Abstract
HIV-1 Vif, an accessory protein in the viral genome, performs an important role in viral pathogenesis by facilitating the degradation of APOBEC3G, an endogenous cellular inhibitor of HIV-1 replication. In this study, intrinsically disordered regions are predicted in HIV-1 Vif using sequence based algorithms. Intrinsic disorder may explain why traditional structure determination of HIV-1 Vif has been elusive, making structure based drug design impossible. To characterize HIV-1 Vif’s structural topology and to map the domains involved in oligomerization we used chemical cross-linking, proteolysis and mass spectrometry. Cross-linking showed evidence of monomer, dimer and trimer species via denaturing gel analysis and an additional tetramer via western blot analysis. We identified 47 unique linear peptides and 24 (13 intramolecular; 11 intermolecular) non-contiguous, cross-linked peptides, among the noncross-linked monomer, cross-linked monomer, cross-linked dimer and cross-linked trimer samples. Almost complete peptide coverage of the N-terminus is observed in all samples analyzed, however reduced peptide coverage in the C-terminal region is observed in the dimer and trimer samples. These differences in peptide coverage or “protections” between dimer and trimer indicate specific differences in packing between the 2 oligomeric forms. Intramolecular cross-links within the monomer suggest that the N-terminus is likely folded into a compact domain, while the C-terminus remains intrinsically disordered. Upon oligomerization, as evidenced by the intermolecular cross-links, the C-terminus of one Vif protein becomes ordered by wrapping back on the N-terminal domain of another. In addition, the majority of the intramolecular cross-links map to regions that have been previously reported to be necessary for viral infectivity. Thus, this data suggests HIV-1 Vif is in a dynamic equilibrium between the various oligomers potentially allowing it to interact with other binding partners.
Keywords: HIV-1 Vif, Oligomerization, Intrinsic Disorder, Mass Spectrometry, Topology
INTRODUCTION
The Human Immunodeficiency Virus type-1 accessory protein, viral infectivity factor or Vif, is a 23 kDa highly basic protein (pI 10.7) that is conserved in all lentiviruses except equine anemia infectious virus. Over the last several years, the function and interactions of HIV-1 Vif have been extensively investigated 1–4. HIV-1 Vif has potential interactions with many viral and cellular macromolecules: APOBEC3G, an endogenous cytidine deaminase 5, 6, along with other family members such as APOBEC3F 7, 8, HIV-1 Gag 9, HIV-1 protease 10, viral RNA 11, 12, and 2 proteins in a cullin-RING ligase complex: Cullin5 13–15 and elongin C 16–18 . When HIV-1 Vif is absent or non-functional, post-infection viral replication and viral production in “non-permissive” primary CD4 T-cells is dramatically reduced. This reduction or inhibition in viral production is likely due to the irreversible effects of the cellular enzyme APOBEC3G found in nonpermissive cells, which inhibits viral replication possibly through its deaminase activity or by preventing the build-up of reverse transcripts 6, 19–23. HIV-1 Vif binds APOBEC3G and targets it for proteosomal degradation through a cullin-RING ligase complex which includes interactions with elongin C and Cullin5 in the Cullin5-elongin BC complex and may also block APOBEC3G’s translation 13, 15, 16, 24–28. Specifically, HIV-1 Vif’s Socs box motif interacts with elongin C 16–18 and an HCCH Zinc binding motif in HIV-1 Vif binds Cullin5 13–15. Therefore, it is likely that HIV-1 Vif’s interactions with other macromolecules are important to its function in suppressing the affects of APOBEC3G.
The lack of or slowing of disease progression to AIDS in HIV-1-infected patients has been correlated with mutations in the HIV-1 Vif gene 29–31. Therefore, blocking HIV-1 Vif’s ability to inhibit APOBEC3G might allow the anti-viral effects of APOBEC3G to prevent the spread of HIV-1 infection. Inhibiting HIV-1 Vif function could therefore potentially suppress viral replication. Thus, HIV-1 Vif is considered a viable therapeutic target either for structure-based inhibitors or as a potential vaccine candidate.
To develop HIV-1 Vif into a viable therapeutic target, a clear understanding is needed of the molecular mechanisms of HIV-1 Vif function, including its oligomerization and interactions with putative functional partners. To date, little biochemical data is available on HIV-1 Vif and, more importantly, no structural data. This lack of data is partially due to an inability to express high levels of soluble recombinant protein using either prokaryotic or baculovirus expression systems. However, as we will show, regions of HIV-1 Vif are very likely to be intrinsically disordered. Intrinsically disordered proteins have extensive regions that lack a fixed tertiary structure 32, and they are characterized by a high net charge and low overall hydrophobicity 33, 34. In addition, proteins with regions known to be disordered tend to bind a large and diverse set of proteins and nucleic acids 34, 35. The above properties of intrinsically disordered proteins are also known characteristics of HIV-1 Vif.
Although a lack of structural or biochemical data for HIV-1 Vif exists, homo-oligomerization has been implicated in its function. A putative oligomerization domain has been found to map to amino acid residues 151–164. If this region is deleted or mutated, HIV-1 Vif function is significantly reduced 36. More specifically, residues 161–164, which map to the PPLP domain of HIV-1 Vif, have been shown to be necessary for HIV-1 Vif oligomerization 37. In addition, peptides corresponding to the region 153–171 drastically reduced the number of HIV-1 Vif oligomers observed. Adding these short peptides also inhibits HIV-1 replication in nonpermissive cells, presumably by competing with the functionally necessary step of oligomerization in HIV-1 Vif 37. Taken together, these data suggest that the oligomerization domain in HIV-1 Vif is residues 151–171.
In this study, intrinsically disordered regions of HIV-1 Vif have been predicted using sequence analyses algorithms. These regions may explain why traditional structure determination, and thus structure based drug design, has been elusive. Mass spectrometry (MS) is an ideal method to obtain structural information at the level of individual amino acids for proteins like HIV-1 Vif, which are difficult to express in large quantities. In contrast to other biophysical and structural techniques, MS uses only nanogram amounts of protein, as opposed to milligrams 38–44. Using a novel approach involving short covalent chemical cross-linkers, proteolysis and various forms of MS techniques, we have identified HIV-1 Vif oligomer (protein-protein) interactions. By determining a series of these interactions, we also obtained a low-resolution structural map of HIV-1 Vif. For the first time, we report data on the structural topology of HIV-1 Vif, map the domains involved in HIV-1 Vif oligomerization, and propose a mechanism in which Vif may bind other proteins.
MATERIALS AND METHODS
HIV-1 Vif
This protein, which had been expressed and purified by Immunodiagnostics, Inc., Woburn, MA, was obtained from the AIDS Research and Reagent Program or directly from Immunodiagnostics, Inc. According to the manufacturer’s product sheet, the HIV-1 Vif protein was strain HXB2, expressed in Escherichia coli with a 6X His tag that is cleaved under native conditions, stored in 50mM Tris, pH 8.0, 150mM sodium chloride, and was >99% pure. Several different lots of HIV-1 Vif protein were obtained and all gave consistent results.
APOBEC3G
This protein, which had been expressed and purified by Immunodiagnostics, Inc., Woburn, MA, was obtained from the AIDS Research and Reagent Program. According to the manufacturer’s product sheet, the protein was expressed in E. coli with a 6X His tag and purified to >95% purity via preparative SDS-PAGE and stored in PBS, 30% glycerol, 0.1% sarcosyl, or it was expressed in baculovirus with a T-tag fusion protein and purified to >95% purity using (NH4)2SO4 fractionation and immuno-affinity chromatography and finally stored in 20mM Tris, pH 8.0, 0.1M sodium chloride, 0.01% sarcosyl.
Co-Immunoprecipitation
HIV-1 Vif was immunocaptured on an EZview™ Red Protein A Affinity gel (Sigma) using an anti-Vif antibody (TG001 obtained from the NIH AIDS Research and Reagent Program). The Protein A Affinity gel with HIV-1 Vif immobilized was washed with 20 mM Tris, pH 8.0, 0.5 M sodium chloride buffer and incubated with APOBEC3G overnight at 4°C. After APOBEC3G incubation, the beads were washed again and the HIV-1 Vif-APOBEC3G complex was eluted from the gel via boiling. The samples were then run on a 16% Tris-Glycine SDS PAGE gel and western blotted using an antibody for APOBEC3G. The same protocol was followed for APOBEC3G, where APOBEC3G is immunocaptured on the Protein A Affinity gel using an anti-APOBEC3G antibody and western blotted using a HIV-1 Vif antibody (TG001).
Cross-linking of HIV-1 Vif
The “zero length” cross-linking agent, EDC (1-ethyl-3-[3-dimethylaminopropyl] carbodiamide; Pierce), and sulfo-NHS (N-hydroxysuccinimide; Pierce) were prepared freshly as 0.1 M stock solutions in deionized water. The cross-linking reaction was performed in solution as described 45. The HIV-1 Vif protein (5 mg/ml) was diluted in 50 µl activation buffer (0.1 M MES, pH 6.0, 0.5 M NaCl) to 0.66 mg/ml, 1 µl of the EDC stock solution was added to give a final EDC concentration of 2 mM, and 2.5 µl of sulfo-NHS stock solution was added to give a final concentration of 5 mM. To avoid random protein-protein interactions, cross-linking was performed in dilute solution. Preliminary experiments determined that the minimal time required for optimal cross-linking in solution was 15 minutes. Thus, the cross-linking reaction was conducted at room temperature for 15 minutes and quenched by adding SDS PAGE gel loading dye with 2-mercaptoethanol. The resulting mixture of HIV-1 Vif oligomers was resolved on a 16% SDS-PAGE gel along with a noncross-linked HIV-1 Vif protein, allowing each species to be individually proteolyzed. Similar cross-linking results were obtained using multiple lots of the HIV-1 Vif protein. The density of the Coomassie blue-stained gel bands was determined using UVP Bio-imaging System EPI Chemi II Dark Room and LabWorks 4.0 software. Cross-linking experiments can only be performed in vitro using purified proteins.
