Abstract
Sphingolipids are membrane components and are involved in cell proliferation, apoptosis and metabolic regulation. In this study we investigated whether de novo sphingolipid biosynthesis in macrophages is regulated by inflammatory stimuli. Lipopolysaccharide (LPS) treatment upregulated Sptlc2, a subunit of serine palmitoyltransferase (SPT), mRNA and protein in Raw264.7 and mouse peritoneal macrophages, but Sptlc1, another subunit of SPT, was not altered. SPT activation by LPS elevated cellular levels of ceramides and sphingomyelin (SM). Pharmacological inhibition of nuclear factor kappa B (NFκB) prevented LPS-induced upregulation of Sptlc2 while transfection of p65 subunit of NFκB upregulated Sptlc2 and increased cellular ceramide levels. In contrast, MAP kinases were not involved in regulation of sphingolipid biosynthesis. Analysis of Sptlc2 promoter and chromatin immunoprecipitation (ChIP) assay showed that NFκB binding sites are located in Sptlc2 promoter region. Our results demonstrate that inflammatory stimuli activate de novo sphingolipid biosynthesis via NFκB and may play a critical role in lipid metabolism in macrophages.
Keywords: inflammation, ceramide, serine palmitoyltransferase, macrophage, NFκB
1. INTRODUCTION
Sphingolipids are important constituents of the plasma membrane in eukaryotes and, as second messengers, modulate apoptosis, cell proliferation and differentiation [1]. Elevation of plasma fatty acids activates de novo biosynthesis of sphingolipids by supplying the substrates, fatty acids [2]. Accumulation of sphingolipid metabolites in tissues is implicated in atherosclerosis, diabetes, and heart failure [3–5]. Plasma ceramide and sphingomyelin (SM) levels correlate with occurrence of human coronary artery disease (CAD) and SM is suggested as an independent CAD risk factor [6].
De novo biosynthesis of sphingolipids is initiated by the serine palmitoyltransferase (SPT) reaction that catalyzes the formation of 3-ketosphinganine from serine and palmitoyl CoA [7]. Mammalian SPT is composed of two different subunits, Sptlc1 and Sptlc2 encoding 53 and 63 kDa proteins, respectively [8]. The amino acid sequence of Sptlc2 has similarity to aminolevulinate synthase, carrying a domain to form a Schiff base with pyridoxal phosphate [9, 10]. In contrast, the role of Sptlc1 protein is not known well and awaits detailed functional studies. Sptlc3, a third subunit of SPT, has been cloned and characterized [10]. The distinct role of Sptlc3 in SPT reaction should be studied further.
Inflammatory diseases and infection alter triglyceride and cholesterol metabolism. In Syrian hamsters, lipopolysaccaride (LPS) increased plasma TG levels and hepatic cholesterol synthesis [11, 12]. It has been reported that sphingolipid biosynthesis is also regulated by inflammatory response in this model [13, 14]. Intraperitoneal LPS injection upregulated SPT mRNA and activity, and activated secretory sphingomyelinase. As a result, ceramide was elevated in liver and plasma [14]. Additionally, LPS upregulated SPT and glucosylceramide synthase resulting in increased glucosylceramide in non-hepatic tissues such as kidney and spleen [15]. Thus, endotoxin activates de novo sphingolipid biosynthesis via cytokines and alters sphingolipid metabolism.
We questioned whether macrophage sphingolipid synthesis was induced in response to inflammatory stimuli. Macrophages require additional lipids for energy and also membranous changes to allow them to engulf and destroy microorganisms or to respond to injury. In this study, we demonstrate that Sptlc2 were activated by endotoxin-induced inflammation; there was no change in Sptlc1. Moreover, we prove that this process is mediated by NFκB.
2. MATERIALS AND METHODS
2.1. Materials
SP600125, SN50 and SB203580 were obtained from Calbiochem (Damstadt, Germany). PD98059 was obtained from Cell Signaling Technology (Beverly, MA). Ceramides (C14:0, C16:0, C18:0, C18:1, C20:0, C24:0, C24:1 ceramides), sphingosine, and sphingomyelin (SM 16:0, C18:0, C18:1) were obtained from Avanti Polar Lipids (Alabaster, AL). Desipramine was obtained from Biomol (Plymouth Meeting, PA).
2.2. Measurement of SPT and SMase activity
Raw264.7 macrophage cells were grown and maintained in DMEM media and 10% FBS (Invitrogen, Carlsbad, CA). Cells were incubated with 1 μg/ml LPS for 8 hrs. Microsomal fraction of cells was isolated by ultracentrifugation at 100,000g for 90 min and maintained in 100 mM HEPES (pH 8.3), 5 mM dithiothreitol, 2.5 mM EDTA (pH 7.4), and 50 μM pyridoxal 5′-phosphate. Measurement of SPT activity was performed as described previously [3]. Briefly, SPT reaction was initiated by adding 200 μM palmitoyl CoA and 1 mM of serine containing L-[14C]-serine (0.5 μCi/reaction) to 100 μg microsomal extracts (GE healthcare, Piscataway, NJ). After incubation for 20 min at 37 °C, the reactions were terminated by adding 0.2 ml of 0.5 N NaOH. Lipid fractions were extracted by addition of chloroform:methanol (2:1) and lower organic layer was isolated and dried under N2 gas. Sphingolipids were separated by silica gel 60 TLC (VWR, Westchester, PA) with the running solvent (Chroroform: Methanol: 2 M NH4OH, 40:10:1) and the amount of produced [14C]-sphinganine was analyzed by autoradiography and scintillation counting.
SMase activity was measured as described previously by Sakata et al [16] and Mintzer et al [17]. Briefly, the cells were harvested and extracted in 1 ml of a lysis buffer containing 10 mM Tris-HCl (pH 7.4) containing 1% Triton X-100 and protease inhibitor cocktails (Roche, Indianapolis, IN) for neutral SMase. For acid SMase, a lysis buffer containing 50 mM sodium acetate buffer (pH5.4) with 1% Triton X-100 and protease inhibitor cocktails. After centrifugation at 17,000g for 10 min at 4°C, the supernatants were used for acid SMase assay and the pellets were for neutral SMase. SMase activity was measured using [14C]-SM (PerkinElmer, Waltham, MA) as substrates. The activity was measured by adding 0.1 M Na Acetate at pH 7.4 or pH 5.4 and chloroform:methanol (2:1) was added to stop the reaction. The reaction mixture was vortexed and centrifuged to separate the phases. The activity was measured by counting released [14C]-choline in aqueous phase by scintillation counter.
2.3. Suppression of Sptlc2 by small interfering RNA
Small interfering RNA (siRNA) duplexes targeting mouse Sptlc2 was obtained from Santa Cruz Biotechnology. RAW 264.7 cells were transfected with 70 pmol mixture of the siRNAs using LipofectAMINE 2000 reagent according to the manufacturer’s instruction (Invitrogen). The cells were cultured for 72 hrs and gene-silencing effect was assessed by immunoblotting with Sptlc2-specific antibody. After Sptlc2 suppression by siRNA, the cells were incubated with 1 μg/ml LPS for 8 hrs and sphingolipid profile was measured by LC/MC/MS as described below.
