Abstract
Post-translational modification of proteins by ubiquitin (Ub) regulates a host of cellular processes including protein quality control, DNA repair, endocytosis and cellular signaling. In the ubiquitination cascade, a thioester-linked conjugate between the Ub C-terminus and the active site cysteine of a ubiquitin-conjugating enzyme (E2) is formed. The E2~Ub conjugate interacts with a ubiquitin ligase (E3) to transfer Ub to a lysine residue on a target protein. The flexibly-linked E2~Ub conjugates have been shown to form a range of structures in solution. In addition, select E2~Ub conjugates oligomerize through a noncovalent “backside” interaction between Ub and E2 components of different conjugates. Additional studies are needed to bridge the gap between the dynamic monomeric conjugates, E2~Ub oligomers and the mechanisms of ubiquitination. We present a new 2.35 Å crystal structure of an oligomeric UbcH5c~Ub conjugate. The conjugate forms a staggered linear oligomer that differs substantially from the “infinite spiral” helical arrangement of the sole previously reported structure of an oligomeric conjugate. Our structure also differs in intra-conjugate conformation from other structurally characterized conjugates. Despite these differences, we find that the backside interaction mode is conserved in different conjugate oligomers and is independent of intra-conjugate relative E2/Ub orientations. We delineate a common intra-conjugate E2-binding surface on Ub. In addition, we demonstrate that an E3 ligase CHIP (carboxyl terminus of Hsp70 interacting protein) interacts directly with UbcH5c~Ub oligomers, not only with conjugate monomers. These results provide insights into the conformational diversity of E2~Ub conjugates and conjugate oligomers, and into their compatibility and interactions with E3 ligases, which have important consequences for the ubiquitination process.
Ubiquitination is a widely utilized posttranslational modification that regulates numerous cellular processes (1-3). In the initial step of a ubiquitination cascade, the ubiquitin (Ub) C-terminus is activated in an ATP-dependent manner by a ubiquitin-activating enzyme (E1). The Ub C-terminus is thereupon linked to the E1 active site cysteine through a thioester bond, and subsequently transferred to the active site cysteine of a ubiquitin-conjugating (E2) enzyme (1). The resulting thioester-linked E2~Ub conjugate interacts with a ubiquitin ligase (E3), which also binds to a target protein. RING- or U-box-type E3s promote the direct transfer of Ub from the E2~Ub conjugate to a target protein lysine, resulting in the formation of an isopeptide bond between the lysine ε-amino group and the Ub C-terminus (4, 5). In contrast, HECT- or RING-in-between-RING- (RBR)-type E3s promote the transfer of Ub to an active-site cysteine of the E3 followed by subsequent isopeptide bond formation between Ub and a substrate lysine (6, 7). In some cases, the substrate N-terminal amine, rather than an internal lysine, is ubiquitinated (8). In addition to target protein lysines, the ε-amino group of any of the seven lysines within Ub or the Ub amino terminus can be ubiquitinated, leading to the formation of linear or branched polyubiquitin chains (9). The well-studied Lys48-linked polyubiquitin chain is utilized for protein degradation by the 26S proteasome (1). Other polyubiquitin chains participate in a variety of cellular pathways; for example, Lys63-linked chains regulate DNA repair and transcription (1, 3).
In general, the topology of an elongating polyubiquitin chain is determined by the identity of both the E2 and the E3 (5, 10-12). The human genome encodes approximately 35 E2s and hundreds of E3s. E3 ligases are known to simultaneously bind the E2~Ub conjugate and target protein, but the mechanism of Ub transfer remains elusive. A number of studies have examined conformational changes within E3s that may bring target lysine residues near the E2~Ub active site (13-16). In addition, E3s are known to increase the rate of Ub release from the E2~Ub conjugate, even in the absence of a target protein (7, 17). E3 enzymes may directly influence the conformations of key catalytic residues in the E2~Ub active site to enhance the Ub release-rate.
While E3s adopt multiple, divergent architectures, E2 enzymes share a conserved α/β topology (18). While structural characterization of conjugates is substantially more challenging than characterization of free E2 enzymes, there are now five structures of E2~Ub conjugates in the PDB. Crystal structures include Ubc13~Ub in complex with Mms2 (PDB 2gmi) (19), UbcH5b~Ub in complex with the NEDD4L-HECT (PDB 3jw0) (20), and an isolated UbcH5b~Ub (PDB 3a33) (21). NMR structural models of E2~Ub conjugates, based on Monte Carlo docking using NMR chemical shift perturbation or cross saturation data, include thioester-linked Ubc1~Ub (PDB 1fxt) (22) and a disulfide linked UbcH8-S-S-Ub (PDB 2kjh) (23). The backbone structures of the E2 and Ub moieties in the conjugates do not deviate significantly from isolated E2 or Ub structures. However, the orientations of Ub relative to the E2 vary distinctly between these structures. The conformational landscape of E2~Ub conjugates in solution has only recently been examined (24). Analysis of UbcH5c~Ub and Ubc13~Ub conjugates by nuclear magnetic resonance (NMR) and small angle X-ray scattering (SAXS) shows that UbcH5c~Ub and Ubc13~Ub conjugates exhibit a range of conformations in solution (24).
Intriguingly, conjugates of a subclass of E2 enzymes form oligomers in solution (25-30). This oligomerization is mediated by the trans-interaction of the Ub of one conjugate with a so-called “backside” interaction surface on the E2 enzyme of another conjugate. Mutagenic disruption of this interaction prevents oligomerization, but also greatly inhibits the ability of these E2 enzymes to generate polyubiquitin chains, suggesting that oligomerization facilitates ubiquitin chain extension (25). Oligomerization may be another mechanism that allows distally located substrate lysines or growing ubiquitin chains to access E2~Ub conjugates. In addition, there is evidence that the backside interaction may modulate the E2 active site and promote catalysis and ubiquitin transfer directly (21, 29). Oligomerization and the “backside” interaction were initially identified and characterized by solution NMR techniques. Subsequently, a crystal structure of a UbcH5b~Ub conjugate (PDB 3a33) exhibited this interaction in the context of the crystal lattice (21). A resulting “infinite spiral” oligomeric arrangement of conjugates in this lattice was proposed to accurately represent the state of the conjugate oligomer in solution. It was unclear as to whether conjugate oligomers exclusively adopt this spiral arrangement.
Additional examinations of the relation between E2~Ub conformations, E2 active site conformation and conjugate oligomerization would provide clues about the activation of E2~Ub conjugates by E3 ligases. and about how substrates access these conjugates. In this report, we describe a new crystal structure of UbcH5c~Ub in space group P1211 at 2.35 Å resolution. The structure shows both a new intra-conjugate conformation and a novel staggered linear oligomeric arrangement mediated through the canonical “backside” interaction. Our UbcH5c~Ub conjugate structure demonstrates that UbcH5~Ub oligomerization does not require a specific relative intra-conjugate Ub orientation. Our structure also suggests that extensive conformational variability is accommodated, and likely present, in conjugate oligomers. Docking of oligomeric conjugates onto E3 ligase structures shows that neither oligomeric arrangement conflicts sterically with even relatively large ligases. By comparing the UbcH5c~Ub conjugate structure to other E2~Ub conjugate structures, we identify a common Ub surface used in intra-conjugate E2/Ub interactions. Finally, we show that the E3 ligase CHIP (carboxyl terminus of Hsp70 interacting protein) interacts directly with UbcH5c~Ub oligomers in solution, showing that E3 ligases may indeed recruit such oligomers to facilitate polyubiquitination.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification and Formation of the UbcH5c~Ub Conjugate
Protein expression in Escherichia coli (BL21 star DE3) was induced by 200 μM IPTG followed by shaking for 18 hours at 16°C. Human UbcH5c (HsUbcH5c) was expressed from the pET28N vector (available from Addgene.org) and purified as previously described (31). Human UbcH5b (HsUbcH5b) and Human full length CHIP (His6-HsCHIP(1-303)) were expressed from pGST-parallel-2 vector and pHis-parallel-2 vectors, respectively, and purified as previously described (32). Reactions to produce UbcH5c~Ub or UbcH5b~Ub conjugate utilized active site Cys-to-Ser (C85S) mutants of the respective E2 enzymes to generate oxyester-linked E2~Ub conjugates. Ser-to-Arg (S22R) mutations were added to the UbcH5 enzymes to produce oligomerization deficient E2~Ub conjugates (25). Conjugation reactions were perfomed by mixing 10 μM human E1, 600 μM mammalian Ub, 300 μM E2 enzyme (HsUbcH5c(C85S), HsUbcH5b(C85S) or HsUbcH5b(C85S/S22R)), 5 mM MgCl2 and 2.5 mM ATP followed by incubation at 30°C for 6 hours. UbcH5~Ub conjugate was purified from conjugation reactions by gel filtration on Superdex 75 resin (G.E. Healthcare) equilibrated with 25 mM HEPES, pH 7, 50 mM NaCl. The purity of UbcH5~Ub gel filtration fractions was assessed by SDS-PAGE. UbcH5~Ub conjugate was concentrated to 0.9 mM prior to use in crystallization trials.
