Abstract
The mammalian target of rapamycin (mTOR) pathway is activated in the majority of human malignancies and thus seems a likely therapeutic target. However, this pathway is genetically complex, complicating studies using pharmacologic mTOR inhibitors. In this issue of the JCI, Hoshii et al. examined mice deficient in one of the mTOR effector complex proteins, Raptor, to elucidate the role of mTORC1 in leukemia. They convincingly demonstrate that Raptor deficiency, with consequent mTORC1 inhibition, blocks differentiation of leukemia cells and prolongs survival, but also allows a population of leukemia-initiating cells to persist in what appears to be a state of dormancy. Translating this new understanding into effective therapeutic strategies will require further study of the molecular mechanisms that underlie these processes.
Introduction
The majority of human cancers are characterized by activation of the serine/threonine kinase mTOR through one or both of its complexes. Given this frequency, the pathway seems a promising therapeutic target, and as of April 2012, ClinicalTrials.gov listed more than 100 cancer clinical trials of mTOR inhibitors, most of which use an mTOR inhibitor in combination with a conventional chemotherapeutic agent and/or another targeted agent. However, it has been challenging to deduce how best to target mTOR therapeutically in human malignancy. Even in benign tumors in tuberous sclerosis complex (TSC) patients (1), in which mTOR activation is generally believed to be the primary driver of tumorigenesis, treatment with mTOR inhibitors yields only partial tumor regression, with regrowth when the agents are stopped (2–4).
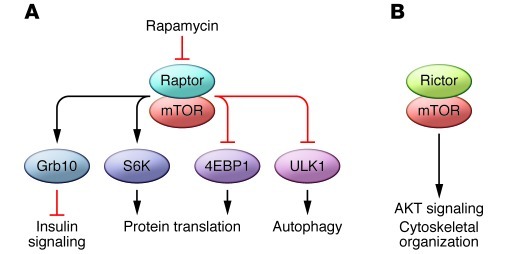
Targeting mTOR in human cancer is complex for many reasons. The two distinct complexes in which mTOR participates have different functions, different sensitivity to mTOR inhibition, and can indirectly regulate each other (5, 6). These two complexes, termed mTOR complex 1 (mTORC1) and mTORC2 are distinguished in part by the mTOR-binding partners, Raptor and Rictor, respectively (Figure 1). The regulation of these complexes, their cellular functions, and their kinase targets are non-overlapping. mTORC1 controls protein translation, cell size, and autophagy through multiple substrates, which include p70 ribosomal protein S6 kinase, eukaryote translation initiation factor 4E binding protein, and ULK1 (7–10). mTORC1 is regulated in part by the Ras homolog Rheb, which is inhibited by the TSC1 and TSC2 proteins. mTORC2 appears to have completely different upstream regulators and downstream kinase targets, which include Akt (5, 11). For example, the TSC proteins, which act via Rheb to inhibit mTORC1, do not directly inhibit mTORC2. Finally, targeting mTOR in cancer can lead to undesired effects on proliferation and survival. For example, feedback loops induced by mTORC1 inhibition can activate kinases such as Akt that drive proliferation, and mTORC1 downregulation induces autophagy, thereby promoting cell survival in some tumors (12–14).
Figure 1. mTOR signaling.
mTOR signaling occurs through two complexes. (A) mTORC1 and (B) mTORC2 utilize the adaptor proteins Raptor and Rictor, respectively. Despite their common components, these two complexes are differentially regulated and have differential effects on cell biology.
The power of genetics
Although mTOR kinase is named for its inhibitory compound (rapamycin), rapamycin and similar agents (often called rapalogs) inhibit only mTORC1, and their inhibition of mTORC1 is incomplete. This is not surprising, since rapamycin actually inhibits mTOR indirectly by binding to FKBP12. Thus mTOR’s other name, FK506 binding protein 12-rapamycin associated protein (FRAP), is perhaps a more accurate designation. Further complexity is introduced by the fact that rapalogs can affect the activity of mTORC2 in some cell types, particularly at higher doses, and because the effects of rapalogs on specific substrates are dynamically regulated. For example, inhibition of S6K phosphorylation is often sustained for longer periods than inhibition of 4EBP1 phosphorylation.
Given these complexities, genetic models have tremendous power to reveal the specific roles of mTORC1 and mTORC2 in disease, as elegantly demonstrated by Hoshii et al. in this issue of the JCI (15). Complete genetic disruption of Raptor results in early embryonic lethality. Previous studies of mice with conditional disruption of Raptor in the adipocyte and muscle lineages had revealed mitochondrial and metabolic defects (9, 10).