2.4. Plasmid construction
Sptlc2 (-2113~-49) promoter cDNA was cloned from mouse genomic DNA (Promega, Madison, WI) by using sense primer 5′-AAA AGG TAC CCA AGA ATG GGT TTC TCT GTG TAG C-3′ and a common anti-sense primer 5′-AAA ACG CGT AGC CAA CTC CCG CTC CGC A -3′. Sptlc2 promoter was incorporated into KpnI and MluI restriction sites in the upstream of pGL3-luciferase (pGL3-luc) vector (Promega, Madison, WI). Additional truncated Sptlc2 promoter inserts were produced by PCR amplification of Sptlc2 full size (-2113) and incorporated into KpnI and MluI restriction sites of pGL3-Luc basic vector (Promega, Madison, WI) with proper primers below. The Sense primers were: 5′-AAA AGG TAC CAA TAA CCT TGC CAA CGC AGT -3′ (-1249); 5′-AAA AGG TAC CCT GGC A AA TGA GTC CAT GAA-3′ (-849); 5′-AAA AGG TAC CAA ACA GAT CAA GTC TCC TGA C -3′ (-749); 5′-AAA AGG TAC CTA GTT CAC AAT GAA GAA CAG T -3′ (-649); 5′-AAA AGG TAC CCT CGG ATT CAA CCT GAG AAC C -3′ (-549); 5′-AAA AGG TAC CTG TTT CTT TCT CCA CAC TCT T-3′ (-449); 5′-AAA AGG TAC CTT GAT GTC TGC AAG CCG CTT -3′ (-300); 5′-AAA GGT ACC GTG GCC CCG CCC TCG CGT GA -3′ (-150). The common antisense primer was used for all PCR reactions. The Sptlc2 promoter (pSptlc2)-pGL3-Luc constructs were verified by DNA sequencing.
2.5. Peritoneal macrophage isolation
After intraperitoneal injection of 2 ml of sterile 3% thioglycollate solution, elicited mouse peritoneal macrophages were isolated from 8 week-old male C57Bl6/J mice by peritoneal lavage with PBS. Isolated macrophages were plated in 12-well plates in RPMI 1640 medium containing 10% FBS, 100 units/ml penicillin and 100 μg/ml streptomycin [18]. All procedures were approved by Gachon University of Medicine and Science Institutional Animal Care and Use Committee (IACUC).
2.6. Transient transfection and luciferase reporter assay
Raw264.7 cells (4 × 105 cells/ml) were cultured in 24-well culture plates to 80% confluency for transfection experiments. Cells were transiently co-transfected with pSptlc2-pGL3-Luc and pcDNA3.1 vector containing p65, wild-type c-Jun amino-terminal kinase (JNKwt), dominant-negative JNK (JNKdn) or IκBα super-repressor (IκBα SR) (S32A, S36A) [19] respectively using FuGENE® HD transfection reagent (Roche, Mannheim, Germany) according to the manufacturer’s instructions. An expression vector for Renilla luciferase, pTK-RL was used to normalize transfection efficiency. After incubation for 24 h, cells were washed with PBS once, lysed in cell lysis buffer, and luciferase and Renilla activity were measured using the Dual-Luciferase Assay System (Promega, Madison, WI) according to the manufacturer’s instructions.
2.7. RNA isolation and real-time RT-PCR
Total RNAs were isolated from Raw264.7 cells using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instruction. First-strand cDNA was synthesized using the iScript™ cDNA Synthesis Kit (Bio-Rad, Hercules, CA). Real-time PCR analysis was performed in ABI7300 equipment (Applied Biosystems Inc, Carlsbad, CA) using SYBR Green Master Mix (Toyobo, Japan). Primers sequences were: sense 5′-GAC GTT GAC ATC CGT AAA G-3′, and anti-sense 5′-CAG TAA CAG TCC GCC T-3′ for β-actin; sense 5′-AGT GGT GGG AGA GTC CCT TT-3′, and anti-sense 5′-CAG TGA CCA CAA CCC TGA TG-3′ for Sptlc1; sense 5′-GGA TAC ATC GGA GGC AAG AA-3′, and anti-sense 5′-ACC TGG TGT TCT CAG CCA AC-3′ for Sptlc2. The PCR condition was 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 15 s and 72 °C for 45 s, and finally extension at 72 °C for 10 min. Expression of genes were expressed as ratio normalized to β-actin.
2.8. Western blot analysis
Whole cell proteins were prepared by using cell lysis buffer containing 20 mM Tris (pH 7.4), 5 mM EDTA (pH 8.0), 10 mM Na4P2O7, 100 mM NaF, 2 mM Na3VO4, 1% NP-40, 1 mM PMSF and protease inhibitors. Thirty micrograms of protein were loaded on 10 % SDS-acrylamide gel by electrophoresis and transferred to PVDF membrane. The blots were blocked with 5% nonfat dried milk in phosphate-buffered saline (PBS) with 0.1% Tween 20 (PBST) and incubated with primary antibodies against Sptlc2 (Abcam, Cambridge, MA), β-actin, JNK, phospho-JNK (Cell Signaling Technology, Danvers, MA), acid SMase (Abcam, Cambridge, MA) or neutral SMase (Santa Cruz Biotechnology, Santa Cruz, CA). After incubation with horseradish peroxidase (HRP)-conjugated Goat anti-Rabbit IgG or Goat anti-Mouse IgG & IgM (Milipore, Bilerica, CA), blots were developed with the enhanced chemiluminescent substrate (Millipore, Bilerica, CA) and detected by a LAS4000 luminescent image analyzer (Fujifilm, Japan).
2.9. Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed according to the manufacturer’s protocol (Millipore, Billerica, MA). Raw264.7 cells were cultured in 10-cm dishes until full confluence. After treatment with or without LPS at 1 μg/ml for 1 h, the DNA-chromatin of cells were cross-linked by the addition of 280 μl of 37% formaldehyde to 10 ml of culture medium for 10 min at room temperature and stopped with glycine buffer. After washed twice with cold PBS, cell pellets were harvested with 1 ml of SDS lysis buffer containing protease inhibitors. The cross-linked protein/DNA was sheared by sonication on ice and then immunoprecipitated with anti-p65 antibody or control IgG. The immune complexes were eluted, reverse cross-linked using 5 M NaCl, and purified by phenol/chloroform extraction. Ethanol-precipitated DNA pellets were redissolved in the ChIP Dilution Buffer. The supernatant of an immunoprecipitation reaction carried out in the absence of antibody was purified and diluted 1:200 as total input DNA control. PCR was carried out on 1 μl of each sample using sense and anti-sense primers against the promoter of interest. PCR products were analyzed on 2 % agarose gels and images were analyzed with a LAS4000 luminescent image analyzer (Fujifilm, Japan).
2.10. Sphingolipid analysis by LC/MS/MS
For quantification of sphingolipids in macrophage, Raw264.7 cells were seeded at a 1 × 106 and incubated at 37 °C for 24 hrs. After LPS treatment or pcDNA3.1-p65 transfection, cells were harvested and lysed using lysis buffer. Known amount of C17:0 -ceramide were added to the cell extracts containing 1 mg of proteins and sphingolipids were extracted with addition of chloroform/methanol (2:1, v/v) containing 0.01% butylated hydroxytoluene. Phospholipids are saponified by adding KOH at 37°C for 2 h. Extracts were neutralized by adding acetic acid and organic phase were separated and dried under N2. Ceramides (C14:0, C16:0, C18:0, C18:1, C20:0, C24:0, C24:1), sphingosine, and SM (C16:0, C18:0, C18:1) were separated by HPLC with C18 column (XTerra C18, 3.5 um, 2.1 * 50 mm) and ionized in positive electrospray ionization mode as described by Yoo et al [20]. [M+]/product ions from corresponding sphingolipid metabolites were monitored for multiple reaction monitoring (MRM) quantification by a bench-top tandem mass spectrometer, API 4000 Q-trap (Applied biosystem, Framingham, MA), interfaced with a electrospray ionization source.