HIV-1 Vif-specific oligomers were identified by western blot analysis using a monoclonal antibody to HIV-1 Vif (TG001) obtained from the NIH AIDS Research and Reagent program. Proteins were transferred from a SDS PAGE gel to nitrocellulose membrane at 200 mAMPS for 2 hours at 4°C. After transfer, the membrane was treated with blocking buffer (10 mM Tris-HCl, pH 8.0, 0.3 M NaCl, 0.25% Tween, and 5% milk) for approximately 4 hours at room temperature. After blocking, the membrane was incubated overnight at 4°C with a 1:20,000 dilution of the HIV-1 Vif monoclonal antibody in blocking buffer. The membrane was then washed 6 times with blocking buffer without milk and incubated at room temperature for 1 hour with goat anti-mouse secondary antibody (1:40,000). The membrane was then washed 6 times and developed using the Pierce Supersignal ECL kit and a Kodak X-Omat machine.
Preparation of Samples for Mass Spectrometry (MS)
Four samples of HIV-1 Vif (the noncross-linked monomer band and the cross-linked monomer, dimer, and trimer) were in-gel digested in preparation for MALDI-TOF MS, LC-ion trap-MS, and LC-QTof-MS analyses in the presence or absence of heavy water (18O) 46, 47. Heavy water (18O) was used in order to label peptides and cross-links. Therefore, peptides digested in the presence of 18O water will be 4 mass units larger than those digested in the presence of 16O water and cross-linked peptides will be 8 mass units larger. Tryptic and chymotryptic in-gel digestion was performed using the Calbiochem ProteoExtract™ All-in-one Trypsin Digestion Kit according to the manufacturer’s protocol or as described 48. Briefly, gel bands containing each oligomer were excised and cut into small pieces. One-half was digested in an Eppendorf tube in the presence of 16O water, and the other half was digested in an Eppendorf tube in the presence of 18O water. The gel pieces were washed 2 times with wash buffer (50 mM ammonium bicarbonate in 50% ethanol) at room temperature. Gel slices were shrunk in 100% ethanol, and incubated for 1 hour at 56°C with 50 mM ammonium bicarbonate containing DTT. After cooling to room temperature, the gel slices were incubated with iodoacetamide for 30 min at room temperature in the dark, washed, shrunk, and dried. Dried gel slices were digested overnight with trypsin (1 µl of 8 ng/µl) at 37°C in the presence of either 16O or 18O water. Peptides were extracted using 50 mM ammonium bicarbonate and 50% N,N-dimethyl formamide and evaporated to dryness using a SpeedVac. Dried peptides were dissolved in 2% acetonitrile, 0.1% TFA. Finally, peptides digested in the presence of 16O water were added to their corresponding half sample digested in the presence of 18O water.
Peptide Analysis using Reflectron MALDI-TOF MS
The peptides extracted from each chosen band were purified using an Omix C18 ZipTip reverse-phase cleanup pipette tip. The ZipTip was prewashed with 50% acetonitrile and equilibrated with 0.1% trifluoroacetic acid (TFA). The peptides were aspirated 5 times through the ZipTip to bind them. The ZipTip was then washed by repeated aspiration of 10 µl of 0.1% TFA, and the peptides were eluted by repeated aspiration of 5 µl of 50% acetonitrile containing 0.1% TFA. The eluate (1 µl) was spotted onto a MALDI-TOF MS target plate with 1 µl of 10 mg/ml α-cyano-4-hydroxycinnamic acid matrix (α-cyano) in 50% acetonitrile, 0.1% TFA, and air dried. Analyses were performed using a Waters MALDI L/R MALDI-TOF mass spectrometer in the reflectron mode to acquire spectra from m/z 400 – 6000. Each spectrum was the sum of 500 laser shots and was lock-mass calibrated using a mixture of synthetic peptides in the lock-mass well. MALDI-TOF heavy water experiments were repeated in triplicate.
Liquid Chromatography Mass Spectrometry (LC-ion trap-MS) of Peptide Fragments
A ThermoFinnigan LTQ linear quadrupole ion trap MS equipped with a Finnigan Surveyor HPLC pumping system was used to perform capillary HPLC nanoelectrospray (NESI) MS and data-dependent MS/MS analyses of the HIV-1 Vif digests. Samples (1–5 µl) were injected into a 35 µl/min flow of 2% acetonitrile in 0.1% formic acid onto a 300 µm × 5 mm C-18 Pepmap trapping column (LC Packings) using a manual NanoPeak injection valve (UpChurch Scientific). A 30 min solvent gradient from 5 to 50% acetonitrile in 0.1% formic acid was then passed in the reverse direction at 200 nl/min through the trapping column. The trapping column eluate was passed through a 75 µm ID × 10 cm ProteoPrep II packed PicoFrit HPLC column/electrospray emitter (New Objectives, Inc.) installed in the NESI source of the mass spectrometer. Positive ion ESI MS were acquired during the elution with one full scan MS followed by data-dependent MS/MS product ion spectra (35% normalized collision energy) of the 10 most intense ions from the full scan MS. The NESI source was operated with the source at 1.8 kV and capillary at 250°C.
LC-QTof-MS Mass Spectrometry of Peptide Fragments (Q-Tof)
A Waters Q-Tof Premier Mass Spectrometer equipped with a Waters CapLC HPLC pumping system was used to perform capillary nanoelectrospray (NESI) MS and data-dependent MS/MS analyses of the HIV-1 Vif digests. Samples (1–5 µl) were injected into a 12 µl/min flow of 2% acetonitrile in 0.1% formic acid onto a CAPTRAP (Michrome Bioresources, Inc.) trapping column using a manual NanoPeak injection valve. A 30-min solvent gradient from 2 to 98% acetonitrile in 0.1% formic acid was then passed through the trapping column in the reverse direction at 200 nl/min. The trapping column eluate was passed through a 75 µm ID × 10 cm ProteoPrep II-packed PicoFrit column/electrospray emitter (New Objectives, Inc.) installed in the NESI source of the mass spectrometer. Data-dependent positive ion NESI MS were acquired during the elution with one full scan MS followed by MS/MS product ion spectra of the 4 most intense ions from the full scan MS lock mass. The NESI source was operated with the source at 2.7 kV and capillary at 200°C.
Data Analysis
Peptides were identified from molecular weight and MS/MS sequence data using SEQUEST and analyzed using PAWS (Protein Analysis Worksheet), GPMAW (General Protein Mass Analysis for Windows), and MassLynx software tools 43. PAWS was used to match peptide MH+ ions observed in the MALDI-TOF MS data to sequences from HIV-1 Vif, and GPMAW was used to similarly match sequences from both normal peptides and cross-linked peptides observed in MALDI-TOF MS and LCMS (ion trap and QTof) analyses. MassLynx was used to analyze MALDI-TOF spectra and to identify 16O and 18O ion pairs from linear and cross-linked peptides. Mass identity assignments of cross-links were made using the following criteria: 1) Trypsin-specific cleavages (R or K) were considered. 2) Since cross-linking reactions form amide bonds between lysine side-chain amine groups and glutamic acid or aspartic acid side-chain carboxyl groups, all identified cross-linked peptides must contain at least one Lys residue in one of the peptides and at least one Glu or Asp residue in the other. 3) Cross-links were assigned only if the ions were present in cross-linked samples and absent in the noncross-linked control. 4) Intramolecular cross-linked peptides were identified by their presence in the monomer cross-link sample. 5) Intermolecular cross-links were inferred when cross-links were only seen in the cross-linked dimer or cross-linked trimer but not in the monomer 49. 6) A crosslink had to be present in at least 2 out of 3 experiments to be accepted. 7) In heavy water experiments when the protein is cleaved with trypsin, 2 atoms of 18O are incorporated into the carboxy terminus of each peptide, resulting in a mass shift of +4 Da for a linear peptide and +8 Da for a cross-linked peptide. Therefore, linear peptides were assigned if a mass shift of +4 Da was observed and cross-linked peptides were assigned if a mass shift of +8 Da was observed 46, 47.
Although high molecular weight cross-links are prone to less accurate mass matches, as the signal-to-noise ratio may affect mass accuracy, particular care was taken to ensure the accuracy of the identified cross-links (Supplemental Table II). These cross-links were only identified if they were reproduced in more than one sample and had the appropriate heavy water label. The majority of the data is within an acceptable parts-per-million (ppm) range; however the few values that diverge slightly meet the other criteria for being cross-linked and are still less than one Dalton different from the theoretical mass.
Structure Predictions
Intrinsically disordered regions of HIV-1 Vif were predicted from PONDR®, Predictors of Natural Disordered Regions, which utilizes sequence based algorithms 50, 51. Access to PONDR® was provided by Molecular Kinetics (6201 Las Pas Trail-Ste 160, Indianapolis, IN 46268; 317-280–8737; main@molecularkinetics.com). VL-XT is copyright©1999 by the WSU Research Foundation all rights reserved. PONDR® is copyright©2004 by Molecular Kinetics, all rights reserved. In addition to the prediction of intrinsic disorder, a consensus secondary structure prediction was obtained from the Pole BioInformatique Lyonnais website using Network Protein Sequence Analysis 52.
RESULTS
Co-Immunoprecipitation of HIV-1 Vif with APOBEC3G
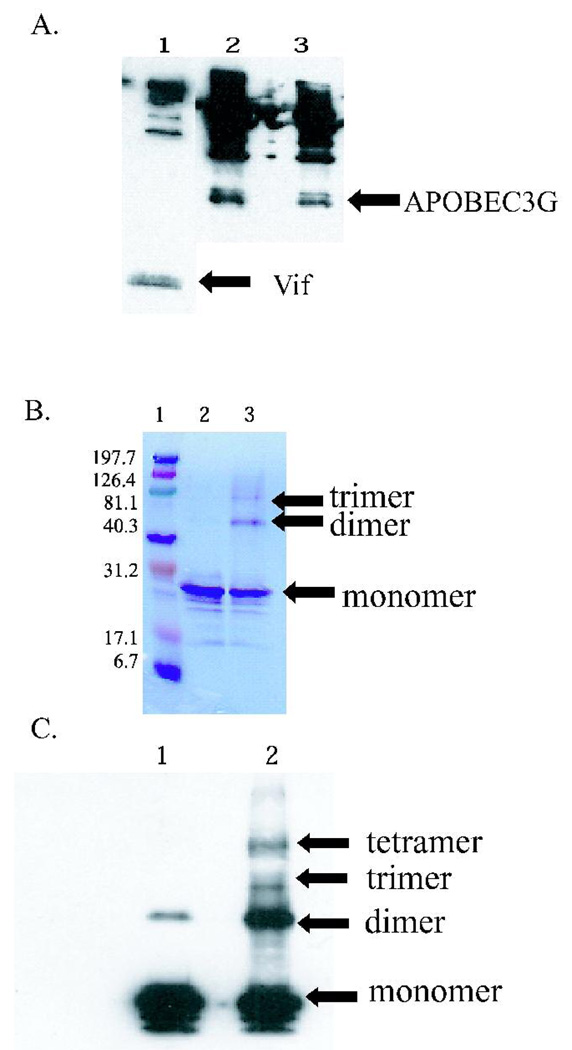
The HIV-1 Vif protein obtained from Immunodiagnostics was determined to be biologically functional by co-immunoprecipitating it with its known binding partner, APOBEC3G. HIV-1 Vif binding was detected both to immobilized APOBEC3G expressed in either E. coli or baculovirus, and APOBEC3G binding to immobilized HIV-1 Vif (1A). These results suggest that the HIV-1 Vif protein obtained was in a biologically relevant form and thus folded and functional.
