Abstract
Oncolytic viral therapy has been explored widely as an option for glioma treatment but its effectiveness has remained limited. CCN1 is an extracellular matrix (ECM) protein elevated in cancer cells that modulates their adhesion and migration by binding cell surface receptors. In this study, we examined an hypothesized role for CCN1 in limiting the efficacy of oncolytic viral therapy for glioma, based on evidence of CCN1 induction that occurs in this setting. Strikingly, we found that exogenous CCN1 in glioma ECM orchestrated a cellular antiviral response that reduced viral replication and limited cytolytic efficacy. Gene expression profiling and real time PCR analysis revealed a significant induction of type-I interferon responsive genes in response to CCN1 exposure. This induction was accompanied by activation of the Jak/Stat signaling pathway, consistent with induction of an innate antiviral cellular response. Both effects were mediated by the binding of CCN1 to the cell surface integrin α6β1, activating its signaling and leading to rapid secretion of interferon-α, which was essential for the innate antiviral effect. Together, our findings reveal how an integrin signaling pathway mediates activation of a type-I antiviral interferon response that can limit the efficacy of oncolytic viral therapy. Further, they suggest therapeutic interventions to inhibit CCN1-integrin α6 interactions to sensitize gliomas to viral oncolysis.
Keywords: CCN1, Cyr61, Glioma, Oncolytic Virus
INTRODUCTION
Glioblastoma multiforme (GBM) is the most common primary brain tumor and despite aggressive therapy involving tumor resection, chemotherapy and radiation treatment, median survival of patients remains less than 15 months from diagnosis (1). Oncolytic viruses (OVs) are biological therapeutics that selectively replicate in and kill tumor cells. These viruses have shown promising results in preclinical models (2), and their safety and efficacy is currently being investigated in clinical trials. Despite these advances, the impact of changes in the tumor microenvironment on OV therapeutic efficacy has not been very well studied.
We have previously described a dose dependent and rapid induction of the secreted angiogenic inducer Cysteine rich 61 (CCN1) in the tumor microenvironment following OV therapy (3). CCN1 is a member of the growth factor inducible immediate early family CCN, named as such for its first three members Cysteine rich 61, connective tissue growth factor (CTGF), and Nephroblastoma-overexpressed (Nov) (4). It is a secreted protein which typically localizes in the extracellular matrix (ECM) and on the cell surface (5), where it binds integrin receptors to modulate a variety of cellular functions including adhesion, migration, and proliferation (6). In brain tumors CCN1 is overexpressed in 68% (27/40) of GBM specimens and in cell lines derived from high-grade gliomas (7). Its increased expression in the mucosa of patients with colorectal cancer has also implicated it in “priming for carcinogenesis” (8) and its oncogenic potential is largely accredited to its activation of integrin-linked kinase-mediated βcatenin-TCF/LEF and AKT (9).
Apart from its induction in glioma cells infected with herpes simplex virus-1 (HSV-1) derived OVs, CCN1 has also been found to be dysregulated in cells after infection with Coxsackievirus B3 (CVB3) and Adenovirus type 12 (Ad12), suggesting that it may play a role in 4 viral infection of mammalian cells (10–11). Here we evaluated the impact of CCN1 expression on OV efficacy. Our findings indicate that CCN1 limits OV replication and cytotoxicity due to its significant activation and enhancement of the innate antiviral type-I interferon (IFN) response in cells. Furthermore, our studies reveal that this IFN response is activated by CCN1 binding to integrin α6β1 on glioma cells, which results in the rapid and early secretion of IFNα and activation of the Jak/Stat signaling pathway. The results from this study demonstrate a novel role for CCN1 and integrin α6β1 in regulating cellular innate defense responses against viral infection and indicate a need for patient selection based on gene expression profiling for therapeutic interventions.
MATERIALS & METHODS
Cells and Viruses
Human LN229, U343, Gli36ΔEGFR-H2B-RFP, U251T2, and U251T3 glioma cell lines are maintained as described (3). EGFR-transduced baby hamster kidney JiEGFR cells are maintained as described (12). Tet-regulated CCN1 expressing clones Cy-1 and Cy-2 were established as described (13). For radiation studies, cells were irradiated at 10gy, using RS-2000 Biological Irradiator. HSV-1-derived OVs, rHSVQ1, rHsvQ1-IE4/5-Luc, and ENVE, have been previously described (14–16).
Animals
All animal experiments were performed in accordance with the Subcommittee on Research Animal Care of The Ohio State University guidelines. Six to eight week old female athymic nude mice were used for all studies.
For CCN1 effects on viral progeny, mice were implanted subcutaneously with 4×106 Cy-1 or LN229 cells into the rear flank and monitored for tumor growth. When tumors reached 100mm3 mice were randomized and fed sucrose±dox (1mg/ml) in drinking water. 2 days post dox treatment initiation, mice were administered rHSVQ1 (1×106 pfu) by direct intratumoral injection and sacrificed 48h post-infection; tumors were harvested for the number of infectious virus particles and analyzed by a standard plaque assay.
For effects of viral progeny on tumor cell growth, mice were implanted subcutaneously with 1.5×107 U251T3 cells into the rear flank. When tumors reached an average of 250mm3 mice were administered ENVE virus by direct intratumoral injection with the indicated dose. Tumor volume was calculated using the following formula: volume=0.5LW2 as described (17).
Antibodies and Reagents
Reagents used in this study were obtained from the following sources: Cilengitide (Merck), Valproic acid & Laminin (Sigma), Fibronectin (Calbiochem), Vitronectin (Promega), CCN1 protein (Cell Sciences). Antibodies were obtained from the following sources: CCN1 (Novus Biologicals), GAPDH & ITGA6 (Abcam), STAT1 & PSTAT1 (Cell Signaling), STAT2, PSTAT2, LM609, P1F6, GoH3, P5D2 & IFNαR2 (Millipore), sheep anti-mouse HRP (GE Healthcare), goat anti-rabbit HRP, IgG negative control (DAKO). IFNα levels were measured from cell supernatants using PBL Interferon Verikine Human IFNα ELISA Kit.
RT-PCR
RNA was isolated using RNeasy Mini Kit (Qiagen). For Quantitative Real-Time PCR, cDNA was made using Superscript First-Strand Synthesis System (Invitrogen). Real time continuous detection of PCR product was achieved using Sybr Green (Applied Biosystems). GAPDH was used as an internal control. Primers were designed using the Primer Express Program (Applied Biosystems) (Supplementary Table S1).
Microarray
Total RNA from Cy-1 cells incubated±dox for 24h was isolated using RNeasy Mini Kit (Qiagen). Samples were then submitted to The Ohio State University Microarray Shared Resource Center for microarray analysis using the Affymetrix GeneChip Analysis. The microarray data from this publication have been submitted to the GEO database (accession number GSE29384).
Statistical Analysis
Results are presented as mean values±standard error of the mean (SEM). Statistical analysis was carried out by unpaired Student’s t-test using GraphPad Prism® 5.01 software. P values <0.05 were considered statistically significant. Affymetrix GeneChip was used for gene expression study. Signal intensities were quantified by Affymetrix software.
