Abstract
Objective and automatic sensor systems to monitor ingestive behavior of individuals arise as a potential solution to replace inaccurate method of self-report. This paper presents a simple sensor system and related signal processing and pattern recognition methodologies to detect periods of food intake based on non-invasive monitoring of chewing. A piezoelectric strain gauge sensor was used to capture movement of the lower jaw from 20 volunteers during periods of quiet sitting, talking and food consumption. These signals were segmented into non-overlapping epochs of fixed length and processed to extract a set of 250 time and frequency domain features for each epoch. A forward feature selection procedure was implemented to choose the most relevant features, identifying from 4 to 11 features most critical for food intake detection. Support vector machine classifiers were trained to create food intake detection models. Twenty-fold cross-validation demonstrated per-epoch classification accuracy of 80.98% and a fine time resolution of 30 s. The simplicity of the chewing strain sensor may result in a less intrusive and simpler way to detect food intake. The proposed methodology could lead to the development of a wearable sensor system to assess eating behaviors of individuals.
Index Terms: Chewing (mastication), food intake detection, monitoring of ingestive behavior (MIB), pattern recognition, wearable sensor
I. Introduction
Overweight and obesity, defined as the abnormal or excessive body fat accumulation, is dramatically expanding from high-income countries to low and middle-income countries, especially in urban settings. The World Health Organization estimated that the overweight adult population would increase from 1.5 billion in 2008 to 2.3 billion in 2015 and that obese adult population would rise from 500 to 700 million worldwide during the same period [1].
The main cause of overweight and obesity is a chronic imbalance between the energy consumed in foods and the energy expended, which is reflected in body weight gain. Environmental factors related to an increased intake of energy-dense food (i.e., fried food) and a decrease in the levels of physical activity due to more sedentary form of life have an important contribution to obesity [2], [3]. Accurate and objective measurement of ingestive behavior (when, how and how much of food is consumed), energy content of ingested food (how many calories were consumed) and energy expenditure (how many calories were expended) are several of the major challenges facing obesity research, which will allow the monitoring of the energy balance to observe and potentially correct behaviors leading to weight gain.
Monitoring of Ingestive Behavior (MIB) and caloric energy intake in free living individuals is arguably the most difficult problem in studying behavioral aspects of obesity. Existing methods such as food recall, food-frequency questionnaires [4], [5], self-report diaries [6], [7] and multimedia diaries [8] suffer from low accuracy as people tend to miscalculate and underreport the food consumed leading to an inaccurate measurement of the daily energy intake [9], [10]. This fact plus the tediousness and lack of robustness of these methods for long-term studies or interventions arise the need for more accurate methods to detect specific patterns of food intake.
A. Food Intake Detection
Objective and automatic methods of MIB based on wearable sensors and/or portable devices were introduced as a potential solution to replace the manual self-reporting methods. MIB methods are being developed to measure periods of food intake with minimal individual’s active participation, which may lead to a better understanding of eating behaviors by improving the accuracy of energy intake estimation, reducing the underreporting and relieving the subject from the recording burden. Incorporation of new technology helped participants to automatically report food consumption [11]. In [12], custom designed software was integrated into a mobile phone with a camera to capture images of foods before and after the meal as well as to include additional food information using voice record. In [13], a similar methodology for mobile phone food record was proposed. A total of 79% of adolescents participating in device evaluation agreed that the software was easy to use. These automatic dietary monitoring methodologies showed to increase the accuracy of food intake but they still rely on individuals taking useful images and self-reporting all consumed foods. In [14], a wearable device that integrates a miniature camera, a microphone and several other sensors (accelerometers, reference lights, etc) for recording food intake was presented. The device is currently under development and evaluation. Another wearable sensor that detected food intake by capturing chewing sounds was presented in [15]. High chewing sound recognition rates were achieved but without considering bite and swallowing sounds. The study was then expanded in [16], where a food recognition system was developed to identify vibration patterns among different foods types. Chewing sounds were captured from 2 subjects by means of a wearable earpad sensor. A classification algorithm discriminated intake of four different food types with an overall accuracy of 86.6%.
