Abstract
The tumor suppressor p53 is arguably the most important transcription factor that safeguards the genome. Although it is clear that the transcriptional activity of p53 is required for its tumor suppressive function, the underlying mechanisms are still largely unknown. In the past several years, genome-wide approaches have provided novel insights into the tumor suppressive functions of p53. This mini-review summarizes recent progress in studying these functions using genome-wide approaches, and offers some perspectives on this rapidly expanding field.
Introduction
As the “guardian” of the genome, p53 is a sequence-specific transcription factor. It is activated in response to cellular stress, such as DNA damage, oncogene over-expression, virus infection and hypoxia. After activation, p53 is stabilized mainly through post-translational modifications. It then binds to chromatin to activate or repress hundreds of downstream targets which are involved in many biological functions [1] (Figure 1). In somatic cells, p53 induces cell cycle arrest, apoptosis or senescence, depending on the type and strength of stresses and the cell types [2]. In fibroblast, activation of p53 by oncogene over-expression leads to senescence. In mouse embryonic stem cells, p53 regulates self-renewal, and in human embryonic stem cells it regulates both, differentiation and apoptosis [3–5].
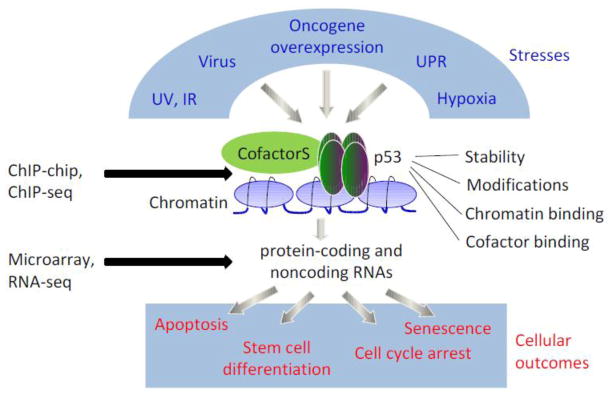
Figure 1. Probing the p53 signaling pathway at a genome wide level.
p53 acts as a hub for many stress signals in cells. Once being activated, p53 is stabilized and subject to post-translational modifications. It also recruits co-factors, binds to chromatin and activates hundreds of downstream targets that include both protein-coding and noncoding RNAs. These downstream targets will elicit various cellular outcomes, such as cell cycle arrest, apoptosis, senescence and stem cell differentiation. A new generation of genome-wide approaches, such as ChIP-seq and RNA-seq, can measure high quality signals of chromatin binding, cofactor recruitment, post-translational modifications and p53-regulated targets.
Despite the fact that much is known about p53 function, many important questions remain. For example, why is there no single p53 target that completely explains the potent tumor suppressive function of p53? Why do most tumors have p53 point mutations rather than p53 deletions? With powerful genome-wide and systems biology approaches, it might be possible to answer some of these questions. This mini-review will give an update of recent progress in using genome-wide approaches to study transcriptional regulation by p53. The basics of some genome-wide platforms, such as chromatin immunoprecipitation based microarray (ChIP-chip), ChIP-sequencing (ChIP-chip), gene expression microarray and RNA sequencing (RNA-seq), have recently been reviewed and will not be detailed here [6].
Location, location, location
Some of the p53 aspects that could benefit from genome-wide studies involve the use of gene expression microarrays to study the downstream targets of p53 [7–8]. To identify p53 downstream targets, Earlier studies using microarrays to detect differentially expressed genes were performed in human bladder carcinoma ECV-304 cells that contain a wild type p53 gene or in human colon carcinoma EB-1 cells that contain an inducible p53 gene [7–8]. Results from these studies suggest that p53 plays much wider roles than previously thought [7]. There are two well-recognized p53 functions: cell cycle arrest and apoptosis. However, it also regulates some relatively under-studied functions, such as the regulation of cytoskeletal structure and cell-cell junctions.
One of the intrinsic limitations of gene expression microarray is that it does not provide specific molecular insights into how p53 regulates its downstream targets. In particular, gene expression microarray does not tell whether the gene expression regulation is the direct effect of transcription factor binding, or an indirect effect on gene expression. There is no doubt that chromatin binding is critical for the tumor suppressive functions of p53 since more than half of human tumors have p53 mutations in the DNA binding domain. So, many interesting questions about p53 interactions with chromatin of are left unanswered by studies with gene expression microarrays. For instance, does stronger chromatin binding correlate with higher expression induction? Is the promoter-proximal region the most important region for p53 to regulate its downstream target? Fueled by these questions, several studies to profile the chromatin binding of p53 in a genome-wide manner were performed.