RESULTS
CCN1 gene expression is upregulated by virus but not by chemotherapy or radiation treatment
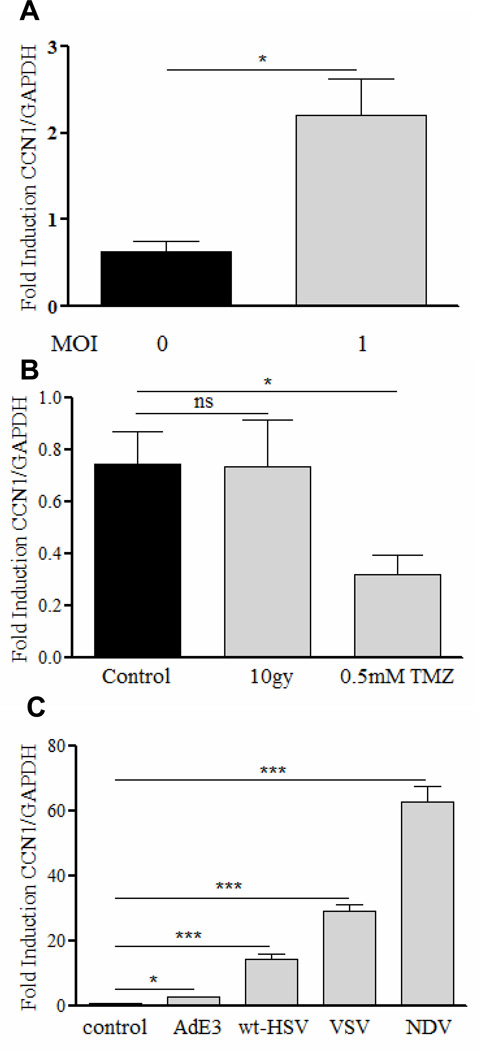
Apart from increased CCN1 gene expression in glioma cells post OV infection, its induction has also been described in H19-7 cells after treatment with etoposide, in UV irradiated human skin fibroblasts, and in HeLa cells infected with Coxsackievirus B3 virus (10, 18–19). Here we tested if induction of CCN1 in glioma cells infected with oncolytic HSV-1 represents a general response to glioma cell killing. Figures 1A and B show that while LN229 glioma cells infected with rHSVQ1 led to a significant increase of CCN1 mRNA, its expression was not increased after radiation or temozolomide treatment. To determine if this response could be generalized to other viruses, we examined changes in its expression in LN229 cells infected with three different viruses in addition to wild type HSV-1: Vesicular stomatitis virus (VSV), Adenovirus (Ad), and Newcastle Disease virus (NDV). Figure 1C shows a significant induction of CCN1 in glioma cells after infection with all the viruses tested indicating that its induction may represent a general response of glioma cells to viral infection.
Figure 1. CCN1 gene expression is upregulated by virus but not by chemotherapy or radiation therapy.
Real time PCR analysis of CCN1 gene expression in LN229 glioma cells treated with (A) rHSVQ1 at MOI=1, (B) 10gy radiation or 0.5mM temozolomide, (C) Vesicular Stomatitis Virus (ts 45) at MOI=0.1, Adenovirus (type 5) at MOI=100, Newcastle Disease Virus at MOI=2, or WtHSV-1 at MOI=1 24h after treatment. Data shown are the mean CCN1 gene expression relative to endogenous GAPDH and error bars are standard error of the mean of at least three replicates and represent at least three independent experiments. ns=not significant, *P<0.05, ***P<0.001
Extracellular CCN1 expression inhibits viral transgene expression, replication, and oncolysis
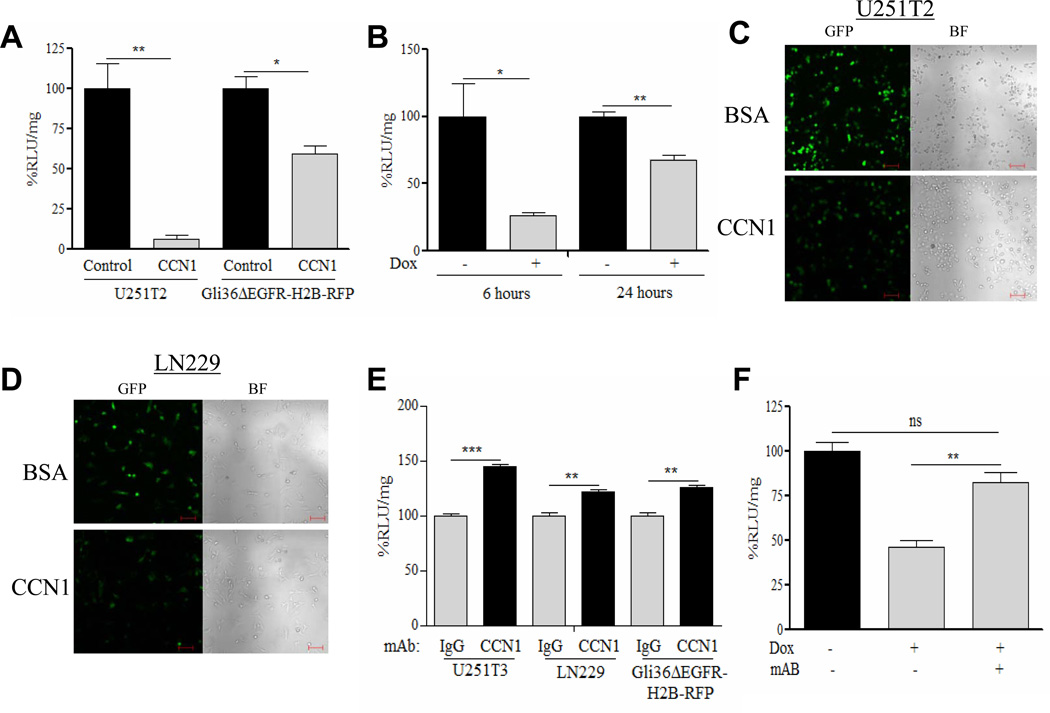
In order to investigate the impact of induction of CCN1 gene expression on viral therapy we analyzed its effect on OV gene expression in glioma cells transiently expressing CCN1 (Gli36ΔEGFR-H2B-RFP and U251T2 cells) and in tet-inducible glioma cells (Cy-1 and Cy-2). Figures 2A & B and Supplementary Figure S1A show a significant reduction in viral transgene expression upon both transient and tet-inducible induction of CCN1 gene expression (Supplementary Figure S1B & C, Supplementary Table 2) and this reduction is dose-dependent (Supplementary Figure S2A). No change was observed in parental LN229 glioma cells treated with dox (Supplementary Figure S2B).
Figure 2. Extracellular CCN1 expression inhibits viral transgene expression, replication, and cell killing.
(A) U251T2 glioma cells and Gli36ΔEGFR-H2B-RFP glioma cells transiently transfected with pcDNA3.1myc-hisB+CCN1 (CCN1) or pcDNA3.1myc-hisB+empty (control), 24h prior to being infected with rHsvQ1-IE4/5-Luc (MOI=1). 24h post infection, virus encoded luciferase activity (relative light units) was measured in infected cell lysates. Data shown are %RLU/mg±SEM relative to control. (B) OV encoded luciferase activity (relative light units: RLU) of Cy-1 tetracycline-inducible glioma cells treated±dox for 24h, prior to infection with rHsvQ1-IE4/5-Luc (MOI=1). Results presented are %RLU/mg±SEM relative to uninduced cells, 6 and 24h post-infection. (C & D) Confocal fluorescent and bright field images of GFP positive infected (C) U251T2 and (D) LN229 glioma cells seeded on plates coated with CCN1/BSA (5ug/ml) infected with rHsvQ1-IE4/5-Luc. (E) Inhibition of endogenous CCN1 increases OV transgene expression in three different glioma cell lines. Quantification of OV encoded luciferase activity of U251T3, LN229, and Gli36ΔEGFR-H2B-RFP glioma cells infected with rHsvQ1-IE4/5-Luc±CCN1 mAb measured 24h post infection. Data shown are %RLU/mg±SEM relative to control. (F) Rescue of CCN1 mediated viral inhibition by CCN1 neutralizing monoclonal antibody. Cy-1 glioma cells treated±dox were infected with rHsvQ1-IE4/5-Luc±CCN1 mAb. Virus encoded luciferase activity was quantified 24h post infection. Results presented are the %RLU/mg±SEM relative to control of at least three different experiments. Scale Bar = 100um, ns=not significant, *P<0.05, **P<0.01, ***P<0.001
To evaluate if the reduction in OV infection/replication was a result of secreted CCN1 in the ECM, we seeded U251T2 and LN229 glioma cells on CCN1/BSA coated plates prior to infection with rHsvQ1-IE4/5-Luc virus. Confocal fluorescent microscopy revealed reduced GFP positive cells when seeded on purified CCN1 compared to BSA (Figure 2C–D). Quantification of OV expressed luciferase indicated a significant reduction of viral transgene expression in cells seeded on CCN1 matrix compared to control (Supplementary Figure S3A-B). To examine the role of endogenous CCN1 on OV replication, we infected glioma cells in the presence or absence of CCN1 neutralizing antibody. Figure 2E shows that inhibition of physiological levels of CCN1 enhances viral transgene expression in three different glioma cell lines. Furthermore, CCN1 mediated reduction in viral transgene expression in dox induced Cy-1 cells was rescued in the presence of CCN1 neutralizing antibody, indicating that CCN1 acting on the cell surface of glioma cells mediates the OV inhibition (Figure 2F).
