Abstract
Streptococcus pneumoniae is the most common cause of severe bacterial meningitis in children, the elderly, and immunocompromised individuals. To identify virulence factors preferentially expressed during meningitis, we conducted niche-specific genome-wide in vivo transcriptomic analysis after intranasal infection of mice with serotype 4 or 6A pneumococci. The expression of 34 bacterial genes was substantially altered in brain tissue of mice infected with either of the 2 strains. Ten upregulated genes were common to both strains, 7 of which were evaluated for their role in the development of meningitis. One previously uncharacterized protein, α-glycerophosphate oxidase (GlpO), was cytotoxic for human brain microvascular endothelial cells (HBMECs) via generation of H2O2. A glpO deletion mutant was defective in adherence to HBMECs in vitro as well as in progression from the blood to the brain in vivo. Mutant bacteria also induced markedly reduced meningeal inflammation and brain pathology compared with wild type, despite similar levels of bacteremia. Immunization of mice with GlpO protected against invasive pneumococcal disease and provided additive protection when formulated with pneumolysin toxoid. Our results provide the basis of a strategy that can be adapted to identify genes that contribute to the development of meningitis caused by other pathogens.
Introduction
Streptococcus pneumoniae is the leading cause of community-acquired pneumonia, bacteremia, and meningitis in children and adults (1). It is the commonest cause of bacterial meningitis in the USA and many countries worldwide (2, 3). Despite effective antimicrobial therapy, pneumococcal meningitis remains highly lethal and has substantial long-term sequelae (4, 5). The availability and use of pneumococcal conjugate vaccines has decreased the incidence of invasive diseases caused by serotypes included in the vaccine formulation. However, this has been offset by substantial increases in both carriage and disease caused by nonvaccine serotypes, which have occupied the niche vacated by the vaccine type pneumococci (6, 7). Therefore, pneumococcal disease continues to be an important health issue worldwide.
Pneumococcal meningitis is preceded by nasopharyngeal colonization, followed by bacteremia and invasion of the CNS (8, 9). However, little is known about the complement of factors required for invasion of, and survival in, the brain by pneumococci. Choline-binding protein A (CbpA, also called PspC or SpsA), pneumolysin (Ply), and neuraminidase A (NanA) are so far the best-studied virulence factors contributing to the development of meningitis (10, 11). Interestingly, immunization of mice with either CbpA, Ply, or both did not afford adequate protection against pneumococcal meningitis (our unpublished observations), suggesting that virulence factors other than CbpA and Ply make an important contribution.
To understand the mechanisms employed by a bacterial pathogen to circumvent the immune system and cause disease, it is critically important to identify the gene products that are specifically required for each stage of the infection process. Several strategies have been employed to screen in vivo–induced genes, including in vivo expression technology (IVET) (12), signature-tagged mutagenesis (STM) (13, 14), differential fluorescence induction (DFI) (15–17), DNA microarray (18, 19), reverse vaccinology (20, 21), antigenome technology (22, 23), and genomic array footprinting (GAF) (24). The availability of complete pathogen genome sequences has stimulated the development and wide-spread application of DNA arrays, which has revolutionized the study of genes that are involved in microbial pathogenesis (25).
Microarray technology is a very powerful tool that provides a global view of integrated cellular processes that are active during infection. This is followed by the combined application of gene cloning, recombinant protein technology, and in vitro functional assays to validate target selection for vaccine development. Protective vaccine components are often derived from proteins that are expressed under specific disease conditions. However, because many virulence factors and antigens are expressed only in vivo, approaches that rely on in vitro–grown bacteria are likely to miss important protective antigens. Hence, the main focus of the present study was to identify factors involved in the pathogenesis of pneumococcal meningitis in order to provide a better understanding of the underlying molecular mechanisms of the disease. We hypothesized that virulence factors that are specifically upregulated during meningitis could serve as potential targets for vaccine development (if surface exposed) and/or therapeutic intervention. To test this, we conducted microarray transcriptomic analysis of bacteria harvested from the blood and brain of mice at various stages of the pathogenic process, examined the role of upregulated genes in pathogenesis, and tested the protective immunogenicity of one of the gene products.
Results
Pathogenesis of pneumococcal meningitis.
In earlier studies, we showed that certain previously characterized pneumococcal virulence genes are differentially expressed in various in vivo niches of CD1 mice (26, 27). In one of those studies, we observed that 2 clinical strains, WCH16 (serotype 6A) and WCH43 (serotype 4), had a high propensity to progress to the brain of mice and proliferate to cause meningitis (27). Therefore, the pathogenesis of the 2 strains was followed at 48, 72, and 96 hours after i.n. infection of CD1 mice in 3 separate experiments (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI45850DS1). The pathogenic profiles clearly showed that stable colonization of the nasopharynx was firmly established over the 48- to 96-hour experimental duration and that invasion of deeper host tissues (particularly the brain) by both strains peaked at 48 to 72 hours after infection.
Differentially expressed genes between blood and brain are identified by microarray analysis.
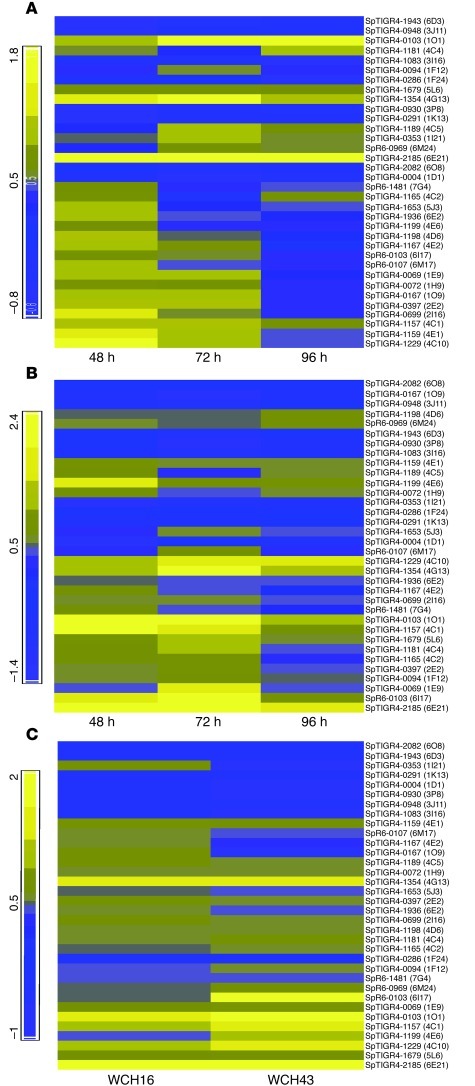
To identify the genes that are differentially expressed by pneumococci in the brain (or attached to the cerebral microvasculature) relative to blood of mice infected with WCH16 and WCH43, total enriched bacterial RNA extracts from the blood and brain of mice infected with the 2 strains at 48, 72, and 96 hours after infection from 3 replicate challenge experiments were subjected to transcriptomic microarray analysis (see Methods). In so doing, only genes that were consistently and reproducibly differentially expressed at all 3 time points, in all 3 replicate experiments, and in the 2 distinct meningitis-causing strains were targeted. Fully annotated microarray data have been deposited in BμG@Sbase (E-BUGS-130; http://bugs.sgul.ac.uk/E-BUGS-130) and also ArrayExpress (E-BUGS-130; http://www.ebi.ac.uk/arrayexpress/experiments/E-BUGS-130). The iterative statistical analyses of the microarray data yielded 34 significantly up- or downregulated genes in the brain of mice infected with either WCH16 (Figure 1A) or WCH43 (Figure 1B). Cross comparison of the 34 regulated genes revealed differentially expressed genes common to both strains (Figure 1C). Further dissection of the genes yielded 10 genes common to both strains (Table 1), 7 of which were chosen for validation by real-time RT-PCR. Commonality of the 7 genes across several serotypes was also confirmed by PCR of genomic DNA using primers flanking the genes in question (not shown). A previous study used microarray analysis to evaluate gene expression in pneumococci harvested from the cerebrospinal fluid of rabbits 4 hours after intracisternal challenge (18). A recent study used DFI to characterize genes involved in the pathogenesis of meningitis (16), while another used GAF to identify pneumococcal genes essential for replication during experimental meningitis (24). However, none of the genes revealed in the current investigation was identified in any of these prior studies. A likely reason for this discrepancy is that the other studies either used in vitro surrogates for given in vivo niches or cross-species comparisons of gene expression between distinct in vivo niches, neither of which are appropriate models for investigating the progression from pneumococcal bacteremia to meningitis. This underscores the uniqueness of the current technique in identifying genes that might be critical to development of meningitis in a model system that mimics progression of invasive pneumococcal disease in humans.
Figure 1. Heat maps showing kinetics of regulated gene expression between blood and brain of mice infected i.n. with WCH16 and WCH43.
(A) Regulated gene expression in WCH16. (B) Regulated gene expression in WCH43. (C) Combined gene expression analyses for WCH16 and WCH43. Yellow to blue scales represent fold difference in mRNA level; yellow, upregulation in brain relative to blood; blue, downregulation in brain relative to blood.
Table 1.
Microarray comparisons of gene expression of S. pneumoniae WCH16 and WCH43 in blood and brain
Genes preferentially upregulated in the brain contribute to pneumococcal meningitis.