Crystallization and X-ray Data Collection
Crystals of human UbcH5c(C85S)~Ub oxyester conjugate were grown by sitting drop vapor diffusion at 293 K in 0.4 μl drops. Sitting drops, consisting of a 1:1 ratio of UbcH5c~Ub (0.9 mM) and a reservoir solution, were prepared using a Crystal Gryphon liquid handling robot (Art Robbins Instruments). Initial screening was conducted using the sparse-matrix crystallization screens JCSG Core I-IV (Qiagen). Optimization of initial screening hits identified an optimal reservoir solution composed of 200 mM tri-potassium citrate and 20% PEG 3350. Resulting single crystals were cryoprotected in LV CryoOil (MiTeGen). X-ray diffraction data was collected with a Cu-Kα source (1.5418 Å wavelength) using a Rigaku MicroMax-007HF generator and a Rigaku Saturn 944+ CCD detector. Data reduction was carried out using d*TREK (33).
Structure Solution and Refinement
Phases for the 2.35 Å dataset were calculated by molecular replacement utilizing the PHASER (34) component of PHENIX (35) using the following search models: PDB ID 3tgd for UbcH5c; PDB ID 1ubq for Ub. A single molecular replacement solution was found in space group P1211. The molecular replacement solution was subjected to automated rebuilding in PHENIX with RESOLVE (36), followed by rounds of iterative refinement (PHENIX) and model building in COOT (37). The refinement protocol used isotropic atomic displacement parameters for all atoms and TLS motion analysis for protein chains (38). All molecular structure figures were prepared with PyMOL (39). The atomic coordinates and structure factors have been deposited in the PDB (accession code 3ugb). Stereochemical and geometric analyses of the UbcH5c~Ub structure were performed using MolProbity (40, 41).
UbcH5~Ub Conjugate Pull-Down Assays
Full-length His6-HsCHIP(1-303) was bound to His Mag Sepharose Ni resin (G.E. Healthcare) by incubating 40 μl of resin suspension with 20 μl of 20 μM His6-HsCHIP(1-303) for 2 hours at 4°C in 25 mM HEPES, pH 7, 150 mM NaCl and 5 mM imidazole. Unbound CHIP was removed by washing the His Mag Sepharose Ni resin with four separate washes of bead wash buffer (500 μl 25 mM HEPES, pH 7, 150 mM NaCl and 5 mM imidazole). Increasing volumes of 20 μM HsUbcH5b(C85S)~Ub or 20 μM HsUbcH5b(C85S/S22R)~Ub were added to tubes containing full-length CHIP bound to His Mag Sepharose Ni resin and incubated for 3 hours at 4°C in bead wash buffer. Unbound UbcH5b~Ub conjugate was removed by washing with 500 μl wash buffer. Protein was eluted from His Mag Sepharose Ni resin by addition of 30 μl 25 mM HEPES, pH 7, 150 mM NaCl and 500 mM imidazole. SDS-PAGE samples were separated on a 12.5% polyacrylamide gel. UbcH5b~Ub conjugate input lanes were loaded with 10 μl of a sample comprised of 20 μl 2x SDS-PAGE buffer (Fisher) and 20 μl of 20 μM UbcH5b~Ub. CHIP input lanes were loaded with 10 μl of a sample comprised of 30 μl 2x SDS-PAGE buffer and 30 μl CHIP eluted from His Mag Sepharose Ni resin. All other lanes were loaded with 10 μl of 30 μl 2x SDS-PAGE buffer and 30 μl CHIP/UbcH5~Ub eluted from His Mag Sepharose Ni resin. Following electrophoresis, gels were stained with IRDye Blue Protein Stain (LI-COR) and imaged using an Odyssey infrared imaging system (LI-COR) with detection at 700 nm. CHIP and UbcH5b~Ub band intensities were quantitated using Image Studio (LI-COR). For each sample, the UbcH5b~Ub band intensity was compared to the intensity of full-length CHIP in the same sample. UbcH5b~Ub and CHIP band intensities were calibrated against inputs of known quantity. Calibration and data plotting were perfomed with Prism (GraphPad).
Cα-Cα Molecular Contact and Sanson-Flamsteed Orientation Plots
Cα-Cα Contact distance plots for intra- and inter-molecular interactions between E2 and Ub chains were calculated for PDB 3ugb (UbcH5c~Ub), PDB 3a33 (UbcH5b~Ub), PDB 3jw0 (UbcH5b~Ub in complex with the NEDD4L-HECT), PDB 2gmi (Ubc13~Ub in complex with Mms2), PDB 1fxt (Ubc1~Ub), PDB 2kjh (UbcH8-S-S-Ub) and PDB 2fuh (UbcH5b/Ub noncovalent complex). For the NMR structures PDB 2fuh, PDB 2kjh and PDB 1fxt, the lowest energy conformer was selected for all analyses. Contact distance plots were calculated using the Bio.PDB module within BioPython (42). Briefly, Cα-Cα distance matrices were generated for select E2 and Ub pairs within each PDB file. These distance matrices were then plotted as a heat map using matplotlib (43) with a distance cutoff of 15 Å.
Sanson-Flamsteed plots were calculated using the Bio.PDB module within BioPython and the base-map toolkit within matplotlib. Briefly, the centers of mass (C.O.M.) for the E2-conjugated ubiquitin chains in PDB 3ugb, PDB 3a33, PDB 3jw0, PDB 2gmi, PDB 1fxt and PDB 2kjh were calculated using the Bio.PDB module. A vector between each Ub C.O.M. and the Ub-conjugated serine-OG or cysteine-SG atom on the respective E2 was calculated using numerical python (numpy). The E2-OG/Ub-C.O.M. vector was converted into spherical polar coordinates using numpy and plotted as a Sanson-Flamsteed projection plot using the basemap toolkit within matplotlib. Sanson-Flamsteed plots for conformers within UbcH5c~Ub and Ubc13~Ub SAXS ensembles were calculated in an identical fashion.
Ub Hot Spot Analysis
Cα-Cα distance matrices generated for each E2~Ub structure using BioPython were utilized to identify the residues most commonly used by Ub to bind E2 through intra-conjugate interactions. A Ub hot spot score was calculated following the procedure described by Winget and Mayor, 2011 (44). For every Ub residue within each E2~Ub structure, the minimum distance to the E2 was calculated. The number of occurrences of minimum Cα-Cα distances less than 10Å were summed across all E2~Ub structures to yield the hot spot score (Supplemental Figure S1). Coloring of the hot spot score was scaled based upon the number of interactions for a given residue.
RESULTS
X-ray Crystal Structure of the Oxyester-Linked UbcH5c~Ub Conjugate
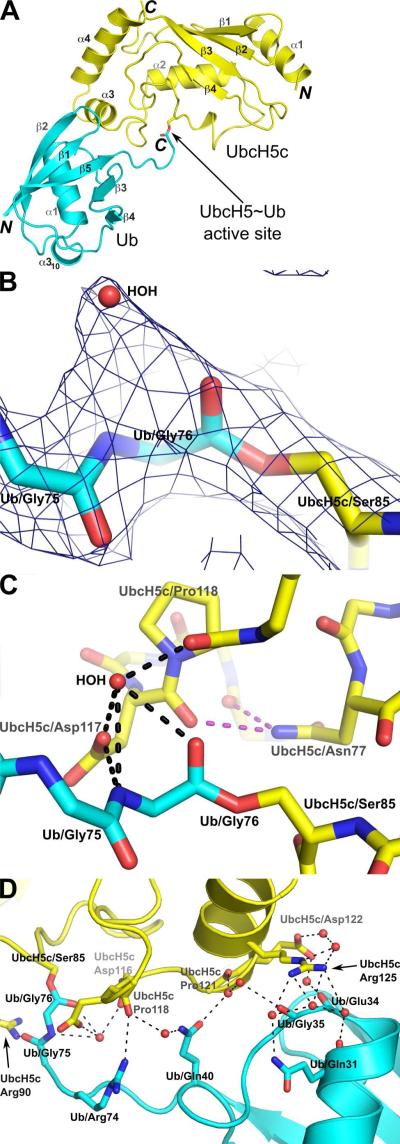
We determined the structure of the oxyester-linked UbcH5c~Ub conjugate by X-ray crystallography in space group P1211 at 2.35 Å resolution (Table 1 and Figure 1A). A simulated-annealing omit map confirms the correct placement of the covalent linkage between the UbcH5c-Ser85 side chain hydroxyl oxygen and the C-terminal Ub-Gly76 carbonyl carbon (Figure 1B). Electron density within a 2Fo-Fc map clearly identifies the UbcH5c~Ub oxyester linkage and surrounding active site residues and bound water molecules (Supplemental Figure S2). Residues surrounding the UbcH5c active site, including the conserved UbcH5c-Asp117 and UbcH5c-Asn77, are poised to stabilize the Ub C-terminus (Figure 1C). UbcH5c-Asn77 does not contact the Ub C-terminus directly, but rather provides a key pair of hydrogen bonds that stabilize the UbcH5c-Asp117/Pro118 loop. In contrast, the UbcH5c-Asp117 side-chain interacts directly with the Ub C-terminus via a water-mediated hydrogen bond network. In addition to interactions between the UbcH5c active site and Ub C-terminus, intra-conjugate contacts between Ub and UbcH5c are mediated by UbcH5c helix α3 residues Pro121, Glu122 and Arg125 and Ub α1-β3 loop residues Glu34, Gly35, Ile36 and Gln40 (Figure 1D).