Our research group is working on the development of methodologies for monitoring and characterization of food intake in free living environment [17]–[20]. In [17] we presented the concept of using chews and swallows as indicators of food intake. In [18], models were created using information from time sequences of both chews and swallows to detect food intake with more that 95% accuracy. In [19], supervised group models and unsupervised individual models were trained to detect food intake by using swallowing alone with 89% recognition accuracy for group models and 93.9% accuracy for individual models. In [20] we presented an acoustical method for detection of swallowing events that could be used as the source of data for methods in [17], [18]. The downside of detecting food intake by monitoring of swallowing alone is the apparent uniqueness of swallowing sound for each individual which results in a need for individual calibration and low accuracy of group recognition models [19]. An appealing alternative may be automatic detection of characteristic jaw motion during chewing. Indeed, previous studies [21] indicate that variations in the jaw motion during chewing differ less between individuals and have well-defined narrow frequency range between 1–2 Hz.
B. Monitoring of Chewing
Ingestion of solid foods can be detected if chewing is present as during food intake a bite is followed by a sequence of chews and swallows, and this process is then repeated throughout an entire meal. Several sensing options are available for monitoring of chewing. Surface Electromyography (EMG) [22] can sense the activation of jaw muscles during mastication by placing electrodes over the skin surface. This measurement technique is obtrusive and may not be suitable for applications under free living conditions. Multi-point sheet-type sensor [23] and strain gauge abutments [24] were also proposed to measure bite and chewing forces. These sensors are placed between the teeth and are likely to produce variations in an individual’s normal mastication patterns. Gold film corrugated strain gauge sensors were designed and tested in [25] to measure the displacement changes associated with jaw motion. The changes in sensor curvature were translated to electrical resistance changes providing and objective and effective method for chewing monitoring. Another sensing option is based on the detection of vibrations produced during food breakdown (chewing sounds). In [16], an earpad sensor was developed to detect those sounds by capturing air-conduced vibrations inside the ear canal.
The goal of this study was to develop a sensor that can non-invasively monitor characteristic jaw motion during chewing as a part of a wearable device and automatic methods for detection of food intake through monitoring of chewing events alone. Detection of jaw motion, rather than the sounds originating from chewing [16] is proposed to achieve a simpler and more accurate sensor system for MIB. A simple methodology is proposed to automatically detect food intake from epoch-divided chewing signal by using a Support Vector Machine (SVM) trained with time and frequency domain features extracted from the captured chewing signal. A forward feature selection procedure was implemented to determine the most relevant set of features representing the data. The optimal size of the epoch was also evaluated along with the most appropriate number of adjacent epochs to be added to the feature vector.