2.11. Statistical analysis
All results are expressed as means ± SEM of at least three experiments with similar results. Data were analyzed by using Student’s t-test and p<0.05 was considered significant.
3. RESULTS
3.1. LPS increases SPT activity and expression of Sptlc2 is upregulated
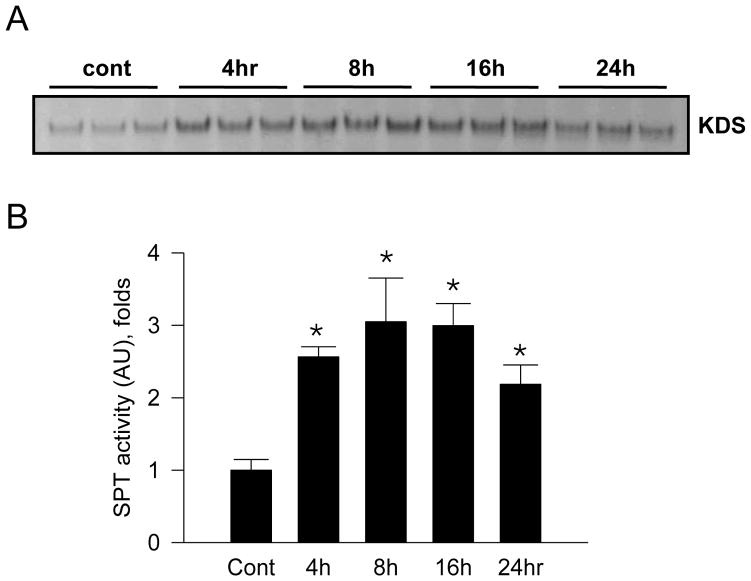
To determine whether endotoxin regulates SPT activity in macrophages, Raw264.7 cells were incubated with 1 μg/ml LPS at various times and SPT activity was measured. Incorporation of [14C]-serine into ketodihydrosphingosine (KDS) was increased by 3 folds at 8 hr (Figure 1).
Figure 1.
Activation of serine palmitoyltransferase (SPT) by LPS in Raw264.7 cells. Raw264.7 macrophage cells were incubated with 1 μg/ml LPS at various times. SPT activity of microsomal fraction (100 μg protein) was measured by using [14C]-serine and palmitoyl CoA as substrates. Lipid fractions were extracted and analyzed by TLC followed by autoradiography (A). The bands indicate ketodihydrosphingosine (KDS). The radioactivity of lipid fractions was measured by scintillation counter (B). n=3, mean ± SEM, *p<0.05 vs. control. Results are representative of two independent experiments.
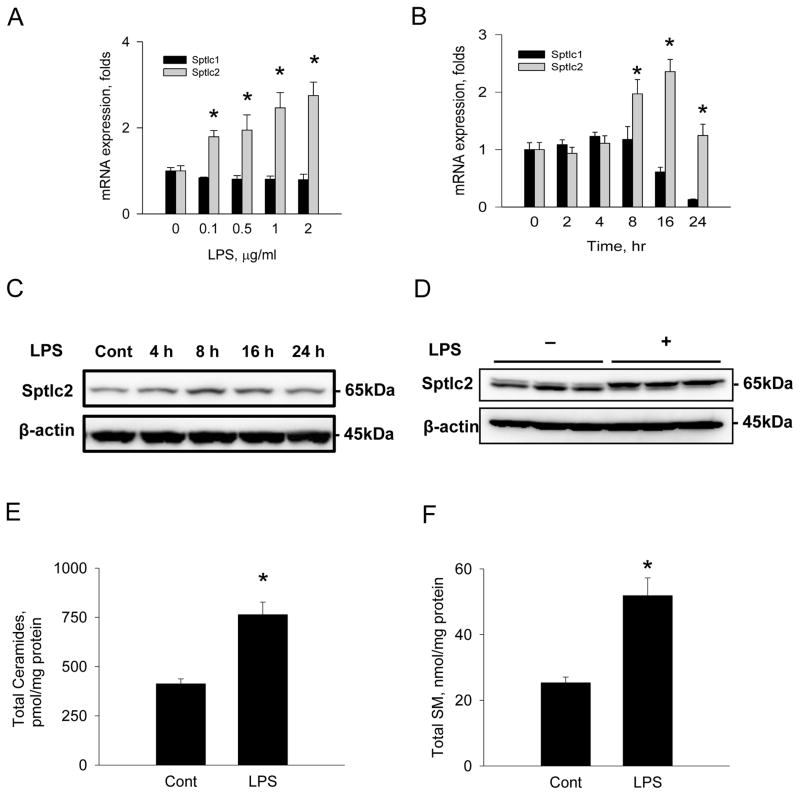
SPT is composed of two major subunits, Sptcl1 and Sptlc2. We measured expression of Sptlc1 and Sptlc2 mRNA after LPS treatment in Raw264.7 cells to examine whether activation of SPT by LPS is due to transcriptional upregulation in macrophage. Cells were incubated with various concentrations of LPS for 8 hrs or incubated with 1 μg/ml for various times and Sptlc1 and Sptlc2 mRNA were measured by RT-PCR. Sptlc2 expression was upregulated in response to LPS in a time- and dose-dependent manner (Figure 2A, B). In contrast to Sptlc2, expression of Sptlc1 was not altered by LPS (Figure 2A, B). Expression of Sptlc2 protein was also elevated in Raw264.7 macrophages and mouse peritoneal macrophages by LPS in a time-dependent manner (Figures 2C, D). Interestingly, Sptlc2 protein from peritoneal macrophage showed two bands, which were not observed in Raw264.7 cells and implied possible post-translational modification of Sptlc2. These results confirm that activation of SPT by LPS is due to transcriptional upregulation of Sptlc2, but not Sptlc1.
Figure 2.
Upregulation of Sptlc2 expression and altered sphingolipid profile by LPS. Raw264.7 macrophage cells were incubated with various concentrations of LPS for 8 hrs (A) and various incubation times with 1 μg/ml LPS (B). mRNA was isolated and expression of Sptlc1 and Sptlc2 were measured by real-time PCR. Raw264.7 cells were incubated with 1 μg/ml LPS for 8 hrs and the cell extracts were analyzed by SDS-PAGE followed by immunoblotting with anti-Sptlc2 antibody (C). Primary peritoneal macrophages were isolated from C57Bl6/J wild-type mice and incubated with 1 μg/ml LPS for 8 hrs (D). Sphingolipids were extracted from Raw264.7 cells incubated with LPS for 8 hrs and analyzed by LC/MS/MS as described in Materials and Methods. Total ceramides (E) and SM (F) were quantified. n =3, Page 37 of 42 mean ± SEM, *p<0.05 vs. control. Results are respresentative of three independent experiments.