Table 1.
Crystallographic data collection and refinement statistics
| HsUJbcH5c(1-147)C85S~HsUb(1-76) | |
|---|---|
| Data Collection | |
| X-ray Source | Rigaku MicroMax-007HF |
| Wavelength (Å) | 1.5418 |
| Space group | P1211 |
| Cell dimensions | |
| a, b, c (Å) | 42.0, 52.5, 53.1 |
| α, β, γ(°) | 90, 101.1, 90 |
| Resolution (Å)a | 52.51-2.35 (2.43-2.35) |
| Rmergeb | 0.048 (0.204) |
| Rr.i.m.c | 0.053 (0.239) |
| 〈I/σTl〉d | 19.2 (5.0) |
| Wilson B factor (Å2) | 41.3 |
| Completeness (%) | 97.1 (78.2) |
| Redundancy | 4.9 (3.1) |
| No. of reflections | 45,575 |
| No. of unique reflections | 9,319 |
| Refinement | |
| Resolution (Å) | 29.57-2.35 |
| No. reflections for refinement | 9,298 |
| Rwork/Rfree | 0.211 / 0.253 |
| Number of atoms | |
| Protein | 1,766 |
| Water | 128 |
| Glycerol | 12 |
| Average B factors | |
| Protein | 45.3 |
| Water | 46.1 |
| Glycerol | 60.0 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.014 |
| Bond angles (°) | 1.071 |
| Ramachandran plot statistics | |
| Favored regions % | 98.2 (214/218) |
| Allowed regions % | 100.0 (218/218) |
| Disallowed regions | 0.0 |
| MolProbity validation statistics | |
| Poor rotamers (%) | 0.0 |
| Cβ deviations >0.25Å | 0 |
| MolProbity clash score | 16.55 |
| MolProbity clash percentile | 76th percentile (N=335, 2.35Å ± 0.25Å) |
| MolProbity score | 1.72 |
| MolProbity score percentile | 98th percentile (N=9,377, 2.35 ± 0.25Å) |
| PDB ID | 3ugb |
Values in parentheses are for the highest resolution shell.
The merging R factor is defined as .
The redundancy-indpendent merging R factor (65) is defined as .
〈I/σI〉 denotes the averaged signal to noise ratio.
Figure 1.
Structure of the UbcH5c~Ub conjugate. (A) Cartoon representation of the UbcH5c~Ub conjugate. The amino- and carboxy-termini of UbcH5c (yellow) and Ub (cyan) are labeled N and C. UbcH5c and Ub secondary structure elements are labeled α or β. (B) A closeup view highlights the oxyester bond between UbcH5c-Ser85 and Ub-Gly76. Water molecules are shown as red spheres and electron density for a simulated annealing omit map (dark blue) is contoured at 1σ. (C) The C-terminal tail of Ub participates in a water mediated hydrogen bond network (black dashes) with the UbcH5c-Asp117 side-chain and UbcH5c-Pro118 backbone. UbcH5c-Asn77 provides supporting hydrogen bonds (purple dashes) that stabilize the UbcH5c-Asp117/Pro118 loop. For clarity, the hydrogen bonded water molecule is labeled “HOH” and only the side-chains of UbcH5c-Asn77, UbcH5c-Ser85, UbcH5c-Asp117 and UbcH5c-Pro118 are shown. (D) Intra-conjugate contacts between UbcH5c and Ub utilize a water mediated hydrogen bond network involving residues UbcH5c-Pro121, UbcH5c-Glu122, UbcH5c-Arg125, Ub-Glu34, Ub-Gly35 and Ub-Gln40.
Orientation of Ubiquitin in E2~Ub Conjugate Structures
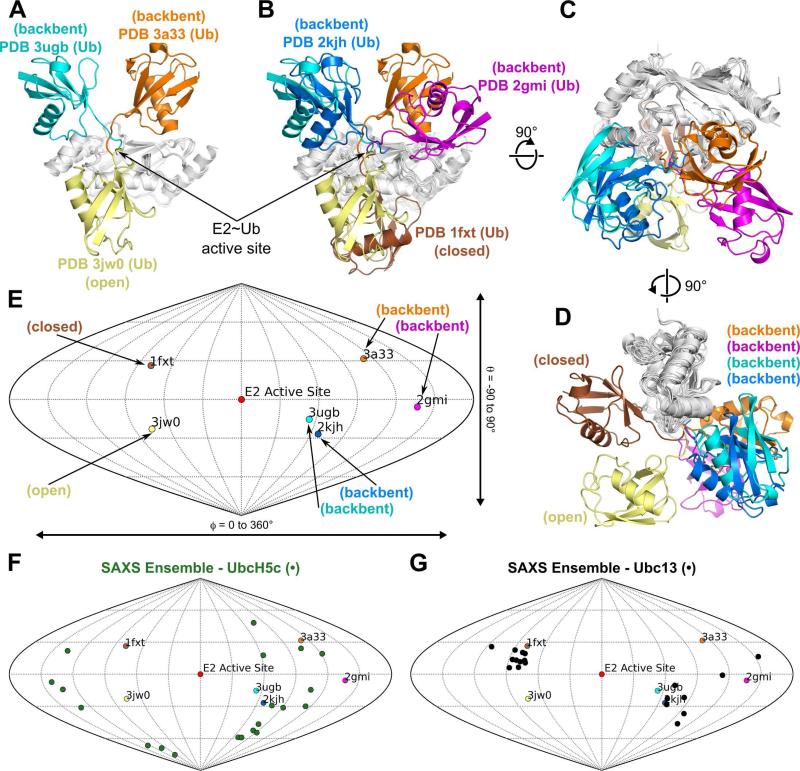
The orientation of Ub within E2~Ub conjugates was previously examined by SAXS and NMR analyses of Ubc13~Ub and UbcH5c~Ub in solution (24). These analyses demonstrated that only a continuum of models, varying between “closed”, “open” and “backbent” states, satisfies the SAXS and NMR data for these conjugates. The “closed” state is defined as an orientation in which the Ub is positioned against the E2 crossover helix α2 (see Figure 1A for secondary structure labeling of E2 enzymes and Ub). In contrast, the “open” state places Ub below the E2 active site in a position with limited E2/Ub contacts. The structure of UbcH5c~Ub presented here (PDB 3ugb) and the two UbcH5b~Ub crystal structures (PDB 3jw0 and PDB 3a33) provide crystallographic evidence for “backbent” (PDB 3a33 and PDB 3ugb) and “open” (PDB 3jw0) states (Figure 2A). The “backbent” state is defined as an orientation that folds the Ub “backwards” against E2 loops 2, 4, 5, or 6. Currently, the UbcH5 subfamily (UbcH5a, UbcH5b and UbcH5c, which are 88% identical and 98% similar in sequence) is the only E2 for which multiple E2~Ub structures are deposited in the PDB. The other conjugate structures Ubc13~Ub (PDB 2gmi), Ubc1~Ub (PDB 1fxt) and UbcH8-S-S-Ub (PDB 2kjh) correspond to “closed” (PDB 1fxt) and “backbent” (PDB 2gmi, PDB 2kjh) states respectively (Figure 2B-D). In addition, a model of the Ube2s~Ub conjugate was reported (45) that places the Ub in the “closed” state (Supplemental Figure S3), similar to Ubc1~Ub (PDB 1fxt).
Figure 2.