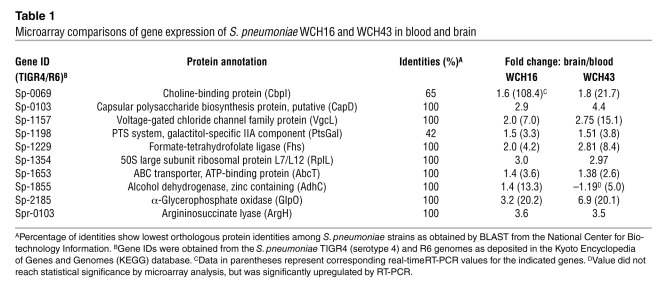
To evaluate the contribution of the genes preferentially upregulated in the brain to pathogenesis, 7 mutants, in which individual target genes were deleted and replaced by a nonpolar antibiotic-resistance cassette, were constructed using overlap PCR (see Methods). A mutation in capD was not constructed because it is a putative capsular biosynthesis protein. A mutant of rplL was also not constructed because of potential lethality issues, being a housekeeping gene. For argH, the sequence is highly variable, with a potential large deletion in WCH16. Therefore, it was not chosen for further study. Mutation of 6 of the 7 genes chosen for further analysis did not adversely affect their growth in vitro in serum broth (SB) (not shown). For the Δfhs mutant, in vitro growth was slightly delayed (∼20 minutes) in the WCH43 background, but its growth in the WCH16 or D39 backgrounds was not affected (not shown).
Initially, the virulence of each mutant relative to that of the isogenic WT strain (WCH43) was assessed in separate i.p. and i.n. infection models. Using this approach, one of the mutants (Δfhs) was completely avirulent in both challenge models (Figure 2, A and B). This mutant was also completely avirulent in both challenge models in WCH16 and D39 backgrounds (not shown). Other single mutants were not significantly attenuated in either challenge model (Figure 2, A and B, and Supplemental Figure 2, A and B). However, we showed previously that while mutagenesis of single mutants may not have a significant impact on pneumococcal virulence, multiple mutations resulted in significant attenuation of virulence (28, 29). Therefore, as a result of the marginal attenuation of virulence observed with the loss of expression of the single genes α-glycerophosphate oxidase (GlpO), PTS system, galactitol-specific IIA component (ptsGal), and voltage-gated chloride channel family protein (vgcl) (denoted ΔglpO, ΔptsGal, and ΔvgcL mutants, respectively) in the i.n. model, the virulence of double mutants (ΔglpO plus ΔptsGal and ΔglpO plus ΔvgcL) relative to WT was investigated. Interestingly, the double mutants were significantly attenuated compared with the WT in the i.n. infection model (Supplemental Figure 2C), indicating additive attenuation of the combined mutants.
Figure 2. Characterization of mutants.
Survival times for mice after (A) i.p. (1 × 105 CFU) and (B) i.n. (2 × 107 CFU) challenge. Each datum point represents 1 mouse. Broken lines denote median survival time for each group. ***P < 0.001, Mann-Whitney U test, 2-tailed. (C) Competition experiments between WT WCH43 and isogenic mutants in mouse brains (n = 14–20) 48 hours after i.n. infection with equal numbers (∼1 × 107 CFU) of WT and mutant. Datum points represent the ratio of mutant bacteria to WT for each animal. The horizontal solid line represents a 1:1 ratio. The horizontal broken line denotes the geometric mean ratio for each comparison. ***P < 0.001, 1 sample t test, 2 tailed. (D) Adherence to HBMECs. Data represent mean ± SEM from 2 independent experiments, each assayed in triplicate. *P < 0.05; ***P < 0.001, Student’s t test, 2 tailed. (E and F) Pathogenesis of WCH43 and its isogenic ΔglpO and complemented mutants. Bacteria were enumerated from the blood and brain of each mouse at 24 hours (E) and 48 hours (F) (n = 16/group per time point). Horizontal line represents geometric mean. *P < 0.05; **P < 0.01, unpaired t test, 2 tailed. BT, back-transformed derivative of ΔglpO mutant.
In order to measure the contribution of each differentially expressed gene to pneumococcal meningitis, we initially carried out separate i.n. infections of mice with WT WCH43 or selected single mutants (ΔglpO and ΔptsGal) to enumerate and compare bacterial load in the blood and brain. For the ΔglpO mutant, bacterial counts in the brain were significantly lower than those of the WT-infected mice (P < 0.05), whereas bacterial counts in the blood of both groups were not significantly different (Supplemental Figure 3). However, for the ΔptsGal mutant, while the overall bacterial load in the brain of WT-infected mice was higher than for those infected with the mutant, the difference did not reach statistical significance, largely due to mouse-to-mouse variation in the levels of infection in the 2 niches (not shown). To circumvent this problem, we evaluated fine differences between WT and mutant by competition experiments in which groups of mice were challenged i.n. with approximately equal numbers of mutant and WT pneumococci. We then calculated the competitive index for each mutant in the various niches at 48 hours after infection. The Δfhs mutant was excluded from this assay because it was completely avirulent in both the i.p. and i.n. challenge models. At 48 hours after infection, ΔadhC, ΔglpO, and ΔptsGal mutants were significantly attenuated in the brain compared with the WT (P < 0.001 in all cases) (Figure 2C). The competitive indices of the mutants (with the exception of the ΔglpO mutant) in the nasopharynx, lungs, and blood varied (Supplemental Figure 4). However, the ΔglpO mutant was significantly attenuated in the nasopharynx and the brain (P < 0.001 in both cases), but not in the lungs or blood (Supplemental Figure 5). A similar scenario was also seen for the ΔglpO mutant in the various niches at 24 and 72 hours after infection (Supplemental Figure 5).
To complement the in vivo findings, the ability of all the mutants to adhere to human brain microvascular endothelial cells (HBMECs) was assessed. In this assay, Δfhs, ΔptsGal, and ΔglpO mutants (especially the ΔglpO mutant) were significantly attenuated for adherence to HBMECs relative to WT bacteria (P < 0.05, P < 0.05, and P < 0.001, respectively) (Figure 2D). The reduced adherence of the ΔglpO mutant to HBMECs was also demonstrated in the WCH16 background (not shown). We also confirmed by real-time RT-PCR analysis that the mutation in glpO did not affect the level of in vitro expression of known pneumococcal virulence genes cbpA, ply, serine protease (prtA), pneumococcal surface protein A (pspA), manganese-dependent superoxide dismutase (sodA), or pyruvate oxidase (spxB) (data not shown).
To clearly define the phenotypic impact of the ΔglpO mutant in WCH43, we carried out a complementation experiment in which the WT glpO gene was back-transformed into the chromosome of the isogenic ΔglpO mutant by homologous recombination (see Methods). The pathogenesis of WT, ΔglpO mutant, and the isogenic back-transformant were then evaluated at 24 and 48 hours after separate i.n. challenge of mice by comparing bacterial load in the blood and brain of the 3 strains. Our results show that bacterial counts in the blood of mice in the 3 groups were not significantly different from each other. However, bacterial counts in the brain of mice infected with the ΔglpO mutant were significantly lower than counts in the brain of mice infected with the WT and glpO–back-transformed strains at 24 and 48 hours after infection (Figure 2, E and F). Together, these assays validate the in vivo transcriptomic approach used to identify the genes being investigated in this study.
GlpO is protective against invasive pneumococcal disease.