Consistent with LPS-mediated upregulation of Sptlc2, cellular levels of ceramides and SM were elevated by 2 folds (Figure 2E, F). Among individual species of ceramides, C16:0, C18:0 and C24:0 ceramide species were elevated by LPS treatment (Table 1). Sphingosine and measured SM species were elevated. Thus, the inflammatory response of macrophages includes changes in sphingolipid metabolism including SPT activation and elevation of cellular ceramides and SM.
Table 1.
Sphingolipid profile in LPS-treated Raw264.7 cells
| Sphingolipids (pmole/mg) | Control | LPS | LPS + Despiramine (20 μM) |
|---|---|---|---|
| C14:0 Cer | 10.1 ± 0.1 | 11.0 ± 0.3 | 11.5 ± 0.2 |
| C16:0 Cer | 77.3 ± 3.8 | 212.7 ± 19.4* | 292.5 ± 22.4 § |
| C18:0 Cer | 0.5 ± 0.1 | 10.7 ± 1.5* | 17.6 ± 1.7*§ |
| C18:1 Cer | 15.4 ± 0.1 | 15.6 ± 0.1 | 14.9 ± 0.1 |
| C24:0 Cer | 231.6 ± 15.8 | 423.3 ± 32.6* | 351.0 ± 41.7*§ |
| C24:1 Cer | 77.9 ± 5.5 | 90.3 ± 10.8 | 98.7 ± 8.0§ |
| SO | 76.5 ± 2.0 | 87.8 ± 1.1* | 67.2 ± 0.1§ |
| SM 16:0 | 25167 ± 1572 | 48973 ± 5033* | 33131 ± 3066§ |
| SM 18:0 | 1401 ± 156 | 2713 ± 380* | 2299 ± 238* |
| SM18:1 | 80.2 ± 5.0 | 154.4 ± 30.3* | 163.5 ± 27.5 |
pmole/mg protein, Cer, ceramide; SO, sphingosine; SM, sphingomyelin. n=3.
p<0.05 vs. Control.
p<0.05 vs. LPS.
3.2. Suppression of Sptlc2 lowers SM levels
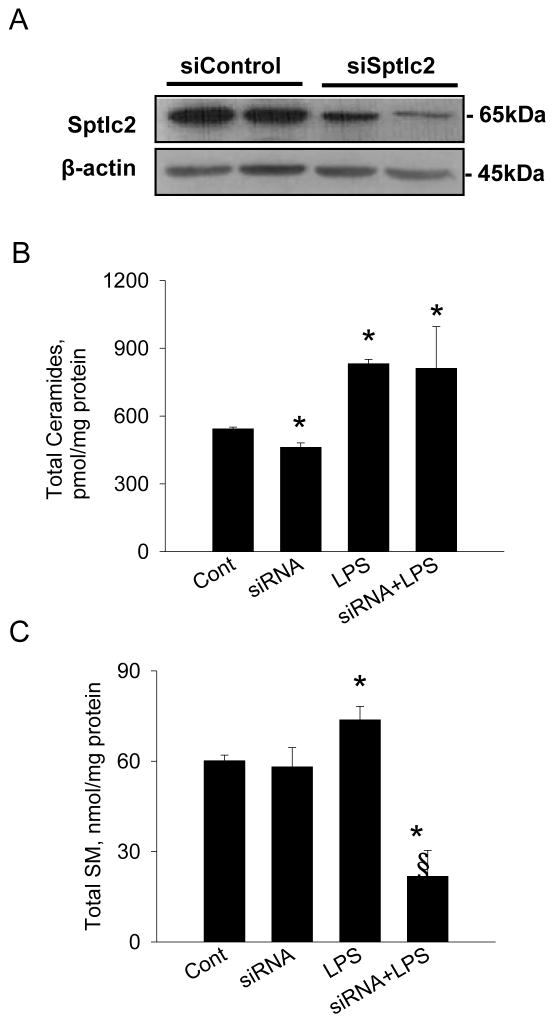
Cellular ceramides are generated by de novo synthesis via SPT as a well as SMase pathways. To determine whether the blockage of de novo sphingolipid biosynthesis abrogates LPS-induced effects on cellular ceramide and SM, we suppressed Sptlc2 expression by Sptlc2-specifc siRNA (Figure 3A). Despite Sptlc2-siRNA lowered cellular ceramides, suppression of Sptlc2 did not alter elevated cellular ceramide by LPS (Figure 3B, Table 2). Instead, SM levels in Sptlc2-siRNA transfected cells were significantly diminished when compared with LPS-treated cells (Figure 3C, Table 2).
Figure 3.
Suppression of Sptlc2 expression by Sptlc2 siRNA and altered sphingolipid profile. Raw264.7 cells were transfected with Sptlc2-siRNA for 72 hrs and Sptlc2 proteins were immunoblotted with Sptlc2-antibody (A). Cells transfected with the control siRNA (siControl) and Sptlc2-siRNA (siSptlc2) were incubated with LPS for 8 hrs. Sphingolipids were extracted and analyzed by LC/MS/MS as described in Materials and Methods. Total ceramides (B) and SM (C) were quantified. n=3, mean ± SEM, *p<0.05 vs. control. § p<0.05 vs. LPS. Results are representative of three independent experiments.
Table 2.
Sphingolipid profile in Raw264.7 cells suppressed by Sptlc2 siRNA
| Sphingolipids (pmole/mg) | Control | Sptlc2 siRNA | LPS | LPS+ Sptlc2 siRNA |
|---|---|---|---|---|
| C14:0 Cer | 11.1± 0.1 | 11.1 ± 0.1 | 11.8 ± 0.1 | 11.9 ± 0.6 |
| C16:0 Cer | 158.7 ± 5.5 | 123.4 ± 5.7* | 299.9 ± 3.8* | 303.4 ± 63.6* |
| C18:0 Cer | 2.5 ± 0.3 | 3.0 ± 0.3 | 8.5 ± 0.5* | 12.4 ± 3.9* |
| C18:1 Cer | 16.7 ± 0.1 | 17.7 ± 0.1* | 16.9 ± 0.2 | 18.4 ± 0.9 |
| C24:0 Cer | 237.4 ± 6.3 | 208.6 ± 9.6 | 391.9 ± 12.0* | 363.7 ± 89.8* |
| C24:1 Cer | 116.9 ± 1.2 | 97.7 ± 5.1* | 102.8 ± 3.8 | 101.3 ± 26.3 |
| SO | 76.5 ± 2.0 | 102.0 ± 1.6* | 106.4 ± 2.6* | 114.0 ± 11.6* |
| SM 16:0 | 58498 ± 1835 | 56563 ± 3599 | 71495 ± 4368* | 21114 ± 8442*§ |
| SM 18:0 | 1515 ± 50.1 | 1476 ± 72.6 | 2108 ± 150.9* | 642 ± 185.5*§ |
| SM18:1 | 120.7 ± 2.5 | 107.5 ± 9.5* | 138.3 ± 5.5 | 40.6 ± 15.4*§ |
pmole/mg protein, Cer, ceramide; SO, sphingosine; SM, sphingomyelin. n=3.
p<0.05 vs. Control.
p<0.05 vs. LPS.