Figure 1. HIV-1 Vif is functional and can form higher order oligomers.
(A) Co-immunoprecipitation of HIV-1 Vif with APOBEC3G. Vif (APOBEC3G) was immobilized on Protein A affinity beads, incubated overnight with APOBEC3G (Vif), and eluted via boiling. Lane 1: Immobilized APOBEC3G interacts with HIV-1 Vif. Lane 2: Immobilized HIV-1 Vif interacts with baculovirus-expressed APOBEC3G. Lane 3: Immobilized HIV-1 Vif interacts with E. coli-expressed APOBEC3G. The higher molecular weight bands observed are the result of heavy chains, light chains, and Protein A background routinely observed in immunoprecipitations. (B) SDS PAGE analysis of HIV-1 Vif cross-links. Lane 1: Molecular weight markers (kDa). Lane 2: Noncross-linked HIV-1 Vif control that is predominately monomer (23 kDa). Lane 3: EDC cross-linked HIV-1 Vif, with evidence for a dimer (46 kDa) and trimer (69 kDa). (C) Western blot of HIV-1 Vif cross-links. Lane 1: A noncross-linked HIV-1 Vif control that is predominantly monomer, with a small amount of dimer. Lane 2: EDC cross-linked HIV-1 Vif, with evidence for dimer, trimer and tetramer forms.
HIV-1 Vif forms higher order oligomers in vitro
To isolate oligomeric forms of HIV-1 Vif, chemical cross-linking experiments were performed. Higher order oligomers of HIV-1 Vif were observed in vitro using the heterobifunctional zero-length cross-linker, EDC. Denaturing PAGE analysis of cross-linked HIV-1 Vif showed the presence of cross-linked monomer (24 kDa), cross-linked dimer (48 kDa), and cross-linked trimer (72 kDa) forms of the HIV-1 Vif protein (Figure 1B, lane 3). The oligomers of HIV-1 Vif were confirmed via western blotting and in addition to the monomer, dimer and trimer forms, a cross-linked tetramer was observed (Figure 1C, lane 2). Densometric analysis of the Coomassie-stained SDS PAGE gel suggests that approximately 46% of the reaction product is cross-linked monomer, 39% is cross-linked dimer, and 15% is cross-linked trimer (Figure 1B, lane 3). A noncross-linked sample of HIV-1 Vif, run as a control, showed a band consistent with a HIV-1 Vif monomer (Figure 1B, lane 2). In the western blot of the noncross-linked monomer, a small amount of HIV-1 Vif dimer is present, which is sometimes observed for oligomeric proteins under reducing and denaturing conditions (Figure 1C, lane 1). The cross-linked monomer, dimer and trimer were analyzed using mass spectrometry to gain insight into HIV-1 Vif’s topology as well as to map the oligomerization domains.
Structure Predictions
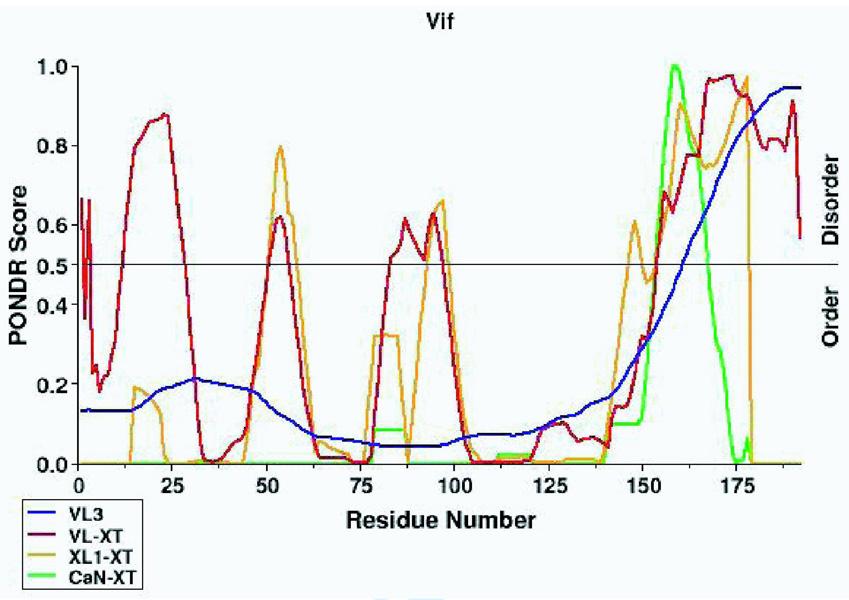
Prior to analysis of the cross-linked HIV-1 Vif samples by mass spectrometry, PONDR®, Predictors of Natural Disordered Regions, was used to determine regions of intrinsic disorder in HIV-1 Vif by submitting the HXB2 HIV-1 Vif sequence 50,51. This program uses four different predictor routines to suggest regions of intrinsic disorder. Only one of the four predictors, VL-XT, predicted the N-terminus of HIV-1 Vif to be disordered, and two of the predictors, VL-XT and XL1-XT, predicted two short regions (possibly extended loops) between residues 50–63 and 87–100 to be disordered. However, all four of the predictors, unanimously predicted the C-terminus to be disordered (Figure 2A). Therefore, this suggests the C-terminus of HIV-1 Vif is intrinsically disordered. In addition to predicting regions of intrinsic disorder, HIV-1 Vif’s secondary structure was predicted using the Pole BioInformatique Lyonnais Network Protein Sequence Analysis (NPS) secondary structure consensus prediction program because intrinsically disordered proteins often do have defined secondary structure (Figure 4A) 52. The majority of HIV-1 Vif’s secondary structure is predicted to be random coil, however, there is predicted secondary structure: the ordered N-terminus is predicted to consistent of mostly beta-sheets and one alpha helix and the disordered c-terminus is predicted to consistent of mostly alpha helixes.
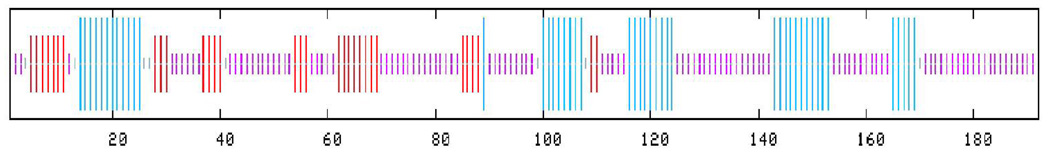
Figure 2. Analysis of HIV-1 Vif using reflectron MALDI-TOF-MS.
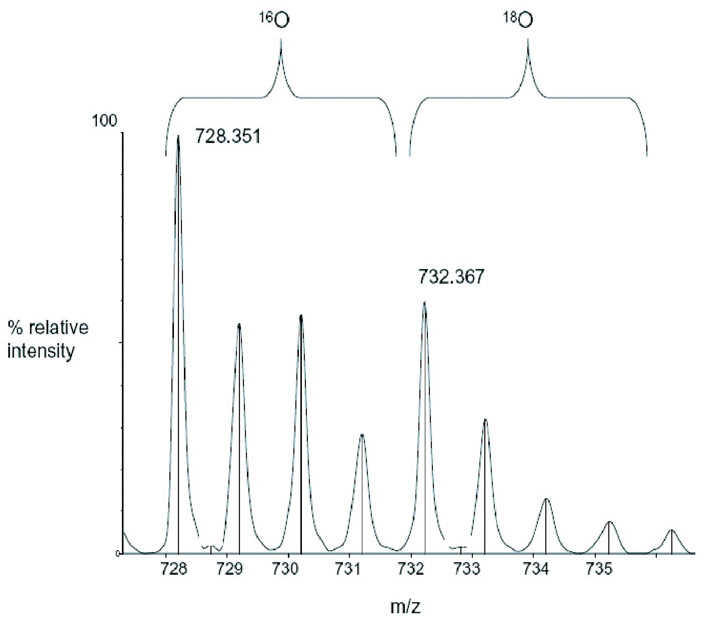
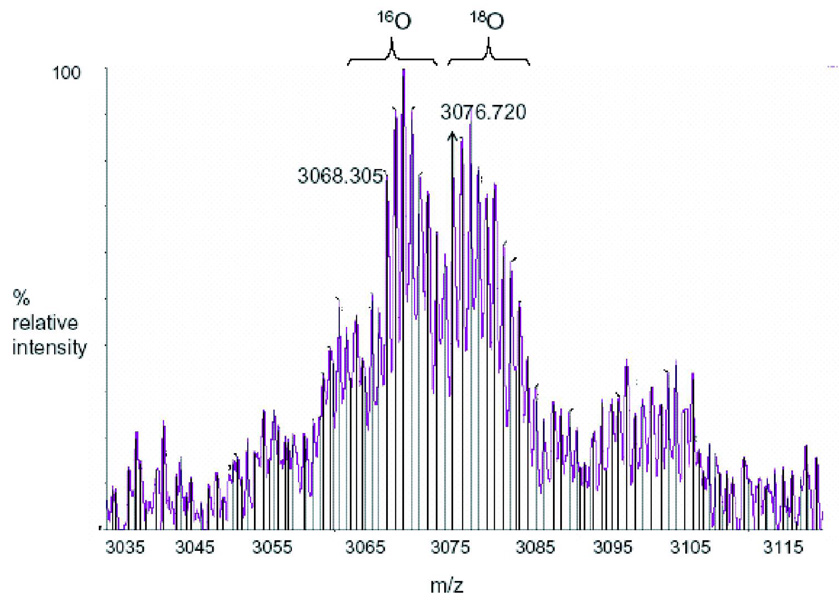
The mass spectrometry data presented here represents an example of the spectra of a peptide (Figure 2B) and a cross-link (Figure 2C) in Tables 1 and 2, respectively. m/z represents the mass-to-charge ratio of the ion and in the case of a singly charged ion represents the peptides MH+ (molecular weight). (A) Predicted regions of intrinsic disorder for HIV-1 Vif using PONDR®, Predictors of Natural Disordered Regions 50,51. The four colors indicate four separate predictor algorithms: blue VL3, red VL-XT, yellow XL1-XT, and green CaN-XT. (B) An unlabelled peptide at m/z 728.351 and its labeled (18O) counterpart 4Da larger at m/z 732.367. This peptide corresponds to residues 37–41 in the protein. (C) An unlabelled intermolecular cross-linked peptide at m/z 3068.305 and its labeled (18O) counterpart 8Da larger at m/z 3076.720. This region corresponds to a cross-link between peptides 51–63 and 159–173 and is only seen in the dimer and trimer. The arrow indicates the beginning of the +8 Da ion series.
