Abstract
Fluorescence molecular tomography is a powerful tool for 3D visualization of molecular targets and pathways in vivo in small animals. Owing to the high degrees of absorption and scattering of light through tissue, the fluorescence tomographic inverse problem is inherently ill-posed. In order to improve source localization and the conditioning of the light propagation model, multiple sets of data are acquired by illuminating the animal surface with different spatial patterns of near-infrared light. However, the choice of these patterns in most experimental setups is ad hoc and suboptimal. This paper presents a systematic approach for designing efficient illumination patterns for fluorescence tomography. Our objective here is to determine how to optimally illuminate the animal surface so as to maximize the information content in the acquired data. We achieve this by improving the conditioning of the Fisher information matrix. We parameterize the spatial illumination patterns and formulate our problem as a constrained optimization problem that, for a fixed number of illumination patterns, yields the optimal set of patterns. For geometric insight, we used our method to generate a set of three optimal patterns for an optically homogeneous, regular geometrical shape and observed expected symmetries in the result. We also generated a set of six optimal patterns for an optically homogeneous cuboidal phantom set up in the transillumination mode. Finally, we computed optimal illumination patterns for an optically inhomogeneous realistically shaped mouse atlas for different given numbers of patterns. The regularized pseudoinverse matrix, generated using the singular value decomposition, was employed to reconstruct the point spread function for each set of patterns in the presence of a sample fluorescent point source deep inside the mouse atlas. We have evaluated the performance of our method by examining the singular value spectra as well as plots of average spatial resolution versus estimator variance corresponding to different illumination schemes.
1. Introduction
Optical imaging techniques have long been popular for generating contrast representing molecular processes in tissue. Over the past decade, their applications have extended from microscopy to macroscopic imaging of deep-tissue optical sources by exploiting the near-infrared (NIR) window where water, oxyhemoglobin, and deoxyhemoglobin, the primary absorbers in tissue, have relatively low absorption coefficients allowing light to penetrate several centimeters inside tissue (Weissleder and Ntziachristos 2003). The non-ionizing nature of NIR light, the availability of a variety of highly specific fluorescent probes, and, finally, low cost offer these methods some leverage over existing radiological techniques for molecular imaging (Hebden et al 1997). However, penetration depths of only a few centimeters are insufficient for most clinical deep-tissue imaging applications. Thus the success of 3D optical imaging methods in clinical diagnostics is restricted to only a few applications. These include NIR spectroscopy and diffuse optical tomography used for mapping brain structure and function in neonates (Koizumi et al 2003, Villringer and Chance 1997, Hebden et al 2002) and optical mammography for breast cancer screening (Ntziachristos and Chance 2001, Culver et al 2003). Optical tomographic methods show great promise in preclinical research, which is a valuable translational tool between in vitro studies and clinical applications. Fluorescence molecular tomography (FMT) and bioluminescence tomography (BLT) have emerged as promising low-cost alternatives to PET and SPECT for functional imaging in small animals, thus greatly impacting diagnostics, drug discovery, and therapeutics (Ntziachristos et al 2005, Gibson et al 2005). Although plagued by tissue autofluorescence, FMT has many advantages over BLT. Compared to most bioluminescent probes, commonly used fluorophores emit light at longer wavelengths (where tissues are less absorbing) consequently offering higher detected signal strengths (Contag and Bachmann 2002, Hielscher 2005). Additionally multiple illumination patterns can be used to generate different mappings from the source space to the detector space making the FMT problem less ill-posed than the BLT problem (Chaudhari et al 2005). The success of FMT can be attributed to a number of breakthroughs.
The availability of a variety of new NIR fluorescent dyes, active or activatable fluorescent biomarkers, and fluorescent proteins expressed by reporter genes has enabled visualization of gene expression and several cellular and subcellular processes in vivo (Massoud and Gambhir 2003, Shu et al 2009).
Advances in instrumentation have led to the development of a range of imaging systems for time-domain, frequency-domain, and continuous-wave (CW) FMT (Kumar et al 2008, Godavarty et al 2005, Zavattini et al 2006). Some of the newer systems feature non-contact tomographic detection using CCD cameras with free-space detection geometries and/or innovative optics for full-surface visualization, thus eliminating the need for optical fibers and matching fluids (Li et al 2009, Graves et al 2003, Patwardhan et al 2005).
Finally, a number of theoretical and computational advances have led to the development of realistic forward models and robust inverse methods. Popular methods employed to model photon propagation through tissue for solving the forward problem include Monte Carlo methods (Hayakawa et al 2001, Boas et al 2002, Chen and Intes 2009) as well as analytical and numerical solutions to the radiative transport equation (Klose et al 2002) and the diffusion equation (Rice et al 2001, Dutta et al 2008, Arridge et al 1993) subject to different boundary conditions (Haskell et al 1994). These forward modeling schemes coupled with fast inversion techniques (Roy and Sevick-Muraca 2001, Ahn et al 2008, Zacharopoulos et al 2009) have made it feasible to reconstruct FMT images accurately and efficiently.
Despite their tremendous potential and increasing popularity, fluorescence tomographic techniques are confounded by high degrees of absorption and scattering of photons propagating through tissue, making the FMT problem ill-posed. One approach for alleviating this problem and improving source localization is to harness spectral variations of tissue optical properties by using multispectral illumination and/or detection (Zacharakis et al 2005, Chaudhari et al 2009). Another approach is to exploit the degree of freedom offered by external illumination in FMT and design a set of spatial illumination patterns that improve the conditioning of the forward model matrix (Dutta et al 2009), and that is the focus of this paper.
FMT setups typically acquire data-sets corresponding to different surface illumination patterns. These patterns generate different excitation fields over the volume, which tune the system matrix. FMT setups available today employ illumination schemes chiefly guided by the availability and simplicity of the light source. Several of these use laser sources with focusing or diffuser lenses to generate point or distributed patch patterns (Graves et al 2003, Zavattini et al 2006, Li et al 2009). Other approaches include raster scanning (Patwardhan et al 2005, Joshi et al 2006) and structured light or spatially modulated illumination patterns (Lukic et al 2009). Most of these approaches are ad hoc. Although various performance metrics could be used to theoretically compare these standard approaches, it is impossible to make an exhaustive set of comparisons, since, for a given number of illumination patterns being used for an experiment, infinitely many designs exist. Therefore, the question we address in this paper is as follows: given a fixed number of illumination patterns, how do we design these patterns so as to maximize the information in the acquired data?
With the availability of Texas Instruments Digital Light Processor (DLP®) chips (Hornbeck 1996, Dudley et al 2003) which work in the near-infrared range and give us precise control over the spatial intensity distribution, it is feasible to generate any set of spatial illumination patterns with grayscale intensity variation (Gardner et al 2010, Bassi et al 2008, Konecky et al 2009, Bélanger et al 2010). The focus of this paper is to compute the set of illumination patterns for CW FMT that maximize the information content in the data by improving the conditioning of the Fisher information matrix. We formulate our problem as a constrained optimization problem that minimizes a cost function derived from the Fisher information matrix and computes the parameterized set of optimal spatial patterns.
Section 2 of this paper provides a description of the CW FMT problem. The formulation of the optimization problem that generates the optimal set of patterns is presented in section 3. In section 4, we describe the methods used to solve the forward and inverse problems, the optimization procedure, and the performance metrics used to evaluate different illumination schemes. Section 5 presents optimal patterns on a cylinder, a cuboidal tissue phantom, and a mouse atlas along with performance comparisons of illumination schemes for the atlas. Finally, a discussion of the results is presented in section 6.