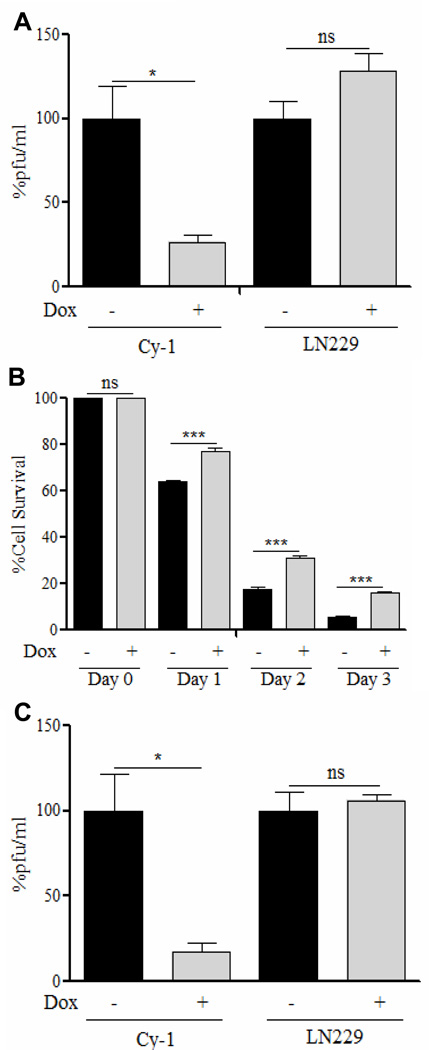
We next evaluated the impact of CCN1 expression on viral replication by measuring the total amount of infectious viral particles released by Cy-1 glioma cells in vitro. Figure 3A shows a significant reduction in viral titers in cells upon CCN1 induction. Consistent with reduced virus replication, we also found a reduction in the ability of OV to kill glioma cells expressing CCN1 (Figure 3B). To test the in vivo relevance of these findings, we examined the impact of CCN1 induction on virus replication in subcutaneous tumors. Mice bearing Cy-1 tumors were fed sucrose water±dox to induce CCN1 expression, two days prior to infection with rHSVQ1. Two days post OV infection, viral progeny was isolated and quantified. We found tumoral expression of CCN1 led to a significant reduction viral progeny by 5.6 fold (Figure 3C); a difference which reduces viral anti-tumor efficacy in vivo (Supplementary figure S4). Collectively, these results demonstrate reduced virus replication and reduced killing of glioma cells with increased levels of CCN1 both in vitro and in vivo.
Figure 3. CCN1 in the ECM limits OV replication and cytotoxicity.
(A) Total infectious virus particles obtained 24h post-infection of Cy-1 or control LN229 cells±dox for 24h prior infection with rHSVQ1 (MOI=2.5). Data shown are fold change in number of virus particles±SEM between control and dox treated cells. (B) Percent surviving cells in infected Cy-1 cells (±dox) relative to uninfected (±dox) cells on days 1, 2, and 3 post infection with rHSVQ1 (MOI=1). Data shown are percent cell survival in infected Cy-1 cells±dox at different time points relative to uninfected cells (day 0). (C) Reduced viral replication in tumors induced to express CCN1. Mice implanted with Cy-1 or LN229 cells fed sucrose±dox were infected intra-tumorally with rHSVQ1 as described in methods. 48h post infection the number of virus particles in each tumor was measured by a standard plaque assay. Data shown are fold change in number virus particles±SEM between control and dox treated cells. ns=not significant, *P<0.05, ***P<0.001.
Transcript profiling uncovered CCN1 mediated induction of type-I IFN response
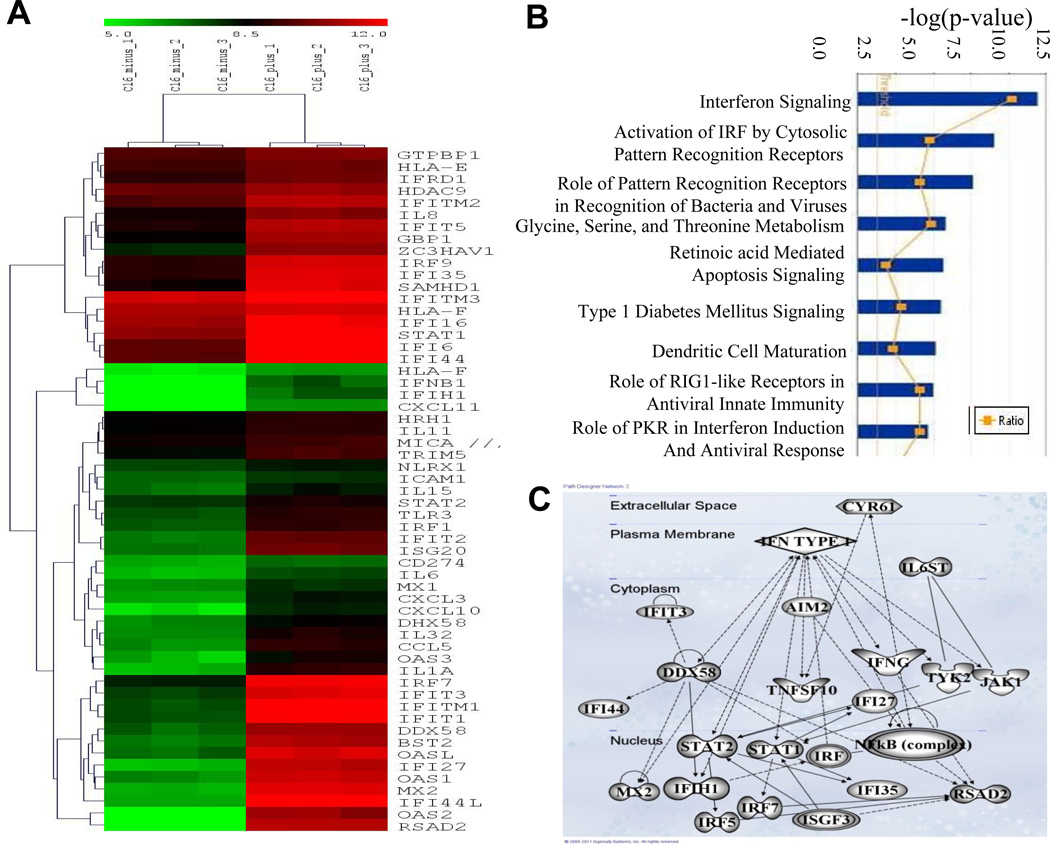
The ECM has been shown to influence cellular gene expression through its interaction with cell surface receptors (20). Transcript profiling of Cy-1 glioma cells induced to express CCN1 revealed a significant induction of the anti-viral type-I IFN pathway (Figure 4A). To identify functional networks and gene ontologies, we analyzed the upregulated gene expression data using Ingenuity Pathway Analysis software. Investigating key biological functions linked to CCN1 gene expression, we found the main functions of genes upregulated with CCN1 were as follows: interferon signaling, activation of interferon regulatory factor (IRF) by cytosolic pattern recognition receptors, and recognition of bacteria and viruses by pattern recognition receptors (Figure 4B). Ingenuity’s Top Network Analysis revealed a highly significant relationship between the genes differentially expressed by CCN1 induction and regulation of the antimicrobial response, inflammatory response, and infection mechanism in glioma cells (Figure 4C). Interestingly both IPA and a detailed PubMed analysis did not reveal a published link between type-I IFN activation and CCN1 expression in ECM.