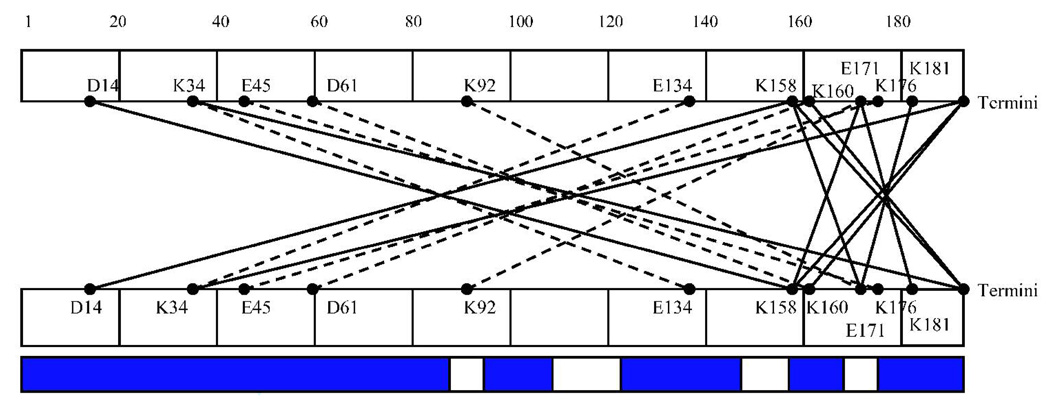
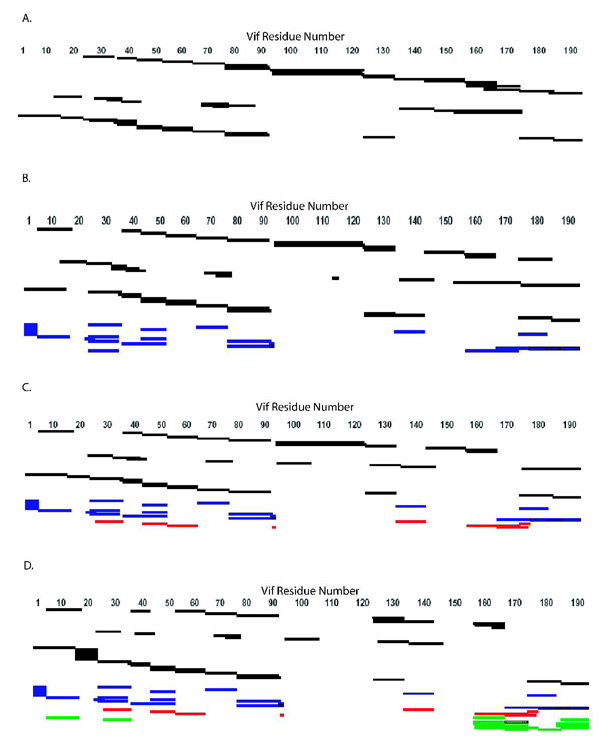
Figure 4. Cross-links of HIV-1 Vif.
Schematic diagram of cross-links observed in different oligomeric states of HIV-1 Vif that were analyzed by MALDI-TOF and heavy water labeling. (A) Consensus secondary structure prediction for HIV-1 Vif 52. Red: beta-sheet. Blue: alpha-helix. Purple: random coil (B) Cross-links observed in the monomer sample. (C) Cross-links observed in the dimer sample and shown going from both the N- to C-terminal and from the C-to N-terminal regions. (D) Cross-links observed in the trimer and shown going from both the N-to C-terminal and from the C- to N-terminal regions. Cross-linking: Dotted lines indicate that cross-links appeared in both the dimer and trimer samples. The blue bars represent peptide coverage for each oligomeric state.
Identification of Noncross-linked Linear Peptides
Detailed examination of the linear peptide coverage can elucidate the regions of HIV-1 Vif that are protected from protease digestion upon folding and oligomerization 53. Because the cross-linking reaction is not 100% efficient, some peptides may be observed with diminished intensity in regions that are important for oligomerization. Noncross-linked HIV-1 Vif, cross-linked monomer, cross-linked dimer, and cross-linked trimer were excised from gels, trypsin digested separately in the presence of 16O and 18O water, and analyzed using reflectron MALDI-TOF MS, LC-ion trap-MS, and LC-QTof-MS. Gel bands were digested in the presence of 18O water in order to label peptides and cross-links for identification. During trypsin cleavage two oxygens are added to the C-terminus of each peptide, therefore peptides digested in the presence of 18O water will be 4 mass units larger than peptides digested in 16O water and cross-links will be 8 mass units larger than their unlabelled counterparts. For example, Figure 2B shows a region of the MALDI-TOF spectrum representing a peptide where an MH+ ion (m/z 728.351) is seen for the unlabelled peptide (16O), and 4 Da higher, an MH+ ion (m/z 732.367) is seen for the labeled peptide (18O). The ion at m/z (mass-to-charge ratio) 728.351 corresponds to a theoretical mass of 728.351 for the peptide 37–41. All the experimental peptides identified via MALDI-TOF were identified in a similar manner to above, and their parts per million and mass difference from the theoretical mass, are shown in Supplemental Table I.
All three mass spectrometry techniques mentioned above and both trypsin and chymotrypsin digests were used to identify peptides that provide 100% sequence coverage for the noncross-linked sample, 99% coverage for the cross-linked monomer, 95% for the dimer and 83% for the trimer (Figure 3 and Figure 4). The difference in coverage (particularly in the C-terminus) between monomer, dimer and trimer suggests that the dimer and trimer samples have more protected areas, which indicates conformational change due to oligomerization. Analysis of the MALDI-TOF MS, LC-ion trap-MS, and LC-QTof-MS data shows complete peptide sequence coverage for all samples analyzed from residues 1–91. The monomer, dimer and trimer samples have an area of protection around residues 92–94 (KKR), which may be due to a conformational change or possibly due to trypsin cleavage at each site, creating single residue peptides. The trimer sample, but not the monomer or dimer sample, has an area of protection from residues 107–121 (IHLYYFDCFDSAIR), suggesting that these residues may be involved in trimerization. This region in the trimer is also well conserved and contains several aromatic residues and one of the 2 cysteines in the protein suggesting this is a functional domain in the trimer. From residues 122–147, there is almost complete peptide coverage in all samples analyzed. The peptide coverage in the C-terminal domain of the dimer and trimer is reduced, or in other words the “protection” is increased, suggesting that this region is involved in oligomerization of the protein, consistent with a previous report that the oligomerization domain is from residues 151–171 36, 37. For example, the trimer sample shows an area of protection from residues 148–157 (LALAALITPK), and a region of protection is seen from 169–173 (LTEDR) in the dimer and trimer. In addition, the sample digested with chymotrypsin contains a peptide in the noncross-linked and cross-linked monomers, corresponding to residues 151–174. This peptide is lost in the dimer and trimer, supporting the importance of this region for oligomerization. The different areas of protection observed between the dimer and trimer samples suggest changes in the details of interactions as the protein oligomeric state transitions from dimer to trimer (Figure 3 and Table I). Finally, the “protection” observed in the C-terminus of the dimer and trimer sample are consistent with these regions becoming more ordered upon oligomerization.
Figure 3. Peptide Sequence Coverage Map from MALDI-TOF, LC-ion trap-MS, and LC-QTof-MS Data.
Coverage maps of tryptic and chymotryptic peptides identified from MALDI-TOF, LC-ion trap-MS, and LC-QTof-MS, and tryptic cross-links identified from MALDI-TOF. Peptides and cross-links were identified via comparison of the experimental molecular weight with a list of theoretical molecular weights calculated from ProteinProspector or GPMAW as well as by a corresponding ion 4Da larger for a peptide and 8Da larger for a cross-link. (A) Noncross-linked. (B) Monomer. (C) Dimer. (D) Trimer. Identified regions are those that are cross-linked to each other and those that are protected. Black: peptides; blue: intramolecular cross-links; red: cross-links in the dimer and trimer; green: trimer cross-links.
Table I. Peptide analysis of HIV-1 Vif.