2. Background
2.1. The forward problem
In continuous wave fluorescence molecular tomography (CW FMT), the 3D biodistribution of the fluorophore inside an animal volume is computed from the steady-state surface fluorescence photon density measured using a CCD camera. With the diffusion approximation, the CW FMT problem can be described by a set of coupled partial differential equations (PDEs) (Hutchinson et al 1995, Milstein et al 2004, Pogue and Patterson 2006):
| (1) |
| (2) |
Here is the diffusion coefficient and μa (r, λ) and , respectively, are the absorption and reduced scattering coefficients at a position r and a wavelength λ. For a surface illumination pattern w(r, λex) at an excitation wavelength λex, the corresponding excitation field, Φ(r, λex), over an animal volume, Ω, can be computed by solving (1). For a known fluorophore distribution q(r) and emission spectral coefficient η(λem), the emission field, Φ(r, λem), can be computed by solving (2) for the previously computed excitation field. In FMT, we assume prior knowledge of the animal surface geometry and tissue optical properties, μa (r, λ) and . The diffusion approximation is valid under the condition . This assumption is valid for most tissue types at near-infrared wavelengths, except near boundaries and in non-scattering parts of some organs (e.g. ventricles in the brain and alveolar sacs in the lungs) (Dehghani et al 1999). Appropriate boundary conditions must be imposed while solving (1) and (2) (Haskell et al 1994, Aronson 1995). The Robin boundary condition, for example, requires that the total inwardly directed photon current at the boundary be zero:
| (3) |
| (4) |
Here ν̂ denotes the outward unit normal at position r on the boundary, ∂Ω, while G depends on the refractive index mismatch at the boundary (Schweiger et al 1995).
The PDEs (1) and (2) can be decoupled and solved subject to conditions (3) and (4), respectively, by replacing the source terms on the right-hand sides of (1) and (2) by point sources (delta functions) at different locations. In a discretized domain with nd detector nodes on the animal surface and ns point source locations distributed inside the volume, the excitation forward model at an excitation wavelength λex can be expressed as a matrix Aex ∈ ℝns×nd, with the j th column of this matrix representing the excitation field caused by illuminating the j th surface node. Similarly the emission forward model at an emission wavelength λem can be expressed as a matrix Aem ∈ ℝnd×ns. The j th column of this matrix represents the surface fluorescence pattern due to the j th internal point source. For a set of p illumination patterns, we discretize the intensity distribution at the surface, w(r, λex), as a set of vectors wk ∈ ℝnd, where k = 1: p. The solution to the forward problem is a system matrix dependent on Aex, Aem, and all the wk vectors.
The block of the system matrix corresponding to the kth illumination pattern can be obtained by diagonally scaling the emission forward model matrix by the excitation intensities at each internal point:
| (5) |
Here is a diagonal matrix representing the excitation field due to wk and is given by
| (6) |
where is the ith component of the vector dk computed from
| (7) |
The full system matrix, A ∈ ℝpnd×ns, is obtained by vertically concatenating the individual forward model matrices for a set of p different illumination patterns:
| (8) |
Here the prime symbol (′) represents matrix transpose.
2.2. The inverse problem
The measured steady-state surface fluorescence patterns corresponding to p different illumination patterns can be stacked up as a single data vector, b ∈ ℝpnd. The unknown fluorophore distribution, q ∈ ℝns, can be computed by solving the following linear system of equations:
| (9) |
CW FMT setups commonly employ one or more mirrors that make the entire surface of the object (animal or phantom) visible in a single CCD camera image (Li et al 2009, Dutta et al 2008, Chaudhari et al 2005). Such setups require CCD cameras with a high dynamic range. Mapping of this data from the CCD image space to the object space requires knowledge of the object surface as well as calibration for various geometric and radiometric mapping parameters (Li et al 2009). The data vector, b, is obtained by interpolating CCD pixel-wise intensity values onto the surface mesh of the animal or phantom being imaged using the calibrated mapping scheme. Its size, therefore, depends on the mesh density.
We pose the image reconstruction problem as an optimization problem that seeks to minimize a regularized least-squares cost function:
| (10) |
Here q̂ is the reconstructed 3D fluorescent source distribution and α is the Tikhonov regularization parameter (Tikhonov and Arsenin 1977). The first term is a data-fitting term, while the second term is a regularization or a penalty term that smooths the reconstructed image. The problem in (10) can be solved analytically using singular value decomposition (SVD) or using a variety of iterative methods, the former being the approach adopted in this paper. Given the SVD, A = UΣV′, of the system matrix, this problem can be solved using the regularized pseudoinverse method:
| (11) |
The system matrix, A, is typically very large in size, with many more rows (pnd) than columns (ns). It is often infeasible to store this large matrix in computer memory. To obtain the solution in (11), it is sufficient to compute A′A and A′b as follows:
| (12) |
| (13) |
Here ‘○’ represents the Hadamard (entrywise) product and and dk can be computed from (6) and (7), respectively. The matrix A′A ∈ ℝns×ns is of the same size irrespective of the number of illumination patterns and/or the number of illumination/detection wavelengths. Its SVD, A′A = VΣ2V′, can be used in (11).
3. Formulation of the optimization problem
According to the Cramér Rao inequality, the inverse of the Fisher information sets a lower bound on the variance of an unbiased estimator (Kay 1993). This establishes a reciprocity between the variance and the information content of an estimator. For a scalar estimator, the Fisher information is a scalar, and the estimator variance can be minimized by maximizing the scalar Fisher information. However, when we are dealing with a vector estimator, the Fisher information is a matrix of elements, and there is no single scalar optimality criterion. In the field of optimal design of experiments, a variety of different functionals of the eigenvalues of the Fisher information matrix (FIM) are traditionally used as optimality criteria. For example, A-optimality minimizes the trace of the inverse of the FIM, D-optimality maximizes the determinant of the FIM, and E-optimality maximizes the smallest eigenvalue of the FIM (Pukelsheim 1993). The approach that we present here is inspired by E-optimality. Our goal is to tune the FIM so that its singular value spectrum approaches that for the perfectly conditioned case. Our method raises the entire spectrum in reference to the largest singular value. Then, for the same regularization parameter, a greater number of singular values (and the corresponding singular vectors) will affect the solution in the optimal case, leading to improved resolution characteristics.
In principle, an unlimited number of illumination patterns can be used. However, this would lead to lengthy data acquisition times (during which the animal needs to remain anesthetized), large data volumes, slow reconstruction, and possible redundancy in collected data, all of which are undesirable in practice. Therefore, we fix the number of patterns and seek to determine the optimal set for this given number.
3.1. Fisher information matrix
For an unknown vector q and an observed data vector b with probability density p(b), the FIM is given by (Kay 1993)
| (14) |
We assume an additive white Gaussian noise model with the noise vector and the data vector b = Aq + n. Without loss of generality, we ignore the constant multiple, , in the covariance. Using (5) and (8), the Fisher information matrix for this system can be expressed as
| (15) |
Using (6), (7), and (15) the FIM can be expressed as a function of the set of illumination patterns, W = [w1w2 ··· wp], where W ∈ ℝnd×p. We denote the ith row vector of Aex by and the ith column vector of Aem by . Then the ijth term of the FIM is given by
| (16) |
where we have used the notations . This implies that the FIM can be conveniently represented as a Hadamard matrix product of two matrices, one dependent on the emission model and the other on the excitation model and the illumination patterns:
| (17) |
Introducing the notation Fem = [(Aem)′Aem] and Gex = [AexWW′(Aex)′], we can rewrite (17) as
| (18) |
3.2. Optimization problem formulation
For the system matrix, A = UΣV′, to have a perfect condition number of 1, the diagonal matrix Σ of singular values should be σ I where σ is a singular value. Note that the singular value σ does not affect the condition number. Then the FIM for the ideal system is
| (19) |
Thus, for perfect conditioning, F should approach Fideal. We would like to find a set of illumination patterns, W, which minimizes the relative difference of the resultant FIM, F (W), and the ideal one, Fideal:
| (20) |
where F (W) is defined in (17) and ||·||F represents the Frobenius norm. The minimizer, (Ŵ, σ̂), of the cost function, Θ(W, σ), with the nonnegativity constraint W ≥ 0 is not unique, since for any γ, Θ(γ Ŵ, γσ̂) = Θ(Ŵ, σ̂). We therefore first solve a simpler optimization problem:
| (21) |
Then we determine the scaling factor by imposing a power constraint, ||W||F = β, on the illumination patterns. The final solution is given by
| (22) |
In practice, the illumination is scaled to exploit the maximum available source power and thus ensure a high signal-to-noise ratio. Since the optimal solution, Ŵ, which minimizes Θ, is simply a scaled version of the solution W opt, we now focus on the simpler problem given in (21).