Figure 4. Transcript profiling of Cy-1 cells induced to express CCN1.
(A) Heat map representing hierarchical clustering of a subset of the differentially regulated genes, plotted using the log 2 values of the genes with p<0.05 (unpaired t test) that are involved in the type-I IFN response. Each column represents a sample plotted in triplicate and each row in the heat map represents a gene that is differentially regulated in that particular comparison of samples. The color scale represents the degree of expression of the gene, green being the lowly expressed (below −3.0) and red being the highly expressed (above +3.0) genes in the sample sets, with black as the center of the scale at ‘0’. (B) Biological functions associated with genes significantly changed by the induction of CCN1. The significance of each canonical pathway is determined based upon the p-values determined using right tailed Fisher’s exact test and with a threshold less than 0.05. The top 9 possible canonical pathways of the genes induced by CCN1 induction are shown. The ratio of number of genes in a given pathway satisfying the cutoff and total number of genes present in that pathway was determined by IPA. (C) Ingenuity Pathway Analysis (IPA) generated pathway associated with type-I interferon responsive genes expressed upon induction of CCN1. Solid lines represent a direct interaction; dotted lines represent an indirect interaction.
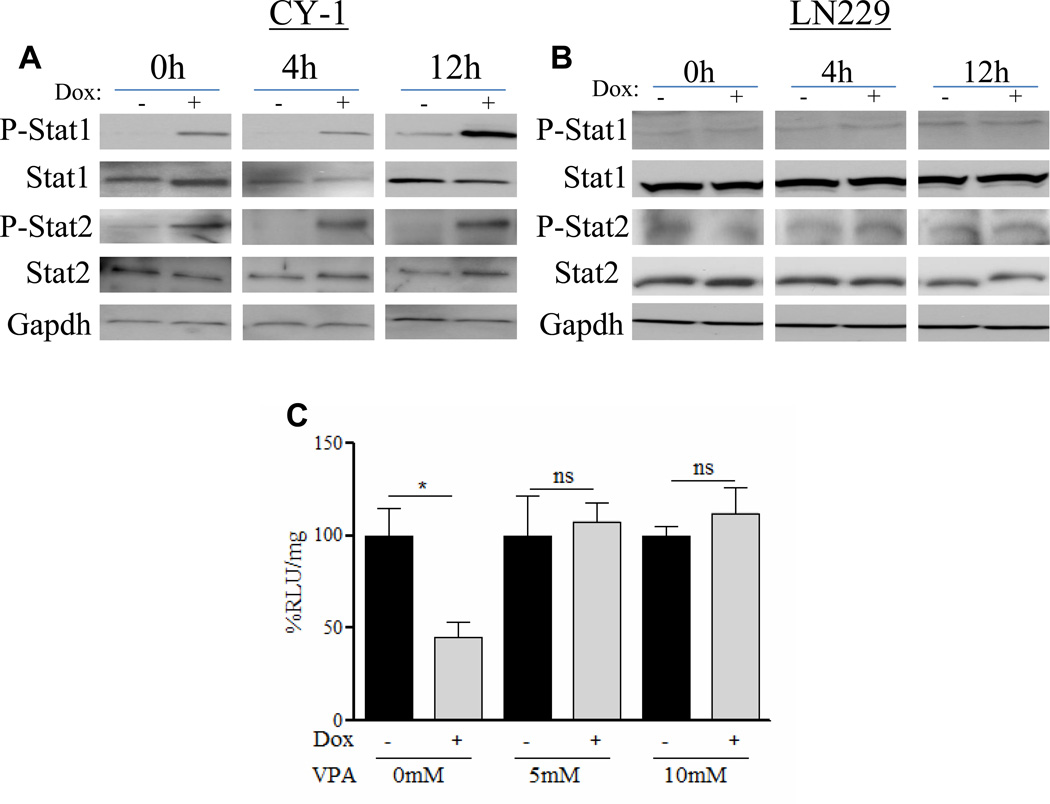
Real time quantitative PCR analysis was utilized to verify induction of a subset of the type-I IFN responsive genes involved in the antiviral defense response in these cells (Table 1). Statistically significant induction of IFNs α and β along with downstream regulatory genes such as signal transducers and activators of transcription 1 and 2 (Stat1 and Stat2), double stranded RNA-dependent protein kinase (PKR), interferon regulatory factors (IRF) 1, 3, and 7, and 2’,5’-oligoadenylate synthetase 2 (OAS2) was observed. These genes were further upregulated in Cy-1 cells expressing CCN1 following infection with rHSVQ1 suggesting an enhanced activation of the type-I IFN response by CCN1 (Table 1). Consistent with this, western blot analysis of Cy-1 cell lysates revealed increased phosphorylation of both Stat1 and Stat2 in cells induced to express CCN1 in the presence and absence of OV infection, suggesting CCN1 both activates and exacerbates the innate cellular antiviral response. No difference was found in phosphorylation status of Stat1 or Stat2 in control LN229 cells treated with dox (Figure 5A–B).
Table 1.
CCN1 induces expression of type-I interferon response gene in the presence and absence of OV
| Gene Name |
Without OV No Dox |
Dox | Fold Induced |
Pvalue | With OV No Dox |
Dox | Fold Induced |
P value |
|---|---|---|---|---|---|---|---|---|
| IFNB | 2.385 | 18.432 | 11.116 | 0.0009 | 22.563 | 36.525 | 1.619 | 0.0302 |
| STAT1 | 1.668 | 13.737 | 8.236 | 0.0001 | 5.556 | 52.442 | 9.439 | <0.0001 |
| IRF7 | 1.748 | 13.748 | 7.865 | 0.0321 | 15.577 | 229.883 | 14.758 | 0.0005 |
| IRF1 | 2.0863 | 12.363 | 5.926 | 0.0001 | 21.151 | 47.901 | 2.265 | 0.0086 |
| PKR | 1.437 | 5.873 | 4.087 | 0.0029 | 2.969 | 25.266 | 8.510 | 0.0001 |
| OAS2 | 16.776 | 62.153 | 3.705 | 0.0094 | 92.739 | 408.286 | 4.402 | <0.0001 |
| IRF9 | 4.922 | 16.379 | 3.281 | 0.0042 | 32.29 | 129.51 | 4.011 | <0.0001 |
| STAT2 | 3.705 | 11.471 | 3.096 | 0.0062 | 11.465 | 76.706 | 6.690 | 0.0041 |
| IFNA | 1.658 | 4.235 | 2.554 | 0.0386 | 20.835 | 32.949 | 1.581 | 0.0366 |
| IRF3 | 1.332 | 2.172 | 1.631 | 0.0101 | 2.863 | 5.693 | 1.988 | 0.0013 |
Abbreviations: IFN, Interferon; STAT, signal transducer and activator; IRF, interferon regulatory factor; PKR, double-stranded RNA-dependent protein kinase; OAS, ’5’-oligoadgenylate synthetase; GAPDH, glyceraldehydes-3-phosphate dehydrogenase; Dox, doxycycline.