Targeting mTORC1
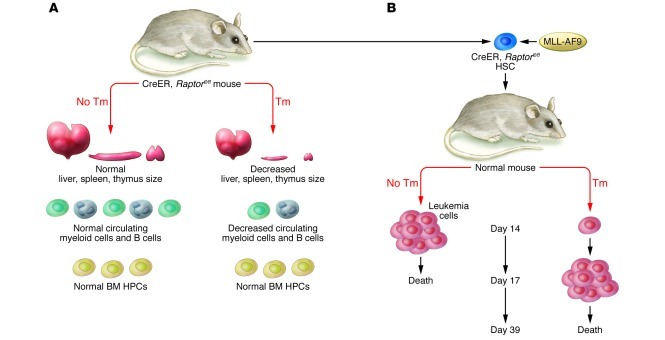
Hoshii et al. focused on mice with Raptor deficiency to elucidate the role of mTORC1 in leukemia. They first studied the impact of Raptor deficiency in hematopoiesis using inducible deletion of Raptor in all tissues. Mice with Raptor deficiency induced by injection of tamoxifen at 8 weeks of age died within 17 days; death was believed to be due to loss of intestinal villi. Importantly, these mice also had fewer white blood cells in peripheral blood and fewer mononuclear cells (MNCs) in bone marrow, and their thymus, spleen, and liver were reduced in size, but hematopoietic progenitor cells (HPC) in the bone marrow were not reduced (Figure 2A). Analysis of blood from these mice revealed decreases in differentiated granulocytes and B cells, which was associated with increased apoptosis in the differentiated cell populations. These data are compelling and support a model in which mTORC1 activity is required for the differentiation and development of B cells and granulocytes, but not for the survival or proliferation of hematopoietic progenitor cells (HPC) in the bone marrow.
Figure 2. Effect of systemic Raptor deficiency on hematopoiesis.
(A) Ablation of Raptor in adult mice resulted in reduced liver, thymus, and spleen size, decreased levels of circulating white blood cells, and fewer MNCs in BM, but normal levels of BM HPCs. (B) Raptor deficiency in leukemia cells prolonged mouse survival in a model of MLL-AF9–driven leukemia. Tm, tamoxifen.
Next, Hoshii et al. used an elegant genetic approach to investigate the role in Raptor in the MLL-AF9–driven model of established acute myelogenous leukemia (AML). They first isolated hematopoietic stem cells from mice carrying the floxed Raptor allele and then retrovirally introduced the MLL-AF9 fusion oncogene to them. When these cells were transplanted into irradiated recipients in the absence of tamoxifen, all of the mice developed leukemia and died within 17 days. However, in the presence of tamoxifen, Raptor deficiency in the leukemia cells reduced the number of leukemia cells in the blood and bone marrow, and the survival of the mice was markedly prolonged (Figure 2B). These data clearly support a critical role of Raptor in leukemia progression. Consistent with the earlier results, in the leukemia model the authors found that Raptor deficiency primarily impacted differentiated cells, allowing undifferentiated AML cells to persist in the bone marrow. Several approaches were used to validate this, including in vivo limiting-dilution transplantation assays and a particularly compelling experiment in which retroviral reintroduction of Raptor restored the ability of the cells to differentiate and induce overt leukemia.
Taken together, the findings of Hoshii et al. convincingly demonstrate in this leukemia model that Raptor deficiency, and consequent mTORC1 inhibition, blocks differentiation of leukemia cells and more than doubles overall survival, but also allows a population of leukemia-initiating cells to persist in what appears to be a dormant state (15). The utilization of a genetic model (Raptor deficiency), rather than a pharmacologic approach, allows a straightforward interpretation of their data.
Therapeutic insight
What are the next steps to translate these findings into improved therapeutic strategies for patients with leukemia (16–18) and other tumors associated with mTORC1 activation (19)? One key knowledge gap involves the molecular mechanisms through which Raptor deficiency induces apoptosis in the differentiated cells but spares the progenitor cells. How do the leukemia “stem cells” proliferate and survive in the absence of mTORC1 activity? Once the mechanisms underlying the persistence of the progenitor population are understood, the next hurdle will be to determine how these mechanisms might be therapeutically targeted. The development of next-generation mTOR inhibitors, including inhibitors of the catalytic activity of mTOR (20), may play a key role in future therapeutic strategies, allowing the activity of mTOR to be more completely blocked.