3.3. Dimensionality reduction
The formulated problem in (21) is fourth-order and non-convex with respect to the patterns W. W is of size nd ×p, which is typically large, and hence solving this problem is computationally intensive (Dutta et al 2008). Additionally, the solutions are not necessarily spatially smooth. Smooth patterns are convenient for experimental implementation since calibration errors in mapping intensities from the light source to the object surface are likely to affect the accuracy of the forward model less. They are also desirable because an optimal set of patterns computed for a mouse atlas can be meaningfully warped to any given mouse shape, as discussed later in section 6. To allow smooth solutions and to enable speedier computation, we parameterize the illumination patterns using a small number of spatially smooth basis functions. The transformation from the basis function domain to the spatial pattern domain is given by
| (23) |
Here yk ∈ ℝm is a vector of the linear coefficients of the basis functions for the kth illumination pattern, L ∈ ℝnd×m is a matrix whose columns are the basis vectors, and Y ∈ ℝm×p with Y = [y1 y2 ··· yp], m being the number of basis vectors used to encode each spatial pattern. Then, the FIM can be represented in terms of Y as follows:
| (24) |
3.4. Modified optimization problem
With the inclusion of basis functions for dimensionality reduction, the modified optimization problem is as follows:
| (25) |
A gradient-based approach can be used to solve the optimization problem in (25). Our cost function as a function of the argument yk, where k = 1: p, is as follows:
| (26) |
where δij is the Kronecker delta function and Ψij is given by
| (27) |
The gradient, ∇k Φ, of the cost function with respect to the kth argument vector, yk, is given by
| (28) |
in (28) can be computed as follows:
| (29) |
Substituting (29) in (28), we have
| (30) |
3.5. Extension to multispectral detection
Our formulation can be extended for application to multispectral detection. We assume that, for each illumination pattern, fluorescence data are collected over s wavelength bins. The fluorescent dye is assumed to have an emission spectral coefficient ηl at the lth wavelength bin. Then the block of the system matrix corresponding to the kth illumination pattern is given by
| (31) |
The Fisher information matrix for this case is
| (32) |
where . The FIM retains the Hadamard product form with the illumination-dependent matrix Gex unchanged. This implies that, once the multispectral emission model-dependent Fem−ms matrix is precomputed, the optimization problem can be solved for the multispectral emission case without any additional computational cost.
4. Methods
The optimization problem formulated in section 3 can be solved to obtain the optimal set of patterns for a given number of patterns, p. This problem is first solved for an optically homogeneous symmetric solid shape—a cylinder—to gather geometric intuition. Next, we solve this problem for an optically homogeneous cuboidal phantom set up in the transillumination mode with the top face illuminated and the bottom face used for detection. Finally, we solve the same problem for an optically inhomogeneous and realistically shaped mouse atlas. We perform source localization studies using the optimal patterns on the mouse atlas and evaluate the performance of the patterns based on appropriate metrics.
4.1. Computation of the forward model
The finite element method (FEM) was used to solve the coupled PDEs (1) and (2) subject to boundary conditions (3) and (4), respectively, to precompute the excitation and emission forward model matrices Aex and Aem.
4.1.1. Tessellated cylinder
The cylinder is chosen since it is a regular geometrical shape without corners and elongated like a real mouse. The idea of this study is to ensure that the numerical scheme for computing the patterns preserves the natural symmetries expected in a regular geometrical shape when no spatial constraints have been imposed. For these simulations, we use a tessellated cylinder of height 40 mm and diameter 20 mm with 2373 tetrahedrons and 588 tessellation nodes. A set of 361 detector nodes on the surface are used for illumination and detection and 1564 point source locations on a uniform, volumetric grid with a 1.5 mm spacing are used for source localization. We use monochromatic excitation at λex = 650 nm and monochromatic detection at λem = 730 nm. Optical properties are assumed to be homogeneously distributed and are assumed to resemble those for muscle tissue (Alexandrakis et al 2005).
4.1.2. Tessellated cuboidal phantom
We simulate a cuboidal phantom of dimensions 40 mm × 30 mm × 20 mm. The phantom is set up in the transillumination mode, a planar imaging mode where the illumination source and the CCD camera are placed on opposite sides of the object (Ntziachristos 2006). The tessellated phantom contains 40 742 tetrahedrons and 7940 nodes. For generating the patterns, we illuminate over the 474 nodes on the top surface (z = 20 mm), detect over the 477 nodes on the bottom surface (z = 0 mm), and use 1155 internal grid points with a uniform 2.5 mm grid spacing for source localization. We use monochromatic excitation at λex = 650 nm and multispectral detection at 710 nm, 730 nm, and 750 nm. We assumed homogeneous optical properties similar to muscle tissue (Alexandrakis et al 2005). We use the emission spectrum of the Alexa Fluor 700 dye, with an emission peak at 719 nm, for computing the forward model.
4.1.3. Digimouse atlas
For more realistic simulations, we use the Digimouse atlas (http://neuroimage.usc.edu/Digimouse.html) (Dogdas et al 2007, Stout et al 2002), a labeled atlas, based on co-registered CT and cryosection images of a 28 g normal male nude mouse. Tissue optical properties are assumed to be spatially inhomogeneous and assigned organ-wise, for 17 different organs, according to published values (Alexandrakis et al 2005). The tessellated atlas volume consists of 306 773 tetrahedrons and 58 244 nodes. For generating the patterns, 810 surface nodes and 1129 internal grid points with a uniform 2.4 mm grid spacing are used. We assume that all surface nodes except those lying on the limbs and the tip of the snout are used for illumination. The Alexa Fluor 700 dye (emission peak 719 nm) is used as the fluorophore. Accordingly, we use an excitation wavelength of λex = 650 nm and perform multispectral detection at 710 nm, 730 nm, and 750 nm.
4.2. Optimization procedure
Since our cost function in (26) is continuous and its gradient is computable using (30), we adopt a gradient-based approach for optimization. We use fmincon, an inbuilt function for constrained optimization in MATLAB® (The Mathworks Inc., Natick, MA, USA). This function chooses the active set method for optimization (Gill et al 1982). The non-convex nature of the cost function implies that several local minima could exist, and our gradient-based optimization approach could get stuck in one of these. Hence, we use a multi-start approach with random initializations. This is a heuristic but straightforward approach to the global minimization problem in which the local minimization algorithm is run from many different starting points, and then the best solution is picked (Liberti and Maculan 2009).
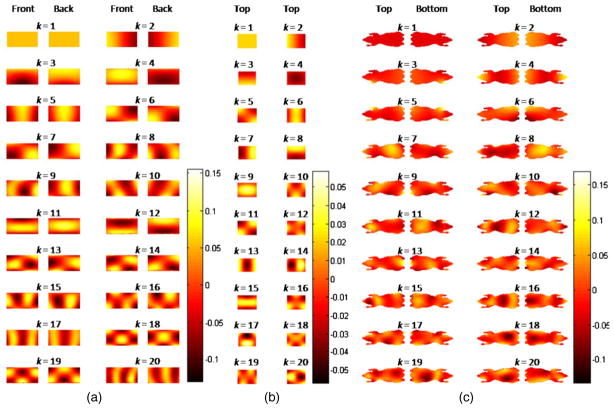
For dimensionality reduction, we define basis functions on the cylinder and atlas surfaces by treating them as smooth 2D manifolds. We use as our basis set eigenfunctions of the Laplace–Beltrami operator (Qiu et al 2006, Joshi 2008, Grenander and Miller 2007). The Laplace–Beltrami operator is a generalization of the Laplace operator on Riemannian and pseudo-Riemannian manifolds. These basis functions allow us to generate a spatially smooth solution. Their orthonormality allows efficient representation of the solution. Since the eigenvalues of a spatial differential operator are measures of the spatial rate of change of the corresponding eigenfunctions, the smaller the eigenvalue the smoother is the corresponding eigenfunction. Therefore, eigenfunctions corresponding to the m = 20 smallest eigenvalues of the operator were used as basis functions. Figure 1 shows front and back views of the tessellated cylinder, top views of the cuboidal phantom, and top and bottom views of the Digimouse atlas with the basis functions displayed on them.