Relative orientations of known E2~Ub structures in the PDB. (A) Three distinct Ub positions within structures of UbcH5~Ub conjugates. PDB 3ugb:UbcH5c~Ub (Ub: cyan, UbcH5c: gray), PDB 3a33:UbcH5b~Ub (Ub: orange, UbcH5c: gray), and PDB 3jw0:UbcH5b~Ub (Ub: yellow, UbcH5b: gray). An arrow denotes the location of the E2~Ub active site. (B) Structures of E2~Ub conjugates exhibit a range of Ub positions. PDB 3ugb:UbcH5c~Ub (Ub: cyan, UbcH5c: gray), PDB 3a33:UbcH5b~Ub (Ub: orange, UbcH5c: gray), PDB 3jw0:UbcH5b~Ub (Ub: yellow, UbcH5b: gray), PDB 2gmi:Ubc13~Ub (Ub: magenta, Ubc13: gray), PDB 1fxt:Ubc1~Ub (Ub:brown, Ubc1: gray) and PDB 2kjh: UbcH8-S-S-Ub (Ub:blue, UbcH8: gray). (C) 90° rotation of (B) about the x-axis. (D) 90° rotation of (C) about the y-axis. (E) Sanson-Flamsteed plot corresponding to the structural overlay in (D) indicating the orientation of the Ub center of mass with respect to the E2 active site Ser-OG or Cys-SG atom in PDB 1fxt (brown), PDB 2gmi (magenta), PDB 2kjh (blue), PDB 3a33 (orange), PDB 3jw0 (yellow) and PDB 3ugb (cyan). (F) Sanson-Flamsteed plot from (E) with the twenty members of the UbcH5c~Ub SAXS ensemble (green) (24). (G) Sanson-Flamsteed plot from (E) with the twenty members of the Ubc13~Ub SAXS ensemble (black) (24).
Sanson-Flamsteed plots have been used to describe tensor orientations for NMR residual dipolar couplings (46, 47) and tilt axis orientations in electron microscopy (48). We used a Sanson-Flamsteed plot of the Ub center of mass with respect to the E2 active site to compare the relative orientations of Ub in different conjugate structures (Figure 2E). Although no two E2~Ub structures have identical Ub positions, some commonalities are present. PDB 3ugb/PDB 2kjh (“backbent” state) and PDB 1fxt/Ube2s~Ub (“closed” state) (Figure 2E and Supplemental Figure S3) exhibit the most similar relative Ub orientations. Furthermore, the Sanson-Flamsteed plot clearly delineates the difference between solution phase SAXS ensembles for UbcH5c~Ub (Figure 2F) and Ubc13~Ub (Figure 2G). While members of the UbcH5c SAXS ensemble occupy a range of Ub positions, members of the Ubc13 ensemble form discrete clusters matching the “closed” and “backbent” states.
Interestingly, the relative orientation of Ub does not correlate with the solvent accessible surface area (SASA) of the E2~Ub active site ester bond (Supplemental Table S1) calculated by WHATIF (49). For example, of the four “backbent” state structures (PDB 2gmi, PDB 2kjh, PDB 3a33 and PDB 3ugb), three can be classified as accessible (PDB 2gmi, PDB 2kjh and PDB 3a33) while the fourth is more occluded (PDB 3ugb). The average active site SASA of the “backbent” state accessible structures is 12.86 Å2 compared to 6.26 Å2 for the single “backbent” state occluded structure. Furthermore, the active site SASA values for the two “closed” structures (PDB 1fxt and Ube2s~Ub) are markedly different. The active site SASA is 11.40 Å2 in PDB 1fxt but is only 6.13 Å2 for Ube2s~Ub. The active site SASA for PDB 3jw0, the only “open” state structure, is 5.37 Å2, within 1 Å2 of the occluded “backbent” state structure PDB 3ugb and the occluded “closed” state model Ube2s~Ub. These results suggest that Ub orientation does not directly dictate the accessibility of the E2~Ub thioester bond.
Specific Interactions Potentiate Ub Orientations
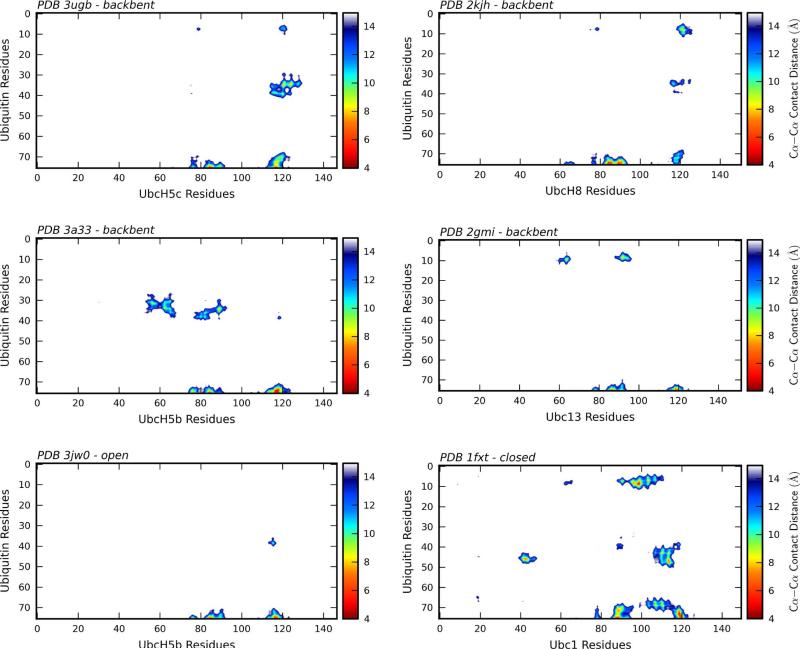
An examination of all six E2~Ub structures using Cα-Cα contact distance plots allows for rapid identification of specific interactions between the E2 and Ub (Figure 3). Although all E2~Ub structures exhibit close contacts between the Ub C-terminus and the E2 active site, each structure also contains additional interactions. An initial analysis of Cα-Cα contact distance plots reveals commonalities that group the structures into four distinct groups.
Figure 3.
Intra-conjugate Cα-Cα contact distance plots. Cα-Cα intra-conjugate contact distance plots for PDB 3ugb (UbcH5c~Ub), 3a33 (UbcH5b~Ub), PDB 3jw0 (UbcH5b~Ub + NEDD4L-HECT), 2gmi (Ubc13~Ub + Mms2), 2kjh (UbcH8-S-S-Ub) and 1fxt (Ubc1~Ub) indicate E2~Ub residue pairs separated by a distance 15 Å or less. For each structure Ub residues are shown on the y-axis and E2 residues are shown on the x-axis. A color bar associated with each plot indicates the Cα-Cα contact distance.
Interactions between Ub residues 8-10 and 35-40 and the E2 helix α3 characterize the first group, comprised of PDB 3ugb (UbcH5c~Ub) and PDB 2kjh (UbcH8-S-S-Ub). The primary difference between the two structures is a slight relative rotation that places the Ub β1-β2 loop closer to the E2 in PDB 2kjh, while the Ub α1-β3 loop is closer in PDB 3ugb. Both PDB 2kjh and PDB 3ugb are examples of the “backbent” state. The second group includes PDB 2gmi (Ubc13~Ub/Mms2) and PDB 3a33 (UbcH5b~Ub), which represent variations on the “backbent” state. Although the Ub orientations are different for PDB 2gmi and PDB 3a33, the E2 interaction surface is nearly identical, with interactions limited to the E2 active site and the E2 β3-β4 loop.
The third group contains a single member, PDB 3jw0 (UbcH5b~Ub/NEDD4L-HECT). Although UbcH5~Ub prefers the “open” state in solution (24), the only crystal structure in an “open” state is the UbcH5b~Ub/NEDD4L-HECT complex (PDB 3jw0), in which the extended position of the Ub is stabilized by interactions with the NEDD4L-HECT. In the absence of a third protein (the HECT domain), such an extended conjugate structure would not form a stable crystal lattice. The PDB 3jw0 Cα-Cα contact distance plot (Figure 3) shows the limited nature of E2/Ub contacts for this “open” state structure. The PDB 3jw0 Cα-Cα contact distance plot indicates residues 39 and 40 near the beginning of Ub strand β3 that most closely approach the E2. Interestingly, these residues are also involved in E2/Ub contacts in the UbcH5c~Ub (PDB 3ugb) and UbcH5b~Ub (PDB 3a33) “backbent” structures.
The single member of the fourth group, PDB 1fxt (Ubc1~Ub), is the only “closed” state structure in the PDB. Intermolecular contacts in this structure and in the Ube2s~Ub model (Supplemental Figure S3), are primarily mediated by the Ub-Ile44 hydrophobic face. Although these contacts are unique to the “closed” state, other E2/Ub contacts involving residues in the Ub β1-β2 loop are similar to the “backbent” state structures PDB 2gmi (Ubc13~Ub) and PDB 2kjh (UbcH8-S-S-Ub).