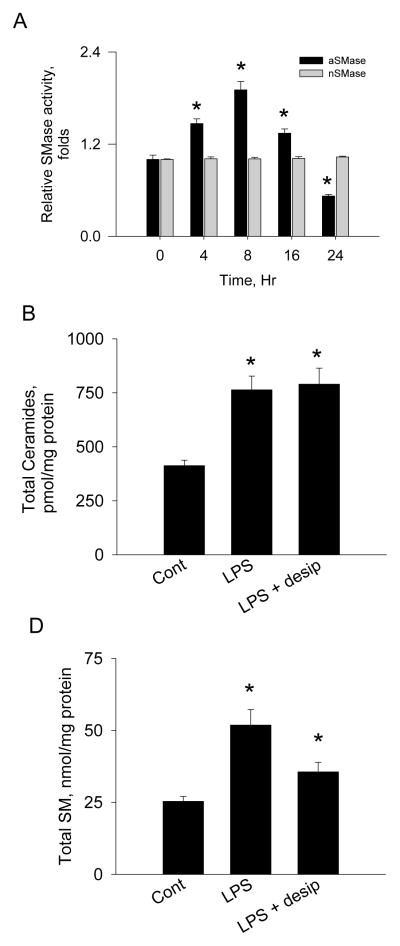
Decreased SM levels and no change in ceramides by siRNA plus LPS imply that SMase pathway is associated with ceramide production when de novo ceramide synthesis is inhibited. To determine whether SMase contributes to LPS-induced elevation of ceramide, the activities of acid SMase and neutral SMase proteins were measured after LPS incubation. We found that acid SMase activity was elevated by 2 folds with no change in neutral SMase (Figure 4A). Inhibition of acid SMase by despiramine and LPS treatment elevated total ceramides (Table 1, Figure 4B) and SM (Table 1, Figure 4C) which is comparable to those of LPS-treated cells. The fact that despiramine did not alter sphingolipid profile suggests that de novo ceramide biosynthesis is the major pathway in response to endotoxin-mediated inflammation. Thus, acid SMase contributes to LPS-induced elevation of cellular ceramides as a compensatory mechanism for de novo ceramide synthesis.
Figure 4.
Activation of acid SMase by LPS. Raw264.7 cells were incubated with LPS at various times. LPS-treated cells were extracted and acid and neutral SMase activities were measured using [14C]-SM as substrates (A). Cells were incubated with LPS and desipramine (desip) for 8 hrs. Sphingolipids were extracted and analyzed by LC/MS/MS as described in Materials and Methods. Total ceramides (B) and SM (C) were quantified. n =3, mean ± SEM, *p<0.05 vs. control. Results are representative of two independent experiments.
3.3. NFκB inhibitor attenuates LPS-induced Sptlc2 expression
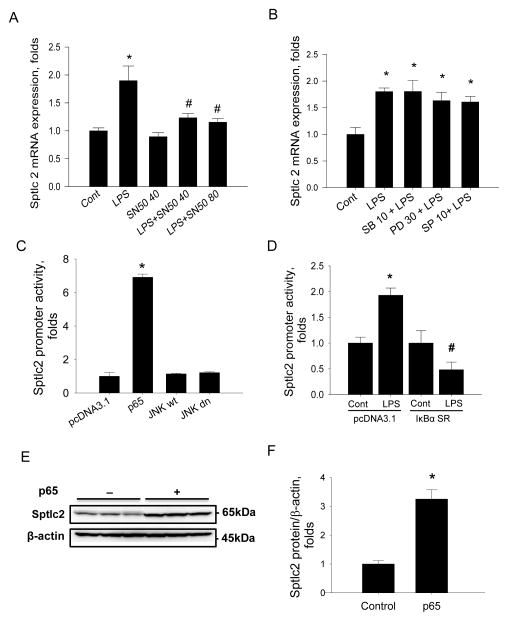
Endotoxin-mediated inflammation activates complex signaling pathways involved in cytokine production and defense mechanisms. To gain further insight into regulation of SPT by inflammation, we inhibited major inflammatory signaling intermediates pharmacologically. Raw264.7 cells were pre-treated with SN50, an inhibitor of NFκB. Then, the cells were treated with LPS and Sptlc2 expression was measured by RT-PCR. Inhibition of NFκB by SN50 suppressed LPS-mediated Sptlc2 upregulation (Figure 5A). In contrast, treatment of SB203580 (an inhibitor of p38 MAP kinase), PD98059 (an inhibitor of MEK1 and ERK1/2 pathway), or SP600125 (an inhibitor of JNK kinase) did not suppress LPS-induced upregulation of Sptlc2 (Figure 5B). These results indicate that NFκB – but not p38, ERK and JNK MAP kinase - pathway regulate Sptlc2 expression in response to inflammation.
Figure 5.
Involvement of inflammatory response pathways in regulation of Sptlc2 expression by LPS in Raw264.7 cells. Cells were incubated with 40 and 80 μM SN50 (A) or 10 μM SB203580 (SB), 30 μM PD98059 (PD), 10 μM SP600125 (SP) (B) respectively in the presence or absence of 1 μg/ml LPS for 8 hrs. Expression of Sptlc2 was measured by RT-PCR. pGL3 reporter construct containing Sptlc2 promoter (-2049~-54) was co-transfected with empty pcDNA3.1, p65, wild-type JNK (JNKwt) or dominant negative JNK (JNKdn) respectively. After 24 hrs incubation, the luciferase activity was measured by Dual-Luciferse Reporter Assay (C). After co-transfection of pcDNA3.1 empty vector or IκBα super-repressor vector (IκBα SR) with Sptlc2 reporter construct and 8 hrs incubation with 1 μM LPS, the activity of luciferase was measured (D). Raw264.7 cells were transfected with pcDNA 3.1 empty vector or pcDNA3.1-p65 vector and incubated for 24 hrs, Sptlc2 protein level were determined by immunoblot (E) and quantified by densitometry (F). Expression of Sptlc2 was normalized by β-actin. n=3, mean ± SEM, *p<0.05 vs. control, #p<0.05 vs LPS with pcDNA3.1 empty vector. Results are representative of two independent experiments.
A luciferase reporter containing the promoter of Sptlc2 (-2049~-54) (pSptlc2 luc reporter) was constructed to elucidate the mechanism of Sptlc2 regulation by NFκB and transiently transfected into Raw264.7 cells. Co-transfection of pSptlc2-luc reporter with pcDNA3.1-p65 DNA, a component of NFκB, increased Sptlc2 promoter activity by 8-fold compared to the control transfected with empty vector, pcDNA3.1 (Figure 5C). In contrast, co-transfection of WT JNK (JNKwt) and dominant negative JNK (JNKdn) DNA did not affect Sptlc2 expression. While Sptlc2 promoter activity increased markedly in LPS-treated cells, co-transfection of IκBα SR (S32A, S36A) prevented LPS-induced elevation of Sptlc2 promoter activity (Figure 5D). Consistently Sptlc2 proteins were increased when cells were transfected with pcDNA3.1-p65 (Figure 5E, F). Sphingolipid profile of pcDNA3.1- p65-transfected cells demonstrated that C16:0, C24:0 and C24:1 ceramide were increased and sphingosine and SM levels were not altered (Table 3). Thus, NFκB is responsible for induction of Sptlc2 expression and elevation of ceramides in Raw264.7 cells.
Table 3.