Figure 1.
(a) Front and back views of a tessellated cylinder (flattened for display purposes with the cylinder axis aligned horizontally), (b) top views of the cuboidal phantom, and (c) top and bottom views of the Digimouse atlas showing eigenfunctions corresponding to the 20 smallest eigenvalues of the Laplace–Beltrami operator. The index k varies from 1 to 20 sorting the eigenvectors in an ascending order of eigenvalues. The basis functions are orthogonal and, therefore, have both positive and negative values at various points.
4.3. Inverse solution and performance metrics
The inverse problem is solved using the regularized pseudoinverse in (11). The illumination patterns on the Digimouse surface are interpolated onto a denser surface mesh with 3234 nodes. For source localization studies, a uniform grid of 9192 point sources with a 1.2 mm spacing is used. Our simulation setup uses surface data from all nodes on the atlas surface except those lying on the limbs and the tip of the snout.
In order to comparatively assess different illumination schemes, we need to establish appropriate performance metrics. We use two approaches for comparing the performance of different illumination patterns. The first approach is to examine the conditioning of the system matrix corresponding to a specific illumination pattern by looking at its singular value distribution. The second approach is to look at average resolution–variance curves to examine the behavior of the inverse solution.
4.3.1. Condition number
The condition number of a matrix is given by the ratio of its largest to its smallest singular value. Although this is a good figure-of-merit for the robustness of the system, the point spread functions (and, hence, the reconstruction results for any source distribution) depend on the singular values as well as the singular vectors (Chaudhari et al 2005). So in addition to looking at the condition number, we analyze properties of the point spread functions corresponding to different source locations inside the animal volume.
4.3.2. Resolution–variance analysis
The mean squared error of an estimator can be decomposed as a sum of the squared bias and the variance. As the regularization parameter is increased, the estimator variance decreases while the estimator bias increases (resolution worsens) and vice versa. Resolution–variance curves, which signify this inherent tradeoff, are commonly used to assess image reconstruction quality (Qi and Leahy 1999, Meng et al 2003, Chaudhari et al 2009). These curves are obtained by sweeping the regularization parameters over a range of values, computing the variance and resolution of the point spread functions, and plotting these two quantities against each other for the different values of the regularization parameter. A lower lying curve implies superior resolution for the same noise variance for the estimator corresponding to an illumination scheme. Since the regularized pseudoinverse operator in (11) is a linear operator, it can be completely characterized in terms of its point spread functions.
When the regularized pseudoinverse method is used for reconstruction, the spatial resolution and noise variance of the estimator can be computed in a closed form, thus eliminating the need for computationally expensive Monte Carlo simulations. For a true source distribution q, the mean value of the estimator for a regularization parameter α can be expressed in terms of the SVD, A = UσV′, of the system matrix as
| (33) |
We would like to compute the spatial resolution from the point spread function, PSFj, for the j th unit point source. For this source, q = ej, where ej is a unit vector with 1 as the j th element. Then PSFj can be computed by substituting q = ej in (33):
| (34) |
The spatial resolution is measured as the full width at half maximum (FWHM) computed from PSFj. Then the average spatial resolution over n different point source locations inside the object is a function of the regularization parameter, α:
| (35) |
For an additive white Gaussian noise model with the covariance , the estimator variance for the j th unit point source, q = ej, is given by
| (36) |
For fair comparison of illumination schemes involving different numbers of illumination patterns, we assume that the total acquisition time is always the same irrespective of the number of patterns used. This implies that, for a scheme with p patterns, the acquisition time per pattern is p times shorter than that for a scheme with 1 pattern. As a result, the individual data-sets are noisier by a factor of p. In other words, the data noise covariance is . With this compensation for acquisition time, the estimator variance averaged over n point source locations is given by
| (37) |
Using (35) and (37), the resolution and the variance for any illumination scheme (and hence any system matrix) averaged over different point source locations inside the volume can be computed for different values of α. The curves of resolution versus variance are obtained by plotting the two quantities against each other for different values of α. Comparison of these curves for different illumination patterns throws light on the relative magnitudes of mean squared error for estimators corresponding to different illumination patterns. It must be noted that (35) and (37) depend on only V and Σ and not on U. Therefore, as described in section 2.2, it is sufficient to compute the SVD A′A = VΣ2V′.
5. Results
By solving the optimization problem described in section 3, one can determine an optimal set of illumination patterns for a given number of patterns, p. We generated optimal sets of patterns for the tessellated cylinder, the cuboidal phantom, and the Digimouse atlas described in section 4.1. Optimal sets of patterns for the atlas were generated for different values of p. The performance of these sets of patterns was then comparatively evaluated using the metrics described in section 4.3.
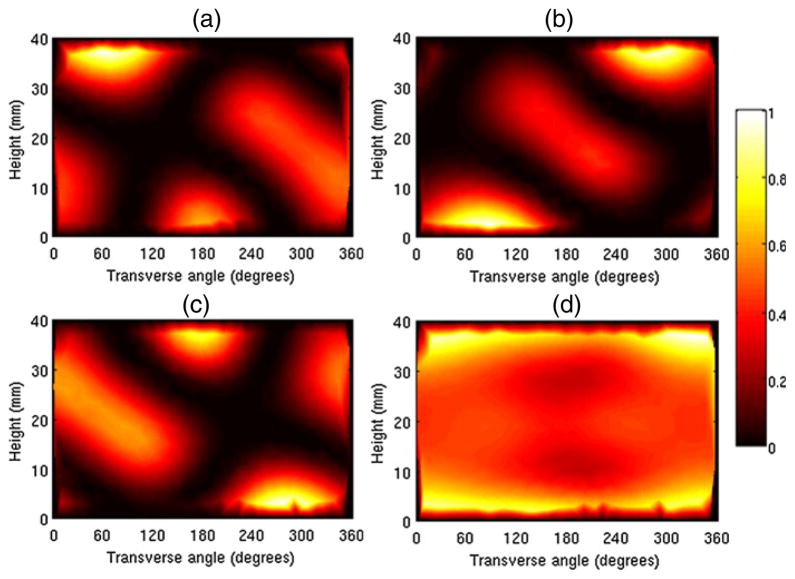
5.1. Patterns on the cylinder
For the study using the tessellated cylinder, we set the number of patterns, p, to 3. The resulting set of three optimal patterns computed for the optically homogeneous tessellated cylinder is shown in figure 2. It is assumed that only the curved surface is used for illumination. The patterns are displayed on the curved surface by laying it out flat. We observe that the pattern (b) can be shifted by 120° (one-third of a full circle) right or left to obtain patterns (a) and (c), respectively. The sum of the three patterns in (d) looks quite uniform overall and additionally indicate rough bilateral symmetry about the horizontal plane cutting the cylinder lengthwise midway. Thus the optimal set of patterns exhibits radial and bilateral symmetries, characteristic of the cylindrical shape.
Figure 2.
A set of three optimal patterns on a tessellated cylinder. The individual patterns are displayed in (a), (b), and (c). The sum of the three patterns, displayed in (d), shows rough overall uniformity of illumination. Only the curved surface of the cylinder was used for illumination. For ease of display, this surface is laid out flat so that the vertical axis represents the height of the cylinder and the horizontal axis represents the transverse angle.