Figure 5. Functional activation of a type-I IFN response by CCN1 mediates OV inhibition.
(A and B) Representative western blots of (A) Cy-1 and (B) LN229 cells treated±dox at 0, 4, and 12h post infection with rHSVQ1 probed for phosphorylated Stat1 and Stat2 in cells in response to CCN1 induction. Total Stat1, Stat2, and GAPDH protein levels were utilized as controls. (C) Cy-1 cells±dox were incubated with valproic acid (VPA) for 16h prior to infection with rHsvQ1-IE4/5-Luc (MOI=1). Virus expressed luciferase activity was quantified. Data shown are %RLU/mg±SEM in dox treated cells relative to uninduced cells. ns=not significant, *P<0.05
To test if the observed CCN1-mediated antiviral effects were dependent on activation of the type-I IFN pathway, we compared viral transgene expression in cells expressing CCN1 in the presence of valproic acid (VPA), an HDAC inhibitor known to interfere with the transcriptional activation of type-I IFN responsive genes (21–22). Figure 5C shows that VPA treatment rescued CCN1 mediated inhibition of viral transgene expression.
CCN1 mediated OV inhibition is dependent on α6β1 integrin receptor-mediated IFNα secretion
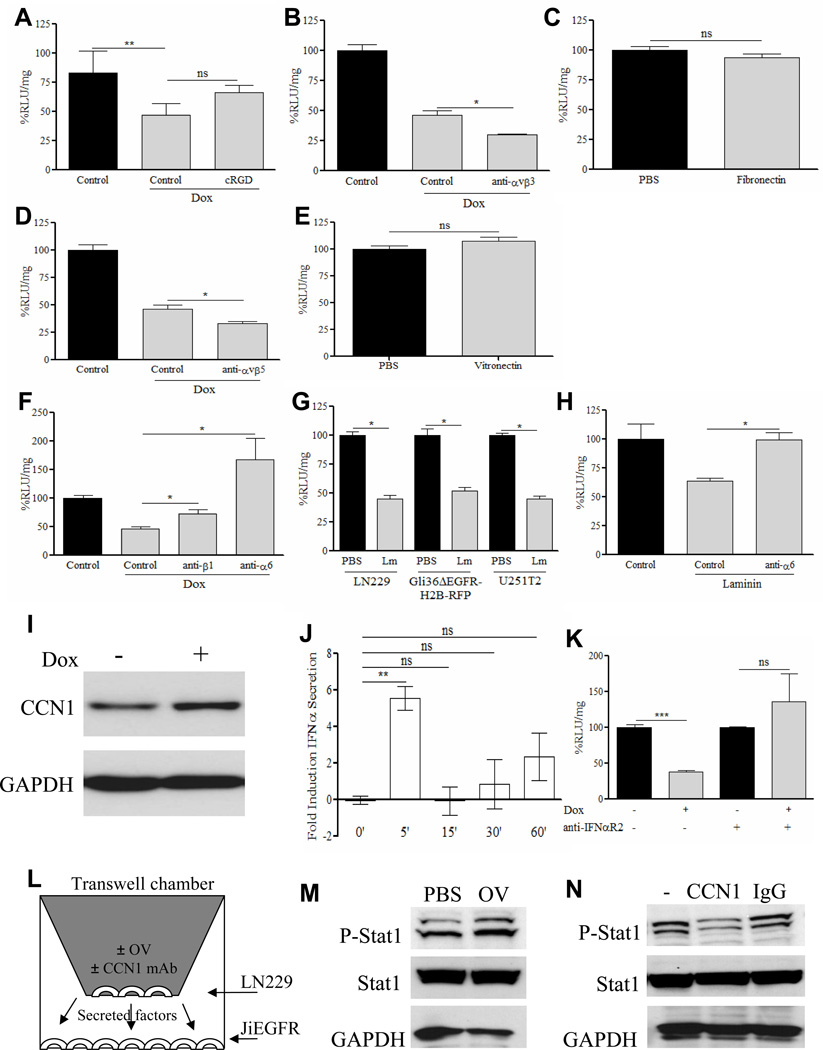
CCN1 is a multifunctional, secreted ECM protein that has been shown to bind to multiple cell surface receptors including integrins αvβ3, αvβ5, and α6β1. In order to determine the cell surface receptor through which CCN1 is mediating its antiviral effects, we investigated the potential contribution of these receptors. We first evaluated the ability of cRGD (Cilengitide; αvβ3 antagonist) and LM609 (a function-blocking monoclonal antibody against αvβ3) to rescue virus inhibition in dox induced Cy-1 cells. Figures 6A–B show that neither agent could rescue CCN1 mediated OV repression. Consistent with this result, LN229 glioma cells plated on fibronectin coated plates (a known αvβ3 activating ligand) also had no effect on OV transgene expression (Figure 6C).
Figure 6. CCN1 mediated OV inhibition is dependent on its interaction with cell surface α6β1 integrin independently of its ability to bind to αvβ3 and αvβ5.
(A&B) Dox induced Cy-1 cells were infected with rHsvQ1-IE4/5-Luc (MOI=0.1), in the presence of (A) Cilengitide (cRGD, 50ug/ml) an αvβ3 antagonist or (B) LM609 a function blocking antibody against anti-αvβ3 (50ug/ml). Viral transgene expression was determined by measuring luciferase activity. Data shown are %RLU/mg relative to control treated cells. (C) LN229 glioma cells were seeded on fibronection (a known αvβ3 agonist) coated plates (5ug/ml) or control plates and infected with rHsvQ1-IE4/5-Luc at MOI=0.1 for 24h. Viral transgene expression was determined by luciferase quantification, normalized to mg protein and represented as % RLU/mg relative to control cells. (D) Dox induced Cy-1 cells were infected with rHsvQ1-IE4/5-Luc (MOI=0.1), in the presence of P1F6, a function blocking antibody against anti- αvβ5 (50ug/ml). Viral transgene expression was determined by luciferase quantification, normalized to mg protein and represented as %RLU/mg relative to control cells. (E) LN229 glioma cells seeded on control or vitronectin (a known agonist for integrin αvβ5) coated plates (5ug/ml) were infected with rHsvQ1-IE4/5-Luc at MOI=0.1 for 24h. Viral expressed luciferase activity was measured and is represented as %RLU/mg relative to control cells. (F) Dox induced Cy-1 cells were infected with rHsvQ1-IE4/5-Luc (MOI=0.1), in the presence of function blocking antibody GoH3 against integrin α6 or P5D2 against integrin β1 (50ug/ml). Viral expressed luciferase activity was measured and is expressed as %RLU/mg relative to un-induced cells. (G) LN229, Gli36ΔEGFR-H2B-RFP, and U251T2 glioma cells were seeded on laminin (a known agonist for α6β1) coated plates (5ug/ml) or non-coated plates and infected with rHsvQ1-IE4/5-Luc at MOI=0.1 for 24h. Viral expressed luciferase activity was measured and is expressed as %RLU/mg relative to control cells. (H) LN229 glioma cells seeded on laminin coated plates (5ug/ml) were infected with rHsvQ1-IE4/5-Luc in the presence or absence of function blocking antibody GoH3 against integrin α6. Viral expressed luciferase activity was quantified, and represented as %RLU/mg relative to control cells. Cy-1 cells were incubated in the presence of dox and harvested at the indicated time points. (I) Representative western blot of Cy-1 cells treated±dox for two minutes indicating CCN1 protein induction at a very early time point. GAPDH protein level was utilized as a control. (J) Supernatants were concentrated and analyzed for changes in IFNα secretion relative to non-treated control cells by ELISA. Data are shown as the mean±SEM of at least three replicates and represent at least three different experiments. (K) Cy-1 cells±dox were incubated with an antibody against the IFNα receptor chain 2 (50μg/ml) prior to infection with rHsvQ1-IE4/5-Luc (MOI=1). Virus expressed luciferase activity (RLU) was quantified. Data shown are %RLU/mg±SEM in dox treated cells relative to uninduced cells. (L) Schematic of experimental setup for 6M–N, showing the culture of JiEGFR cells with infected or uninfected LN229 cells. (M) Western blot for Stat1 phosphorylation of JiEGFR cells, cultured in the presence of secreted medium from infected or uninfected LN229 cells. Total Stat1 and GAPDH were used as controls. (N) Western blot for Stat1 phosphorylation of JiEGFR cells, cultured in the presence of secreted medium from infected LN229 cells, cultured in the presence or absence of CCN1 neutralizing antibodies. Total Stat1 and GAPDH were used as controls. ‘=minute, ns=not significant, *P<0.05, **P<0.01, ***P<0.001
We next assessed the potential role of integrin αvβ5 in CCN1 mediated OV inhibition. Figure 6D shows that treatment of glioma cells with P1F6 (an αvβ5 function blocking antibody) did not rescue CCN1 mediated reduction of OV. Moreover, activation of cell surface αvβ5 by vitronectin (a known αvβ5 activating ligand), also did not affect OV transgene expression (Figure 6E).