The patterns of Cα-Cα contacts across different E2 enzymes and variable intra-conjugate relative Ub orientations indicate that a relatively limited subset of Ub residues mediate interaction with the E2. To highlight the key interacting residues, we converted the E2~Ub Cα-Cα contact distance plots (Figure 3) into a Ub hot spot score, following the protocol of Winget and Mayor (44). A heatmap produced from the Ub hot spot score was used to color residues on Ub (Figure 4 A, B and Supplemental Figure S1). Residues 8-11, 36, 39, 71 and 73-76 are the Ub epitopes most commonly involved in intra-conjugate interactions. These residues map to a single face of Ub formed by loops 1 and 3 and the C-terminal tail. A closer examination of the E2~Ub structures emphasizes that each E2~Ub structure uses nearly the same face of Ub to interact with the E2.
Figure 4.
Ubiquitin intra-conjugate E2-interaction hot spot mapping. (A) Cartoon representation of Ub residues (PDB ID 1ubq) are colored by the Ub hot spot score as determined according to degree of participation in intra-conjugate interaction with the E2 for structures PDB 1fxt, PDB 2gmi, PDB 2kjh, PDB 3a33, PDB 3jw0 and PDB 3ugb. The side-chain atoms of residues most commonly participating in intra-conjugate interactions with the E2 are shown as sticks. (B) Surface representation of (A). A color bar indicates coloring used to represent the hot spot score in (A) and (B).
UbcH5 and the Backside Interaction
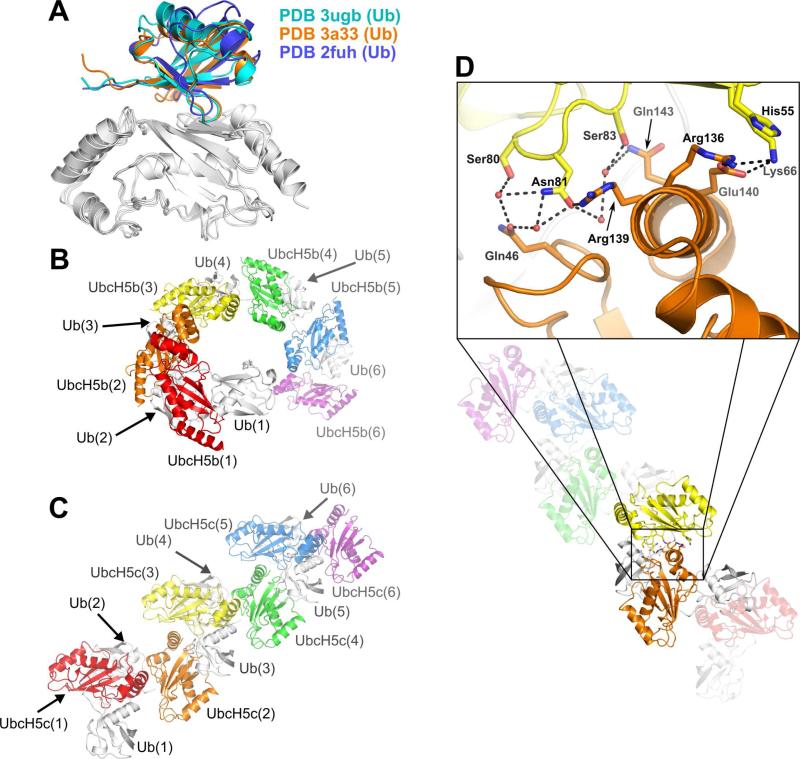
The UbcH5/Ub backside interaction was first reported by Brzovic et al. (25). The NMR structure of the non-covalent UbcH5c/Ub complex (PDB 2fuh) identified intimate contacts between the UbcH5c β-sheet and the Ub-Ile44 hydrophobic face and β4-α310 loop (Figure 5A). E2/Ub backside interactions are present in models of non-covalent Ube2g2/Ub and Ubc2b/Ub complexes (50) (51) and in a crystal structure of a non-covalent (unconjugated) UbcH5a/Ub complex (PDB 3ptf) (52). Backside-mediated contacts have also been observed between the E2 Ubc9 and the small ubiquitin like modifier (SUMO) (26, 29). The first example of the backside interaction in the context of E2~Ub conjugates was observed in the crystal structure of UbcH5b~Ub in space group P6122 (PDB 3a33) (21). Our new structure of the UbcH5c~Ub conjugate in space group P1211 provides additional crystallographic evidence of the backside interaction in an E2~Ub conjugate (Figure 5A). Cα-Cα contact distance plots for the UbcH5/Ub backside interaction within both UbcH5~Ub structures demonstrate that each structure utilizes a nearly identical pattern of E2/Ub contacts (Supplemental Figure S4).
Figure 5.
The canonical UbcH5/Ub backside interaction and relative intra-conjugate Ub orientation determine the geometry of UbcH5~Ub oligomers. (A) The positions of ubiquitin chains participating in a backside interaction with UbcH5 (white) are nearly identical for PDB 3ugb (cyan), PDB 3a33 (orange) and PDB 2fuh (blue). (B) The relative orientations of UbcH5b and Ub in PDB 3a33 in combination with the UbcH5/Ub backside interaction produce an infinite spiral of UbcH5b~Ub molecules. (C) The relative orientations of UbcH5c and Ub in PDB 3ugb in combination with the UbcH5/Ub backside interaction create an infinite staggered linear array of UbcH5c~Ub molecules. (D) Residues mediating contacts between UbcH5c molecules from neighboring conjugates in the staggered linear array are shown as sticks. Water molecules are shown as red spheres. Hydrogen bonds and salt bridges are shown as dashed lines (black). For panels (B-D), UbcH5 molecules are colored red, orange, yellow, green, blue or purple and all Ub molecules are colored white.
Structural Variability and Diversity in UbcH5~Ub Conjugate Oligomerization
Extensive line broadening observed in NMR spectra of UbcH5~Ub conjugates (25) suggested that the UbcH5/Ub backside interaction causes UbcH5~Ub conjugates to form higher-order oligomers. The first crystal structure of an oligomerized conjugate (UbcH5b~Ub) (21) revealed an infinite spiral oligomer (Figure 5B). Our structure presented here (PDB 3ugb) demonstrates a second architecture for UbcH5~Ub oligomers, in which the Ub orientation results in a staggered linear array oligomer (Figure 5C). The UbcH5~Ub structures PDB 3a33 and PDB 3ugb demonstrate that the UbcH5/Ub backside interaction is compatible with multiple relative Ub orientations. Additionally, the positions of UbcH5 molecules within the staggered linear array result in E2/E2 interactions (Figure 5D). Such interactions were predicted in the original UbcH5~Ub backside interaction study (25) but are not present in the infinite spiral oligomer (PDB 3a33). The E2/E2 interaction surface within the staggered linear array comprises a surface area of 330 Å2 as calculated by PISA (53). The interactions at this E2/E2 interface include a water mediated hydrogen bond network, a salt bridge between Glu140 and Lys66 and a cation-π interaction between Arg136 and His55 (Figure 5D). Despite the addition of the E2/E2 interface in the staggered linear UbcH5~Ub oligomers, the two architectures result in similar total buried surface areas, due the different orientations of Ub in the two oligomers. A staggered linear UbcH5c~Ub dimer buries 1725 Å2 of surface area while an infinite spiral UbcH5b~Ub dimer buries a surface area of 1700 Å2.
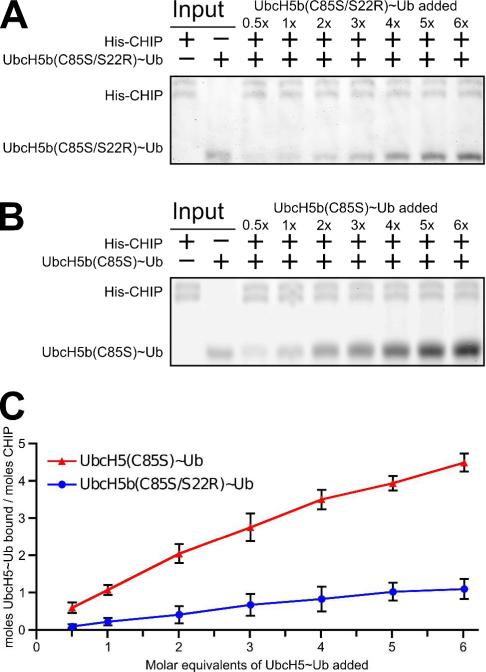
E3 Ubiquitin Ligase CHIP Interacts with UbcH5~Ub Conjugate Oligomers
Existing structures of UbcH5 in complex with both HECT- and RING-/U-box-type E3 enzymes do not occlude the UbcH5 residues involved in backside-mediated oligomerization. While E3s should be able to bind to directly to such oligomers, this interaction has not been experimentally verified. We therefore performed pull-down assays with the U-box-type ligase CHIP and UbcH5b~Ub conjugates. We first bound His6-tagged CHIP to Ni-coated magnetic beads and probed with increasing concentrations of oligomerization-deficient UbcH5b(C85S/S22R)~Ub conjugate. The UbcH5b-S22R mutation prevents backside-mediated oligomerization, resulting in a monomeric UbcH5~Ub species, but does not perturb the interaction between CHIP and UbcH5b. The pull-down assays (Figure 6A) indicate that CHIP interacts with the monomeric conjugate in a saturable manner (Figure 6C). In contrast, CHIP pulls down a higher quantity of oligomerization-competent UbcH5b(C85S)~Ub that increases linearly with the addition of conjugate and does not saturate within the range examined (Figure 6B). These results demonstrate that CHIP interacts directly with an oligomerized UbcH5~Ub conjugate. In our experiment, based on a comparison of the intensities of the respective UbcH5b(C85S)~Ub and UbcH5b(C85S/S22R)~Ub bands to the intensities of input controls (data not shown), each CHIP-bound conjugate oligomer is comprised of four UbcH5~Ub conjugates on average. The trend seen in Figure 6C suggests that UbcH5~Ub oligomers bound to CHIP may contain even higher numbers of conjugate molecules.