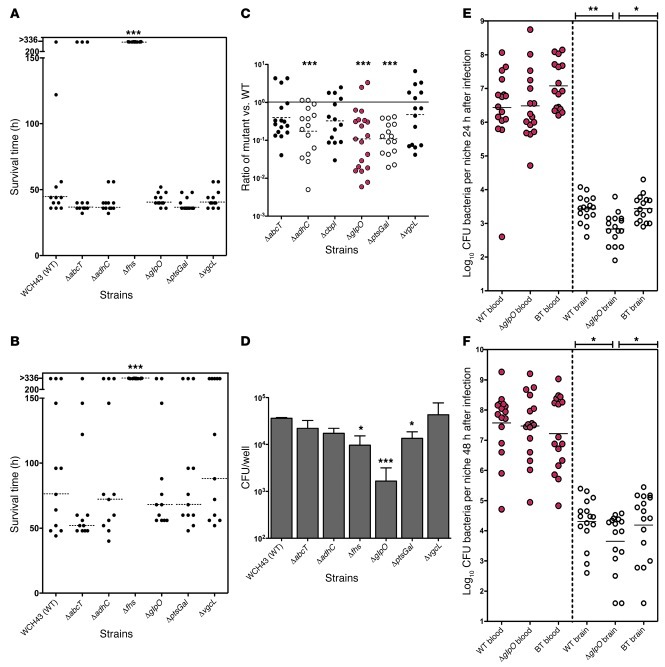
In view of the consistent phenotype observed for the ΔglpO mutant as well as predicted expression of the encoded protein (GlpO) on the pneumococcal surface, the mechanism or mechanisms by which the protein contributes to development of meningitis were further evaluated. First, recombinant GlpO was expressed and purified as a His6-tagged fusion protein in E. coli, as detailed in Methods. Localization of GlpO on the pneumococcal surface was demonstrated by flow cytometry using polyclonal anti-GlpO serum on WT WCH43 (Figure 3A) and WCH16 (not shown), while the isogenic ΔglpO mutant was not labeled. The specificity of the anti-GlpO serum was confirmed by Western blot analysis of whole-cell lysates of WT WCH43 and its otherwise isogenic ΔglpO mutant and glpO–back-transformed strains. This showed clear labeling of a 67-kDa species in the WT and back-transformed strains, but not in the ΔglpO mutant (Figure 3B). Second, mice immunized with recombinant GlpO were significantly protected against i.p. challenge with WCH43 compared with the placebo group, as judged by median survival time (P = 0.0171; Mann-Whitney U test) (Figure 3C). Protection was similar to that achieved by immunization with the nontoxic Ply derivative (PdT). Even stronger protection was achieved using GlpO+PdT (P = 0.0052; Mann-Whitney U test). The overall survival rate for mice in this group was superior to that for mice immunized with either PdT alone (P = 0.0373; Fisher’s exact test) or the placebo group (P = 0.0013; Fisher’s exact test). These results suggest an additive protective effect of the GlpO+PdT combination. Furthermore, individual antibody titers in sera collected 1 week before challenge for 4 surviving mice immunized with GlpO were significantly higher than those for the 4 mice that died (P = 0.04; Student’s t test). This suggested that high GlpO antibody titers correlated with protection, as was shown previously for another Ply toxoid, PdB (30). Third, we tested the functional property of anti-GlpO serum in pathogenesis by passive i.p. immunization of mice followed by i.p challenge with WT WCH43. As a control, serum derived from alum-immunized mice was used. At 30 hours after infection, bacterial counts in the brain of mice passively immunized with anti-GlpO serum were significantly lower than counts in the brain of the sham-immunized mice (P < 0.05), while bacterial counts in the blood of both groups were not significantly different (Figure 3D). Furthermore, the ratio of bacterial numbers in the brain versus the blood in the anti–GlpO-treated mice was significantly lower than that for the control mice (P < 0.01) (Figure 3E). These results suggest that anti-GlpO could be acting to block progression of the bacteria from the blood to the brain by impeding GlpO’s biological activity. We also determined whether anti-GlpO could promote clearance of pneumococci, by measuring deposition of complement on the bacterial surface. FACS analysis showed that WCH43 incubated with anti-GlpO fixed significantly more C3 on their surface than bacteria incubated with nonimmune serum (Figure 3F).
Figure 3. GlpO-mediated immunity.
(A) Flow cytometry of WT WCH43 and ΔglpO using polyclonal anti-GlpO or nonimmune serum. Representative scan from 1 of 2 experiments is shown. (B) Western blot analysis of lysates of WT WCH43 (WT), ΔglpO, and BT, using anti-GlpO serum to show specific labeling of 68-kDa GlpO, with PsaA (36 kDa) labeled by coadministered polyclonal anti-PsaA as a loading control. (C) Active i.p. immunization with alum, GlpO, PdT, and GlpO+PdT and i.p. challenge with 3 × 104 CFU of WCH43. Data represent survival time. *P < 0.05; **P < 0.01, Mann-Whitney U test, 2 tailed. (D) Passive immunization with anti-GlpO and i.p. challenge with 5 × 104 CFU of WCH43, using serum from alum-immunized mice as a control. Bacteria in blood and brain were enumerated at 30 hours; horizontal lines indicate geometric mean CFU. *P < 0.05, unpaired t test (1 tailed). (E) Ratios of bacteria in the brain versus blood compared for alum- and anti-GlpO–immunized mice, as in D above. **P < 0.01, unpaired t test, 1 tailed. (F) C3 deposition on the surface of WCH43 treated with anti-GlpO or nonimmune serum was determined by FACS. Representative scan from 1 of 3 experiments is shown.
GlpO is cytotoxic toward HBMECs.
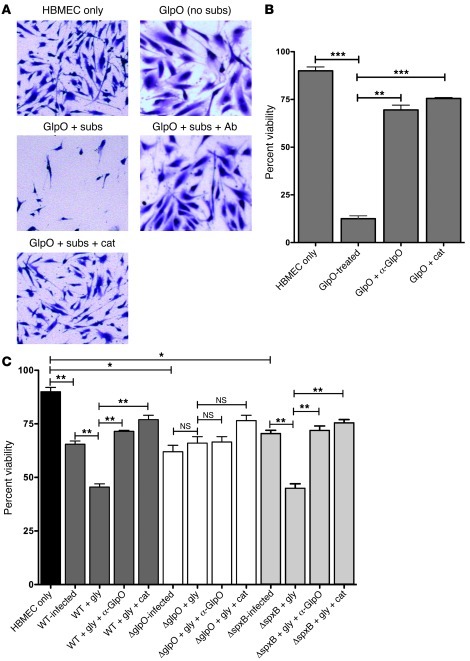
Interestingly, an ortholog of pneumococcal GlpO from Mycoplasma mycoides subsp. mycoides Small Colony has been reported to be cytotoxic for embryonic calf nasal epithelial cells (31, 32). We thus hypothesized that GlpO could contribute to pathogenesis of meningitis by release of H2O2 from metabolism of α-glycerophosphate present in the brain. Therefore, we investigated the ability of purified GlpO to mediate cytotoxicity toward HBMECs. Since the pneumococcal glycerol metabolic pathway is absent, assays were carried out in the presence of the actual GlpO substrates, ATP and glycerol-3-phosphate, and the cofactor flavin adenine dinucleotide (FAD). Cytotoxicity was assessed microscopically (Figure 4A) as well as with a quantitative cell viability assay (Figure 4B). At a dose of 0.1 μg/ml, GlpO almost totally destroyed the monolayer, with viability reduced to 13% compared with 90% for untreated HBMECs (P = 0.001). Highly significant neutralization of GlpO-mediated HBMEC cytotoxicity was achieved by addition of either polyclonal anti-GlpO mouse serum or catalase, with viability increasing to 70% and 75%, respectively (P = 0.003 and P = 0.001, respectively).
Figure 4. HBMEC cytotoxicity and cell viability assays.
(A) HBMEC monolayers were treated as indicated on each panel and stained with crystal violet. (B and C) Cell viability assays (mean ± SEM of 6 replicates) of HBMECs treated with purified GlpO protein (B) or WCH43 and mutants thereof (C). *P < 0.05; **P < 0.01; ***P < 0.001, unpaired t test (2 tailed). WT, WT WCH43; gly, glycerol; subs, substrate (glycerol-3-phosphate +FAD + ATP); cat, catalase; Ab (or α-GlpO), polyclonal anti-GlpO serum.
Cytotoxicity for HBMECs was also investigated using WT WCH43 and its isogenic ΔglpO and ΔspxB mutants, with and without exogenous glycerol to provide a substrate for GlpO. HBMEC monolayers exposed to WT WCH43 exhibited marked cytotoxicity, as evident from cellular swelling, increased cell detachment (Supplemental Figure 6A), and significant loss of viability (P = 0.01) (Figure 4C). The extent of cytotoxicity was significantly enhanced by addition of glycerol (P = 0.01). On the other hand, there was significant neutralization of cytotoxicity by polyclonal anti-GlpO mouse serum, while addition of catalase was even more protective (P = 0.004 and P = 0.006, respectively, relative to WT cells with glycerol) (Figure 4C). In the absence of glycerol, the ΔglpO mutant induced HBMEC cytotoxicity similar to that of the WT WCH43, indicating that some of the cytotoxicity is attributable to other factors such as Ply or SpxB (the cell culture medium contains glucose, enabling SpxB to generate H2O2 from pyruvate). As expected, addition of glycerol did not increase cytotoxicity of the ΔglpO mutant, nor was cytotoxicity neutralized by anti-GlpO. However, there was reduction in cytotoxity in the presence of catalase, consistent with neutralization of SpxB-generated H2O2. The behavior of the ΔspxB mutant was similar to that of WT WCH43 in all experimental conditions (Figure 4C and Supplemental Figure 6A). Results of cytotoxicity assays using the ΔglpO back-transformant were essentially the same as those of the WT strain (not shown). These findings confirm that HBMEC cytotoxicity of WCH43 is largely mediated by GlpO-dependent generation of H2O2 using glycerol as substrate.
GlpO induces H2O2 release from glycerol metabolism in vitro.
H2O2 production was also assayed in cultures of WCH43 and the various mutants (Supplemental Figure 6B). The ΔglpO mutant produced much less H2O2 than WCH43 in the presence of glycerol, but produced high levels of H2O2 in the presence of glucose. In contrast, the ΔspxB mutant produced moderate levels of H2O2 in the presence of glycerol, but much less H2O2 when glucose was used as the carbon source. Thus, GlpO is largely responsible for H2O2 generation by the pneumococcus when glycerol is the available carbon source, while SpxB is largely responsible in the presence of glucose.
GlpO enhances meningeal inflammation.