CCN1 binds to and activates integrin α6β1 on fibroblasts, vascular smooth muscle cells, and vascular endothelial cells (23–25). More recently, glioblastoma stem cells were also found to express the integrin α6 chain of this heterodimeric receptor (26). To investigate if CCN1 mediated OV inhibition was due to the activation of integrin α6β1 on glioma cells we measured the impact of function-blocking monoclonal antibodies against α6 and β1 on viral infection. Figure 6F shows that the inhibition in OV transgene expression observed when Cy-1 cells express CCN1 is rescued in the presence of function-blocking monoclonal antibodies against either α6 or β1. Consistent with this, glioma cells plated on laminin (a known α6β1 activating ligand) leads to a significant inhibition of OV transgene expression (Figure 6G). This ability of laminin to inhibit viral transgene expression is rescued in the presence of a function-blocking antibody against integrin α6, indicating that CCN1 mediated activation of integrin α6β1 on glioma cells leads to the induction of an anti-viral defense response (Figure 6H). Supplementary figure S5A shows presence of integrin α6 on all glioma cell lines tested.
Integrin mediated cell-matrix interactions are known to play a role in protein secretion (27–29), and among these, integrin α6β1 has been shown to mediate insulin secretion in primary rat β-cells (30–31). In order to further delineate the underlying mechanism behind integrin α6β1 activation of the type-I IFNs we performed an ELISA looking for changes in the IFNα secretion pattern in the presence of CCN1. A time course analysis with Cy-1 cells induced to express CCN1 indicated that this protein is induced within minutes after treatment with dox (Figure 6I). Interestingly, minutes after protein induction, we observed a rapid burst in the secretion of IFNα (Figure 6J) independent of its gene expression (Supplementary Figure S5B) and independent of dox treatment (data not shown). This suggests that CCN1 protein induction mediates a rapid type-I IFN secretion in glioma cells. Additionally, CCN1 mediated OV transgene inhibition was rescued by IFNα2 receptor blocking antibody indicating that secreted IFNα was required for this antiviral effect in vitro (Figure 6K). Consistent with this, CCN1 did not have an antiviral effect on U87ΔEGFR cells, which have a homozygous deletion of the entire IFNA/IFNW gene cluster and of the IFNB1 gene (Supplementary Figure S5C) (32–34).
To examine if CCN1 induced by OV infection could activate this antiviral response in adjacent uninfected cells, we cultured JiEGFR cells, which are resistant to HSV infection (12) (Supplementary Figure S6), in the presence of LN229 cells infected with OV. Figure 6L–M shows increased phosphorylation of Stat1 in JiEGFR cells. More significantly, this increased phosphorylation is rescued in the presence of CCN1 neutralizing antibodies (Figure 6N) indicating that endogenous CCN1 induced after OV infection could activate Jak/Stat signaling in adjacent uninfected cells.
Collectively, these results indicate that increased expression of CCN1 in the tumor microenvironment leads to the activation of integrin α6β1 on glioma cells, resulting in the secretion of IFNα and activation of an antiviral response in the tumor microenvironment which ultimately limits OV infection and replication (Supplementary Figure S7).
DISCUSSION
CCN1 is a pleiotropic ECM molecule which binds several cell surface receptors, and modulates cell signaling events affecting diverse cellular functions including proliferation, adhesion, and migration. In the current study, we report the induction of CCN1 gene expression in glioma cells infected with several different viruses. We further show that CCN1 in the tumoral ECM binds to cell surface α6β1 integrin receptors to activate an innate anti-viral defense response by the secretion of IFNα. Collectively, these results suggest that secretion of CCN1 upon infection orchestrates an “alarm signal” in the tumor microenvironment which activates an antiviral state in adjacent uninfected cells leading to increased resistance to viral infection/replication (Figure 6L–N). To our knowledge, this is the first report linking integrin binding and activation by extracellular CCN1 to secretion of IFNα and activation of the antiviral type-I IFN response. Although CCN1’s role as a pro-inflammatory molecule is beginning to be realized (35), its effect on the type-I IFN response is quite novel and this is the first study linking an inhibitory role of CCN1 to OV therapy. This study has several implications for biological therapies and viral infections.
CCN1’s role in tumor biology has been extensively studied, and depending on the tissue type has been found both pro- and anti-tumorigenic (36). Apart from negatively modulating the cellular response to OV therapy, the expression of CCN1 protein in breast, prostate, and ovarian cancer correlates with a poor prognosis (37–39). Conversely, its expression in lung, endometrial, and gastric cancer has been associated with a better prognosis and outcome (40–42). Though the reason underpinning CCN1’s opposing effect in different tissue has not been elucidated, it may depend in part on the context in which CCN1 is expressed differing by the presence of co-activators and repressors, and the receptor expression profiles present in different tissues.
Here we show that CCN1 activates a type-I IFN pro-inflammatory cascade in glioma cells by binding to and activating the α6β1 integrin receptor and inducing secretion of IFNα. We show that CCN1 expression not only upregulates the type-I IFNs α and β, but also several downstream mediators of the type-I IFN response known to play key roles in the cellular antiviral defense response such as PKR and OAS (43–44). These results suggest that while expression of CCN1 leads to increased angiogenesis and invasion in the tumors, it also interferes with oncolytic viral therapy and inhibition of this pathway may provide opportunities to enhance OV anti-tumor efficacy.
Recently, integrin α6 has been recognized as an enrichment marker for glioblastoma stem cells (GSCs) (26), and was found to be coexpressed with CD133 (a widely accepted glioma stem cell marker) in GBM biopsies. Apart from increased tumorigenicity, glioma stem cells have been shown resistant to both radiation and chemotherapy (45). In U87 glioma cells, it has been shown that stable cell surface expression of integrin α6β1 leads to both enhanced proliferation and decreased apoptosis in vitro and in vivo (46). The results from our study indicate that CCN1 mediated activation of integrin α6 contributes to the reduced efficacy of viral oncolytic therapy and it will be interesting to understand how the CCN1-integrin α6 interaction plays a role in glioma therapeutic resistance.