Sphingolipid profile in p65-transfected Raw264.7 cells
| Sphingolipids (pmole/mg) | Control pcDNA3.1 | pcDNA3.1-p65 |
|---|---|---|
| C14:0 Cer | 4.1 ± 0.1 | 4.3 ± 0.1 |
| C16:0 Cer | 72.9 ± 1.2 | 104.7 ± 5.6* |
| C18:0 Cer | <0 | <0 |
| C18:1 Cer | 4.4 ± 0.2 | 4.5 ± 0.3 |
| C24:0 Cer | 178.9 ± 13.5 | 391.4 ± 36.8* |
| C24:1 Cer | 68.4 ± 3.4 | 133.3 ± 14.3* |
| SO | 40.3 ± 2.2 | 47.8 ± 3.2 |
| SM 16:0 | 1526.7 ± 254.4 | 1734.8 ± 131.9 |
| SM 18:0 | 152.2 ± 30.6 | 154.6 ± 10.4 |
| SM18:1 | 1.4 ± 1.2 | 2.6 ± 0.6 |
pmole/mg protein, Cer, ceramide; SO, sphingosine; SM, sphingomyelin. n=3.
p<0.05 vs. Control.
3.4. NFκB subunit p65 binds to the promoter of Sptlc2
Sptlc2 promoter (-2113~-49) was truncated to different sizes and then incorporated into pGL3 in order to identify the possible binding location for NFκB subunit p65. After co-transfection of pSptlc2-luc reporter and p65 for 24 h, the luciferase activity in Raw264.7 cells was measured. We found that truncated promoter of Sptlc2 from -2049 to -849 showed high luciferase activity by p65. The promoter region from -649 to -449 had a moderate effect, significantly lower than that of full size promoter. The promoter containing between -300 and -150 showed the lowest effect, close to that of empty pGL3 vector (Figure 6A). After treatment with or without LPS for 8 h, the protein/DNA complex was fragmented and immunoprecipitated by anti-p65 antibody. The primers specific for detection of Sptlc2 promoter -449~-250 and -849~-650 were designed and PCR was performed using the immunoprecipitated fragments as template. As shown in Figure 6B, the gene sequence of both Sptlc2 promoter -449~-250 and -849~-650 were detected in the chromatin DNA fragments immunoprecipitated by anti-p65 antibody after LPS treatment. Therefore there are at least two distinct binding sites for NFκB in the Sptlc2 promoter which is involved in up-regulation of Sptlc2 gene expression under LPS stimulation.
Figure 6.
Promoter analysis of Sptlc2 by NFκB in Raw264.7 cells. Sptlc2 reporter construct with various lengths of Sptlc2 promoter were co-transfected with pcDNA2.1 empty vector or pcDNA2.1-p65 vector and the luciferase activity was measured (A). Raw264.7 cells were incubated with 1 μg/ml LPS for 8 hrs and chromatin was immunoprecipitated by anti-p65 antibody. Immunoprecipitaed chromatin DNA was fragmented by sonication to produce 0.2~0.4 kb DNA fragments. PCR was performed to measure the promoter region of -449~-250 and -845~-650 (B). n=3, mean ± SEM. §p<0.05 vs. empty control. #p<0.05 vs. -849 promoter. *p<0.05 vs. -449 promoter. Results are respresentative of two independent experiments.
4. DISCUSSION
Sphingolipids play important roles in cell biology [21]. Imbalanced sphingolipid metabolism leads to development of neurological dysfunction [22, 23], and is associated with other generalized metabolic diseases such as diabetes and CAD [3, 4, 6, 24]. To determine whether inflammatory response is closely associated with regulation of sphingolipid biosynthesis, we investigated regulation of sphingolipid biosynthesis by endotoxin in macrophages. In this study, we demonstrated the following: 1) Inflammatory response elicited by LPS increases SPT activity via upregulation of Sptlc2 in macrophages, 2) acid SMase is involved in LPS-induced ceramide increase, 3) upregulation of Sptlc2 by LPS is caused by the NFκB pathway not by other MAP kinase pathways. 4) NFκB-specific response elements exist in the promoter region of Sptlc2.
De novo biosynthesis of sphingolipids starts with condensation of serine and palmitoyl CoA to produce 3-ketosphinganine by SPT enzyme. SPT is composed of two subunits, Sptlc1 and Sptlc2 [8]. These subunits are assumed to form a heterodimer [8]. Recently, a third SPT subunit, Sptlc3 has been reported [10]. While overexpression of Sptlc1 in HEK293 cells had no effect on SPT activity, Sptlc2- or Sptlc3-overexpressing cells showed increased SPT activity by 2- to 3-fold [10]. Sptlc1 must not be rate limiting in the regulation of SPT; it is likely to be produced in excess and act as an anchor for other SPT subunit in the endoplasmic reticulum [10]. Possibly when the other subunits are expressed at greater levels, they have a readily available partner. Therefore, the stoichiometry of Sptlc1 vs. Sptlc2 and identification of other possible binding partners regulating SPT activity deserve further study.
Sptlc2 has 68% homology with Sptlc3 and it is speculated that Sptlc3 is an isoform of Sptlc2, each binding to Sptlc1 independently. Depending on the tissue distribution and the balance of Sptlc2 and Sptlc3 could be a cellular mechanism to regulate sphingolipid pools. In this study, we detected Sptlc3 expression at very low levels in Raw264.7 macrophage (data not shown) and focused on Sptlc1 and Sptlc2. Although various physical and biochemical agents upregulate Sptlc2 mRNA, such as ultraviolet irradiation [25, 26], inflammatory cytokines [14, 15] and endotoxin [14, 15], relatively little is known about the regulatory mechanism of SPT in de novo sphingolipid biosynthesis in macrophages.
The response to infection and inflammation is associated with hepatic synthesis of acute-phase inflammatory proteins [27]. Administration of LPS, a component of the gram-negative bacterial cell wall, increased hepatic SPT activity and de novo sphingolipid biosynthesis resulting in lipoproteins enriched in ceramide and SM [14]. Additionally, ceramide and glucosylceramide (GlcCer) are accumulated in kidney and spleen when SPT and GlcCer synthase are upregulated [15]. In response to infection, macrophages are major inflammatory cells with an ability to produce inflammatory cytokines, such as TNFα, IL-1β, and IL-6. Moreover, these cells are active participants in phagocytosis of microbes and damaged tissues. Our results demonstrated that LPS-induced SPT activation is occurring in macrophages. As a result, elevated levels of cellular ceramide and SM were found. Sptlc2, but not Sptlc1 was upregulated by endotoxin. These data confirm that Sptlc2 is the major limiting subunit responsible for catalytic function of SPT [10].