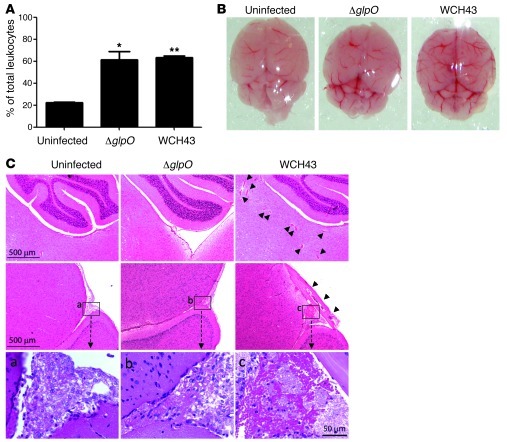
Analysis of blood 48 hours after infection revealed that WT WCH43– or ΔglpO-challenged mice showed a similar significant increase in percentage of neutrophils in the leukocyte population, relative to that in uninfected mice (Figure 5A). Macroscopic (Figure 5B) and histopathological examination (Figure 5C) of the brains of mice at 48 hours after infection with WT WCH43 showed gross inflammation and extensive blood vessel dilation, microhemorrhages and microthrombi in brain parenchyma, and meningeal inflammation. However, damage was much less marked in the brains of ΔglpO-challenged mice (Figure 5, B and C). Evans blue (EB) dye extravasation was then used to assess whether the difference in meningeal inflammation had an impact on blood-brain barrier (BBB) permeability (see Methods). Macroscopic examination showed that the perfused brains of 4 of the 6 WCH43-challenged mice were markedly more pigmented than those of uninfected mice, while only 1 brain from the 6 ΔglpO-challenged mice exhibited any evidence of increased pigmentation, consistent with an overall reduction in dye extravasation and BBB permeability in this latter group (Supplemental Figure 7).
Figure 5. Pathological investigations of mice challenged with WCH43 and ΔglpO.
(A) Neutrophil percentages determined from differential counts of leukocytes in Giemsa-stained mouse peripheral blood smears at 48 hours after i.n. challenge. Data represent mean ± SEM from 2 experiments. *P < 0.05; **P < 0.01, Student’s t test (relative to uninfected mice). (B). Macroscopic views of freshly removed mouse brains at 48 hours after i.n. challenge. demonstrating surface vessel dilations, in WCH43- and ΔglpO-challenged mice. (C) Microscopic views of H&E-stained mouse brain sections at 48 hours after challenge. The upper panels are from the mid-brain/cerebellum region, showing small vessel dilations, microhemorrhages, and microthrombi in the parenchyma (arrowheads) in WCH43-challenged mice. The middle panels are from the boundary between the mid-brain and cerebral cortex, showing meningeal inflammation, particularly in WCH43-challenged mice (arrowheads). Meningeal regions labeled a, b, and c are enlarged in the bottom panels. Scale bars are as indicated. Views presented are representative of 6 sections derived from 2 different mice for each challenge strain.
Increased GlpO expression in meningitis-prone S. pneumoniae strains.
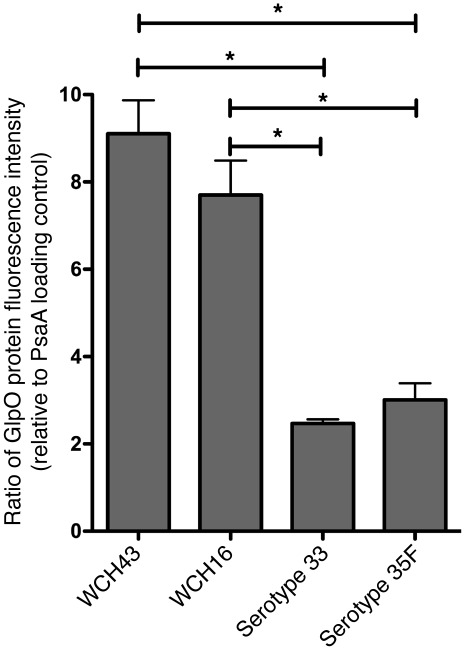
Given the unequivocal contribution of GlpO to progression of meningitis in meningitis-prone strains WCH16 and WCH43, we determined whether there were differences in baseline levels of GlpO expression between these and 2 other pneumococcal strains (belonging to serotypes 33 and 35F) known not to be associated with meningitis. Expression levels were determined by quantitative Western blotting. In this assay, the non–meningitis-associated strains exhibited approximately 3-fold lower GlpO expression levels than either of the meningitis-prone strains (P < 0.05 in all cases) (Figure 6).
Figure 6. GlpO expression.
GlpO expression in lysates of meningitis-prone strains (WCH16 and WCH43) and non–meningitis-associated strains (serotypes 33 and 35F) was determined by quantitative Western blotting using anti-GlpO, as described in Methods. Data represent fluorescence intensity of the anti-GlpO–reactive species, relative to those of the PsaA loading control for each strain (mean ± SEM of quadruplicate experiments). *P < 0.05, unpaired t test, 2 tailed.
Discussion
S. pneumoniae continues to be responsible for high global morbidity and mortality from pneumonia, bacteremia, and meningitis.
While major efforts have recently been directed toward the development of cheap and effective vaccines against pneumococcal pneumonia and bacteremia, similar measures toward the prevention of pneumococcal meningitis are lacking. This could in part be because protection against meningitis is assumed for vaccines that prevent bacteremia. Nevertheless, pneumococcus is now the commonest cause of bacterial meningitis in children, the elderly, and adults with specific underlying medical conditions and accounts for the most severe form of bacterial meningitis in terms of the degree of morbidity and mortality (33). Indeed, in spite of the widespread use of the 7-valent pneumococcal conjugate vaccine (Prevnar), S. pneumoniae is still the commonest cause of bacterial meningitis in children in the USA, mostly due to nonvaccine serotypes (3). Therefore, the development of effective, multivalent vaccines providing non–serotype-dependent protection represents the best prospect for managing pneumococcal diseases, including meningitis, in the 21st century. However, the critical determinants that enable certain strains of pneumococci to cross the BBB to cause meningitis are still largely unknown. We hypothesized that progression from the blood to the brain will require niche-specific alterations in virulence gene expression and that surface-exposed virulence factors that are upregulated during this transition could be protective immunogens against meningitis.
Until recently, progress on the systematic examination of pneumococcal gene expression patterns in distinct host niches was hampered by technical difficulties associated with isolating sufficient quantities of pure and intact bacterial RNA from infected tissues to perform accurate, quantitative mRNA analyses. These difficulties might explain why previous studies had used suboptimal in vivo models or in vitro surrogates to estimate pneumococcal gene expression patterns in distinct host niches. However, in this study, the technical difficulties of in vivo–derived RNA extraction, enrichment, and linear amplification were overcome and further refined, permitting the first microarray comparison and analysis of transcription kinetics between distinct host niches of the same animal, including sites such as the nasopharynx, where pneumococci exist in very low numbers. This provides a superior model system for analysis of pneumococcal gene expression during pathogenesis of invasive disease.
The present study focused on identifying S. pneumoniae proteins that are important for the development of meningitis. First, we compared in vivo–derived transcriptomes of pneumococci simultaneously harvested from the blood and brain of infected mice. Second, the role of key genes was investigated at the molecular level, using targeted mutagenesis, mouse models of disease, and functional assays. Using this approach, several genes encoding proteins that could potentially contribute to meningitis were identified, foremost among which was formate-tetrahydrofolate ligase (Fhs). Bioinformatic analysis showed that Fhs is a metabolic enzyme that is not predicted to be surface exposed or secreted and hence is a suboptimal vaccine target. Nevertheless, the mechanism or mechanisms whereby this protein contributes to virulence is the subject of a separate investigation.
Another gene of interest identified in our screen was glpO, encoding GlpO, which catalyzes the oxidation of α-glycerophosphate (an intermediate in glycerol metabolism), to dihydroxyacetone phosphate, with the concomitant reduction of oxygen to H2O2 (34). Since the pneumococcus lacks catalase, it produces excessive amounts of H2O2 as a by-product of aerobic metabolism, with deleterious effects on both the host and competing nasopharyngeal microflora (35, 36). In M. mycoides subsp. mycoides SC, GlpO is involved in production and translocation of toxic H2O2 into host cells, causing inflammation and cell death (32). Our in vitro studies using a ΔglpO mutant as well as purified protein have shown that pneumococcal GlpO has a potent cytotoxic effect on HBMECs and that this is a consequence of H2O2 generation from glycerol metabolism. The ΔglpO mutant also exhibited markedly reduced adherence to HBMECs. Previous studies have shown that generation of H2O2 by, or in response to, pneumococci has major deleterious effects on the brain, triggering apoptosis in microglia and neurons (37, 38), and may also mediate peroxidation of brain lipids (39). However, GlpO is not the only pneumococcal enzyme capable of generating H2O2. The others are SpxB, which has been implicated in pneumococcal meningitis (albeit using a rat intrathecal challenge model that bypasses the BBB) (40), and the NADH oxidase (Nox), shown to be important in respiratory tract and otitis media infection models (41). It is unlikely that SpxB plays the major role in H2O2-mediated damage in pneumococcal meningitis. The spxB gene is not upregulated in the brain, as its substrate is pyruvate produced from glucose metabolism. Our in vitro studies showed that a ΔspxB mutant produced significant amounts of H2O2 in the presence of glycerol, but not glucose, whereas the ΔglpO mutant produced high H2O2 in the presence of glucose, but not glycerol. Glucose is more abundant in blood than in the brain, while glycerol is more abundant in the brain than in the blood (42, 43). Thus, although SpxB may play a significant role in colonization and sepsis, as has been suggested by others (10, 44, 45), GlpO is expected to be the major pneumococcal generator of H2O2 in the brain.