In a somewhat limited whole-genome manner, using tiling microarray or chromatin immunoprecipitation paired end tag (ChIP-PET), the chromatin binding of p53 was mapped [9–12]. From these early studies, it appeared that the promoter region was not the only part of these locations bound by p53. The majority of p53 binding sites were found within the gene bodies and/or at the downstream regions [9, 11–12]. These results raised more questions: if the non-promoter binding sites of p53 are functional, how do they contribute to transcription mediated by the basal transcription machinery at the promoter region? It is highly possible that chromatin looping is involved in this signal transmission. Using recently developed Chromosome Conformation Capture Carbon Copy (5C) [13], this hypothesis can now be tested on a genome-wide scale. Recently, genome-wide chromatin looping mediated by estrogen receptor alpha has been mapped using chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) [14]. A similar approach could be used to probe the role of chromatin looping in mediating p53 function.
Did we find the right target?
A long-standing question on p53 function is whether there is a single downstream target that can mimic its tumor suppressor activities. So far, no such target gene has been identified. One of the most well studied downstream targets, Cdkn1a (also called p21 or Waf1), is critical for p53-mediated cell cycle arrest [15–16]. However, Cdkn1a knockout (KO) mice do not display the dramatic tumor phenotypes at seven months of age as observed in p53 KO mice [15–16]. Another p53 target, Bbc3 (also called Puma), is reported to be essential for p53-mediated apoptosis [17]. Similar to Cdkn1a KO mice, Bbc3 KO mice do not develop tumors at six month age [17]. So, why don’t Cdkn1a and Bbc3 KO mice phenocopy p53 KO mice? There are at least two possible answers to this question. The first possibility is that we simply have not found the correct target. A recent genome-wide report has provided evidence to support this possibility [18]. In that study, the authors generated a mouse with p53 mutations at amino acids 25 and 26 [called p53 (25, 26)]. This p53 mutant is defective in the induction of cell cycle arrest genes, such as Cdkn1a and Mdm2, as well as the pro-apoptotic gene, Bbc3 [18]. However, the p53 (25, 26) mice have a similar tumor incidence as wild type mice [18]. These results suggest that these canonical p53 targets are dispensable for the tumor suppressive functions of p53 [18]. Intriguingly, the authors identified dozens of new p53 targets that phenocopy p53 in tumor suppression in a K-Ras driven non-small cell lung cancer model [18]. Using other cancer models, such as central nervous system tumors and B-cell lymphomas, the same group observed similar results [19]. These results strongly indicate that the previously believed tumor suppressive roles of Cdkn1a and Bbc3 need be re-evaluated [18–19].
A second possibility is that a subset of p53 targets cooperatively mediates the tumor suppressive function. Each target carries only a partial p53-mediated function. This concept is particularly attractive given that tumorigenesis is a multi-step process [20]. This hypothesis could be tested by generating mice with several deletions in p53 targeted genes. However, this is a technically challenging task since the possible number of multi-gene knockout strains increases exponentially as the number of genes of interest increases. To guide the design of multi-gene knockout experiments, it is helpful to generate a picture of the p53 signaling network using the combination of genome-wide and systems biology approaches. Gene-set based approaches can then be used to dissect the roles of subsets of p53 regulated genes in tumor suppression [10, 21]. Key nodes for each pathway can be identified and functionally tested. By doing so, we could reduce the apparent complexity of p53-mediated transcriptional network and make the second possibility testable. Novel therapeutic strategies might also be developed to treat cancer by simultaneously targeting multiple pathways that are dis-regulated upon loss of p53.
Can we outsmart cancer by studying only cancer cells?