Figure 6.
Interaction of UbcH5~Ub conjugate oligomers with the U-box-type E3 CHIP. (A) Full-length His6-CHIP, bound to Ni-coated magnetic beads, was used to pull down backside-mediated oligomerization deficient HsUbcH5b(C85S/S22R)~Ub (A) or backside-mediated oligomerization competent HsUbcH5b(C85S)~Ub (B) at the indicated molar equivalents. Eluted CHIP and UbcH5b~Ub in (A) and (B) were analyzed by SDS-PAGE. Gels were stained with IRDye Blue Protein Stain (LI-COR) and detected by fluorescence imaging at 700nm. (C) LI-COR quantified UbcH5~Ub band intensities were compared to the eluted CHIP intensity within the same lane, calibrated against input bands and plotted as a function of the molar equivalents of UbcH5b~Ub added. Data points represent the average of 3 separate trials with error bars indicating the standard deviation.
DISCUSSION
E2~Ub conjugates play a central role in the ubiquitination cascade. The structure of UbcH5c~Ub adds to a small but growing library of E2~Ub conjugate structures and allows for insights into the effect of Ub orientation on active site accessibility, organization and intra- and inter-conjugate interactions. Here we find that UbcH5~Ub conjugates are not limited to the previously reported infinite spiral arrangement but can also adopt a linearly staggered arrangement. The UbcH5/Ub backside interaction is maintained in both oligomers and is not constrained by the variable intra-conjugate Ub orientation.
Are extended oligomers that result from iteration of the UbcH5/Ub backside interaction compatible with E3 ubiquitin ligases? We found direct evidence for an interaction between a U-box-type E3 and UbcH5~Ub oligomers (Figure 6). We examined the compatibility of the two structurally characterized UbcH5~Ub conjugate oligomers with the UbcH5-interacting domains of representative E3 ubiquitin ligases. We docked both the staggered linear array and infinite spiral UbcH5~Ub oligomers onto U-box- and HECT- domains using the structure of UbcH5a in complex with the U-box domain of CHIP (32) and the structure of UbcH5b~Ub in complex with the HECT domain of NEDD4L (20) (Supplemental Figure S5). The staggered linear array and infinite spiral UbcH5~Ub oligomers are tolerated in both docked models without any steric clashes. We extended the docking analysis by using the full-length structures of CHIP (54) and the Cul1-Rbx1-Skp1-Skp2 SCF ligase, a multisubunit RING-type ligase (55). Both architectures are accommodated by full length CHIP and SCF ligases without any steric clashes (Supplemental Figure S6). Both oligomers extend away from the E3s, and no E3/conjugate interactions are evident other than the E2/E3 interaction surface between the E3 RING/U-box domains and the “initiating” E2s of the oligomers that are present in the original crystal structures.
Throughout the staggered linear array and infinite spiral UbcH5~Ub oligomeric chains, the RING/U-box E3 binding site is unobstructed, The extended oligomers thus contain many potentially accessible RING/U-box E3 binding sites, suggesting that more than one E3 may bind to a UbcH5~Ub oligomeric assembly. While our pulldown experiment does not allow us to distinguish between this possibility and the possibility that the E3 binds only to the “initiating” conjugate in the oligomer, a closer examination of modeled E3/UbcH5~Ub complexes (Supplemental Figures S5 and S6) provides insight. HECT domains bind Ub in an orientation that does not allow for a simultaneous backside interaction, as the non-covalently interacting E2 would clash with the HECT domain. Most likely, HECT-type ligases therefore bind only to the “initiating” conjugate. Such a steric restriction does not apply to the CHIP or SCF ligases, suggesting that these RING/U-box E3s are able to bind at multiple sites along an oligomer. However, Deshaies and coworkers showed that formation of a transient “closed” state is required for E3-mediated ubiquitination by the E2 enzyme Cdc34, and likely also by UbcH5 (56). In the “closed” state, the canonical Ub-Ile44 binding surface is occluded by the E2 and cannot participate in the backside interaction. However, the E2 backside binding surface of a “closed” state conjugate is unobstructed and competent to bind ubiquitin. This suggests that the “closed” state could be allowed for the initiating conjugate but not for other conjugates in the oligomer. Thus, if ubiquitination is to proceed, the mechanistic requirement of a “closed” state Ub orientation may limit RING/U-box E3s to binding only the “initiating” conjugate.
Solution phase studies indicate that the Ub orientation is dynamic (24, 51), implying that UbcH5~Ub oligomers may be dynamic as well. Models of CHIP and SCF ligases in complex with UbcH5~Ub oligomers (Supplemental Figure S6) suggest that a wide range of conformationally dynamic UbcH5~Ub oligomers are accommodated by RING/U-box E3 ubiquitin ligases. The range of possible UbcH5~Ub oligomers can be represented by combining the UbcH5/Ub backside interaction with intra-conjugate Ub orientations that are compatible with UbcH5~Ub oligomerization. Unlike the “closed” state, “open” and “backbent” state Ub orientations are compatible with the UbcH5~Ub backside interaction and could occur anywhere within an E2~Ub conjugate oligomer. Combinations of UbcH5~Ub conjugates with intra-conjugate Ub orientations from the UbcH5b~Ub (PDB 3a33) and UbcH5c~Ub (PDB 3ugb) structures could produce many distinct oligomers (Supplemental Figure S7A). For example, a chain assembly of four UbcH5~Ub conjugates utilizing the two Ub orientations from PDB 3a33 and PDB 3ugb could form 8 (23) different isomers. However, the topology of conjugate oligomers is likely even less constrained than suggested by the two current oligomeric crystal structures. An ensemble of twenty E2~Ub orientations was required to fully reproduce the experimental SAXS curve for UbcH5c~Ub (24). If we limit the available UbcH5~Ub orientations to those found in current E2~Ub structures that are compatible with backside-mediated oligomerization (5 structures: PDB 2gmi, PDB 2kjh, PDB 3a33, PDB 3jw0 and PDB 3ugb), an oligomer comprised of four UbcH5~Ub conjugates could form as many as 125 (53) different isomers (Supplemental Figure S7B-F). The diversity of possible E2~Ub oligomer topologies, in combination with the conformational dynamics observed by SAXS and NMR, suggest that that UbcH5~Ub oligomers are likely to be conformationally diverse and dynamic. While the staggered linear array and infinite spiral architectures are currently the only structurally characterized UbcH5~Ub oligomers, these architectures probably represent only two of many UbcH5~Ub conjugate oligomers.
The continuum of transiently occupied “open”, “closed,” and “backbent” states identified by SAXS and NMR analyses (24) and by the E2~Ub structures discussed here may have important functional consequences apart from their effects on oligomerization. For one, with the exception of the “closed” state model PDB 1fxt (Ubc1~Ub), it is significant that none of the structures utilize the canonical Ub-Ile44 recognition surface for intra-conjugate interactions. The I44 surface is engaged by the majority of known ubiquitin-binding domains (57). The use of a noncanonical Ub recognition surface could allow the exposed Ub-Ile44 surface to participate in additional interactions in larger ternary complexes, such as those suggested by models of the interactions between Ub, the ZNF216 A20-like zinc finger ubiquitin binding domain, and the ubiquitin associated (UBA) domain of p62 (58). Additionally, exposure of the Ub-Ile44 surface in a E2~Ub conjugate would allow for engagement with ubiquitin binding domains on proteins that undergo self-ubiquitination even in the absence of E3 ligases (59).