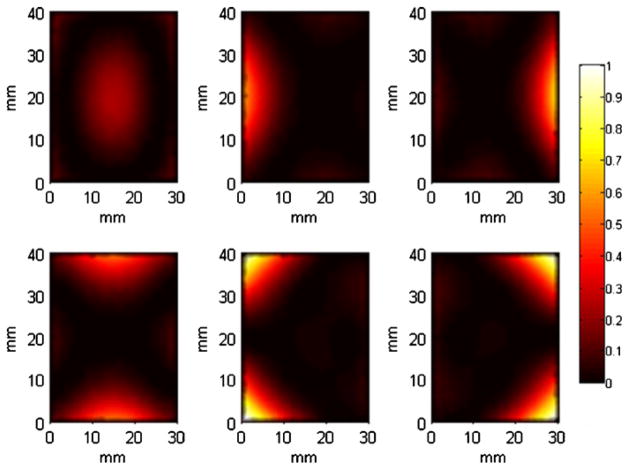
5.2. Patterns on the cuboidal phantom
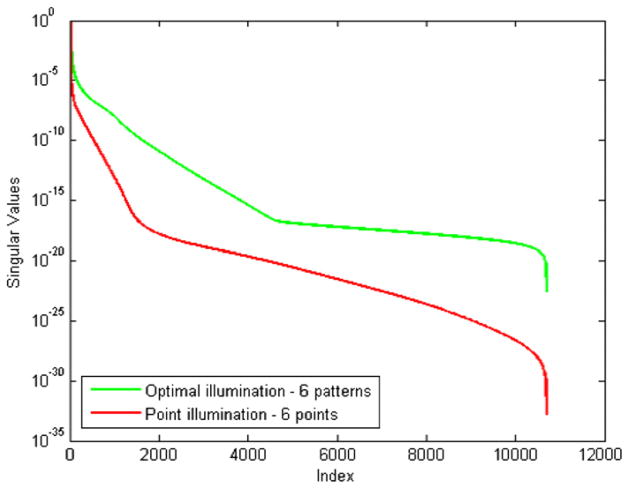
We generated a set of p = 6 patterns for the cuboidal phantom placed in the transillumination mode with illumination from the top and detection from the bottom. The resulting set of optimal illumination patterns on the top surface of the phantom is shown in figure 3. For reference, we generated another transillumination setup where 6 evenly spaced points on the top face (z = 20 mm) are illuminated. The point locations are (10, 10, 20), (20, 10, 20), (10, 20, 20), (20, 20, 20), (10, 30, 20), and (20, 30, 20), where all positions are in millimeters. The singular value spectra for the optimal scheme and the point illumination scheme compared in figure 4 show that the former is more well conditioned than the latter.
Figure 3.
A set of six optimal patterns on the top surface (z = 20 mm) of a tessellated cuboidal phantom set up in the transillumination mode.
Figure 4.
Singular value spectra for the optimal illumination scheme and evenly spaced point illumination scheme for the cuboid in the transillumination mode. Both schemes use a set of six illumination patterns.
5.3. Patterns on the Digimouse atlas
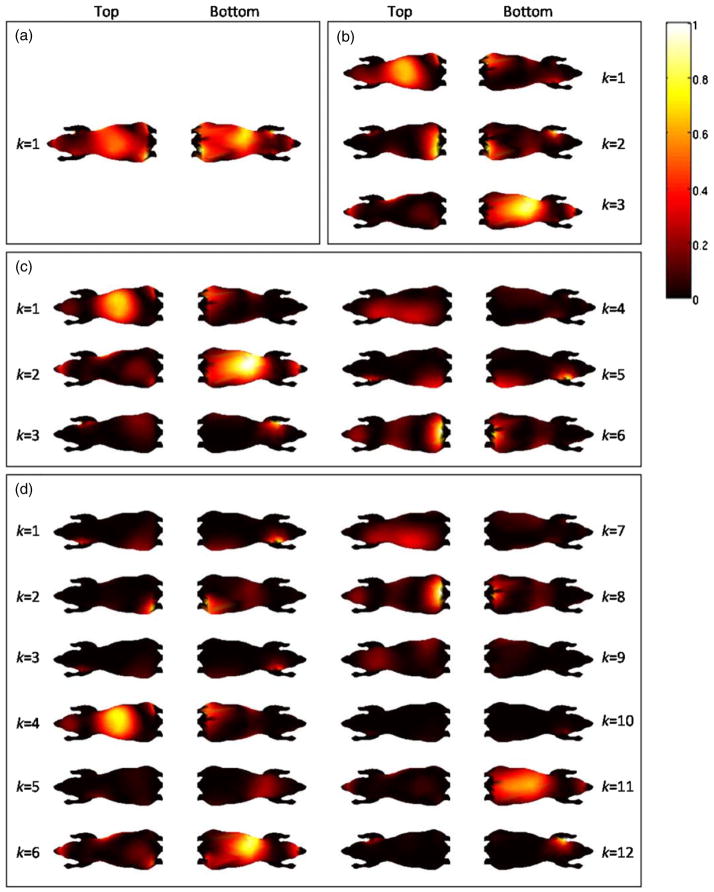
Optimal sets of patterns for the Digimouse atlas were generated for different numbers of patterns, p. We picked the values p = 1, 3, 6, and 12 and looked at the corresponding optimal sets of patterns. The average computation times for sets of 1, 3, 6, and 12 patterns were 5 min, 16 min, 2 h, and 20 h, respectively, on a 2.33 GHz quad-core Intel Xeon® processor. The optimal illumination schemes for the chosen numbers of patterns are shown in figure 5. For each set of patterns, the intensities are normalized with respect to the maximum intensity value for that set for display purposes. Unlike the cylinder, the mouse atlas is of an optically inhomogeneous, irregular shape. Accordingly, the patterns do not exhibit any clear symmetry.
Figure 5.
Projections of the top and bottom surfaces of the Digimouse atlas showing optimal patterns for (a) p = 1, (b) p = 3, (c) p = 6, and (d) p = 12.
5.3.1. Point source localization
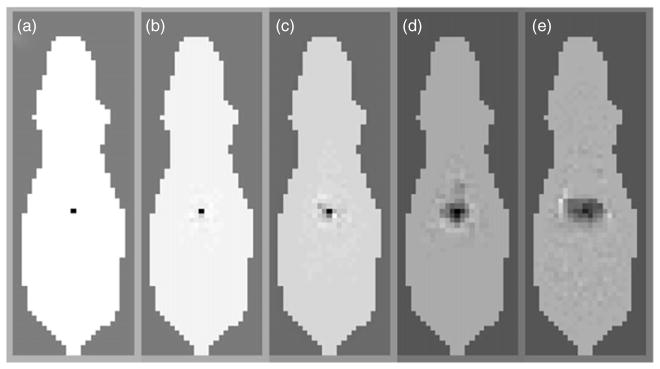
We reconstructed a sample deep source inside the trunk of the mouse at a depth of 10.4 mm below the top surface and 7.6 mm from the bottom surface. The PSFs were generated using (34) for the optimal sets of patterns computed for p = 1, 3, 6, and 12. Figure 6 shows coronal slices of the Digimouse atlas displaying the point spread functions for the optimal illumination schemes for different values of p. For reference, we have studied a set of p = 3 patterns each of which uniformly illuminates one of three different longitudinal sections of the mouse. To generate these PSFs, we picked values of the regularization parameter that equalize the estimator variances as computed using (37) for the different illumination schemes. These values were 4.90×10−11, 3.26×10−12, 1.23×10−13, and 8.35×10−16, respectively, for optimal illumination schemes with p = 1, 3, 6, and 12, and 10−13 for the p = 3 uniform illumination scheme. We observe improvement in the measured FWHM from the cases p = 1 through 12. The FWHMs of the PSFs for the different cases are provided in table 1. Also, figure 6 and table 1 indicate that the reconstructed point source for the p = 3 optimal case appears to have better resolution than the p = 3 uniform case for the same estimator variance.