II. Methods
A. Jaw Motion Sensor
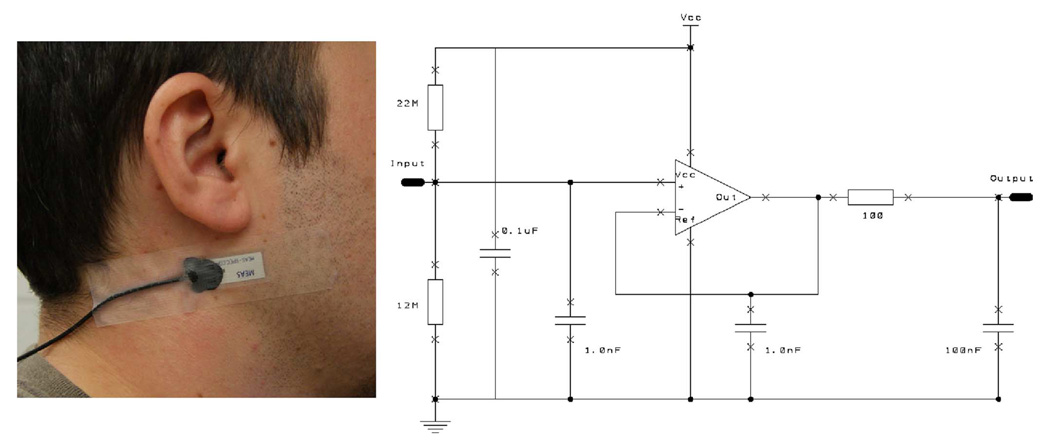
The purpose of this sensor is to detect characteristic jaw motion during chewing while being non-invasive, non-obtrusive and socially acceptable. In [17] we determined that the best sensor location is immediately below the outer ear (Fig. 1, left) where the jaw motion can be detected by monitoring changes in skin curvature due to changes in distance between the mandible (jaw) and temporal/occipital bones of the skull during chewing. The changes in skin curvature can be detected by a strain sensor. Testing of various foil gauges including a corrugated thin-film sensor [25] produced reasonable results in monitoring of the skin curvature but unacceptably high energy consumption due to low electrical resistance. Low power consumption is a major requirement as the chewing sensor is envisioned as a part of a wearable sensor system. To address the power consumption requirement, dynamic skin strain was monitored by an off-the-shelf piezoelectric film strain gauge sensor. The selected sensor was the LDT0-028K vibration sensor manufactured by Measurement Specialties (http://www.meas-spec.com/). The piezoelectric film element is a PVDF polymer of 28 µm thickness with screen-printed Ag-ink electrodes and laminated with an acrylic coating. This laminated film element develops high voltage output when flexed and can generate up to 7 V with a tip deflection of 2 mm, however, sensor response is frequency dependent. Testing of the sensor has demonstrated response in the range −0.05 V to 0.05 V when flexed at frequencies < 1 Hz, thus allowing for a simple interface through a buffering and level shifting non-inverting op-amp circuit. The strain sensor was attached by medical tape to the area below the outer ear (Fig. 1, left). Such attachment satisfied the requirement of non-invasiveness and partially satisfied the requirements of non-obtrusiveness and social acceptance. In the future, the sensor can be further miniaturized, made as a wireless “band-aid” sensor and thus improve in these categories. The high impedance of the piezoelectric film was buffered with unity gain by an ultra-lower power TLV-2452 operational amplifier (Texas Instruments) with 1 GOhm differential input resistance. Fig. 1 (right) illustrates the buffer circuit used. A voltage divider was added to the input stage to set a DC offset. Capacitors were connected at input, output and feedback stages to remove high frequency RF noise and eliminate high-voltage spikes that may be created by accidental high frequency excitation. The buffered signal was sampled at 100 Hz and quantized with 16 bits by a data acquisition module USB-1608FS (Measurement Computing). Fig. 2 (top) shows an example of the signal obtained after buffering.
Fig. 1.
Left: strain sensor attached below the outer ear to detect movement of the lower jaw. Right: sensor amplifier.