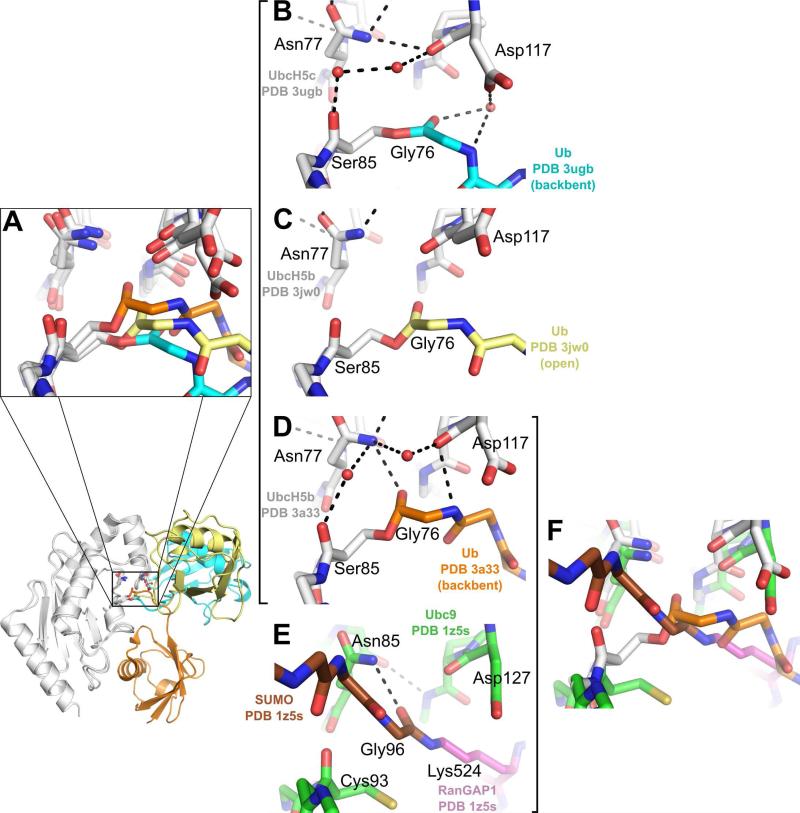
The transiently occupied states of the conjugate may also have more direct implications for catalysis and ubiquitin transfer. Our comparison of active site SASA suggests that the relative orientation of Ub does not dictate the accessibility of the active site thioester bond. However, solvent accessibility is not the sole requirement for active site competency. Previous studies have shown that both a conserved asparagine (UbcH5-Asn77)(60) and a conserved aspartate (UbcH5-Asp117)(7, 60) play critical roles in Ub transfer to lysines. Although the specific roles of these two residues remain to be experimentally confirmed, it has been proposed that that UbcH5-Asn77 stabilizes the oxyanion and hydrogen bonds to the Ub-Gly76 carbonyl prior to Ub transfer, while UbcH5-Asp117 is thought to act as a general base, both deprotonating and lowering the pKa of the incoming lysine. Interestingly, the positions and orientations of the Ub backbone at the thioester and the key active site residues UbcH5-Asn77, UbcH5-Asp117, and UbcH5-Ser85 (Figure 7A) differ in the three available UbcH5~Ub structures (PDB 3a33, PDB 3jw0 and PDB 3ugb).
Figure 7.
Comparison of molecular environments surrounding the E2~Ub active site. (A) Comparison of E2~Ub active site environments for PDB 3ugb (UbcH5c:white, Ub:cyan), PDB 3a33 (UbcH5b:white, Ub:orange) (21) and PDB 3jw0 (UbcH5b: white, Ub: yellow) (20). A zoomed-out cartoon view highlights the location of the E2~Ub active site. (B) E2~Ub active site environment for PDB 3ugb (UbcH5c: white, Ub: cyan). (C) E2~Ub active site environment for PDB 3jw0 (UbcH5b: white, Ub: yellow). (D) E2~Ub active site environment for PDB 3a33 (UbcH5b: white, Ub: orange). (E) E2~Ub active site environment for PDB 1z5s (Ubc9: green, SUMO: brown, RanGAP1: purple) (61). (F) Comparison of E2~Ub active site environments for PDB 3a33 (UbcH5b: white, Ub: orange) and PDB 1z5s (Ubc9: green, SUMO: brown, RanGAP1: purple).
In our UbcH5c~Ub structure (PDB 3ugb, Figure 7B), the Ub backbone participates in a water-mediated hydrogen bond to the UbcH5-Asp117 side-chain. UbcH5-Asn77 stabilizes the UbcH5-Asp117-Pro118 loop and is not in position to form a hydrogen bond with the carbonyl of Ub-Gly76. UbcH5-Asn77-mediated stabilization of the Asp117-Pro118 loop is observed in both PDB 3ugb (UbcH5c~Ub) and PDB 2gmi (Ubc13~Ub/Mms2) E2~Ub structures. In contrast, the structure of UbcH5b~Ub in complex with NEDD4L-HECT exhibits a different active site organization. In this structure (PDB 3jw0, Figure 7C) UbcH5-Asp117 is disengaged from the Ub backbone, allowing rotation of the Ub backbone towards UbcH5-Asn77. The UbcH5b~Ub structure (PDB 3a33, Figure 7D) presents a third, distinct active site organization. In this structure the Ub-Gly76 backbone carbonyl is hydrogenbonded to the UbcH5-Asn77 side-chain, a position that may allow the UbcH5-Asn77 to stabilize the oxyanion upon attack by a lysine. Furthermore, UbcH5-Asp117 does not interact with the Ub C-terminus but may be poised for interaction with an incoming lysine. While there are no transition state structures, or post-Ub transfer structures for the UbcH5/Ub system, a structure of Ubc9 in complex with SUMOylated RanGAP1 (61) (Figure 7E) provides clues to the organization of the E2 active site after Ub transfer. Intriguingly, the active site organizations for the UbcH5b~Ub structure (PDB 3a33) and the Ubc9/RanGAP1~SUMO complex (PDB 1z5s) are remarkably similar (Figure 7F). In both structures, the Ub-Gly76 (SUMO-Gly96) is in position to interact with the E2-Asn77 side-chain. Additionally, the position of the E2-Asp117 side-chain is similarly disengaged from the Ub or SUMO backbone in both structures.
The difference in active site orientations among the UbcH5~Ub structures thus raises an interesting question. What is the relationship between relative Ub orientation, active site organization and active site activation? Although the “closed” state Ub orientation has been shown to be important for activating Ub transfer by aminolysis (56), the absence of a UbcH5~Ub or E2~Ub “closed” state structure in which side chain orientations have been explicitly determined prevents a direct comparison to the existing UbcH5~Ub structures. The relationship between E2~Ub active site organization and Ub orientation may be important for E2 enzymes that interact with different families of E3 enzymes. For example, the UbcH5 family of E2 enzymes participates in Ub transfer with RING-, U-box-, and HECT-type E3s with widely divergent architectures. It is unlikely that interactions with RING-/U-box-type versus HECT-type E3 enzymes results in identical or even similar relative Ub orientations. The E2~Ub active site must nevertheless be activated or made competent for activation, to allow for E3-mediated nucleophilic attack and ubiquitin transfer. The ability to tune the E2~Ub active-site reactivity by altering Ub orientation may enable the E2~Ub to be competent for Ub transfer via aminolysis or transthiolation, depending upon the identity of the E3 ligase.
The number of E2~Ub conjugate structures deposited in the PDB is now beginning to reach a critical mass that allows for insights into the mechanisms underlying ubiquitination. Nevertheless, additional structures of E2~Ub conjugates are still needed. In particular, E2~Ub conjugate structures in complex with a range of E3 enzymes may show whether the mechanisms of ubiquitination are specific to each class of E3 enzymes, or whether more general mechanisms exists. Furthermore, structures of E2~Ub conjugates from additional E2 families will shed light on the role of both intra- and inter-conjugate E2/Ub interactions. For the UbcH5 family of E2~Ub conjugates, transition state or pre- and post-transition state structures would contribute significantly to our understanding of the molecular mechanisms of ubiquitination. Although a transition-state structure including the oxyanion intermediate poses a difficult challenge to structural biology, recent advances in synthetically modified Ub moieties (62-64) may provide potential avenues of approach to characterize pre- and post-transition state structures. While much work remains to be completed on the road to characterizing the mechanisms of E3-mediated ubiquitination, the structure of the UbcH5c~Ub conjugate presented here provides valuable insights into intra-conjugate E2/Ub interactions, the organization of the E2~Ub active site, and the dynamic, versatile architecture of E2~Ub conjugate oligomers.
Supplementary Material
Acknowledgments
FUNDING SOURCE STATEMENT
The authors acknowledge financial support from the US National Institutes of Health (grant RO1-GM080271 to S.M. and grant RO1-GM088055 to R.E.K.). R.C.P. was partly supported by U.S. Public Health Service (PHS) National Research Service Award (NRSA) postdoctoral fellowship T32 HL007914. J.N.P. was supported by U.S. PHS NRSA predoctoral fellowship T32 GM008268.