Although activation of macrophages is associated with greater ceramide levels, the pathways responsible for this are not well defined. While SPT is responsible for de novo synthesis of ceramide, SMase produces ceramide from SM supplied from plasma lipoproteins or endogenous sources. Treatment with LPS or cytokines such as IL1β and TNFα activates serum acid SMase in WT mice [28]. In addition, TNFα-mediated activation of lysosomal acid SMase increased cellular ceramide. Subsequently, elevated levels of ceramide activate NFκB producing inflammatory cytokines in HL-60 leukemia cells and THP-1 macrophages [16, 29]. This vicious cycle accelerates the systemic inflammatory response by producing more cytokines and it implies that ceramide acts as a signaling molecule in inflammation. Our results demonstrated that LPS-induced elevation of ceramide did not involve neutral SMase in Raw264.7 cells. Instead, the activity of acid SMase was elevated by LPS. Thus, ceramide production in response to LPS was caused by activation of SPT and acid SMase. While suppression of Sptlc2 by siRNA decreased cellular ceramides but not SM, LPS elevated cellular ceramides and diminished SM significantly in Sptlc2-siRNA transfected cells. This observation implies that generated ceramides were produced from SM by action of acid SMase when de novo synthesis was suppressed. In contrast, despiramine, an inhibitor of acid SMase, in the presence of LPS elevated ceramide and SM probably due to less contribution of acid SMase to ceramide production. These results indicate that LPS-induced activation of de novo sphingolipid biosynthesis contributes to elevation of ceramides and acid SMase is a compensatory substitute to maintain basal ceramide levels.
Endotoxin-mediated inflammation activates various mitogen-activated protein kinase (MAPKs). Activated MAPKs such as ERK, JNK and p38 subsequently phosphorylate transcription factors for production of cytokines and inflammatory proteins [30]. We found that pharmacological inhibition of ERK, JNK and p38 pathways did not suppress LPS-induced upregulation of Sptlc2. Instead, inhibition of NFκB prevented LPS-induced upregulation of Sptlc2. Overxpression of p65, an active subunit of NFκB, upregulated Sptlc2 and the blockage of NFκB pathway by IκBα prevented Sptlc2 upregulation (Figure 5). However, the overexpression of JNK and dominant negative mutant JNK did not affect Sptlc2 expression in macrophages (Figure 5C). Despite it has been reported that elevation of ceramide activates JNK in myocytes [31–33], activated JNK did not affect ceramide biosynthesis in macrophages.
We demonstrated that NFκB regulates Sptlc2 transcription by direct binding to the Sptlc2 promoter. NFκB binding sites are located in the promoter regions of Sptlc2 (-449~-300 and -845~-649). Sequence analysis of Sptlc2 promoter found three consecutive putative NFκB response elements (NKRE) in - 725~-686 region (GGAAAGTCTTCAAAGGAAATTGCCCTAGGAAAACCCGG). However, we could not find any putative NKRE in the region of -449~-300. Sptlc2 promoter has been characterized previously by Linn et al. regarding the proximal 335 bp initiator and several downstream promoter elements [34]. Thus, endotoxin activates NFκB [29], which then activates de novo sphingolipid biosynthesis by upregulation of Sptlc2 in macrophages.
In conclusion, this study shows that SPT is activated by inflammatory response via upregulation of Sptlc2, a subunit of SPT. Elevated ceramide levels are due to endotoxin-mediated activation of de novo biosynthesis and acid SMase. Among the inflammatory signaling pathways, NFκB is involved in endotoxin-mediated Sptlc2 upregulation by binding to the specific regions of Sptlc2 promoter. Thus, NFκB is a major transcription factor regulating de novo sphingolipid biosynthesis in macrophages.
RESEARCH HIGHLIGHTS
Inflammatory response by LPS increases SPT activity via upregulation of Sptlc2 in macrophages
Upregulation of Sptlc2 by LPS is caused by the NFκB pathway not by other MAP kinase pathways.
NFκB-specific response elements exist in the promoter region of Sptlc2.
Acknowledgments
This study was supported by a grant of the Korea Healthcare technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea. (A090614).
Abbreviations
- LPS
lipopolysaccharide
- SPT
Serine palmitoyltransferase
- NFκB
nuclear factor kappa B
- ChIP
chromatin immunoprecipitation
- SM
sphingomyelin
- SMase
sphingomyelinase
- CAD
coronary artery disease
- JNK
c-Jun amino-terminal kinase
- dn
dominant negative
- SR
super-repressor
- ERK
extracellular-signal-related kinase
- MAPK
Mitogen-activated protein kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pettus BJ, Chalfant CE, Hannun YA. Ceramide in apoptosis: an overview and current perspectives. Biochim Biophys Acta. 2002;1585:114–25. doi: 10.1016/s1388-1981(02)00331-1. [DOI] [PubMed] [Google Scholar]
- 2.Chavez JA, Summers SA. Characterizing the effects of saturated fatty acids on insulin signaling and ceramide and diacylglycerol accumulation in 3T3-L1 adipocytes and C2C12 myotubes. Arch Biochem Biophys. 2003;419:101–9. doi: 10.1016/j.abb.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 3.Park TS, Panek RL, Mueller SB, Hanselman JC, Rosebury WS, Robertson AW, et al. Inhibition of sphingomyelin synthesis reduces atherogenesis in apolipoprotein E-knockout mice. Circulation. 2004;110:3465–71. doi: 10.1161/01.CIR.0000148370.60535.22. [DOI] [PubMed] [Google Scholar]
- 4.Holland WL, Brozinick JT, Wang LP, Hawkins ED, Sargent KM, Liu Y, et al. Inhibition of ceramide synthesis ameliorates glucocorticoid-, saturated-fat-, and obesity-induced insulin resistance. Cell Metab. 2007;5:167–79. doi: 10.1016/j.cmet.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Park TS, Hu Y, Noh HL, Drosatos K, Okajima K, Buchanan J, et al. Ceramide is a cardiotoxin in lipotoxic cardiomyopathy. J Lipid Res. 2008;49:2101–12. doi: 10.1194/jlr.M800147-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang XC, Paultre F, Pearson TA, Reed RG, Francis CK, Lin M, et al. Plasma sphingomyelin level as a risk factor for coronary artery disease. Arterioscler Thromb Vasc Biol. 2000;20:2614–8. doi: 10.1161/01.atv.20.12.2614. [DOI] [PubMed] [Google Scholar]
- 7.Miyake Y, Kozutsumi Y, Nakamura S, Fujita T, Kawasaki T. Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem Biophys Res Commun. 1995;211:396–403. doi: 10.1006/bbrc.1995.1827. [DOI] [PubMed] [Google Scholar]
- 8.Weiss B, Stoffel W. Human and murine serine-palmitoyl-CoA transferase--cloning, expression and characterization of the key enzyme in sphingolipid synthesis. Eur J Biochem. 1997;249:239–47. doi: 10.1111/j.1432-1033.1997.00239.x. [DOI] [PubMed] [Google Scholar]
- 9.Dickson RC, Lester RL, Nagiec MM. Serine palmitoyltransferase. Methods Enzymol. 2000;311:3–9. doi: 10.1016/s0076-6879(00)11060-2. [DOI] [PubMed] [Google Scholar]