Figure 6.
Point spread functions for (a) optimal patterns for p = 12, (b) optimal patterns for p = 6, (c) optimal patterns for p = 3, (d) uniform patterns for p = 3, and (e) optimal pattern for p = 1.
Table 1.
Full width at half maximum of point spread functions in figure 6.
| Pattern type | FWHM (mm) |
|---|---|
| Optimal, p = 12 | 1.20 |
| Optimal, p = 6 | 1.20 |
| Optimal, p = 3 | 1.63 |
| Uniform, p = 3 | 3.02 |
| Optimal, p = 1 | 5.91 |
5.3.2. Performance evaluation
We used the performance metrics described in section 4.3 to investigate the benefit of having larger numbers of patterns in the optimal illumination scheme and to compare the optimal scheme for a set of three patterns with the uniform illumination scheme described in section 5.3.1.
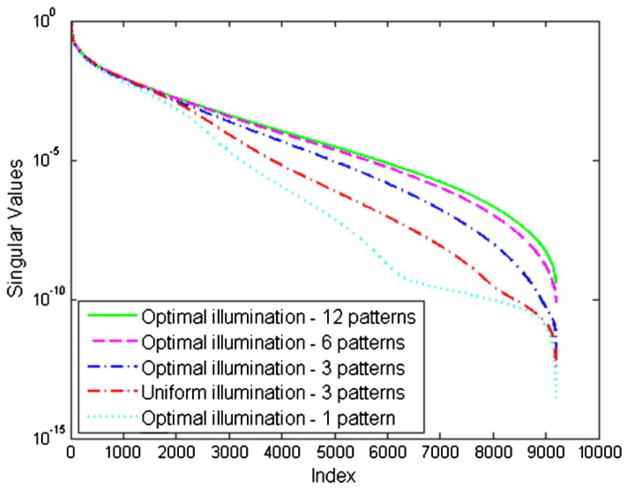
Figure 7 shows the singular value distributions for the different illumination schemes. The singular values are plotted in descending order and normalized with respect to the largest singular value. The condition numbers of the system matrices are provided in table 2. The condition numbers clearly improve as p is increased from 1 through 12. The optimal illumination pattern for p = 3 is observed to generate a better conditioned system matrix than the p = 3 uniform illumination scheme.
Figure 7.
Singular value distributions for different illumination schemes.
Table 2.
Condition numbers of the system matrix for different illumination schemes.
| Pattern type | Condition number |
|---|---|
| Optimal, p = 12 | 2.5575 × 109 |
| Optimal, p = 6 | 1.3466 × 1010 |
| Optimal, p = 3 | 7.1619 × 1011 |
| Uniform, p = 3 | 2.8376 × 1012 |
| Optimal, p = 1 | 3.3014 × 1013 |
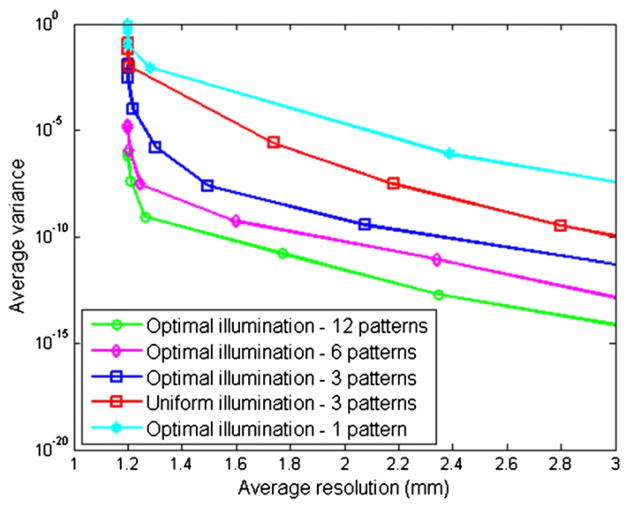
The plots of average resolution versus average variance for the different illumination schemes are shown in figure 8. These are obtained by averaging resolution and variance for n = 250 randomly picked sources on the uniform source grid for different values of the regularization parameter. The regularization parameter was swept over values within the range 10−30–10−4. The best possible FWHM is 1.2 mm, which is the grid spacing. As explained in section 4.3.2, the idea underlying these curves is the trade-off between resolution and variance, which implies that, a decrease in estimator variance is accompanied by an increase in FWHM and vice versa. The intensities of the different sets were normalized to ensure that all sets of patterns have the same average intensity. As expected, as p goes from 1 to 12, the curves lie lower. In other words, the resolution–variance properties improve. Also, figure 8 indicates that, for the same average variance, the p = 3 optimal illumination pattern offers better average resolution than the p = 3 uniform illumination scheme.
Figure 8.
Average resolution versus variance curves for different illumination schemes.
6. Discussion
We have developed an optimization framework for generating optimal spatial illumination patterns for CW FMT based on an approach that seeks to improve the condition number of the Fisher information matrix. We formulated our problem as a constrained optimization problem which, for a given number of patterns, can be solved to compute the set of optimal patterns which maximize the information content in the acquired data. Our formulation assumes an i.i.d. Gaussian noise model. The fourth-order, non-convex cost function was minimized in the presence of a non-negativity constraint using a gradient-based approach. A multi-start method with random initializations was used to ensure global convergence. The dimensionality of the optimization problem was reduced using a set of geometrical basis functions defined on the 2D manifold representing the surface of the object being imaged. Eigenfunctions of the Laplace–Beltrami operator were used as basis functions owing to their spatial smoothness and orthogonality. To ensure that this numerical approach is geometrically meaningful, we looked at optimal patterns generated for an optically homogeneous tessellated cylinder and observed radial and bilateral symmetries that are intrinsic to the cylindrical shape. We then applied this method to compute optimal patterns for an optically homogeneous cuboidal phantom in the transillumination mode. We compared this scheme with an evenly spaced point illumination scheme by examining the singular value spectra and showed that the optimal scheme is more well-conditioned than the point illumination scheme. Finally, we used our method to compute optimal patterns for the optically inhomogeneous and realistically shaped Digimouse atlas. The obtained patterns look smooth and physically realizable. For the same number of patterns, p = 3, the optimal set was shown to perform better than a uniform illumination scheme on the basis of condition number and average resolution versus variance curves. Since typical FMT experimental setups use a larger number of illumination patterns, we have generated optimal patterns for up to p = 12 and presented their singular value spectra and average resolution–variance characteristics. For fair comparison, the variance was computed based on the assumption of equality of total data acquisition times for different values of p.
In our simulations, we have assumed that the entire surface of the mouse except for the limbs and the tip of the snout is being illuminated. However, in an experimental setting, it may not be feasible to illuminate the entire mouse surface at one go. For such applications, our formulation can be easily modified to introduce spatial constraints on the illumination patterns. Another useful modification of our formulation would be its extension to include multispectral excitation. Owing to the larger variability of tissue optical properties at the excitation wavelengths of commonly used fluorescent dyes and proteins, for the same number of wavelength bins, multispectral excitation typically leads to a better conditioned system matrix than multispectral detection (Chaudhari et al 2009). We, therefore, believe that optimal illumination coupled with multispectral excitation will allow us to pack more information into the acquired data for a given number of illumination patterns and a given number of wavelengths.
Currently, the main limitation of our method is the large computation time required to generate large numbers of patterns. We, therefore, would like to explore more efficient numerical implementations that will enable us to compute larger sets of patterns. This will also allow us to investigate the benefits of using large (say > 20) sets of patterns and, in the process, determine the minimum number of patterns that can be used without making the acquired data overly redundant.
The framework for generating optimal patterns assumes prior knowledge of mouse surface topography and tissue optical properties and requires the forward problem to be solved prior to the optimization procedure. It might not be feasible to repeat the optimization procedure for each animal in between the surface profiling and fluorescence data acquisition steps of an experiment. An alternative approach would be to use the atlas as a surrogate and to warp its surface to match that of the target mouse being imaged (Joshi et al 2009), and, in doing so, we can warp the optimal patterns onto the surface of the target.