ABBREVIATIONS
- CHIP
carboxyl terminus of Hsp70 interacting protein
- E1
ubiquitin-activating enzyme
- E2
ubiquitinconjugating enzyme
- E3
ubiquitin ligase
- HECT
homologous to E6AP carboxy terminus
- NMR
nuclear magnetic resonance
- RING
really interesting new gene
- SAXS
small angle X-ray scattering
- Ub
ubiquitin
Footnotes
SUPPORTING INFORMATION AVAILABLE
Molecular modeling protocol to generate the Ube2s~Ub conjugate, Ube2s~Ub Cα-Cα contact plot, comparison of Ube2s~Ub to E2~Ub structures deposited in the PDB, sequence scoring chart for Ub hot spot analysis, 2Fo-Fc electron density map for the UbcH5c~Ub oxyester linkage, Cα-Cα contact plots and structural alignments of UbcH5/Ub structures deposited in the PDB that contain the backside interaction, modeled complexes between UbcH5~Ub oligomers and CHIP-U-box, NEDD4L-HECT, full length CHIP and Cul1-Rbx1-Skp1-Skp2 SCF ligases and models of variable topology E2~Ub oligomers. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Cyr DM, Hohfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 5.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 6.Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci U S A. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciechanover A, Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.David Y, Ziv T, Admon A, Navon A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J Biol Chem. 2010;285:8595–8604. doi: 10.1074/jbc.M109.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim HC, Huibregtse JM. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol Cell Biol. 2009;29:3307–3318. doi: 10.1128/MCB.00240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 13.Capili AD, Lima CD. Taking it step by step: mechanistic insights from structural studies of ubiquitin/ubiquitin-like protein modification pathways. Curr Opin Struct Biol. 2007;17:726–735. doi: 10.1016/j.sbi.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 16.Verdecia MA, Joazeiro CA, Wells NJ, Ferrer JL, Bowman ME, Hunter T, Noel JP. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol Cell. 2003;11:249–259. doi: 10.1016/s1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- 17.Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci U S A. 2005;102:18890–18895. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wenzel DM, Stoll KE, Klevit RE. E2s: structurally economical and functionally replete. Biochem J. 2011;433:31–42. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 20.Kamadurai HB, Souphron J, Scott DC, Duda DM, Miller DJ, Stringer D, Piper RC, Schulman BA. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol Cell. 2009;36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakata E, Satoh T, Yamamoto S, Yamaguchi Y, Yagi-Utsumi M, Kurimoto E, Tanaka K, Wakatsuki S, Kato K. Crystal structure of UbcH5b~ubiquitin intermediate: insight into the formation of the self-assembled E2~Ub conjugates. Structure. 2010;18:138–147. doi: 10.1016/j.str.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Hamilton KS, Ellison MJ, Barber KR, Williams RS, Huzil JT, McKenna S, Ptak C, Glover M, Shaw GS. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9:897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 23.Serniwka SA, Shaw GS. The structure of the UbcH8-ubiquitin complex shows a unique ubiquitin interaction site. Biochemistry. 2009;48:12169–12179. doi: 10.1021/bi901686j. [DOI] [PubMed] [Google Scholar]
- 24.Pruneda JN, Stoll KE, Bolton LJ, Brzovic PS, Klevit RE. Ubiquitin in motion: structural studies of the ubiquitin-conjugating enzyme~ubiquitin conjugate. Biochemistry. 2011;50:1624–1633. doi: 10.1021/bi101913m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Capili AD, Lima CD. Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J Mol Biol. 2007;369:608–618. doi: 10.1016/j.jmb.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi YS, Jeon YH, Ryu KS, Cheong C. 60th residues of ubiquitin and Nedd8 are located out of E2-binding surfaces, but are important for K48 ubiquitin-linkage. FEBS Lett. 2009;583:3323–3328. doi: 10.1016/j.febslet.2009.09.034. [DOI] [PubMed] [Google Scholar]
- 28.Das R, Mariano J, Tsai YC, Kalathur RC, Kostova Z, Li J, Tarasov SG, McFeeters RL, Altieri AS, Ji X, Byrd RA, Weissman AM. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol Cell. 2009;34:674–685. doi: 10.1016/j.molcel.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK. Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J. 2007;26:2797–2807. doi: 10.1038/sj.emboj.7601711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W, Tu D, Li L, Wollert T, Ghirlando R, Brunger AT, Ye Y. Mechanistic insights into active site-associated polyubiquitination by the ubiquitin-conjugating enzyme Ube2g2. Proc Natl Acad Sci U S A. 2009;106:3722–3727. doi: 10.1073/pnas.0808564106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D, 3rd, Fukuda M, Ohta T, Klevit R. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitinligase complex. Proc Natl Acad Sci U S A. 2003;100:5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Z, Kohli E, Devlin KI, Bold M, Nix JC, Misra S. Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct Biol. 2008;8:26. doi: 10.1186/1472-6807-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 34.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terwilliger TC. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 39.DeLano W. The PyMol Molecular Graphics System. 2002.
- 40.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunter JD. Matplotlib: A 2D Graphics Environment. Computing in Science & Engineering. 2007;9:90–95. [Google Scholar]
- 44.Winget JM, Mayor T. The diversity of ubiquitin recognition: hot spots and varied specificity. Mol Cell. 2010;38:627–635. doi: 10.1016/j.molcel.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 45.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitz C, Stanton-Cook MJ, Su XC, Otting G, Huber T. Numbat: an interactive software tool for fitting Deltachi-tensors to molecular coordinates using pseudocontact shifts. J Biomol NMR. 2008;41:179–189. doi: 10.1007/s10858-008-9249-z. [DOI] [PubMed] [Google Scholar]
- 47.Valafar H, Prestegard JH. REDCAT: a residual dipolar coupling analysis tool. J Magn Reson. 2004;167:228–241. doi: 10.1016/j.jmr.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 48.Winkler H, Zhu P, Liu J, Ye F, Roux KH, Taylor KA. Tomographic sub-volume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J Struct Biol. 2009;165:64–77. doi: 10.1016/j.jsb.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990;8:52–56. 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 50.Bocik WE, Sircar A, Gray JJ, Tolman JR. Mechanism of polyubiquitin chain recognition by the human ubiquitin conjugating enzyme Ube2g2. J Biol Chem. 2011;286:3981–3991. doi: 10.1074/jbc.M110.189050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miura T, Klaus W, Gsell B, Miyamoto C, Senn H. Characterization of the binding interface between ubiquitin and class I human ubiquitin-conjugating enzyme 2b by multidimensional heteronuclear NMR spectroscopy in solution. J Mol Biol. 1999;290:213–228. doi: 10.1006/jmbi.1999.2859. [DOI] [PubMed] [Google Scholar]
- 52.Bosanac I, Phu L, Pan B, Zilberleyb I, Maurer B, Dixit VM, Hymowitz SG, Kirkpatrick DS. Modulation of K11-linkage formation by variable loop residues within UbcH5A. J Mol Biol. 2011;408:420–431. doi: 10.1016/j.jmb.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 53.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 54.Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 55.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 56.Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol Cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garner TP, Strachan J, Shedden EC, Long JE, Cavey JR, Shaw B, Layfield R, Searle MS. Independent interactions of ubiquitin-binding domains in a ubiquitin-mediated ternary complex. Biochemistry. 2011;50:9076–9087. doi: 10.1021/bi201137e. [DOI] [PubMed] [Google Scholar]
- 59.Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, Bonifacino JS, Hurley JH. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat Struct Mol Biol. 2006;13:264–271. doi: 10.1038/nsmb1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu PY, Hanlon M, Eddins M, Tsui C, Rogers RS, Jensen JP, Matunis MJ, Weissman AM, Wolberger C, Pickart CM. A conserved catalytic residue in the ubiquitinconjugating enzyme family. EMBO J. 2003;22:5241–5250. doi: 10.1093/emboj/cdg501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geurink PP, El Oualid F, Jonker A, Hameed DS, Ovaa H. A General Chemical Ligation Approach Towards Isopeptide-Linked Ubiquitin and Ubiquitin-Like Assay Reagents. Chembiochem. 2011. [DOI] [PMC free article] [PubMed]
- 63.Castaneda C, Liu J, Chaturvedi A, Nowicka U, Cropp TA, Fushman D. Non-enzymatic assembly of natural polyubiquitin chains of any linkage composition and isotopic labeling scheme. J Am Chem Soc. 2011;133:17855–17868. doi: 10.1021/ja207220g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.El Oualid F, Merkx R, Ekkebus R, Hameed DS, Smit JJ, de Jong A, Hilkmann H, Sixma TK, Ovaa H. Chemical synthesis of ubiquitin, ubiquitin-based probes, and diubiquitin. Angew Chem Int Ed Engl. 2010;49:10149–10153. doi: 10.1002/anie.201005995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weiss M. Global indicators of X-ray data quality. Journal of Applied Crystallography. 2001;34:130–135. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.