Most genome-wide studies reported so far have been performed in established cancer cell lines, such as HCT116 (human colon carcinoma) and U2OS (human osteosarcoma) [10, 12]. It is worth noting that cells with a wild type p53 gene do not necessarily have a normal p53 signaling pathway. In the case of glioblastoma, around 87% of tumors have an abnormality in the p53 signaling pathway, while only about 35% of these tumors have a mutated p53 gene [22]. This observation suggests that around 52% of glioblastomas have a wild type p53 gene, but the p53 signaling pathway in these tumors is disrupted. Therefore, caution needs to be taken when interpreting results of p53 studies obtained from HCT116, U2OS, and other tumor-derived cell lines. For example, Shaked et al. compared the chromatin binding of p53 in normal human cells versus cancer cell lines using a custom array [23]. In primary (non-tumorigenic) cells, p53 has limited chromatin binding in the absence of stress [23]. Cellular stresses greatly increase the occupancy of p53 to chromatin [23]. This is in contrast to the behavior of p53 in established cancer cell lines [24–25]. In established cancer cell lines, stress have little effect on p53 binding to chromatin [25]. Thus, the unique cellular environment in cancer cell lines can change the behavior of p53. Because of the low coverage of the array, it is difficult to assess whether these conclusions will still hold when the genome-wide binding of p53 is measured.
In addition to the concern of using cancer cell lines to study p53 signaling, prolonged culturing in vitro is also a factor that needs to be considered. Long-term culturing in vitro brings about new epigenetic markers [26]. Epigenetic modifications have been shown to alter the chromatin binding of p53 [27–28]. With these caveats, translating the results from in vitro cultured cancer cells into the tumor suppressive functions of p53 is extremely challenging [29]. Mapping of p53 signaling in low-passage primary cells or fresh tissue samples will greatly help in obtaining a more physiologically-relevant landscape of the p53 signaling pathway, which will serve as a standard for studying p53 signaling in normal versus cancer cells.
Cell type-specific p53 signaling, an epigenetic story?
Another interesting observation is that different cell types have different p53 dependent responses to stress, as exemplified by ionizing irradiation [30]. In addition, the differences in response to p53 inactivation in different tissues may lead to development of different tumor types. For example, mice harboring p53 loss are more likely to develop lymphomas, while mice with p53 hotspot mutations, such as R172H and R270H, are prone to carcinomas [31–32]. Moreover, tumors arising from different tissues respond to p53 reactivation differently [33–34]. Mouse liver carcinoma cells are subject to senescence upon p53 reactivation [34], while mouse lymphoma cells undergo apoptosis [33]. The cell type-specific p53 response partially results from differential p53 activation in different tissues [30]. Whether and how epigenetic events play a role in cell type-specific p53 responses has not been fully explored. One of the approaches for addressing the epigenetic influence on cellular outcomes is to use the combination of genome-wide and systems biology approaches to decipher how the epigenetic landscapes and cell type-specific factors shape the response to p53 in different cell types. For an initial pilot study, mouse embryonic stem (mES) cells, primary embryonic fibroblast (MEF) cells and neural progenitor cells (NPC) could be suitable models. The p53 signaling pathway in mES stem cells has been profiled using an integrative genome-wide approach [4]. This study revealed a surprising connection between p53 and the Wnt signaling pathway. Interestingly, the induction of the Wnt signaling pathway by p53 is specific to mES cells because the induction is greatly diminished in MEF or NPC cells, although p53 activation is comparable in all cells (Li and Huang, unpublished results). These results suggest that epigenetic events play a role in mES specificity to p53 dependent induction of Wnt. It is worth mentioning that this epigenetic event restricts the p53/Wnt axis to an early developmental stage, as observed in mES cells [35]. At later developmental stages the connection between p53 and Wnt is lost, perhaps as a mechanism for dissociating p53 from the oncogenic effects of the Wnt signaling. A comparison of p53 signaling pathways during ES cell differentiation and among these model cell types will facilitate the understanding why different cell types respond differently to p53 activation or loss of function.
Not every function of p53 has a desirable effect. For example, p53-mediated acute apoptosis of intestinal and hair follicle cells during chemo- or radiotherapy represent a significant side effect that limits the therapeutic potential [36]. A greater understanding of the mechanisms underlying the cell type-specific p53 signaling offers potential opportunities for the development of useful strategies to maximize the tumor killing effect of the therapy while minimizing the side effects.