- 10.Hornemann T, Richard S, Rutti MF, Wei Y, von Eckardstein A. Cloning and initial characterization of a new subunit for mammalian serine-palmitoyltransferase. J Biol Chem. 2006;281:37275–81. doi: 10.1074/jbc.M608066200. [DOI] [PubMed] [Google Scholar]
- 11.Feingold KR, Hardardottir I, Memon R, Krul EJ, Moser AH, Taylor JM, et al. Effect of endotoxin on cholesterol biosynthesis and distribution in serum lipoproteins in Syrian hamsters. J Lipid Res. 1993;34:2147–58. [PubMed] [Google Scholar]
- 12.Feingold KR, Staprans I, Memon RA, Moser AH, Shigenaga JK, Doerrler W, et al. Endotoxin rapidly induces changes in lipid metabolism that produce hypertriglyceridemia: low doses stimulate hepatic triglyceride production while high doses inhibit clearance. J Lipid Res. 1992;33:1765–76. [PubMed] [Google Scholar]
- 13.Lightle S, Tosheva R, Lee A, Queen-Baker J, Boyanovsky B, Shedlofsky S, et al. Elevation of ceramide in serum lipoproteins during acute phase response in humans and mice: role of serine-palmitoyl transferase. Arch Biochem Biophys. 2003;419:120–8. doi: 10.1016/j.abb.2003.08.031. [DOI] [PubMed] [Google Scholar]
- 14.Memon RA, Holleran WM, Moser AH, Seki T, Uchida Y, Fuller J, et al. Endotoxin and cytokines increase hepatic sphingolipid biosynthesis and produce lipoproteins enriched in ceramides and sphingomyelin. Arterioscler Thromb Vasc Biol. 1998;18:1257–65. doi: 10.1161/01.atv.18.8.1257. [DOI] [PubMed] [Google Scholar]
- 15.Memon RA, Holleran WM, Uchida Y, Moser AH, Grunfeld C, Feingold KR. Regulation of sphingolipid and glycosphingolipid metabolism in extrahepatic tissues by endotoxin. J Lipid Res. 2001;42:452–9. [PubMed] [Google Scholar]
- 16.Sakata A, Ochiai T, Shimeno H, Hikishima S, Yokomatsu T, Shibuya S, et al. Acid sphingomyelinase inhibition suppresses lipopolysaccharide-mediated release of inflammatory cytokines from macrophages and protects against disease pathology in dextran sulphate sodium-induced colitis in mice. Immunology. 2007;122:54–64. doi: 10.1111/j.1365-2567.2007.02612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mintzer RJ, Appell KC, Cole A, Johns A, Pagila R, Polokoff MA, et al. A novel high-throughput screening format to identify inhibitors of secreted acid sphingomyelinase. J Biomol Screen. 2005;10:225–34. doi: 10.1177/1087057104272546. [DOI] [PubMed] [Google Scholar]
- 18.Chang ZQ, Lee JS, Gebru E, Hong JH, Jung HK, Jo WS, et al. Mechanism of macrophage activation induced by beta-glucan produced from Paenibacillus polymyxa JB115. Biochem Biophys Res Commun. 2010;391:1358–62. doi: 10.1016/j.bbrc.2009.12.064. [DOI] [PubMed] [Google Scholar]
- 19.Lee YR, Kweon SH, Kwon KB, Park JW, Yoon TR, Park BH. Inhibition of IL-1beta-mediated inflammatory responses by the IkappaB alpha super-repressor in human fibroblast-like synoviocytes. Biochem Biophys Res Commun. 2009;378:90–4. doi: 10.1016/j.bbrc.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Yoo HH, Son J, Kim DH. Liquid chromatography-tandem mass spectrometric determination of ceramides and related lipid species in cellular extracts. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;843:327–33. doi: 10.1016/j.jchromb.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 21.Worgall TS. Sphingolipids: major regulators of lipid metabolism. Curr Opin Clin Nutr Metab Care. 2007;10:149–55. doi: 10.1097/MCO.0b013e328028fda3. [DOI] [PubMed] [Google Scholar]
- 22.Penno A, Reilly MM, Houlden H, Laura M, Rentsch K, Niederkofler V, et al. Hereditary sensory neuropathy type 1 is caused by the accumulation of two neurotoxic sphingolipids. J Biol Chem. 2006;285:11178–87. doi: 10.1074/jbc.M109.092973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCampbell A, Truong D, Broom DC, Allchorne A, Gable K, Cutler RG, et al. Mutant SPTLC1 dominantly inhibits serine palmitoyltransferase activity in vivo and confers an age-dependent neuropathy. Hum Mol Genet. 2005;14:3507–21. doi: 10.1093/hmg/ddi380. [DOI] [PubMed] [Google Scholar]
- 24.Hojjati MR, Li Z, Zhou H, Tang S, Huan C, Ooi E, et al. Effect of myriocin on plasma sphingolipid metabolism and atherosclerosis in apoE-deficient mice. J Biol Chem. 2005;280:10284–9. doi: 10.1074/jbc.M412348200. [DOI] [PubMed] [Google Scholar]
- 25.Grether-Beck S, Timmer A, Felsner I, Brenden H, Brammertz D, Krutmann J. Ultraviolet A-induced signaling involves a ceramide-mediated autocrine loop leading to ceramide de novo synthesis. J Invest Dermatol. 2005;125:545–53. doi: 10.1111/j.0022-202X.2005.23782.x. [DOI] [PubMed] [Google Scholar]
- 26.Farrell AM, Uchida Y, Nagiec MM, Harris IR, Dickson RC, Elias PM, et al. UVB irradiation up-regulates serine palmitoyltransferase in cultured human keratinocytes. J Lipid Res. 1998;39:2031–8. [PubMed] [Google Scholar]
- 27.Baumann H, Gauldie J. The acute phase response. Immunol Today. 1994;15:74–80. doi: 10.1016/0167-5699(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 28.Wong ML, Xie B, Beatini N, Phu P, Marathe S, Johns A, et al. Acute systemic inflammation up-regulates secretory sphingomyelinase in vivo: a possible link between inflammatory cytokines and atherogenesis. Proc Natl Acad Sci U S A. 2000;97:8681–6. doi: 10.1073/pnas.150098097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z, Costanzo M, Golde DW, Kolesnick RN. Tumor necrosis factor activation of the sphingomyelin pathway signals nuclear factor kappa B translocation in intact HL-60 cells. J Biol Chem. 1993;268:20520–3. [PubMed] [Google Scholar]
- 30.Rao KM. MAP kinase activation in macrophages. J Leukoc Biol. 2001;69:3–10. [PubMed] [Google Scholar]
- 31.Chavez JA, Knotts TA, Wang LP, Li G, Dobrowsky RT, Florant GL, et al. A role for ceramide, but not diacylglycerol, in the antagonism of insulin signal transduction by saturated fatty acids. J Biol Chem. 2003;278:10297–303. doi: 10.1074/jbc.M212307200. [DOI] [PubMed] [Google Scholar]
- 32.Powell DJ, Turban S, Gray A, Hajduch E, Hundal HS. Intracellular ceramide synthesis and protein kinase Czeta activation play an essential role in palmitate-induced insulin resistance in rat L6 skeletal muscle cells. Biochem J. 2004;382:619–29. doi: 10.1042/BJ20040139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitz-Peiffer C, Craig DL, Biden TJ. Ceramide generation is sufficient to account for the inhibition of the insulin-stimulated PKB pathway in C2C12 skeletal muscle cells pretreated with palmitate. J Biol Chem. 1999;274:24202–10. doi: 10.1074/jbc.274.34.24202. [DOI] [PubMed] [Google Scholar]
- 34.Linn SC, Andras LM, Kim HS, Wei J, Nagiec MM, Dickson RC, et al. Functional characterization of the promoter for the mouse SPTLC2 gene, which encodes subunit 2 of serine palmitoyltransferase. FEBS Lett. 2006;580:6217–23. doi: 10.1016/j.febslet.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]