In conclusion, this is the first study to reveal the effect of a secreted matricellular integrin binding protein on the initiation of an innate type-I IFN cellular defense response to virus infection. This study suggests that therapeutic interventions which inhibit the CCN1-integrin α6 interaction may sensitize glioma to chemo and radiation therapies and viral oncolysis. Future studies will evaluate the extent to which expression of CCN1 and/or integrin α6 receptor on tumors can serve as a predictor of patient response to oncolytic viral therapy.
Supplementary Material
Acknowledgments
Financial Support: This study was supported by the NIH grants: RO1NS064607-01, RO1CA150153-01, and P30NS045758.
GRANT SUPPPORT: RO1NS064607-01, RO1CA150153-01, and P30NS045758.
REFERENCES
- 1.Stupp R, Hegi ME, Gilbert MR, Chakravarti A. Chemoradiotherapy in malignant glioma: standard of care and future directions. J Clin Oncol. 2007 Sep 10;25(26):4127–4136. doi: 10.1200/JCO.2007.11.8554. [DOI] [PubMed] [Google Scholar]
- 2.Kaur B, Cripe TP, Chiocca EA. "Buy one get one free": armed viruses for the treatment of cancer cells and their microenvironment. Curr Gene Ther. 2009 Oct;9(5):341–355. doi: 10.2174/156652309789753329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurozumi K, Hardcastle J, Thakur R, Shroll J, Nowicki M, Otsuki A, et al. Oncolytic HSV-1 infection of tumors induces angiogenesis and upregulates CYR61. Mol Ther. 2008 Aug;16(8):1382–1391. doi: 10.1038/mt.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Du XY. Functional properties and intracellular signaling of CCN1/Cyr61. J Cell Biochem. 2007 Apr 15;100(6):1337–1345. doi: 10.1002/jcb.21194. [DOI] [PubMed] [Google Scholar]
- 5.Yang GP, Lau LF. Cyr61, product of a growth factor-inducible immediate early gene, is associated with the extracellular matrix and the cell surface. Cell Growth Differ. 1991 Jul;2(7):351–357. [PubMed] [Google Scholar]
- 6.Chen CC, Lau LF. Functions and mechanisms of action of CCN matricellular proteins. Int J Biochem Cell Biol. 2009 Apr;41(4):771–783. doi: 10.1016/j.biocel.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie D, Yin D, Wang HJ, Liu GT, Elashoff R, Black K, et al. Levels of expression of CYR61 and CTGF are prognostic for tumor progression and survival of individuals with gliomas. Clin Cancer Res. 2004 Mar 15;10(6):2072–2081. doi: 10.1158/1078-0432.ccr-0659-03. [DOI] [PubMed] [Google Scholar]
- 8.Hong Y, Ho KS, Eu KW, Cheah PY. A susceptibility gene set for early onset colorectal cancer that integrates diverse signaling pathways: implication for tumorigenesis. Clin Cancer Res. 2007 Feb 15;13(4):1107–1114. doi: 10.1158/1078-0432.CCR-06-1633. [DOI] [PubMed] [Google Scholar]
- 9.Xie D, Yin D, Tong X, O'Kelly J, Mori A, Miller C, et al. Cyr61 is overexpressed in gliomas and involved in integrin-linked kinase-mediated Akt and beta-catenin-TCF/Lef signaling pathways. Cancer Res. 2004 Mar 15;64(6):1987–1996. doi: 10.1158/0008-5472.can-03-0666. [DOI] [PubMed] [Google Scholar]
- 10.Kim SM, Park JH, Chung SK, Kim JY, Hwang HY, Chung KC, et al. Coxsackievirus B3 infection induces cyr61 activation via JNK to mediate cell death. J Virol. 2004 Dec;78(24):13479–13488. doi: 10.1128/JVI.78.24.13479-13488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorn A, Zhao H, Granberg F, Hosel M, Webb D, Svensson C, et al. Identification of specific cellular genes up-regulated late in adenovirus type 12 infection. J Virol. 2005 Feb;79(4):2404–2412. doi: 10.1128/JVI.79.4.2404-2412.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uchida H, Chan J, Goins WF, Grandi P, Kumagai I, Cohen JB, et al. A double mutation in glycoprotein gB compensates for ineffective gD-dependent initiation of herpes simplex virus type 1 infection. J Virol. 2010 Dec;84(23):12200–12209. doi: 10.1128/JVI.01633-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaur B, Brat DJ, Devi NS, Van Meir EG. Vasculostatin, a proteolytic fragment of brain angiogenesis inhibitor 1, is an antiangiogenic and antitumorigenic factor. Oncogene. 2005 May 19;24(22):3632–3642. doi: 10.1038/sj.onc.1208317. [DOI] [PubMed] [Google Scholar]
- 14.Terada K, Wakimoto H, Tyminski E, Chiocca EA, Saeki Y. Development of a rapid method to generate multiple oncolytic HSV vectors and their in vivo evaluation using syngeneic mouse tumor models. Gene Ther. 2006 Apr;13(8):705–714. doi: 10.1038/sj.gt.3302717. [DOI] [PubMed] [Google Scholar]
- 15.Wakimoto H, Fulci G, Tyminski E, Chiocca EA. Altered expression of antiviral cytokine mRNAs associated with cyclophosphamide's enhancement of viral oncolysis. Gene Ther. 2004 Jan;11(2):214–223. doi: 10.1038/sj.gt.3302143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoo JY, Haseley A, Bratasz A, Chiocca EA, Zhang J, Powell K, et al. Antitumor Efficacy of 34.5ENVE: A Transcriptionally Retargeted and "Vstat120"-expressing Oncolytic Virus. Mol Ther. 2011 Oct 25; doi: 10.1038/mt.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoo JY, Kim JH, Kim J, Huang JH, Zhang SN, Kang YA, et al. Short hairpin RNA-expressing oncolytic adenovirus-mediated inhibition of IL-8: effects on antiangiogenesis and tumor growth inhibition. Gene Ther. 2008 May;15(9):635–651. doi: 10.1038/gt.2008.3. [DOI] [PubMed] [Google Scholar]
- 18.Kim KH, Min YK, Baik JH, Lau LF, Chaqour B, Chung KC. Expression of angiogenic factor Cyr61 during neuronal cell death via the activation of c-Jun N-terminal kinase and serum response factor. J Biol Chem. 2003 Apr 18;278(16):13847–13854. doi: 10.1074/jbc.M210128200. [DOI] [PubMed] [Google Scholar]
- 19.Quan T, Qin Z, Xu Y, He T, Kang S, Voorhees JJ, et al. Ultraviolet irradiation induces CYR61/CCN1, a mediator of collagen homeostasis, through activation of transcription factor AP-1 in human skin fibroblasts. J Invest Dermatol. 2010 Jun;130(6):1697–1706. doi: 10.1038/jid.2010.29. [DOI] [PubMed] [Google Scholar]
- 20.Spencer VA, Xu R, Bissell MJ. Extracellular matrix, nuclear and chromatin structure, and gene expression in normal tissues and malignant tumors: a work in progress. Adv Cancer Res. 2007;97:275–294. doi: 10.1016/S0065-230X(06)97012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suh HS, Choi S, Khattar P, Choi N, Lee SC. Histone deacetylase inhibitors suppress the expression of inflammatory and innate immune response genes in human microglia and astrocytes. J Neuroimmune Pharmacol. 2010 Dec;5(4):521–532. doi: 10.1007/s11481-010-9192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otsuki A, Patel A, Kasai K, Suzuki M, Kurozumi K, Chiocca EA, et al. Histone deacetylase inhibitors augment antitumor efficacy of herpes-based oncolytic viruses. Mol Ther. 2008 Sep;16(9):1546–1555. doi: 10.1038/mt.2008.155. [DOI] [PubMed] [Google Scholar]