Peptides fragments were identified by mass spectrometry (MALDI-TOF-MS, LC-ion trap-MS, or LC-QTof-MS [Q-Tof] indicated by MS) from uncross-linked (UnX), cross-linked monomer (Mono), dimer, or trimer and digested with either trypsin (T) or chymotrypsin (C). Italics indicate identification by Q-Tof; bold indicates identification by both Q-Tof and LC-ion trap-MS.
| UnX-T | UnX-T- MS |
UnX-C | Mono- T |
Mono-T- MS |
Mono-C | Dimer-T | Dimer-T- MS |
Dimer- C |
Trimer-T | Trimer-T- MS |
Trimer- C |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1–15 | 1–15 | 1–15 | 1–15 | ||||||||
| 5–19 | 5–19 | 5–19 | |||||||||
| 12–21 | 12–21 | ||||||||||
| 16–22 | 16–22 | 16–22 | |||||||||
| 23–26 | 23–26 | 23–26 | 23–26 | ||||||||
| 22–30 | 22–30 | 22–30 | |||||||||
| 23–34 | |||||||||||
| 23–36 | 23–36 | 23–36 | 23–36 | ||||||||
| 27–41 | |||||||||||
| 30–39 | |||||||||||
| 31–39 | 31–39 | ||||||||||
| 31–40 | 31–40 | ||||||||||
| 35–41 | 35–41 | 35–41 | 35–41 | ||||||||
| 37–41 | 37–41 | 37–41 | 37–41 | 37–41 | 37–41 | 37–41 | 37–41 | ||||
| 39–44 | 39–44 | 39–44 | 39–44 | ||||||||
| 42–50 | 42–50 | 42–50 | 42–50 | 42–50 | 42–50 | 42–50 | |||||
| 42–63 | 42–63 | 42–63 | 42–63 | ||||||||
| 51–63 | 51–63 | 51–63 | 51–63 | 51–63 | 51–63 | 51–63 | 51–63 | ||||
| 64–77 | 64–77 | 64–77 | 64–77 | 64–77 | 64–77 | 64–77 | 64–77 | ||||
| 70–79 | 70–79 | 70–79 | 70–79 | ||||||||
| 70–89 | |||||||||||
| 71–79 | 71–79 | 71–79 | |||||||||
| 78–90 | 78–90 | 78–90 | 78–90 | 78–90 | 78–90 | 78–90 | 78–90 | ||||
| 78–91 | 78–91 | 78–91 | 78–91 | ||||||||
| 78–92 | |||||||||||
| 93–122 | |||||||||||
| 94–121 | 94–121 | 94–121 | |||||||||
| 94–122 | 94–122 | 94–122 | |||||||||
| 95–106 | 95–106 | ||||||||||
| 112– 115 |
|||||||||||
| 122–132 | 122–132 | 122–132 | |||||||||
| 123– 132 |
123–132 | 123– 132 |
123–132 | 123– 132 |
123–132 | 123–132 | 123–132 | ||||
| 126– 135 |
126–135 | ||||||||||
| 133–157 | |||||||||||
| 136–147 | 136– 147 |
136– 147 |
136–147 | ||||||||
| 142–157 | 142–157 | 142–157 | |||||||||
| 148–174 | |||||||||||
| 151–174 | 151– 174 |
||||||||||
| 158–168 | 158–168 | 158–168 | 158–168 | ||||||||
| 159–168 | 159–168 | 159–168 | 159–168 | ||||||||
| 159–173 | |||||||||||
| 161–168 | 161–168 | ||||||||||
| 161–173 | |||||||||||
| 174– 184 |
174–184 | 174– 184 |
174–184 | 174– 184 |
174–184 | ||||||
| 175– 192 |
175– 192 |
||||||||||
| 182–192 | |||||||||||
| 185– 192 |
185– 192 |
185– 192 |
185–192 | ||||||||
Identification of Cross-linked Peptides
In addition to the linear peptides, cross-linked peptides were observed using reflectron MALDI-TOF MS labeled with heavy water (18O). Cross-linked peptides were identified via MALDI-TOF MS by the presence of peaks not seen in the noncross-linked sample and a +8 Da mass shift obtained from the incorporation of four 18Os in the two cross-linked peptides. All the experimental cross-links identified via MALDI-TOF, with their parts per million and mass difference from the theoretical mass, are shown as supplemental data (Supplemental Table II). For example, Figure 2C shows a region of the MALDI-TOF spectrum from an intermolecular cross-link seen only in the dimer and trimer, where an ion (m/z 3068.305) in the trimer sample is seen for the unlabelled (16O) cross-link and 8 mass units away, an ion (m/z 3076.720) is seen for the labeled (18O) cross-link. For this high molecular weight range there is a good correlation, less than half a Dalton, between the m/z ion at 3068.305 and a theoretical m/z ion of 3068.710 for the cross-linked peptides 51–63 linked to 159–173.
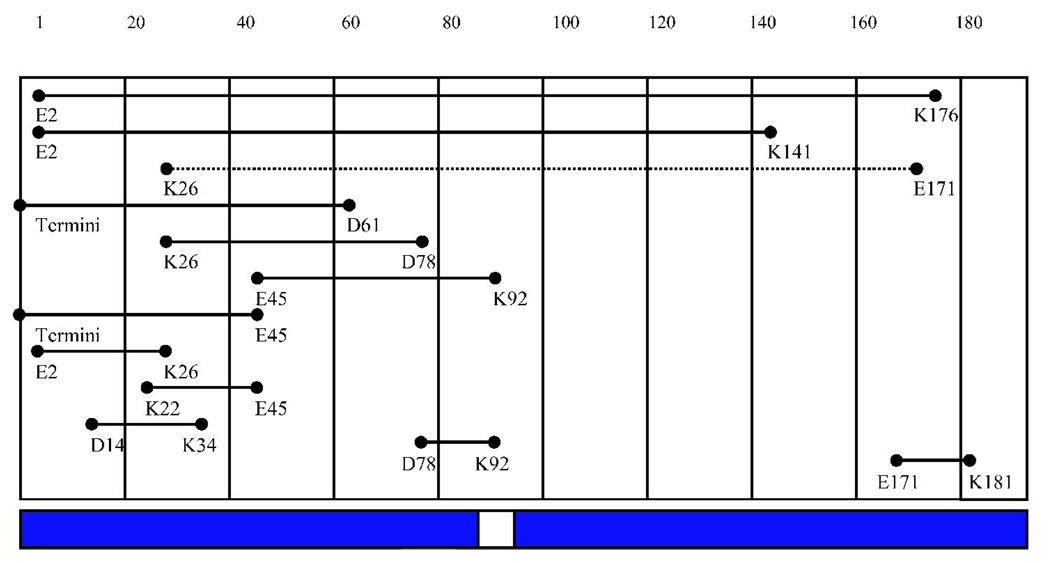
Thirteen intramolecular cross-links were identified in all cross-linked samples but not in the noncross-linked control (Table II), in which two of these intramolecular cross-links map to the same residues, and so appear only once in Figure 4B (E45 to K92). This core of intramolecular cross-links seen in all 3 samples are between distinct and distant residues separated in sequence, indicating that HIV-1 Vif is compactly folded as cross-linking suggests these residues are close to each other in 3-dimensional space. In addition, one cross-linked peptide, 23–34 linked to 159–172, is specific to the monomer sample, suggesting a possible conformational change upon oligomerization (Table II and Figure 4B).
Table II.
Cross-links of HIV-1 Vif. Cross-linked residues identified by MALDI-TOF-MS and heavy water labeling of trypsin-digested monomeric, dimeric or trimeric forms of HIV-1 Vif. The cross-linking agent EDC specifically links lysine (K) to either aspartic acid (D) or glutamic acid (E). If more than one lysine or acid is within an identified fragment, all are listed. Linking residues are listed sequentially. Cross-linked residues in bold appear in Figure 4. Italics indicate that experimental molecular weights were consistent with theoretical molecular weights of both cross-links.
| Monomer | Dimer | Trimer | |||
|---|---|---|---|---|---|
| K26, K34 | E171, D172 | ||||
| Termini | E45 | Termini | E45 | Termini | E45 |
| Termini | D61, E76 | Termini | D61, E76 | Termini | D61, E76 |
| E2 | K26, K34 | E2 | K26, K34 | E2 | K26, K34 |
| E54, D61 | K176, K179 | E54, D61 | K176, K179 | E54, D61 | K176, K179 |
| E2 | K141 | E2 | K141 | E2 | K141 |
| E2 | K176, K179, K181 | E2 |
K176, K179, K181 |
E2 | K176, K179, K181 |
| D14 | K26, K34 | D14 | K26, K34 | D14 | K26, K34 |
| K22, K26 | E45 | K22, K26 | E45 | K22, K26 | E45 |
| K26, K34 | D78, E88 | K26, K34 | D78, E88 | K26, K34 | D78, E88 |
| E45 | K91, K92 | E45 | K91, K92 | E45 | K91, K92 |
| E45 | K92 | E45 | K92 | E45 | K92 |
| D78, E88 | K91, K92 | D78, E88 | K91, K92 | D78, E88 | K91, K92 |
| E171, D172 | K181 | E171, D172 | K181 | E171, D172 | K181 |
| K34 | E134 | K34 | E134 | ||
| E45 | K176, K179 | E45 | K176, K179 | ||
| D61 | K160, K168 | D61 | K160, K168 | ||
| K92 | E171, D172 | K92 | E171, D172 | ||
| D14 | K158, K160, K168 | ||||
| K34 | Termini | ||||
| K158, K160, K168 | E171, D172 | ||||
| K158, K160, K168 | Termini | ||||
|
K160, K168, E171, D172 |
Termini | ||||
|
K160, K168, E171, D172, K176, K179 |
Termini | ||||
| E171, D172 | K181 |
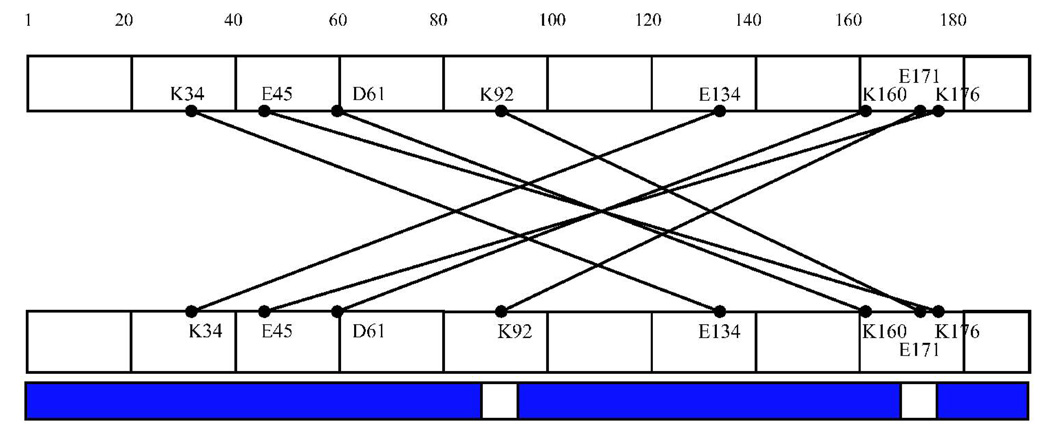
Intermolecular cross-links were observed in the cross-linked dimer and trimer samples (Table II and Figure 4C, D). These intermolecular cross-links were identified by their presence in the dimer and/or trimer sample but not in the noncross-linked or cross-linked monomer samples, and by the presence of a +8 Da mass unit shift. Seven cross-links specific to the trimer and 4 in both the dimer and trimer were identified. The majority of cross-links observed in the cross-linked dimer and trimer mapped to the N- and C-termini of the protein. The trimer also contained a set of cross-links that mapped to the C-terminus of each respective monomer, suggesting that along with an N- to C-terminus interaction in the trimer, there is also a C- to C-terminus interaction.