Pneumococcal GlpO clearly plays a significant role in the pathogenesis of pneumococcal meningitis. In an i.n. challenge model that mimics the natural route and course of infection in humans, numbers of the ΔglpO mutant in the brain compartment were significantly lower than the WT in both separate challenge and competition experiments, whereas numbers in the blood were not significantly different. Moreover, passive immunization with anti-GlpO reduced the numbers of WT WCH43 entering the brain, but did not affect the level of bacteremia. There was also markedly lower meningeal inflammation and histological damage in the brains of ΔglpO-challenged mice, compared with those challenged with WT WCH43. The increased pathology in the latter would be expected to increase BBB permeability, and this is consistent with the apparent increase in extravasation of EB dye in mice infected with WCH43 versus ΔglpO. However, even though the ΔglpO mutant was less virulent than the isogenic WT strain, there was still residual tissue damage, suggesting that other factors also play an important role. A strong candidate is Ply, which has been shown to be cytotoxic to a wide range of eukaryotic cells and also elicits host inflammatory responses capable of generating ROS production (46, 47).
To our knowledge, this study provides the first direct evidence that GlpO plays a significant role in pneumococcal meningitis. The underlying mechanism most likely involves in situ generation of H2O2 from glycerol metabolism as well as promotion of adherence to the cerebrovascular endothelium, collectively promoting penetration of the BBB and generation of meningeal inflammation. GlpO was also an effective vaccine antigen in its own right and provided additive protection when coadministered with Ply toxoid. We showed that anti-GlpO serum significantly neutralizes H2O2 production and the associated cytotoxic activity of GlpO. We also used flow cytometry to show that incubation of WT WCH43 with anti-GlpO promotes deposition of C3 on the bacterial surface. Thus, the mechanism underlying the protection elicited by immunization with GlpO is likely to include direct antibody-mediated neutralization of GlpO activity in vivo (thereby attenuating penetration of the BBB) as well as promotion of opsonophagocytic clearance.
This study also identified other proteins that could potentially contribute substantially to the disease process, and their roles are currently under investigation. Nevertheless, the demonstration by active and passive immunization-challenge experiments that GlpO is protective against invasive pneumococcal disease and contributes to the development of meningitis strongly suggest that it might be a suitable candidate for future neuroimmunoprotective strategies against pneumococcal meningitis. Moreover, the in vivo transcriptomic strategy described in this paper can be adapted to the discovery of virulence genes that contribute to the development of meningitis caused by other pathogens such as Haemophilus influenzae type b and Neisseria meningitidis.
Methods
Bacterial strains and growth conditions.
The pneumococcal strains used in this study were serotype 2 (D39), serotype 4 (WCH43), and serotype 6A (WCH16) (Supplemental Table 1). Serotype-specific capsule production was confirmed by quellung reaction, as described previously (28). Opaque-phase variants of the 3 strains, selected on Todd-Hewitt broth supplemented with 1% yeast extract–catalase (THY-catalase) plates (48), were used in all animal experiments. Before infection, the bacteria were grown statically at 37°C in SB to A600 of 0.16 (equivalent to approximately 5 × 107 CFU/ml).
Challenge (i.n.) of mice and harvesting of bacteria for gene expression analyses.
Outbred 5- to 6-week-old female CD1 (Swiss) mice were used in all experiments. For WCH16 and WCH43 challenges, groups of 40 mice were used. The mice were anesthetized by i.p. injection of pentobarbital sodium (Nembutal; Rhone-Merieux) at a dose of 66 mg/g of body weight and challenged i.n. with 50 μl of bacterial suspension containing approximately 1 × 107 CFU in SB. The challenge dose was confirmed retrospectively by serial dilution and plating of the inocula on blood agar. At each of 48, 72, and 96 hours after challenge, at least 12 mice were sacrificed by CO2 asphyxiation, and blood and brain samples were processed essentially as described previously (26, 49). This included extensive perfusion with ice-cold PBS between collection of blood and dissection of the brain. Before processing, brain samples were further rinsed in 25 ml sterile, ice-cold PBS before homogenization to remove contaminating blood from the surface. A 40-μl aliquot of each sample was serially diluted in SB and plated on blood agar to enumerate pneumococci present in each niche. Blood plates were incubated at 37°C in 5% CO2 overnight. Samples were then stored at –80°C until further processing was done. The experiment was performed 3 times for each strain.
Extraction, enrichment, and amplification of total bacterial RNA from mouse tissues.
RNA for microarray experiments was isolated and purified from either blood-borne bacteria or a mixture of bacteria that had either crossed over to the brain or were still in transition (attached to the microvascular endothelium) with acid phenol:chloroform:isoamyl alcohol (125:24:1; pH 4.5; Ambion). Extracted RNA samples were checked for purity and integrity as described previously (26, 27). Bacterial RNA samples from a minimum of 5 mice that satisfied these criteria from a specific niche were pooled. The RNA was then purified further using a QIAGEN RNeasy Mini Kit. Samples were further enriched for prokaryotic RNA using the MicrobEnrich Kit (Ambion); bacterial mRNA from brain samples was further enriched using the MicrobExpress Kit (Ambion). The amount of RNA recovered following purification/enrichment was determined by A260/280 measurements.
Microarray analysis of bacterial RNA.
Microarray experiments were performed on whole-genome S. pneumoniae PCR arrays obtained from the Bacterial Microarray Group at St. George’s Hospital Medical School, London, United Kingdom (BμG@S). The array was designed using TIGR4 base strain annotation (50) and extra target genes from strain R6 (51). The array design is available at BμG@Sbase (A-BUGS-14; http://bugs.sgul.ac.uk/A-BUGS-14) and also ArrayExpress (A-BUGS-14; http://www.ebi.ac.uk/arrayexpress/).
Microarray probes were generated using the 3DNA Array 900 MPX Labeling Kit (Genisphere) following the manufacturer’s guidelines. Total amplified RNA of S. pneumoniae obtained from either mouse blood or brain from the 2 strains was used, and pair-wise comparisons were made between blood and brain RNA samples from the 48-, 72-, and 96-hour time points. RNA samples were reverse transcribed using Superscript III (Invitrogen) and then labeled with either Alexa Fluor 546 or Alexa Fluor 647 dye. The fluorescently labeled cDNAs for each pair-wise comparison were then combined and hybridized to the surface of the microarray, essentially as described (52, 53).
Slides were scanned at 10 μM resolution using a GenePix 4000B Scanner (Molecular Devices). Detector PMT voltages were adjusted individually for each slide so that the total red (Alexa Fluor 647) and green (Alexa Fluor 546) fluorescence signals for each channel were approximately equal, while the total number of features with signal above the maximum detectable was minimized. Foreground and background mean pixel intensity values were extracted from the scanned images for both channels (Alexa Fluor 546, Alexa Fluor 647) using the Spot plugin (CSIRO) within the R statistical software package ( http://www.R-project.org). The Limma plugin for R (54) was used for data processing and statistical analysis. After background subtraction, the foreground intensities were log2 transformed, and a single ratio (Alexa Fluor 647/Alexa Fluor 546) value was obtained for each probe. Ratio values were normalized using the print-tip Loess normalization routine (55). The replicate arrays were normalized to each other to give similar ranges of mRNA expression values. For each probe across the arrays, a linear model was fitted to determine a final expression value for each mRNA probed and associated statistics (56). These statistics were used to rank the mRNAs from those most likely to be differentially expressed to the least likely using false discovery rate values of P < 0.05. Microarray analysis examining RNA from infected blood and brains was performed on a total of 9 independent hybridizations from 3 separate assays, including 1 dye reversal per comparison for each strain.
Relative quantitation real-time RT-PCR.
For a subset of selected pneumococcal genes that were significantly upregulated in the brain by microarray analysis, gene expression was validated using a 1-step RT-PCR kit (Invitrogen) in a LightCycler 480 II (Roche) as described previously (27). The relative gene expression was analyzed using the 2–ΔΔCT method (57). The reference gene was 16S rRNA. The primer pairs used for gene expression analysis are listed in Supplemental Table 2. All data were obtained from 3 biological replicates.
Construction of mutants and assessment of bacterial growth in vitro.
Defined, nonpolar mutants of genes of interest were constructed in WCH43 (serotype 4) and, in some cases, also in WCH16 and D39. Mutants were constructed by overlap extension PCR as described previously (58) and validated by PCR and sequencing to be in-frame deletion mutation replacements. All PCR procedures were performed with the Phusion High Fidelity Kit (FINNZYMES). The primer pairs used for construction and validation of the mutants are listed in Supplemental Table 2. In order to evaluate the growth rate of the mutants in comparison with the WT, bacterial strains were grown in SB and A600 monitored overnight on a Spectramax M2 spectrophotometer (Millennium Science).
Virulence assessment of mutants.
To assess the virulence potential of mutants, groups of 12 mice were challenged i.p. or i.n with either mutant or WT bacteria. The survival of mice was monitored 4 times daily for the first 5 days, twice daily for the next 5 days, and then daily until 14 days after challenge. Differences in median survival times for mice between groups were analyzed by the Mann-Whitney U test (1 and 2 tailed).
Pathogenesis and competition experiments.