It is worth noting that mice harboring R172H or R270H mutations in the p53 gene have a different tumor spectrum compared to mice with p53 knockout, suggesting a gain of function of mice with p53 mutations [31–32]. Given the fact that most tumors have p53 mutations rather than p53 deletions, the study of the gain of function of p53 mutants is very relevant to tumor biology. However, the underlying mechanisms of this gain of function are largely unknown. A genome-wide study comparing p53 signaling in wild type, p53 null and p53 mutant cells may provide insights into these mechanisms.
Decoding p53 regulated transcriptome
Early genome-wide studies suggest that p53 does not only regulates the expression of protein-coding RNAs, but also changes the expression of non-coding RNAs [9, 37–39]. These non-coding RNAs include microRNAs (miRNAs) and large intergenic noncoding RNAs (lincRNAs). Using microarrays, the miR-34 family of miRNAs have been shown to be activated by p53 and involved in cell cycle arrest [37, 40–41]. The first identified p53 regulated lincRNA is lincRNA-p21 which is involved in the transcriptional repression mediated by p53 [38]. More recently, another lincRNA called PANDA, located between lincRNA-p21 and the Cdkn1a gene, has been identified as a p53 regulated non-coding RNA [39]. PANDA binds to NF-YA transcriptional factor and inhibits its pro-apoptotic function. Therefore, PANDA is a p53-regulated pro-survival RNA. According to the latest annotation from GENCODE [42], the number of non-coding RNAs has bypassed 10,000. Therefore, it is expected that more non-coding RNAs will be identified as p53 regulated targets in the years to come, as the deep sequencing technology starts to generate more comprehensive transcriptome.
Future directions
Two key requirements for implementation of systems biology are the scale and quality of the measurements [43]. As various genome-wide technologies gradually evolve, the timing for systems biology in p53 function is also evolving. In addition, single-cell imaging technology and mathematical models are being adopted for p53 studies [44]. Because p53 signaling is a complex network, true signals underlying p53 functions are mixed with cellular “noises”. Using the combination of systems biology and genome-wide studies, we should be able to extract the true signals which are required for developing more effective therapeutic strategies in cancer treatment.
Another area that needs additional effort is the study of cell type and tissue specific roles of p53 in a more mechanistic and genome-wide manner. Conventional studies have generated valuable information about cell type-specific roles of p53. However, they provide little mechanistic insight into how different cell types respond to p53 activation. Since different cell types carry the same DNA sequence, epigenetic events may play a role. The NIH Roadmap Epigenomics project has already started to map DNA methylation and histone modifications in various cell types [45]. It will be interesting to see how epigenetic landscape and chromatin structure shape p53 signaling in different cell types. Knowledge gained from these studies may explain how tumors containing the wild type p53 gene escape from p53-mediated surveillance system.
Non-coding RNAs also play important roles in p53-mediated cell cycle arrest, apoptosis and tumorigenesis [39, 46–47]. However, compared to protein-coding RNAs, the regulation of non-coding RNAs by p53 has not been fully explored. As more powerful genome-wide approaches, such as RNA sequencing, are adopted for p53 research, there will likely be great increase in the discovery of p53 regulated non-coding RNAs in the next few years.
Highlights.
Mechanisms underlying the tumor suppressive functions of p53 are still unclear
Binding of p53 at non-promoter regions is not fully understood
Dissection of p53 regulated transcriptome is challenging
Noncoding RNAs join the family of p53 targets
Epigenetic regulation of chromatin binding of p53 has not been fully explored
Acknowledgments
We thank Nan Roche and Lyuba Varticovski for their critical reading and useful comments. J.H.’s laboratory is supported by an intramural fund at the Center of Cancer Research, the National Cancer Institute and the National Institute of Health. The authors would like to apologize to their colleagues whose studies cannot be covered in this short review because of the page limit.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 3.Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 4.Lee KH, Li M, Michalowski AM, Zhang X, Liao H, Chen L, Xu Y, Wu X, Huang J. A genomewide study identifies the Wnt signaling pathway as a major target of p53 in murine embryonic stem cells. Proc Natl Acad Sci U S A. 2010;107:69–74. doi: 10.1073/pnas.0909734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qin H, Yu T, Qing T, Liu Y, Zhao Y, Cai J, Li J, Song Z, Qu X, Zhou P, Wu J, Ding M, Deng H. Regulation of apoptosis and differentiation by p53 in human embryonic stem cells. J Biol Chem. 2007;282:5842–5852. doi: 10.1074/jbc.M610464200. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X, Huang J. Integrative genome-wide approaches in embryonic stem cell research. Integr Biol (Camb) 2010;2:510–516. doi: 10.1039/c0ib00068j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 8.Maxwell SA, Davis GE. Differential gene expression in p53-mediated apoptosis-resistant vs. apoptosis-sensitive tumor cell lines. Proc Natl Acad Sci U S A. 2000;97:13009–13014. doi: 10.1073/pnas.230445997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, Wheeler R, Wong B, Drenkow J, Yamanaka M, Patel S, Brubaker S, Tammana H, Helt G, Struhl K, Gingeras TR. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 10.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q, Ng HH, Ruan Y. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 11.Smeenk L, van Heeringen SJ, Koeppel M, Gilbert B, Janssen-Megens E, Stunnenberg HG, Lohrum M. Role of p53 serine 46 in p53 target gene regulation. PLoS One. 2011;6:e17574. doi: 10.1371/journal.pone.0017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smeenk L, van Heeringen SJ, Koeppel M, van Driel MA, Bartels SJ, Akkers RC, Denissov S, Stunnenberg HG, Lohrum M. Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res. 2008;36:3639–3654. doi: 10.1093/nar/gkn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RK, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 16.Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 17.Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, McKinnon PJ, Cleveland JL, Zambetti GP. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 18.Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, Kenzelmann Broz D, Basak S, Park EJ, McLaughlin ME, Karnezis AN, Attardi LD. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang D, Brady CA, Johnson TM, Lee EY, Park EJ, Scott MP, Attardi LD. Full p53 transcriptional activation potential is dispensable for tumor suppression in diverse lineages. Proc Natl Acad Sci U S A. 2011;108:17123–17128. doi: 10.1073/pnas.1111245108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Mizuno H, Spike BT, Wahl GM, Levine AJ. Inactivation of p53 in breast cancers correlates with stem cell transcriptional signatures. Proc Natl Acad Sci U S A. 2010;107:22745–22750. doi: 10.1073/pnas.1017001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaked H, Shiff I, Kott-Gutkowski M, Siegfried Z, Haupt Y, Simon I. Chromatin immunoprecipitation-on-chip reveals stress-dependent p53 occupancy in primary normal cells but not in established cell lines. Cancer Res. 2008;68:9671–9677. doi: 10.1158/0008-5472.CAN-08-0865. [DOI] [PubMed] [Google Scholar]
- 24.Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 25.Kaeser MD, Iggo RD. Chromatin immunoprecipitation analysis fails to support the latency model for regulation of p53 DNA binding activity in vivo. Proc Natl Acad Sci U S A. 2002;99:95–100. doi: 10.1073/pnas.012283399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soria C, Estermann FE, Espantman KC, O’Shea CC. Heterochromatin silencing of p53 target genes by a small viral protein. Nature. 2010;466:1076–1081. doi: 10.1038/nature09307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka T, Ohkubo S, Tatsuno I, Prives C. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell. 2007;130:638–650. doi: 10.1016/j.cell.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 30.MacCallum DE, Hupp TR, Midgley CA, Stuart D, Campbell SJ, Harper A, Walsh FS, Wright EG, Balmain A, Lane DP, Hall PA. The p53 response to ionising radiation in adult and developing murine tissues. Oncogene. 1996;13:2575–2587. [PubMed] [Google Scholar]
- 31.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, El-Naggar AK, Lozano G. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 33.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 34.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li M, Huang J. A new puzzling role of p53 in mouse embryonic stem cells. Cell Cycle. 2010;9:1669–1670. doi: 10.4161/cc.9.9.11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, Gudkov AV. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 37.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007 doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Kong B, Langerod A, Borresen-Dale AL, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ, Chang HY. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 42.Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, Rossier C, Ucla C, Hubbard T, Antonarakis SE, Guigo R. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl 1):S4, 1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macilwain C. Systems biology: evolving into the mainstream. Cell. 2011;144:839–841. doi: 10.1016/j.cell.2011.02.044. [DOI] [PubMed] [Google Scholar]
- 44.Loewer A, Batchelor E, Gaglia G, Lahav G. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell. 2010;142:89–100. doi: 10.1016/j.cell.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, Farnham PJ, Hirst M, Lander ES, Mikkelsen TS, Thomson JA. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:R152–161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]