Acknowledgments
This work was supported by the National Cancer Institute under grants R01CA121783 and R44CA138243.
References
- Ahn S, Chaudhari AJ, Darvas F, Bouman CA, Leahy RM. Fast iterative image reconstruction methods for fully 3D multispectral bioluminescence tomography. Phys Med Biol. 2008;53:3921–42. doi: 10.1088/0031-9155/53/14/013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrakis G, Rannou FR, Chatziioannou AF. Tomographic bioluminescence imaging by use of a combined optical-pet (OPET) system: a computer simulation feasibility study. Phys Med Biol. 2005;50:4225–41. doi: 10.1088/0031-9155/50/17/021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson R. Boundary conditions for diffusion of light. J Opt Soc Am A. 1995;12:2532–9. doi: 10.1364/josaa.12.002532. [DOI] [PubMed] [Google Scholar]
- Arridge SR, Schweiger M, Hiroaka M, Delpy DT. A finite element approach for modeling photon transport in tissue. Med Phys. 1993;20:299–309. doi: 10.1118/1.597069. [DOI] [PubMed] [Google Scholar]
- Bassi A, D’Andrea C, Valentini G, Cubeddu R, Arridge S. Temporal propagation of spatial information in turbid media. Opt Lett. 2008;33:2836–8. doi: 10.1364/ol.33.002836. [DOI] [PubMed] [Google Scholar]
- Bélanger S, Abran M, Intes X, Casanova C, Lesage F. Real-time diffuse optical tomography based on structured illumination. J Biomed Opt. 2010;15:016006. doi: 10.1117/1.3290818. [DOI] [PubMed] [Google Scholar]
- Boas D, Culver J, Stott J, Dunn A. Three dimensional Monte Carlo code for photon migration through complex heterogeneous media including the adult human head. Opt Express. 2002;10:159–70. doi: 10.1364/oe.10.000159. [DOI] [PubMed] [Google Scholar]
- Chaudhari AJ, Ahn S, Levenson R, Badawi RD, Cherry SR, Leahy RM. Excitation spectroscopy in multispectral optical fluorescence tomography: methodology, feasibility and computer simulation studies. Phys Med Biol. 2009;54:4687–704. doi: 10.1088/0031-9155/54/15/004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari AJ, Darvas F, Bading JR, Moats RA, Conti PS, Smith DJ, Cherry SR, Leahy RM. Hyperspectral and multispectral bioluminescence optical tomography for small animal imaging. Phys Med Biol. 2005;50:5421–41. doi: 10.1088/0031-9155/50/23/001. [DOI] [PubMed] [Google Scholar]
- Chen J, Intes X. Time-gated perturbation Monte Carlo for whole body functional imaging in small animals. Opt Express. 2009;17:19566–79. doi: 10.1364/OE.17.019566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contag CH, Bachmann MH. Advances in in vivo bioluminescence imaging of gene expression. Annu Rev Biomed Eng. 2002;4:235–60. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
- Culver JP, Choe R, Holboke MJ, Zubkov L, Durduran T, Slemp A, Ntziachristos V, Chance B, Yodh AG. Three-dimensional diffuse optical tomography in the parallel plane transmission geometry: evaluation of a hybrid frequency domain/continuous wave clinical system for breast imaging. Med Phys. 2003;30:235–47. doi: 10.1118/1.1534109. [DOI] [PubMed] [Google Scholar]
- Dehghani H, Delpy DT, Arridge SR. Photon migration in non-scattering tissue and the effects on image reconstruction. Phys Med Biol. 1999;44:2897. doi: 10.1088/0031-9155/44/12/303. [DOI] [PubMed] [Google Scholar]
- Dogdas B, Stout D, Chatziioannou A, Leahy RM. Digimouse: a 3D whole body mouse atlas from CT and cryosection data. Phys Med Biol. 2007;52:577–87. doi: 10.1088/0031-9155/52/3/003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley D, Duncan WM, Slaughter J. Emerging digital micromirror device (DMD) applications. Proc SPIE. 2003;4985:14–25. [Google Scholar]
- Dutta J, Ahn S, Joshi AA, Leahy RM. Optimal illumination patterns for fluorescence tomography. IEEE Int. Symp. on Biomedical Imaging: From Nano to Macro, 2009 (ISBI ’09); 2009. pp. 1275–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta J, Ahn S, Leahy RM. Optimized illumination patterns for fluorescence tomography. World Molecular Imaging Conf., 2008 (WMIC’08) Abstract Book; Nice, France. 2008. [Google Scholar]
- Dutta J, Ahn S, Li C, Chaudhari AJ, Cherry SR, Leahy RM. Computationally efficient perturbative forward modeling for 3D multispectral bioluminescence and fluorescence tomography. Proc SPIE. 2008;6913:69130. [Google Scholar]
- Gardner C, Dutta J, Mitchell G, Ahn S, Li C, Harvey P, Gershman R, Sheedy S, Mansfield J, Cherry SR, Leahy RM, Levenson R. Improved in vivo fluorescence tomography and quantitation in small animals using a novel multiview, multispectral imaging system. Proc Biomed Optics (BIOMED) Topical Meeting (OSA) 2010:BTuF1. [Google Scholar]
- Gibson AP, Hebden JC, Arridge SR. Recent advances in diffuse optical imaging. Phys Med Biol. 2005;50:R1–43. doi: 10.1088/0031-9155/50/4/r01. [DOI] [PubMed] [Google Scholar]
- Gill PE, Murray W, Wright MH. Practical Optimization. New York: Academic; 1982. [Google Scholar]
- Godavarty A, Sevick-Muraca EM, Eppstein MJ. Three-dimensional fluorescence lifetime tomography. Med Phys. 2005;32:992–1000. doi: 10.1118/1.1861160. [DOI] [PubMed] [Google Scholar]
- Graves EE, Ripoll J, Weissleder R, Ntziachristos V. A submillimeter resolution fluorescence molecular imaging system for small animal imaging. Med Phys. 2003;30:901–11. doi: 10.1118/1.1568977. [DOI] [PubMed] [Google Scholar]
- Grenander U, Miller M. Pattern Theory: From Representation to Inference. New York: Oxford University Press; 2007. [Google Scholar]
- Haskell RC, Svaasand LO, Tsay TT, Feng TC, McAdams MS, Tromberg BJ. Boundary conditions for the diffusion equation in radiative transfer. J Opt Soc Am A. 1994;11:2727–40. doi: 10.1364/josaa.11.002727. [DOI] [PubMed] [Google Scholar]
- Hayakawa CK, Spanier J, Bevilacqua F, Dunn AK, You JS, Tromberg BJ, Venugopalan V. Perturbation Monte Carlo methods to solve inverse photon migration problems in heterogeneous tissues. Opt Lett. 2001;26:1335–7. doi: 10.1364/ol.26.001335. [DOI] [PubMed] [Google Scholar]
- Hebden JC, Arridge SR, Delpy DT. Optical imaging in medicine: I. Experimental techniques. Phys Med Biol. 1997;42:825–40. doi: 10.1088/0031-9155/42/5/007. [DOI] [PubMed] [Google Scholar]
- Hebden JC, Gibson A, Yusof RM, Everdell N, Hillman EMC, Delpy DT, Arridge SR, Austin T, Meek JH, Wyatt JS. Three-dimensional optical tomography of the premature infant brain. Phys Med Biol. 2002;47:4155–66. doi: 10.1088/0031-9155/47/23/303. [DOI] [PubMed] [Google Scholar]
- Hielscher AH. Optical tomographic imaging of small animals. Curr Opin Biotechnol. 2005;16:79–88. doi: 10.1016/j.copbio.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Hornbeck LJ. Digital light processing and mems: reflecting the digital display needs of the networked society. Proc SPIE. 1996;2783:2–13. [Google Scholar]