For initial pathogenesis experiments, mutant and WT strains were grown separately in SB to A600 = 0.16 (approximately 5 × 107 CFU/ml). For pathogenesis experiments, 10 mice were challenged i.n. with 50 μl suspension containing approximately 1 × 107 CFU of either WT or the isogenic ΔglpO mutant. At 48 hours after challenge, mice from each separate infection experiment were sacrificed, bacteria were enumerated from the blood and brain (as described previously), and data were analyzed using unpaired t test (1 tailed). For competition experiments, mutant and WT bacteria were mixed at an input ratio of 1:1, and 20 mice were challenged i.n. with 50 μl bacterial suspension containing approximately 1 × 107 CFU. At 24, 48, and 72 hours after challenge, 5 mice from each mixed infection experiment were sacrificed, and samples from the nasopharynx, lungs, blood, and brain were processed as described previously. A 40 μl aliquot of each sample was serially diluted in SB and plated on blood agar and blood agar with a selective antibiotic to determine the ratio of mutant to WT bacteria. The experiment was repeated at least twice, and data from all experiments were pooled. Competitive indices were calculated as the ratio of mutant to WT bacteria recovered from each niche adjusted by the input ratio, using the 1 sample t test, 1 or 2 tailed.
Complementation experiments.
In order to clearly define the phenotypic effect of the ΔglpO mutant, the mutation was complemented by back-transforming the WT glpO gene into the chromosome of the ΔglpO mutant by homologous recombination. The otherwise isogenic back-transformants were selected on the loss of antibiotic (spectinomycin) resistance by replica plating on plain blood agar and blood agar containing spectinomycin. The pathogenesis of WT, ΔglpO mutant, and the isogenic back-transformant were then evaluated at 24 and 48 hours after separate i.n. challenge of mice as described above and analyzed using unpaired t test (2 tailed).
Adherence assay.
HBMECs (obtained from ScienCell Research Laboratories) were grown in F-12 medium supplemented with 10% (v/v) heat-inactivated FCS, 25 mM Hepes, and 100 U/ml penicillin-streptomycin. Cells were incubated at 37°C in 5% CO2. WT and mutant pneumococci were tested for their adherence to HBMECs after incubation for 2 hours, as described previously (59). Assays were performed in triplicate from 3 independent experiments. Differences between means were analyzed using the unpaired Student’s t test (2 tailed).
Cloning and expression of His6-tagged GlpO proteins.
The glpO and glpOΔFAD open reading frames were PCR amplified from WCH43 genomic DNA with forward and reverse primers (Supplemental Table 2), which incorporate SphI and SalI restriction sites. The PCR fragments were digested with the same enzymes and cloned into the corresponding restriction sites in pQE-31 (QIAGEN) to generate recombinant plasmids. High-level expression in a lipid A (lpxM) mutant of E. coli BL21 (DE3) (60) transformed with pREP4 (QIAGEN) was induced with 2 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 3 hours. The cells were harvested by centrifugation at 6,000 g for 10 minutes and lysed in a French pressure cell; the expressed protein was then purified on a nickel–nitrilotriacetic acid column, essentially as described previously (61). GlpO and GlpOΔFAD were purified under denaturing conditions and refolded by dialysis with buffer exchange, according to Qiagen recommendations. Each protein was judged to be more than 98% pure by SDS-PAGE and staining with Coomassie Brilliant Blue R-250. Recombinant GlpOΔFAD was used for immunization, as it has been shown that the FAD fragment is not essential for immunogenicity (60). However, the full recombinant GlpO protein containing the FAD fragment was used for cytotoxicity assays.
Active and passive immunization-challenge experiments.
For active immunization challenge, groups of 5- to 6-week-old female CD1 mice (12 per group) were immunized i.p. with GlpOΔFAD, Ply toxoid (PdT), or GlpOΔFAD+PdT, in alum adjuvant (Imject Alum no. 77161; Pierce). Each mouse received 3 doses of 10 μg of each antigen in 100 μg alum at 14-day intervals. Mice given the placebo received an identical course of saline plus alum. Sera were collected from individual mice by retroorbital bleeding 1 week after the third immunization. Two weeks after the third immunization, mice were challenged with approximately 3 × 104 CFU of WCH43. Mice were closely monitored for survival over 14 days. Differences in survival of mice between groups were analyzed by the Mann-Whitney U test (2 tailed). Overall survival rates of mice between groups were compared by Fisher’s exact test (2 tailed).
For passive immunization challenge with anti-GlpO serum, ELISA titer for anti-GlpO serum was determined from pooled sera from GlpO-immunized mice, as described previously (61). For challenge, 10 mice were immunized i.p. with high titer (32,000) anti-GlpO serum 1 hour prior to i.p. challenge with 5 × 104 CFU of WCH43. Serum from alum-immunized mice was injected i.p. into another group of 10 mice 1 hour before challenge, as a control. Bacteria were then enumerated from blood and brain of mice from each group at 30 hours after challenge, and ratios of bacteria in the brain to blood were determined. Data were analyzed using the unpaired t test (1 tailed).
Cytotoxicity to HBMECs, cell viability assay, and H2O2 production.
Cytotoxicity assays were performed essentially as described for M. mycoides subsp. mycoides SC (31). The various pneumococcal strains (with or without the indicated treatments) were used to infect confluent HBMEC monolayers in 8-well chamber tissue culture slides (Thermo Scientific) at a multiplicity of infection of 100 bacteria per cell, for 2 hours. Optimum concentration of purified recombinant GlpO protein for analysis was initially determined prior to the assay by titrating at 30, 10, 1, and 0.1 μg/well, and 0.1 μg/well was chosen. Viability of HBMEC monolayers after cytotoxicity assay was analyzed using the Countess Cell Counter System (Invitrogen) according to the manufacturer’s recommendations.
For in vitro assays for H2O2 production, strains were grown in complete C+Y medium (62) to A600 = 0.25. Cells were washed once in the same medium and then resuspended in twice the starting culture volume of fresh medium, consisting of either 1% glucose or 1% glycerol. Control tubes for each treatment contained either catalase or anti-GlpO serum. Aliquots of each sample were then immediately withdrawn and placed on ice (zero time point). Cells were then incubated for 1 hour, after which supernatants from all treatments were collected by centrifugation at 14,000 g for 3 minutes and placed on ice. H2O2 production by the pneumococcal strains (with or without the indicated treatments) was then carried out by a modification of the method described previously (35).
Complement deposition assay and surface localization studies.
Detection of complement C3 deposition on pneumococci was performed essentially as described previously (63) Briefly, approximately 5 × 106 opaque phase WT WCH43 grown in SB to mid-log phase was washed twice in PBS and preincubated with a 1/200 dilution of polyclonal mouse anti-GlpO serum for 30 minutes at 37°C. Thereafter, bacteria were washed twice in PBS and resuspended in 20 μl of pooled fresh naive mouse serum and incubated for 30 minutes at 37°C. Fresh bacteria (not preincubated with anti-GlpO) were also resuspended in 20 μl of pooled fresh naive mouse serum and incubated for 30 minutes at 37°C. Opsonized bacteria were then washed 3 times in PBS and resuspended in PBS containing a 1:100 dilution of FITC-conjugated goat anti-mouse C3 antibody (ICN) and incubated on ice for 30 minutes. After incubation, cells were washed and resuspended in PBS for analysis by flow cytometry (BD FACSCanto; BD Biosciences). Untreated bacteria labeled with the FITC-conjugated goat anti-mouse C3 antibody were used as control.
For surface localization studies, WT and ΔglpO mutant WCH43 and WCH16 were grown as described above. After washing twice in PBS, bacteria were incubated with a 1/200 dilution of either polyclonal anti-GlpO or serum from sham-immunized mice for 1 hour at 37°C. Bacteria were then washed 3 times in PBS, resuspended in PBS containing a 1:200 dilution of Alexa Fluor 488–conjugated goat anti-mouse C3 antibody (Invitrogen), and incubated on ice for 1 hour. After incubation, cells were washed 3 times in PBS and resuspended in PBS for analysis by flow cytometry (BD FACSCanto; BD Biosciences). Untreated bacteria labeled with Alexa Fluor 488–conjugated goat anti-mouse C3 antibody (Invitrogen) were used as control.
Western blotting.
Bacteria were grown to mid-exponential phase (A600 = 0.16 in SB), pelleted by centrifugation, washed once in PBS, and then resuspended in PBS to ×10 concentrations. Thereafter, the cultures were lysed by addition of 0.1% sodium deoxycholate at 37°C for 30 minutes and total cellular protein determined by A280 measurements on a NanoDrop 1000 Spectrophotometer (Thermo Scientific). Approximately 50 μg of whole-cell lysates of each sample was separated on a 4%–12% Bis Tris NuPAGE gel and transferred onto an iBlot Dry Blotting (Nitrocellulose) System (Life Technologies; Invitrogen). Electroblotted samples were then processed either for relative quantification of GlpO between strains using the Odyssey Infrared Imaging System and associated software (LI-COR Biosciences) essentially, as described previously (64), or for presence or absence of GlpO using alkaline phosphatase–conjugated secondary antibody, as described previously (65).
Examination of blood and brain samples of infected mice.
At 48 hours after i.n. infection of mice with either WT WCH43 or its isogenic ΔglpO mutant, blood samples were taken via cardiac puncture and collected into lithium heparin tubes. Blood smears were made from fresh mouse blood, fixed in 100% methanol, and stained with Giemsa solution (BDH 35086.2500) and then examined by light microscopy (Axiophot; ZEISS); digital images were taken with a CCIZ Soft Imaging System and analySIS LS Research software. Blood leukocytes were extracted by lysing red blood cells with a hypotonic shock, fixed in 1% paraformaldehyde, and then analyzed with FACSCanto and CellQuest Pro software (BD). Mouse brains were removed and placed in petri dishes containing ice-cold PBS for macroscopic examination. For histological examination, the brains were fixed in 4% formaldehyde overnight at 4°C. They were then embedded in paraffin, sectioned, and stained with H&E, essentially as described previously (66). The tissue sections were examined using light microscopy, and digital images were taken as described above. For each of these analyses, uninfected mice were used as controls. Data are presented as mean ± SEM, and differences between uninfected and infected mice were analyzed using Student’s t test. P < 0.05 was considered significant.