Overall, cross-link sequence coverage was observed in 62.5% of the monomer, 69.3% of the dimer, and 69.8% of the trimer. Complete cross-link sequence coverage is not expected since cross-linked fragments are likely to exist in digested dimers and trimers above 4000 Da. Molecular weights greater than 4000 are difficult to uniquely identify, given the current technology. The differences in levels of coverage observed between cross-links in the monomer and those in the dimer and trimer suggest a conformational change that allows the C-terminus to become more accessible to cross-linking, and therefore suggesting the region becomes more ordered upon higher order oligomerization. The redundancy in intermolecular cross-links indicates that oligomerization occurs in a unique, specific manner that is likely functionally significant and not due to nonspecific aggregation. The intramolecular cross-links observed give insight into the tertiary fold of the HIV-1 Vif protein, whereas the intermolecular cross-links give insight into the quaternary fold of the homo-oligomer (Figure 4, Figure 5 and Table II).
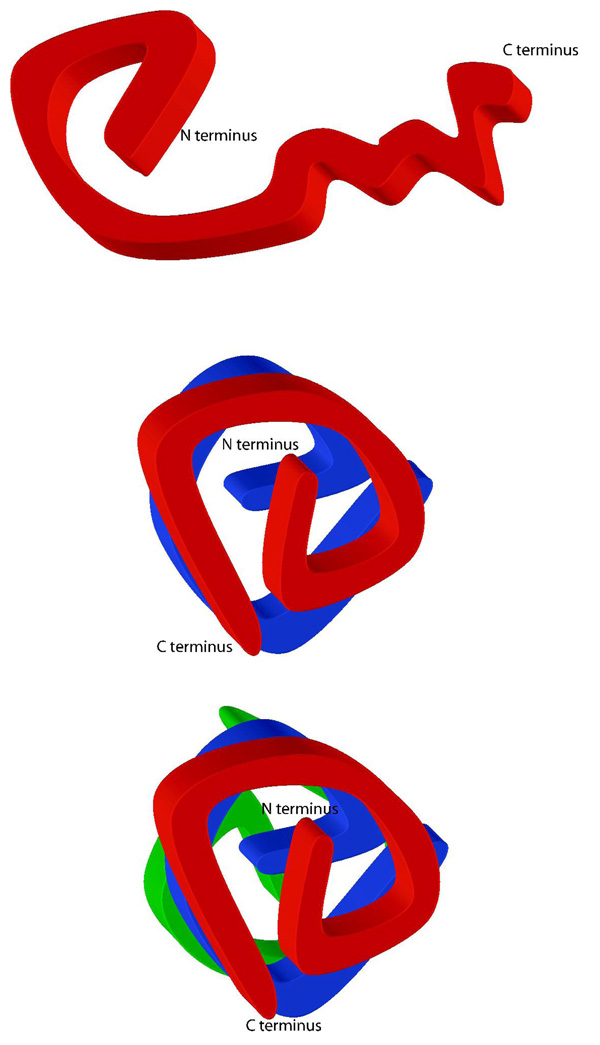
Figure 5. Topology and Multimerization of HIV-1 Vif.
(A) Intramolecular cross-links suggest that the N-terminus is folded into a compact domain and the C-terminus is less structured. The HIV-1 Vif monomer is globular in shape. (B and C) Schematic of how the HIV-1 Vif dimer and trimer may fold. The carboxyl terminus becomes more ordered upon oligomerization.
DISCUSSION
HIV-1 Vif interacts directly or indirectly with APOBEC3G, an endogenous cellular inhibitor of HIV-1 replication, and targets it for proteosomal degradation through a cullin-RING ligase complex, thus allowing the virus to replicate. The proteosomal degradation of APOBEC3G depends on HIV-1 Vif binding members of the cullin-RING ligase complex such as elongin C and Cullin5. HIV-1 Vif binds elongin C through its SOCS box domain which appears to be regulated by phosphorylation of the SOCS box 6, 16, 24, 26, and HIV-1 Vif binds Cullin5 through a HCCH zinc binding motif 13–15. This function of HIV-1 Vif makes it a critical factor in HIV-1 infectivity and viral spread, and hence provides an ideal target for antiviral agents. In addition, to APOBEC3G, elongin C and Cullin5, Vif has also been shown to bind a host of other proteins including HIV-1 NCp7 (Gag) and HIV-1 viral RNA9, 12, 54, 55. High resolution structural data would help to identify residues involved in protein folding and in interactions with other proteins at the atomic level. This data would also provide insight into the protein interfaces and residues that are potential targets for antiviral strategies, similar to the insights used for exploring structure-based inhibition of HIV-1 protease 56. However, to date no structural information exists on HIV-1 Vif at the atomic level or any biophysical information at the single amino acid level. This lack of information is partly due to the inability to express soluble HIV-1 Vif at amounts needed for conventional high-resolution structural techniques. Because of this inability to express large quantities of soluble HIV-1 Vif, experiments such as gel filtration or analytical centrifugation are also not possible. We, therefore, utilized techniques that require small amounts of protein to map the topology of HIV-1 Vif. Specifically we used cross-linking, proteolysis and state-of-the-art mass spectrometry (LC-QTof-MS, LC-ion trap-MS and MALDI-TOF MS) techniques 38, 40, 43, 46, 47 to obtain data on the topology and oligomerization of HIV-1 Vif.
The cross-linking analysis of HIV-1 Vif resulted in the elucidation of a variety of oligomeric states: monomer, dimer and trimer. Proteolytic digestion and comparison of the resulting peptides from these cross-linked oligomers and a noncross-linked sample allowed the determination of many key aspects of the tertiary and quaternary structure of HIV-1 Vif. Linear peptides were identified that cover the entire protein for both the noncross-linked and cross-linked monomers of HIV-1 Vif. In the dimer and trimer, however, certain regions of the oligomer are not observed, indicating that cross-linking has “protected” them from proteolysis. Such protected linear peptides are observed in the dimer and trimer at residues 169–173 and in the trimer alone at 107–121 and 148–157. The difference in protection between the dimer and trimer indicates specific differences in packing between the two oligomers, likely reflecting an increase in order upon oligomerization.
In addition to linear peptides, 24 specific intra- and intermolecular cross-links were also identified in the various oligomeric states (Figure 4). These observed cross-links are not due to nonspecific aggregation since only a limited number of lysines and aspartic/glutamic acid residues in HIV-1 Vif are involved. In fact, only 9 of 14 (64.29%) lysines are involved in cross-links and 7 of 16 (43.75%) acidic residues are involved in cross-links, where the combination of possible cross-links is 224 if all lysines randomly interacted with all possible acids. Since the “zero-length” cross-linking agent, EDC, only links groups that are within 5 Å, the cross-links that are observed are specific and not the result of non-specific aggregation. Multiple, essentially redundant cross-links, are also observed between similar regions of the protein particularly in the N-terminus, indicating a specific fold. Thirteen cross-links were observed in the cross-linked monomer, dimer and trimer, and are therefore intramolecular. Ten of these intramolecular cross-links are within the amino-terminal half of HIV-1 Vif, indicating that this region is likely folded into a compact domain. Only one cross-link is observed between residues 171 and 181 in the carboxy-terminal half of HIV-1 Vif, indicating that this region is likely less ordered in the monomer. Between the two domains there are only three cross-links (between residues: 2 and 141; 2 and 176; and 26 and 171) and none in the last 16 residues of the protein (Figure 4B), indicating minimal interactions between N- and C-terminal regions of the protein within the monomer.
Additional specific intermolecular cross-links were observed in the dimer and trimer of HIV-1 Vif. Four cross-links observed in the dimer are also observed in the trimer, where they link residues in the amino half of the protein, between 34 and 92, with residues in the carboxyl half of the protein, between 134 and 176. An additional 6 cross-links are observed in the trimer. Two of these cross-links involve more extensive interactions between the two termini of the protein, and the remaining 4 intermolecular cross-links occur within the carboxy quarter of the protein, between residues 158 to 192. This observation suggests that, in contrast to its disordered structure as a monomer, the carboxy-terminal domain becomes more ordered upon dimerization and trimerization.
Proteins that contain regions of intrinsic disorder are characterized by having regions with poorly defined tertiary structure when studied in isolation. These proteins become more ordered upon oligomerizing or binding to other biological macromolecules 32, 35. In addition, intrinsically disordered proteins have high net charge and low overall hydrophobicity 33, 34. Interestingly, both of these characteristics are attributable to HIV-1 Vif. The propensity for being intrinsically disordered can be successfully predicted from a protein’s sequence utilizing analysis programs such as PONDR® in the Database of Protein Disorder (DisProt) 50, 51. In fact, PONDR VL-XT successfully predicted that regions of p53 and Mdm2 had intrinsic disorder 57. In p53 PONDR VL-XT predicts the middle portion of the protein is ordered, while both the N-and C-termini are predominantly disordered, which was experimentally confirmed by NMR. In addition “disorder-to-order transitions” have been observed in p53 to occur both upon tetramerization and upon other proteins binding. Similarly in Mdm2, the p53 binding domain has been predicted and confirmed to be ordered while other regions are confirmed to undergo “disorder-to-order transitions” 57. Interestingly, Mdm2 targets p53 for proteosomal degradation in an analogous fashion to HIV-1 Vif and APOBEC3G.
When the HIV-1 VifHXB2 sequence is submitted to PONDR® 51 all four predictors score the C-terminus as being highly disordered (Figure 2A). Only one of the four predictors suggests that the N-terminus might also be disordered. However, our mass spectrometry data, with ten internal cross-links within the N-terminus strongly supports that it is predominantly ordered unlike the C-terminus where only one internal cross-link was observed (Figure 4B). Thus, the C-terminus of monomeric HIV-1 Vif is likely intrinsically disordered and may become more ordered upon oligomerization. This model is supported by our cross-linking data, since four new cross-links (Figure 4C) and another six additional cross-links (Figure 4D) were obtained in the C-terminus for the dimer and trimer, respectively. Figure 5 shows a 2-dimensional schematic of how HIV-1 Vif may homo-oligomerize based on observed areas of protection and cross-links from the mass spectrometry analysis: with the carboxy termini becoming more structured as oligomerization occurs. This is consistent with the secondary structure prediction (Figure 4A) where the twists and turns correspond to areas predicted to be random coils. The ordering of HIV-1 Vif upon oligomerization is consistent with the behavior of other intrinsically disordered proteins that become more ordered upon binding other biological macromolecules 32, 35.