- Hutchinson C, Lakowicz J, Sevick-Muraca E. Fluorescence lifetime-based sensing in tissues: a computational study. Biophys J. 1995;68:1574–82. doi: 10.1016/S0006-3495(95)80330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA. PhD Thesis. University of Southern California; Los Angeles: 2008. Geometric methods for image registration and analysis. [Google Scholar]
- Joshi AA, Chaudhari A, Li C, Shattuck D, Dutta J, Leahy RM, Toga AW. Posture matching and elastic registration of a mouse atlas to surface topography range data. IEEE Int. Symp. on Biomedical Imaging: From Nano to Macro, 2009 (ISBI ’09); 2009. pp. 366–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Bangerth W, Sevick-Muraca EM. Non-contact fluorescence optical tomography with scanning patterned illumination. Opt Express. 2006;14:6516–34. doi: 10.1364/oe.14.006516. [DOI] [PubMed] [Google Scholar]
- Kay SM. Fundamentals of Statistical Signal Processing: Estimation Theory. Englewood Cliffs, NJ: Prentice Hall; 1993. [Google Scholar]
- Klose AD, Netz U, Beuthan J, Hielscher AH. Optical tomography using the time-independent equation of radiative transfer: Part 1. Forward model. J Quant Spectrosc Radiat Transfer. 2002;72:691–713. [Google Scholar]
- Koizumi H, Yamamoto T, Maki A, Yamashita Y, Sato H, Kawaguchi H, Ichikawa N. Optical topography: practical problems and new applications. Appl Opt. 2003;42:3054–62. doi: 10.1364/ao.42.003054. [DOI] [PubMed] [Google Scholar]
- Konecky SD, Mazhar A, Cuccia D, Durkin AJ, Schotland JC, Tromberg BJ. Quantitative optical tomography of sub-surface heterogeneities using spatially modulated structured light. Opt Express. 2009;17:14780–90. doi: 10.1364/oe.17.014780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Raymond S, Dunn A, Bacskai B, Boas D. A time domain fluorescence tomography system for small animal imaging. IEEE Trans Med Imaging. 2008;27:1152–63. doi: 10.1109/TMI.2008.918341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Mitchell GS, Dutta J, Ahn S, Leahy RM, Cherry SR. A three-dimensional multispectral fluorescence optical tomography imaging system for small animals based on a conical mirror design. Opt Express. 2009;17:7571–85. doi: 10.1364/oe.17.007571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti L, Maculan N. Global Optimization: From Theory to Implementation. New York: Springer; 2009. [Google Scholar]
- Lukic V, Markel VA, Schotland JC. Optical tomography with structured illumination. Opt Lett. 2009;34:983–5. doi: 10.1364/ol.34.000983. [DOI] [PubMed] [Google Scholar]
- Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–80. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- Meng L, Rogers W, Clinthorne N, Fessler J. Feasibility study of compton scattering enchanced multiple pinhole imager for nuclear medicine. IEEE Trans Nucl Sci. 2003;50:1609–17. doi: 10.1109/NSSMIC.2002.1239548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein AB, Stott JJ, Oh S, Boas DA, Millane RP, Bouman CA, Webb KJ. Fluorescence optical diffusion tomography using multiple-frequency data. J Opt Soc Am A. 2004;21:1035–49. doi: 10.1364/josaa.21.001035. [DOI] [PubMed] [Google Scholar]
- Ntziachristos V. Fluorescence molecular imaging. Annu Rev Biomed Eng. 2006;8:1–33. doi: 10.1146/annurev.bioeng.8.061505.095831. [DOI] [PubMed] [Google Scholar]
- Ntziachristos V, Chance B. Probing physiology and molecular function using optical imaging: applications to breast cancer. Breast Cancer Res. 2001;3:41–6. doi: 10.1186/bcr269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos V, Ripoll J, Wang L, Weissleder R. Looking and listening to light: the evolution of whole-body photonic imaging. Nat Biotechnol. 2005;23:313–20. doi: 10.1038/nbt1074. [DOI] [PubMed] [Google Scholar]
- Patwardhan S, Bloch S, Achilefu S, Culver J. Time-dependent whole-body fluorescence tomography of probe bio-distributions in mice. Opt Express. 2005;13:2564–77. doi: 10.1364/opex.13.002564. [DOI] [PubMed] [Google Scholar]
- Pogue BW, Patterson MS. Review of tissue simulating phantoms for optical spectroscopy, imaging and dosimetry. J Biomed Opt. 2006;11:041102. doi: 10.1117/1.2335429. [DOI] [PubMed] [Google Scholar]
- Pukelsheim F. Optimal Design of Experiments (Wiley Series in Probability and Mathematical Statistics) New York: Wiley; 1993. [Google Scholar]
- Qi J, Leahy R. A theoretical study of the contrast recovery and variance of map reconstructions from pet data. IEEE Trans Med Imaging. 1999;18:293–305. doi: 10.1109/42.768839. [DOI] [PubMed] [Google Scholar]
- Qiu A, Bitouk D, Miller M. Smooth functional and structural maps on the neocortex via orthonormal bases of the Laplace–Beltrami operator. IEEE Trans Med Imaging. 2006;25:1296–306. doi: 10.1109/tmi.2006.882143. [DOI] [PubMed] [Google Scholar]
- Rice BW, Cable MD, Nelson MB. In vivo imaging of light-emitting probes. J Biomed Opt. 2001;6:432–40. doi: 10.1117/1.1413210. [DOI] [PubMed] [Google Scholar]
- Roy R, Sevick-Muraca EM. Three-dimensional unconstrained and constrained image-reconstruction techniques applied to fluorescence, frequency-domain photon migration. Appl Opt. 2001;40:2206–15. doi: 10.1364/ao.40.002206. [DOI] [PubMed] [Google Scholar]
- Schweiger M, Arridge SR, Hiraoka M, Delpy DT. The finite element method for the propagation of light in scattering media: boundary and source conditions. Med Phys. 1995;22:1779–92. doi: 10.1118/1.597634. [DOI] [PubMed] [Google Scholar]
- Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, Tsien RY. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–7. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout D, Chow P, Silverman R, Leahy RM, Lewis X, Gambhir S, Chatziioannou A. Creating a whole body digital mouse atlas with PET, CT and cryosection images. Mol Imag Biol. 2002;4:S27. [Google Scholar]
- Tikhonov AN, Arsenin VY. Solutions of Ill-Posed Problems. Washington, DC: VH Winston and Sons; 1977. [Google Scholar]
- Villringer A, Chance B. Non-invasive optical spectroscopy and imaging of human brain function. Trends Neurosci. 1997;20:435–42. doi: 10.1016/s0166-2236(97)01132-6. [DOI] [PubMed] [Google Scholar]
- Weissleder R, Ntziachristos V. Shedding light onto live molecular targets. Nature Med. 2003;9:123–8. doi: 10.1038/nm0103-123. [DOI] [PubMed] [Google Scholar]
- Zacharakis G, Kambara H, Shih H, Ripoll J, Grimm J, Saeki Y, Weissleder R, Ntziachristos V. Volumetric tomography of fluorescent proteins through small animals in vivo . Proc Natl Acad Sci USA. 2005;102:18252–7. doi: 10.1073/pnas.0504628102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharopoulos AD, Svenmarker P, Axelsson J, Schweiger M, Arridge SR, Andersson-Engels S. A matrix-free algorithm for multiple wavelength fluorescence tomography. Opt Express. 2009;17:3042–51. doi: 10.1364/oe.17.003042. [DOI] [PubMed] [Google Scholar]
- Zavattini G, Vecchi S, Mitchell G, Weisser U, Leahy RM, Pichler BJ, Smith DJ, Cherry SR. A hyperspectral fluorescence system for 3d in vivo optical imaging. Phys Med Biol. 2006;51:2029–43. doi: 10.1088/0031-9155/51/8/005. [DOI] [PubMed] [Google Scholar]