We also carried out BBB permeability measurement by EB dye extravasation using a modification of the method described previously (67). Briefly, at 48 hours after i.n. infection, groups of mice challenged with either WT WCH43 or its isogenic ΔglpO mutant were injected i.p. with 100 μl of 2% (w/v) EB dye (in PBS) for 2 hours. Mice were then euthanized followed by transcardial perfusion with PBS. Uninfected mice (as controls) were similarly treated. The brains of the mice were then harvested and photographed.
Study approval.
The Animal Ethics Committee of the University of Adelaide approved all animal experiments. The study was conducted in compliance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes (7th Edition, 2004) and the South Australian Animal Welfare Act, 1985.
Supplementary Material
Acknowledgments
We wish to thank Claudia Trappetti, Antonio Focareta, and Jan Cook for help with animal experiments, and Charles Plumptre for help with flow cytometry. We thank Lindsay Dent for the gift of anti-mouse C3 antibody. This work was supported by the Meningitis Research Foundation (UK) (802.0 to A.D. Ogunniyi, J.C. Paton, and L.K. Mahdi) and the National Health and Medical Research Council of Australia (NHMRC) (627142 to J.C. Paton and A.D. Ogunniyi; 565526 to J.C. Paton). We acknowledge BμG@S (the Bacterial Microarray Group at St George’s, University of London) for supply of the microarray slides and advice and The Wellcome Trust for funding the multicollaborative microbial pathogen microarray facility under its Functional Genomics Resources Initiative. J.C. Paton is a NHMRC Australia Fellow.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(6):2208–2220. doi:10.1172/JCI45850.
References
- 1.Lynch JP 3rd, Zhanel GG. Streptococcus pneumoniae: epidemiology and risk factors, evolution of antimicrobial resistance, and impact of vaccines. . Curr Opin Pulm Med. 2010;16(3):217–225. doi: 10.1097/MCP.0b013e3283385653. [DOI] [PubMed] [Google Scholar]
- 2.Sakata H, et al. Results of a multicenter survey of diagnosis and treatment for bacterial meningitis in Japan. J Infect Chemother. 2010;16(6):396–406. doi: 10.1007/s10156-010-0064-6. [DOI] [PubMed] [Google Scholar]
- 3.Nigrovic LE, Kuppermann N, Malley R. Children with bacterial meningitis presenting to the emergency department during the pneumococcal conjugate vaccine era. Acad Emerg Med. 2008;15(6):522–528. doi: 10.1111/j.1553-2712.2008.00117.x. [DOI] [PubMed] [Google Scholar]
- 4.Dery MA, Hasbun R. Changing epidemiology of bacterial meningitis. Curr Infect Dis Rep. 2007;9(4):301–307. doi: 10.1007/s11908-007-0047-7. [DOI] [PubMed] [Google Scholar]
- 5.Jit M. The risk of sequelae due to pneumococcal meningitis in high-income countries: A systematic review and meta-analysis. J Infect. 2010;61(2):114–124. doi: 10.1016/j.jinf.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Singleton RJ, et al. Invasive pneumococcal disease caused by nonvaccine serotypes among alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. JAMA. 2007;297(16):1784–1792. doi: 10.1001/jama.297.16.1784. [DOI] [PubMed] [Google Scholar]
- 7.Hicks LA, et al. Incidence of pneumococcal disease due to non-pneumococcal conjugate vaccine (PCV7) serotypes in the United States during the era of widespread PCV7 vaccination, 1998–2004. J Infect Dis. 2007;196(9):1346–1354. doi: 10.1086/521626. [DOI] [PubMed] [Google Scholar]
- 8.Meli DN, Christen S, Leib SL, Tauber MG. Current concepts in the pathogenesis of meningitis caused by Streptococcus pneumoniae. . Curr Opin Infect Dis. 2002;15(3):253–257. doi: 10.1097/00001432-200206000-00007. [DOI] [PubMed] [Google Scholar]
- 9.Koedel U, Scheld WM, Pfister HW. Pathogenesis and pathophysiology of pneumococcal meningitis. Lancet Infect Dis. 2002;2(12):721–736. doi: 10.1016/S1473-3099(02)00450-4. [DOI] [PubMed] [Google Scholar]
- 10.Orihuela CJ, Gao G, Francis KP, Yu J, Tuomanen EI. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J Infect Dis. 2004;190(9):1661–1669. doi: 10.1086/424596. [DOI] [PubMed] [Google Scholar]
- 11.Uchiyama S, et al. The surface-anchored NanA protein promotes pneumococcal brain endothelial cell invasion. J Exp Med. 2009;206(9):1845–1852. doi: 10.1084/jem.20090386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng JP, et al. Identification of Streptococcus pneumoniae genes specifically induced in mouse lung tissues. . Can J Microbiol. 2008;54(1):58–65. doi: 10.1139/W07-117. [DOI] [PubMed] [Google Scholar]
- 13.Blomberg C, et al. Pattern of accessory regions and invasive disease potential in Streptococcus pneumoniae. . J Infect Dis. 2009;199(7):1032–1042. doi: 10.1086/597205. [DOI] [PubMed] [Google Scholar]
- 14.Polissi A, et al. Large-scale identification of virulence genes from Streptococcus pneumoniae. . Infect Immun. 1998;66(12):5620–5629. doi: 10.1128/iai.66.12.5620-5629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marra A, et al. Differential fluorescence induction analysis of Streptococcus pneumoniae identifies genes involved in pathogenesis. . Infect Immun. 2002;70(3):1422–1433. doi: 10.1128/IAI.70.3.1422-1433.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu WC, et al. In vivo characterization of Streptococcus pneumoniae genes involved in the pathogenesis of meningitis by differential fluorescence induction. . Saudi Med J. 2010;31(4):382–388. [PubMed] [Google Scholar]
- 17.Bartilson M, Marra A, Christine J, Asundi JS, Schneider WP, Hromockyj AE. Differential fluorescence induction reveals Streptococcus pneumoniae loci regulated by competence stimulatory peptide. . Mol Microbiol. 2001;39(1):126–135. doi: 10.1046/j.1365-2958.2001.02218.x. [DOI] [PubMed] [Google Scholar]
- 18.Orihuela CJ, Radin JN, Sublett JE, Gao G, Kaushal D, Tuomanen EI. Microarray analysis of pneumococcal gene expression during invasive disease. Infect Immun. 2004;72(10):5582–5596. doi: 10.1128/IAI.72.10.5582-5596.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grifantini R, et al. Previously unrecognized vaccine candidates against group B meningococcus identified by DNA microarrays. Nat Biotechnol. 2002;20(9):914–921. doi: 10.1038/nbt728. [DOI] [PubMed] [Google Scholar]
- 20.Rappuoli R. Reverse vaccinology. Curr Opin Microbiol. 2000;3(5):445–450. doi: 10.1016/S1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- 21.Rappuoli R. Reverse vaccinology, a genome–based approach to vaccine development. Vaccine. 2001;19(17–19):2688–2691. doi: 10.1016/S0264-410X(00)00554-5. [DOI] [PubMed] [Google Scholar]
- 22.Meinke A, Henics T, Hanner M, Minh DB, Nagy E. Antigenome technology: a novel approach for the selection of bacterial vaccine candidate antigens. Vaccine. 2005;23(17–18):2035–2041. doi: 10.1016/j.vaccine.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Giefing C, et al. Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J Exp Med. 2008;205(1):117–131. doi: 10.1084/jem.20071168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molzen TE, et al. Genome-wide identification of Streptococcus pneumoniae genes essential for bacterial replication during experimental meningitis. . Infect Immun. 2011;79(1):288–297. doi: 10.1128/IAI.00631-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conway T, Schoolnik GK. Microarray expression profiling: capturing a genome-wide portrait of the transcriptome. Mol Microbiol. 2003;47(4):879–889. doi: 10.1046/j.1365-2958.2003.03338.x. [DOI] [PubMed] [Google Scholar]
- 26.LeMessurier KS, Ogunniyi AD, Paton JC. Differential expression of key pneumococcal virulence genes in vivo. Microbiology. 2006;152(pt 2):305–311. doi: 10.1099/mic.0.28438-0. [DOI] [PubMed] [Google Scholar]
- 27.Mahdi LK, Ogunniyi AD, LeMessurier KS, Paton JC. Pneumococcal virulence gene expression and host cytokine profiles during pathogenesis of invasive disease. Infect Immun. 2008;76(2):646–657. doi: 10.1128/IAI.01161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berry AM, Paton JC. Additive attenuation of virulence of Streptococcus pneumoniae by mutation of the genes encoding pneumolysin and other putative pneumococcal virulence proteins. . Infect Immun. 2000;68(1):133–140. doi: 10.1128/IAI.68.1.133-140.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogunniyi AD, et al. Contributions of pneumolysin, pneumococcal surface protein A (PspA), and PspC to pathogenicity of Streptococcus pneumoniae D39 in a mouse model. . Infect Immun. 2007;75(4):1843–1851. doi: 10.1128/IAI.01384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogunniyi AD, et al. c-di-GMP is an effective immunomodulator and vaccine adjuvant against pneumococcal infection. Vaccine. 2008;26(36):4676–4685. doi: 10.1016/j.vaccine.2008.06.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pilo P, et al. A metabolic enzyme as a primary virulence factor of Mycoplasma mycoides subsp. mycoides small colony. . J Bacteriol. 2005;187(19):6824–6831. doi: 10.1128/JB.187.19.6824-6831.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bischof DF, Vilei EM, Frey J. Functional and antigenic properties of GlpO from Mycoplasma mycoides subsp. mycoides SC: characterization of a flavin adenine dinucleotide–binding site deletion mutant. . Vet Res. 2009;40(4):35. doi: 10.1051/vetres/2009018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swartz MN. Bacterial meningitis--a view of the past 90 years. N Engl J Med. 2004;351(18):1826–1828. doi: 10.1056/NEJMp048246. [DOI] [PubMed] [Google Scholar]