The mechanisms through which leukemia progenitor populations in this MLL-AF9 model resist mTORC1 inhibition may include feedback activation of the PI3K/Akt pathway and induction of autophagy (14, 21). Deciphering these pathways and determining whether they are relevant in human leukemia will almost certainly involve years of work. Furthermore, even if the observed differential sensitivity between differentiated and undifferentiated cells holds true in other leukemia models, the resistance mechanisms within progenitor cells in any one leukemia model may not be broadly translatable to others, or to other neoplastic drivers. For example, does the regrowth of tumors in TSC patients following discontinuation of mTORC1 inhibitors reflect a similar reduction in the differentiated cells with persistence of a progenitor cell population? If the answer is yes, will the underlying mechanisms be similar to those in leukemia models? Do these models and mechanisms apply to solid tumors as well?
Toward an integrated understanding
Mouse models and clinical trials will continue to provide critical insights into our understanding of mTOR functions, the list of agents with which mTOR inhibitors can be combined is growing, and clinical successes are emerging. How can we optimize the utilization mTORC1-targeted therapies in a timeframe that will benefit patients living with cancer today? In my opinion, a new data set must be added to the equation: those patients whose tumors exhibit unusually significant, “outlier” responses to mTORC1 inhibition, either as a single agent or in combination. These response outliers represent the “finish line,” and are pivotal to deducing the clusters of germline and somatic genetic features that predict response. When we combine these data with the starting line of animal models and clinical trials, we will accelerate the discovery of determinants of response that would otherwise take years to deduce. Working from both the start and the finish of the race to identify effective combinatorial targeted therapeutics will facilitate the rapid integration of mTORC1-targeted therapies into strategies that induce durable responses in human malignancy.
Footnotes
Conflict of interest: The author has declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(6):1970–1972. doi:10.1172/JCI64227
See the related article beginning on page 2114.
References
- 1.Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355(13):1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- 2.McCormack FX, et al. Efficacy and safety of sirolimus in lymphangioleiomyomatosis. N Engl J Med. 2011;364(17):1595–1606. doi: 10.1056/NEJMoa1100391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krueger DA, et al. Everolimus for subependymal giant–cell astrocytomas in tuberous sclerosis. . N Engl J Med. 2010;363(19):1801–1811. doi: 10.1056/NEJMoa1001671. [DOI] [PubMed] [Google Scholar]
- 4.Bissler JJ, et al. Sirolimus for angiomyolipoma in tuberous sclerosis complex or lymphangioleiomyomatosis. N Engl J Med. 2008;358(2):140–151. doi: 10.1056/NEJMoa063564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40(2):310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guertin DA, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt–FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11(6):859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Jacinto E, et al. SIN1/MIP1 maintains rictor–mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127(1):125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 9.Polak P, Cybulski N, Feige JN, Auwerx J, Ruegg MA, Hall MN. Adipose–specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 2008;8(5):399–410. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Bentzinger CF, et al. Skeletal muscle–specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8(5):411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Oh WJ, Jacinto E. mTOR complex 2 signaling and functions. Cell Cycle. 2011;10(14):2305–2316. doi: 10.4161/cc.10.14.16586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu J, Parkhitko A, Henske EP. Autophagy: an ‘Achilles’ heel of tumorigenesis in TSC and LAM. Autophagy. 2011;7(11):1400–1401. doi: 10.4161/auto.7.11.17652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330(6009):1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkhitko A, et al. Tumorigenesis in tuberous sclerosis complex is autophagy and p62/sequestosome 1 (SQSTM1)-dependent. Proc Natl Acad Sci U S A. 2011;108(30):12455–12460. doi: 10.1073/pnas.1104361108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoshii T, et al. mTORC1 is essential for leukemia propagation but not stem cell self-renewal. J Clin Invest. 2012;122(6):2114–2129. doi: 10.1172/JCI62279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martelli AM, et al. Targeting the translational apparatus to improve leukemia therapy: roles of the PI3K/PTEN/Akt/mTOR pathway. Leukemia. 2011;25(7):1064–1079. doi: 10.1038/leu.2011.46. [DOI] [PubMed] [Google Scholar]
- 17.Guo D, Teng Q, Ji C. NOTCH and phosphatidylinositide 3–kinase/phosphatase and tensin homolog deleted on chromosome ten/AKT/mammalian target of rapamycin (mTOR) signaling in T–cell development and T–cell acute lymphoblastic leukemia. Leuk Lymphoma. 2011;52(7):1200–1210. doi: 10.3109/10428194.2011.564696. [DOI] [PubMed] [Google Scholar]
- 18.Altman JK, Sassano A, Platanias LC. Targeting mTOR for the treatment of AML. New agents and new directions. Oncotarget. 2011;2(6):510–517. doi: 10.18632/oncotarget.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dazert E, Hall MN. mTOR signaling in disease. Curr Opin Cell Biol. 2011;23(6):744–755. doi: 10.1016/j.ceb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov. 2011;10(11):868–880. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- 21.Alers S, Loffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32(1):2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]