- 23.Leu SJ, Lam SC, Lau LF. Pro-angiogenic activities of CYR61 (CCN1) mediated through integrins alphavbeta3 and alpha6beta1 in human umbilical vein endothelial cells. J Biol Chem. 2002 Nov 29;277(48):46248–46255. doi: 10.1074/jbc.M209288200. [DOI] [PubMed] [Google Scholar]
- 24.Chen CC, Chen N, Lau LF. The angiogenic factors Cyr61 and connective tissue growth factor induce adhesive signaling in primary human skin fibroblasts. J Biol Chem. 2001 Mar 30;276(13):10443–10452. doi: 10.1074/jbc.M008087200. [DOI] [PubMed] [Google Scholar]
- 25.Grzeszkiewicz TM, Lindner V, Chen N, Lam SC, Lau LF. The angiogenic factor cysteine-rich 61 (CYR61, CCN1) supports vascular smooth muscle cell adhesion and stimulates chemotaxis through integrin alpha(6)beta(1) and cell surface heparan sulfate proteoglycans. Endocrinology. 2002 Apr;143(4):1441–1450. doi: 10.1210/endo.143.4.8731. [DOI] [PubMed] [Google Scholar]
- 26.Lathia JD, Gallagher J, Heddleston JM, Wang J, Eyler CE, Macswords J, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010 May 7;6(5):421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quan WY, Ko JA, Yanai R, Nakamura Y, Nishida T. Integrin-mediated inhibition of interleukin-8 secretion from human neutrophils by collagen type I. J Leukoc Biol. 2009 Mar;87(3):487–491. doi: 10.1189/jlb.0209098. [DOI] [PubMed] [Google Scholar]
- 28.Mousa SA, Lorelli W, Campochiaro PA. Role of hypoxia and extracellular matrix-integrin binding in the modulation of angiogenic growth factors secretion by retinal pigmented epithelial cells. J Cell Biochem. 1999 Jul 1;74(1):135–143. [PubMed] [Google Scholar]
- 29.Hammar E, Tomas A, Bosco D, Halban PA. Role of the Rho-ROCK (Rho-associated kinase) signaling pathway in the regulation of pancreatic beta-cell function. Endocrinology. 2009 May;150(5):2072–2079. doi: 10.1210/en.2008-1135. [DOI] [PubMed] [Google Scholar]
- 30.Bosco D, Meda P, Halban PA, Rouiller DG. Importance of cell-matrix interactions in rat islet beta-cell secretion in vitro: role of alpha6beta1 integrin. Diabetes. 2000 Feb;49(2):233–243. doi: 10.2337/diabetes.49.2.233. [DOI] [PubMed] [Google Scholar]
- 31.Parnaud G, Hammar E, Rouiller DG, Armanet M, Halban PA, Bosco D. Blockade of beta 1 integrin-laminin-5 interaction affects spreading and insulin secretion of rat beta-cells attached on extracellular matrix. Diabetes. 2006 May;55(5):1413–1420. doi: 10.2337/db05-1388. [DOI] [PubMed] [Google Scholar]
- 32.Alain T, Lun X, Martineau Y, Sean P, Pulendran B, Petroulakis E, et al. Vesicular stomatitis virus oncolysis is potentiated by impairing mTORC1-dependent type I IFN production. Proc Natl Acad Sci U S A. 2010 Jan 26;107(4):1576–1581. doi: 10.1073/pnas.0912344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wollmann G, Robek MD, van den Pol AN. Variable deficiencies in the interferon response enhance susceptibility to vesicular stomatitis virus oncolytic actions in glioblastoma cells but not in normal human glial cells. J Virol. 2007 Feb;81(3):1479–1491. doi: 10.1128/JVI.01861-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olopade OI, Jenkins RB, Ransom DT, Malik K, Pomykala H, Nobori T, et al. Molecular analysis of deletions of the short arm of chromosome 9 in human gliomas. Cancer Res. 1992 May 1;52(9):2523–2529. [PubMed] [Google Scholar]
- 35.Kular L, Pakradouni J, Kitabgi P, Laurent M, Martinerie C. The CCN family: A new class of inflammation modulators? Biochimie. 2011 Mar;93(3):377–388. doi: 10.1016/j.biochi.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Dhar A, Ray A. The CCN family proteins in carcinogenesis. Exp Oncol. Mar;32(1):2–9. [PubMed] [Google Scholar]
- 37.Xie D, Miller CW, O'Kelly J, Nakachi K, Sakashita A, Said JW, et al. Breast cancer. Cyr61 is overexpressed, estrogen-inducible, and associated with more advanced disease. J Biol Chem. 2001 Apr 27;276(17):14187–14194. doi: 10.1074/jbc.M009755200. [DOI] [PubMed] [Google Scholar]
- 38.Sun ZJ, Wang Y, Cai Z, Chen PP, Tong XJ, Xie D. Involvement of Cyr61 in growth, migration, and metastasis of prostate cancer cells. Br J Cancer. 2008 Nov 18;99(10):1656–1667. doi: 10.1038/sj.bjc.6604712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gery S, Xie D, Yin D, Gabra H, Miller C, Wang H, et al. Ovarian carcinomas: CCN genes are aberrantly expressed and CCN1 promotes proliferation of these cells. Clin Cancer Res. 2005 Oct 15;11(20):7243–7254. doi: 10.1158/1078-0432.CCR-05-0231. [DOI] [PubMed] [Google Scholar]
- 40.Chen PP, Li WJ, Wang Y, Zhao S, Li DY, Feng LY, et al. Expression of Cyr61, CTGF, and WISP-1 correlates with clinical features of lung cancer. PLoS One. 2007;2(6):e534. doi: 10.1371/journal.pone.0000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chien W, Kumagai T, Miller CW, Desmond JC, Frank JM, Said JW, et al. Cyr61 suppresses growth of human endometrial cancer cells. J Biol Chem. 2004 Dec 17;279(51):53087–53096. doi: 10.1074/jbc.M410254200. [DOI] [PubMed] [Google Scholar]
- 42.Maeta N, Osaki M, Shomori K, Inaba A, Kidani K, Ikeguchi M, et al. CYR61 downregulation correlates with tumor progression by promoting MMP-7 expression in human gastric carcinoma. Oncology. 2007;73(1–2):118–126. doi: 10.1159/000121000. [DOI] [PubMed] [Google Scholar]
- 43.Thomis DC, Samuel CE. Mechanism of interferon action: evidence for intermolecular autophosphorylation and autoactivation of the interferon-induced, RNA-dependent protein kinase PKR. J Virol. 1993 Dec;67(12):7695–7700. doi: 10.1128/jvi.67.12.7695-7700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cassady KA, Gross M, Roizman B. The second-site mutation in the herpes simplex virus recombinants lacking the gamma134.5 genes precludes shutoff of protein synthesis by blocking the phosphorylation of eIF-2alpha. J Virol. 1998 Sep;72(9):7005–7011. doi: 10.1128/jvi.72.9.7005-7011.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altaner C. Glioblastoma and stem cells. Neoplasma. 2008;55(5):369–374. [PubMed] [Google Scholar]
- 46.Delamarre E, Taboubi S, Mathieu S, Berenguer C, Rigot V, Lissitzky JC, et al. Expression of integrin alpha6beta1 enhances tumorigenesis in glioma cells. Am J Pathol. 2009 Aug;175(2):844–855. doi: 10.2353/ajpath.2009.080920. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.