- 34.Claiborne A. Studies on the structure and mechanism of Streptococcus faecium L-alpha-glycerophosphate oxidase. . J Biol Chem. 1986;261(31):14398–14407. [PubMed] [Google Scholar]
- 35.Pericone CD, Overweg K, Hermans PW, Weiser JN. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. . Infect Immun. 2000;68(7):3990–3997. doi: 10.1128/IAI.68.7.3990-3997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pericone CD, Park S, Imlay JA, Weiser JN. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the fenton reaction. . J Bacteriol. 2003;185(23):6815–6825. doi: 10.1128/JB.185.23.6815-6825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bermpohl D, et al. Bacterial programmed cell death of cerebral endothelial cells involves dual death pathways. J Clin Invest. 2005;115(6):1607–1615. doi: 10.1172/JCI23223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braun JS, et al. Pneumococcal pneumolysin and H(2)O(2) mediate brain cell apoptosis during meningitis. J Clin Invest. 2002;109(1):19–27. doi: 10.1172/JCI12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffmann O, et al. Interplay of pneumococcal hydrogen peroxide and host-derived nitric oxide. Infect Immun. 2006;74(9):5058–5066. doi: 10.1128/IAI.01932-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffmann OM, Becker D, Weber JR. Bacterial hydrogen peroxide contributes to cerebral hyperemia during early stages of experimental pneumococcal meningitis. J Cereb Blood Flow Metab. 2007;27(11):1792–1797. doi: 10.1038/sj.jcbfm.9600474. [DOI] [PubMed] [Google Scholar]
- 41.Yu J, et al. Characterization of the Streptococcus pneumoniae NADH oxidase that is required for infection. . Microbiology. 2001;147(pt 2):431–438. doi: 10.1099/00221287-147-2-431. [DOI] [PubMed] [Google Scholar]
- 42.Reinstrup P, Stahl N, Mellergard P, Uski T, Ungerstedt U, Nordstrom CH. Intracerebral microdialysis in clinical practice: baseline values for chemical markers during wakefulness, anesthesia, and neurosurgery. Neurosurgery. 2000;47(3):701–709. doi: 10.1097/00006123-200009000-00035. [DOI] [PubMed] [Google Scholar]
- 43.Kiyoshima A, Kudo K, Nishida N, Ikeda N. HPLC simultaneous determination of glycerol and mannitol in human tissues for forensic analysis. Forensic Sci Int. 2002;125(2–3):127–133. doi: 10.1016/S0379-0738(01)00624-7. [DOI] [PubMed] [Google Scholar]
- 44.Spellerberg B, et al. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. . Mol Microbiol. 1996;19(4):803–813. doi: 10.1046/j.1365-2958.1996.425954.x. [DOI] [PubMed] [Google Scholar]
- 45.Regev-Yochay G, Trzcinski K, Thompson CM, Lipsitch M, Malley R. SpxB is a suicide gene of Streptococcus pneumoniae and confers a selective advantage in an in vivo competitive colonization model. . J Bacteriol. 2007;189(18):6532–6539. doi: 10.1128/JB.00813-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paton JC, Andrew PW, Boulnois GJ, Mitchell TJ. Molecular analysis of the pathogenicity of Streptococcus pneumoniae: the role of pneumococcal proteins. . Annu Rev Microbiol. 1993;47:89–115. doi: 10.1146/annurev.mi.47.100193.000513. [DOI] [PubMed] [Google Scholar]
- 47.Hirst RA, Sikand KS, Rutman A, Mitchell TJ, Andrew PW, O’Callaghan C. Relative roles of pneumolysin and hydrogen peroxide from Streptococcus pneumoniae in inhibition of ependymal ciliary beat frequency. . Infect Immun. 2000;68(3):1557–1562. doi: 10.1128/IAI.68.3.1557-1562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiser JN, Austrian R, Sreenivasan PK, Masure HR. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect Immun. 1994;62(6):2582–2589. doi: 10.1128/iai.62.6.2582-2589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogunniyi AD, Giammarinaro P, Paton JC. The genes encoding virulence-associated proteins and the capsule of Streptococcus pneumoniae are upregulated and differentially expressed in vivo. . Microbiology. 2002;148(pt 7):2045–2053. doi: 10.1099/00221287-148-7-2045. [DOI] [PubMed] [Google Scholar]
- 50.Tettelin H, et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. . Science. 2001;293(5529):498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 51.Hoskins J, et al. Genome of the bacterium Streptococcus pneumoniae strain R6. . J Bacteriol. 2001;183(19):5709–5717. doi: 10.1128/JB.183.19.5709-5717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCluskey J, Hinds J, Husain S, Witney A, Mitchell TJ. A two-component system that controls the expression of pneumococcal surface antigen A (PsaA) and regulates virulence and resistance to oxidative stress in Streptococcus pneumoniae. . Mol Microbiol. 2004;51(6):1661–1675. doi: 10.1111/j.1365-2958.2003.03917.x. [DOI] [PubMed] [Google Scholar]
- 53.Ogunniyi AD, et al. Central role of manganese in regulation of stress responses, physiology, and metabolism in Streptococcus pneumoniae. . J Bacteriol. 2010;192(17):4489–4497. doi: 10.1128/JB.00064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smyth GK. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. 2005. Limma: linear models for microarray data. In: Gentleman R, Carey S, Dudoit R, Irizarry W, eds. pp. 397–420. New York, New York, USA: Springer; [Google Scholar]
- 55.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31(4):265–273. doi: 10.1016/S1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 56. Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004. 3 (article 3). [DOI] [PubMed] [Google Scholar]
- 57.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT Method. . Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 58.Horton RM, Ho SN, Pullen JK, Hunt HD, Cai Z, Pease LR. Gene splicing by overlap extension. Methods Enzymol. 1993;217:270–279. doi: 10.1016/0076-6879(93)17067-f. [DOI] [PubMed] [Google Scholar]
- 59.Talbot UM, Paton AW, Paton JC. Uptake of Streptococcus pneumoniae by respiratory epithelial cells. . Infect Immun. 1996;64(9):3772–3777. doi: 10.1128/iai.64.9.3772-3777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cognet I, et al. Expression of recombinant proteins in a lipid A mutant of Escherichia coli BL21 with a strongly reduced capacity to induce dendritic cell activation and maturation. . J Immunol Methods. 2003;272(1–2):199–210. doi: 10.1016/S0022-1759(02)00506-9. [DOI] [PubMed] [Google Scholar]
- 61.Ogunniyi AD, Folland RL, Briles DE, Hollingshead SK, Paton JC. Immunization of mice with combinations of pneumococcal virulence proteins elicits enhanced protection against challenge with Streptococcus pneumoniae. . Infect Immun. 2000;68(5):3028–3033. doi: 10.1128/IAI.68.5.3028-3033.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lacks S, Hotchkiss RD. A study of the genetic material determining an enzyme in Pneumococcus. Biochim Biophys Acta. 1960;39(3):508–518. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 63.Brown JS, et al. The classical pathway is the dominant complement pathway required for innate immunity to Streptococcus pneumoniae infection in mice. . Proc Natl Acad Sci U S A. 2002;99(26):16969–16974. doi: 10.1073/pnas.012669199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harvey RM, Ogunniyi AD, Chen AY, Paton JC. Pneumolysin with low hemolytic activity confers an early growth advantage to Streptococcus pneumoniae in the blood. . Infect Immun. 2011;79(10):4122–4130. doi: 10.1128/IAI.05418-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ogunniyi AD, Grabowicz M, Briles DE, Cook J, Paton JC. Development of a vaccine against invasive pneumococcal disease based on combinations of virulence proteins of Streptococcus pneumoniae. . Infect Immun. 2007;75(1):350–357. doi: 10.1128/IAI.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H, Paton JC, Paton AW. Pathologic changes in mice induced by subtilase cytotoxin, a potent new Escherichia coli AB5 toxin that targets the endoplasmic reticulum. . J Infect Dis. 2007;196(7):1093–1101. doi: 10.1086/521364. [DOI] [PubMed] [Google Scholar]
- 67.Schaper M, Leib SL, Meli DN, Brandes RP, Tauber MG, Christen S. Differential effect of p47phox and gp91phox deficiency on the course of Pneumococcal Meningitis. Infect Immun. 2003;71(7):4087–4092. doi: 10.1128/IAI.71.7.4087-4092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.