Thus, in the absence of high resolution structural data, we have successfully obtained information on the oligomeric state and molecular topology of HIV-1 Vif using state-of-the-art cross-linking and mass spectroscopy techniques that have been extensively used with other proteins 38, 40, 43, 46, 47. This data both from identification of peptide protection and identification of specific cross-links complements and expands previous reports on the identification of important residues and regions of HIV-1 Vif required for HIV-1 infectivity. Data from single or multiple site directed mutagenesis experiments showed that 20 residues either individually or in combination reduced viral infectivity by greater than 85% and that another 9 residues reduced infectivity greater than 50% 58. Our mass spectrometry data overlaps with 27 of these 29 sites that have been identified as being important for infectivity. Three variants [(Q105A, L106A, I107V); (Y111A, F112A); (C114S)] containing 6 of the 27 sites of mutation that reduce infectivity by greater than 85% 58 overlap directly with a protected peptide we observed in the trimer (107–121). This protected peptide contains H108, C114 and I120 which are important in Cullin5 binding 13–15. Thus the fact that 107–121 is protected in the trimer may imply trimerization is essential for HIV-1 Vif activity, perhaps by facilitating Cullin5 binding by the displacement of one or more HIV-1 Vif monomers. Another 21 of these important residues map near two separate cross-links that only occur upon dimer and trimer formation. Five of these 21 residues, in three variants [(P161A, P162A, L163A); (P164A); (S165A)], map adjacent to a cross-link of K160 to D61. The remaining sixteen of these 21 residues map to the region contiguous with K34 cross-linking with E134 where 4 variants [(W38A, F39A, Y40A); (H43A, Y44A); (C133S); (S144A, L145A, Q146A)] affect infectivity by greater than 85% and three variants [(M29A, Y30A, I31V); (Y135A, Q136A); (N140A, K141A)] affect infectivity by greater than 50% 58. Our cross-linking data of K34 linking with E134 indicates that these essential regions, 29–44 and 133–146, are close together in three-dimensional space. This implies in the dimer and trimer these regions likely come together to form a single molecular surface creating a “hotspot” for biological activity possibly facilitating Cullin5 binding through C133 of the HCCH zinc binding motif 13–15.
The HIV-1 viral RNA binding site in HIV-1 Vif, residues 1–64, further supports the proposed “hot spot” for biological activity since it overlaps with “hot spot” residues 29–44. RNA binding activity was significantly reduced by the following HIV-1 Vif mutants: W11A, Y30A, and Y40A 55, which in the case of Y30A and Y40A, are adjacent to the proposed “hotspot” for biological activity. In addition, HIV-1 Vif’s binding to viral RNA appears to occur in a cooperative manner resulting from HIV-1 Vif multimers binding to the viral RNA 12.
Peptide protection data in the dimer and/or trimer of residues 148–157 and 169–173 and cross-links at residues 158, 160, and 170 are also consistent with the previously identified oligomerization domain, 151–171 36, 37. In fact, 148–157 overlaps with the SOCS box, residues 145–169, where HIV-1 Vif interacts with elongin C in the elongin BC complex. This is similar to the trimer specific peptide protection of residues 107–121 corresponding with Cullin5 binding. In addition, the oligomerization domain is also adjacent to and overlapping with the putative HIV-1 NCp7 (Gag) binding site, residues 157–179 54. Therefore, our protection and cross-linking data not only is consistant with the previously reported oligomerization domain, but the overlap of this region with the NCp7 (Gag) binding site may suggest that Gag is important for HIV-1 Vif function. A possible molecular mechanism is that the Cullin5-elonginBC complex displaces one (or more) of the HIV-1 Vif monomers thereby inducing a conformational change that may also facilitate the binding and targeting of APOBEC3G for degradation. Therefore, the structurally important regions identified from our mass spectrometry analysis in HIV-1 Vif likely undergo a “disorder-to-order” transition upon oligomerization or binding to other macromolecules such as Cullin5, elongin C, viral RNA, and HIV-1 NCp7 (Gag).
Although HIV-1 Vif is not highly conserved the majority of our cross-links and peptide protections occur at residues that are conserved among HIV-1 subtypes. Among those residues in cross-links, E2, D14, K22, K26, and E34 in the N-terminus and E134, K141, K158, K160, K171, and K176 in the C-terminus cluster as two regions of high sequence conservation. Conservation of these residues along with cross-linking data is consistent with residues 29–44 and 133–146 being a “hotspot” for HIV-1 Vif’s biological activity. The sequence of HIV-1 Vif is not conserved among other lentiviral Vifs. However, like HIV-1 Vif, PONDR® predicts that the C-terminus of HIV-2 and SIV Vif are both disordered. Therefore, these lentiviral Vifs may undergo a similar “disorder-to-order” transition upon oligomerization and binding of other macromolecules similar to the one proposed for HIV-1 Vif.
This first study of HIV-1 Vif’s molecular structure allows us to develop the following hypothesis: HIV-1 Vif monomers are likely in dynamic equilibrium between various homo-oligomers (dimer and trimer), Cullin5-elonginBC complex, APOBEC3G and possibly other biological macromolecules such as HIV-1 NCp7 (Gag) and HIV-1 viral RNA. The HIV-1 Vif monomer will likely only exist transiently with an N-terminal region that has a defined topology while the C-terminus is intrinsically disordered. This disorder may facilitate the dynamic equilibrium of the various complexes. Upon binding other macromolecules the structure of the C-terminus of the HIV-1 Vif monomer becomes more defined, which is what we observed in molecular detail upon the formation of HIV-1 Vif dimers and trimers in the C-terminal region (Figures 4 and 5). Specifically, this increase in structure, or “disorder-to-order” transition, is likely to be critical to HIV-1 Vif’s function in viral infectivity, as regions where we observed specific cross-links and protected peptides correspond to previously identified regions critical to viral infection. Further structure determination and biochemical analysis of these complexes must be performed in order to better elucidate the specifics of these interactions as future inhibitor design will involve disruption of one or more of these critical interactions involved in HIV-1 Vif function.
Acknowledgements
We thank Jinal Patel (UMass Proteomics Mass Spectrometry Facility) for help with collecting and analyzing the mass spec data; Jennifer Saporita for help with determining the complete density of gel bands; Melissa Grabowski for critically reading the manuscript; Mary Munson, William Kobertz, and C. Robert Matthews for helpful discussions; the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH and Dr. Opendra Sharma. This work was supported by a grant from the NIH (R21 AI056935 and R21 AI067021) and a UMass CFAR grant (5P3O AI42845).
Glossary
The abbreviations used are
- HIV
human immunodeficiency virus
- Vif
virion infectivity factor
- MALDI-TOF
matrix-assisted laser desorption/ionization-time of flight
- LC-ion trap-MS
ion trap liquid chromatography mass spectrometry
- LC-QTof-MS
quadrupole time of flight liquid chromatography mass spectrometry
- m/z
mass-to-charge ratio
REFERENCES
- 1.Baraz L, Kotler M. The Vif protein of human immunodeficiency virus type 1 (HIV-1): enigmas and solutions. Current Medical Chemistry. 2004;11:221–231. doi: 10.2174/0929867043456124. [DOI] [PubMed] [Google Scholar]
- 2.Lake J-a, Carr J, Feng F, Mundy L, et al. The role of Vif during HIV-1 infection: interaction with novel host cellular factors. Journal of Clinical Virology. 2003;26:143–152. doi: 10.1016/s1386-6532(02)00113-0. [DOI] [PubMed] [Google Scholar]
- 3.Navarro F, Landau NR. Recent insights into HIV-1 Vif. Current Opinion in Immunology. 2004;16:477–482. doi: 10.1016/j.coi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Rose KM, Marin M, Kozak SL, Kabat D. The viral Infectivity factor (Vif) of HIV-1 unveiled. TRENDS in Molecular Medicine. 2004;10(6):291–297. doi: 10.1016/j.molmed.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Madani N, Kabat D. Cellular and viral specificities of human immunodeficiency virus type 1 vif protein. Journal of Virology. 2000;74:5982–5987. doi: 10.1128/jvi.74.13.5982-5987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002 August 8;418(6898):646–650. doi: 10.1038/nature00939. 2002. [DOI] [PubMed] [Google Scholar]
- 7.Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. The Embo Journal. 2004;23(12):2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu B, Sarkis PTN, Luo K, Yu Y, et al. Regulation of Apobec3F and Human Immunodeficiency Virus Type 1 Vif by Vif-Cul5-ElonB/C E3 Ubiquitin Ligase. Journal of Virology. 2005;79(15):9579–9587. doi: 10.1128/JVI.79.15.9579-9587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bardy M, Gay B, Pebernard S, Chazal N, et al. Interaction of human immunodeficiency virus type 1 Vif with Gag and Gag-Pol precursors: co-encapsidation and interference with viral protease-mediated Gag processing. Journal of General Virology. 2001;82:2719–2733. doi: 10.1099/0022-1317-82-11-2719. [DOI] [PubMed] [Google Scholar]
- 10.Hutoran M, Britan E, Baraz L, Blumenzweig I, et al. Abrogation of Vif function by peptide derived from the N-terminal region of the human immunodeficiency virus type 1 (HIV-1) protease. Virology. 2004;330:261–270. doi: 10.1016/j.virol.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 11.Cancio R, Spadari S, Maga G. Vif is an auxiliary factor of the HIV-1 reverse transcriptase and facilitates abasic site bypass. Journal of Biochemistry. 2004;383(3):475–482. doi: 10.1042/BJ20040914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henriet S, Richer D, Bernacchi S, Decroly E, et al. Cooperative and Specific Binding of Vif to the 5' Region of HIV-1 Genomic RNA. Journal of Molecular Biology. 2005;354:55–72. doi: 10.1016/j.jmb.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 13.Mehle A, Thomas ER, Rajendran KS, Gabuzda D. A zinc-binding region in Vif binds Cul5 and determines Cullin selection. Journal of Biological Chemistry. 2006;281(25):17259–17265. doi: 10.1074/jbc.M602413200. [DOI] [PubMed] [Google Scholar]
- 14.Xiao Z, Ehrlich E, Yu Y, Luo K, et al. Assembly of HIV-1 Vif-Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology. 2006;349(2):290–299. doi: 10.1016/j.virol.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Luo K, Xiao Z, Ehrlich E, Yu Y. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl. Acad. Sci USA. 2005;102(32):11444–11449. doi: 10.1073/pnas.0502440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehle A, Goncalves J, Santa-Marta M, McPike M, et al. Phosphoryylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes & Development. 2004;18(23):2861–2866. doi: 10.1101/gad.1249904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu X, Yu Y, Liu B, Luo K, et al. Induction of APOBEC3G Ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF Complex. Science. 2003;302(5647):1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 18.Yu Y, Xiao Z, Ehrlich ES, Yu X, et al. Selective assembly of HIV-1 vif-Cul5 ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes & Development. 2004;18:2867–2872. doi: 10.1101/gad.1250204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabuzda DH, Lawrence K, Langhoff E, Terwilliger E, et al. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. Jounal of Virology. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakai H, Shibata R, Sakuragi J, Sakuragi S, et al. Cell-dependent requirement of human immunodeficiency virus type 1 Vif protein for maturation of virus particles. Journal of Virology. 1993;67(3):1663–1666. doi: 10.1128/jvi.67.3.1663-1666.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sova P, Volsky DJ. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. Journal of Virolgoy. 1993;67:6322–6326. doi: 10.1128/jvi.67.10.6322-6326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.vonSchwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. Journal of Virology. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bishop KN, Holmes RK, Malim MH. Antiviral Potency of APOBEC Proteins Does Not Correlate with Cytidine Deamination. Journal of Virology. 2006;80(17):8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif Protein binds the editing enzyme APOBEC3G and induces its degradation. Nature Medicine. 2003;9(11):1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 25.Sheehy AM, Gaddis NC, Malim MH. The Antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nature Medicine. 2003;9(11):1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 26.Stopak K, Noronha Cd, Yonemoto W, Greene WC. HIV-1 Vif Blocks the Antiviral Activity of APOBEC3G by Impairing Both Its Translation and Intracellular Stability. Molecular Cell. 2003;12(3):591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi M, Takaori-Kondo A, Miyauchi Y, Iwai K, et al. Ubiquitination of APOBEC3G by an HIV-1 Vif-Cullin5-ElonginB-ElonginC complex is essential for Vif function. Journal of Biological Chemistry. 2005;280(19):18573–18578. doi: 10.1074/jbc.C500082200. [DOI] [PubMed] [Google Scholar]
- 28.Shirakawa K, Takaori-Kondo A, Kobayashi M, Tomonaga M, et al. Ubiquitination of APOBEC3 proteins by the Vif-Cullin5-ElonginB-ElonginC complex. Virology. 2006;344(2):263–266. doi: 10.1016/j.virol.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 29.Hassaine G, Agostini I, Candotti D, Bessou G, et al. Characterization of Human Immunodeficiency Virus Type 1 vif Gene in Long-Term Asymptomatic Individuals. Virology. 2000;276:169–180. doi: 10.1006/viro.2000.0543. [DOI] [PubMed] [Google Scholar]
- 30.Farrow MA, Somasundaran M, Zhang C, Gabuzda D, et al. Nuclear Localization of HIV Type 1 Vif Isolated from a Long-Term Asymptomatic Individual and Potential Role in Virus Attenuation. AIDS Res Hum Retroviruses. 2005;21(6):565–574. doi: 10.1089/aid.2005.21.565. [DOI] [PubMed] [Google Scholar]
- 31.Sakurai A, Jere A, Yoshido A, Yamada T, et al. Functional analysis of HIV-1 vif genes derived from Japanese long-term nonprogressors and progressors for AIDS. Microbes and Infection. 2004;6(9):799–805. doi: 10.1016/j.micinf.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Romero PR, Zaida S, Fang YY, Uversky VN, et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. PNAS. 2006;103(22):8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dyson HJ, Wright PE. Intrinsically Unstructured Proteins and Their Functions. Nature Reviews. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 34.Fink AL. Natively unfolded proteins. Current Opinion in Structural Biology. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Dunker A, Cortese M, Romero P, Iakoucheva L, et al. Flexible nets. The roles of intrinisic disorder in protein interaction networks. FEBS. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 36.Yang S, Sun Y, Zhang H. The Multimerization of Human Immunodeficiency Virus Type 1 Vif Protein. The Journal of Biological Chemistry. 2001;276(7):4889–4893. doi: 10.1074/jbc.M004895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang B, Li L, Lu Z, Fan X, et al. Potent Suppression of Viral Infectivity by the Peptides That Inhibit Multimerization of Human Immunodeficiency Virus Type 1 (HIV-1) Vif Proteins. The Journal of Biological Chemistry. 2003;278(8):6596–6602. doi: 10.1074/jbc.M210164200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Back JW, Jong Ld, Muijsers AO, Koster CGd. Chemical Cross-linking and Mass Spectrometry for Protein Structural Modeling. Journal Molecular Biology. 2003;331:303–313. doi: 10.1016/s0022-2836(03)00721-6. [DOI] [PubMed] [Google Scholar]
- 39.Eyles SJ, Kaltashov IA. Methods to study protein dynamics and folding by mass spectrometry. Methods. 2004;34:88–99. doi: 10.1016/j.ymeth.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 40.Farmer TB, Caprioli RM. Determination of Protein-Protein Interactions by Matrix-assisted Laser Desorption/Ionization Mass Spectrometry. Journal of Mass Spectrometry. 1998;33:697–704. doi: 10.1002/(SICI)1096-9888(199808)33:8<697::AID-JMS711>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 41.Kalkhof S, Ihling C, Mechtler K, Sinz A. Chemical Cross-linking and High-Performance Fourier Transform Ion Cyclotron Resonance Mass Spectrometry for Protein Interaction Analysis: Application to a Calmodulin/Target Peptide Complex. Analytical Chemistry. 2005;77(2):495–503. doi: 10.1021/ac0487294. [DOI] [PubMed] [Google Scholar]
- 42.Schulz DM, Ihling C, Clore GM, Sinz A. Mapping the Topology and Determination of a Low-Resolution Three Dimensional Structure of the Calmodulin-Melittin Complex by Chemical Cross-linking and High-Resolution FTICRMS: Direct Demonstration of Multiple Binding Modes. Biochemistry. 2004;43:4703–4715. doi: 10.1021/bi036149f. [DOI] [PubMed] [Google Scholar]
- 43.Sinz A. Chemical Cross-linking and mass spectrometry for mapping three-dimensional structures of proteins and protein complexes. Journal of Mass Spectrometery. 2003;38:1225–1237. doi: 10.1002/jms.559. [DOI] [PubMed] [Google Scholar]
- 44.Trester-Zedlitz M, Kamada K, Burley SK, Fenyo D, et al. A Modular Cross-Linking Approach for Exploring Protein Interactions. Journal of American Chemical Society. 2003;125:2416–2425. doi: 10.1021/ja026917a. [DOI] [PubMed] [Google Scholar]
- 45.Grabarek Z, Gergely J. Zero-length crosslinking procedure with the use of active esters. Analytical Biochemistry. 1990;185:131–135. doi: 10.1016/0003-2697(90)90267-d. [DOI] [PubMed] [Google Scholar]
- 46.Yao X, Freas A, Ramirez J, Demirev P, et al. Proteolytic 18O labeling for comparative proteomics: model studies with two serotypes of adenovirus. Anal Chem. 2001;73(13):2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 47.Back J, Notenboom V, de Koning L, Muijsers A, et al. Identification of cross-linked peptides for protein interaction studies using mass spectrometry and 18O labeling. Anal Chem. 2002;74(17):4417–4422. doi: 10.1021/ac0257492. [DOI] [PubMed] [Google Scholar]
- 48.Soskic V, Godovac-Zimmerman J. Improvement of an in-gel tryptic digestion method for matrix-assisted laser desorption/ionization-time of flight mass spectrometry peptide mapping by use of volatile solubilizing agents. Proteomics. 2001;1(11):1364–1367. doi: 10.1002/1615-9861(200111)1:11<1364::AID-PROT1364>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 49.Davidson WS, Hilliard GM. The Spatial Organization of Apolipoprotein A–I on the Edge of Discoidal High Density Lipoprotein Particles. The Journal of Biological Chemistry. 2003;278(29):27199–27207. doi: 10.1074/jbc.M302764200. [DOI] [PubMed] [Google Scholar]
- 50.Romero P, Obradovic Z, Li X, Garner EC, et al. Sequence Complexity of Disordered Protein. PROTEINS: Structure, Function, and Genetics. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 51.Vucetic S, Obradovic Z, Vacic V, Radivojac P, et al. DisProt: A Database of Protein Disorder. Bioinformatics. 2005;21:137–140. doi: 10.1093/bioinformatics/bth476. [DOI] [PubMed] [Google Scholar]
- 52.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: Network Protein Sequence Analysis. TIBS. 2000;25(3):147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 53.Strochlic L, Cartaud A, Labas V, Hoch W, et al. MAGI-1C: A Synaptic MAGUK Interacting with MuSK at the Vertebrate Neuromuscular Junction. Journal of Cell Biology. 2001;153(5):1127–1132. doi: 10.1083/jcb.153.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bouyac M, Courcoul M, Bertoia G, Baudat Y, et al. Human Immunodeficiency Virus Type 1 Vif Protein Binds to the Pr55Gag Precursor. Journal of Virology. 1997;71(12):9358–9365. doi: 10.1128/jvi.71.12.9358-9365.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang H, Pomerantz RJ, Dornadula G, Sun Y. Human Immunodeficiency Virus Type 1 Vif Protein Is an Integral Component of an mRNP Complex of Viral RNA and Could Be Involved in the Viral RNA Folding and Packaging Process. The Journal of Virology. 2000;74(18):8252–8261. doi: 10.1128/jvi.74.18.8252-8261.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.King N, Prabu-Jeyabalan M, Nalivaika E. Combating susceptibility to drug resistance: lessons from HIV-1 protease. Chemical Biology. 2004;11(10):1333–1338. doi: 10.1016/j.chembiol.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 57.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, et al. Intrinsic Disorder in Cell-signaling and Cancer-associated Proteins. Journal of Molecular Biology. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 58.Simon JHM, Sheehy AM, Carpentier EA, Fouchier RAM, et al. Mutational Analysis of the Human Immunodeficiency Virus Type 1 Vif Protein. Journal of Virology. 1999;73(4):2675–2681. doi: 10.1128/jvi.73.4.2675-2681.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]