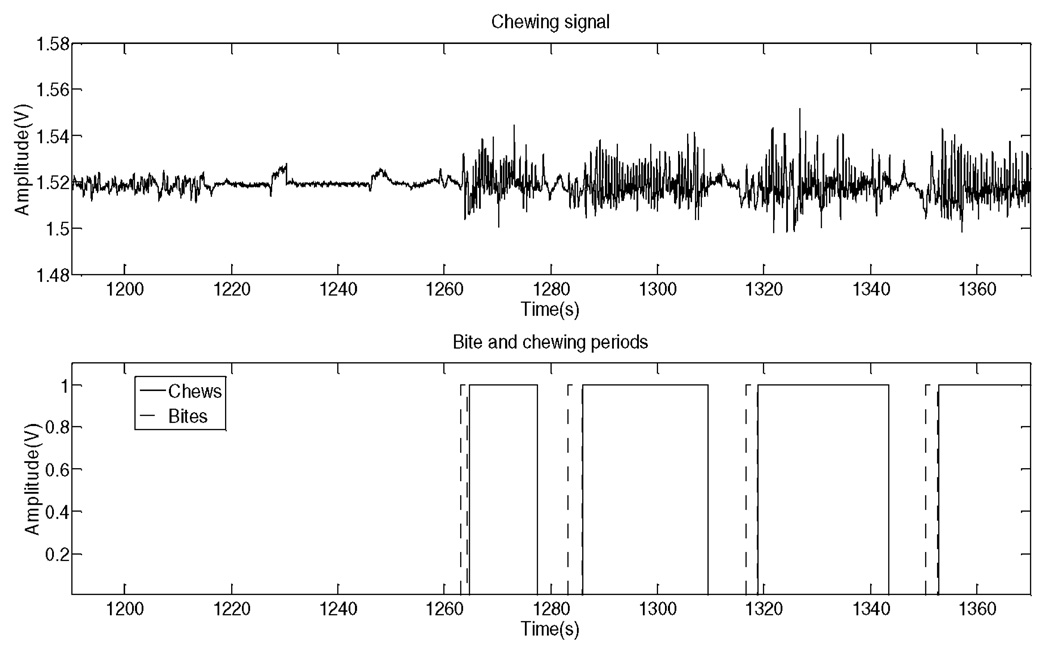
Fig. 2.
Top: Strain sensor signal collected during resting and food ingestion. Bottom: Bites and chews marked during annotation of food intake by a human rater.
B. Data Collection
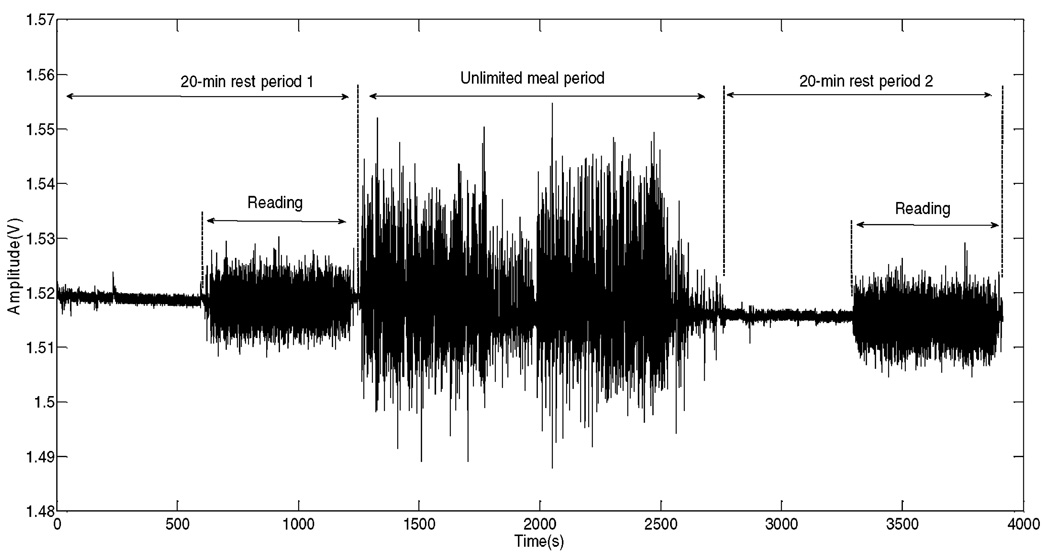
Data used in this study were collected during a human study described in [17]. A total of 20 healthy volunteers (11 males and 9 females, age ranged from 18 to 57 years) who did not present any medical condition that would interfere with normal food intake were recruited. The average body mass index (BMI) was 29.0 ± 6.4 kg/m2, to verify that the proposed sensors may be implemented in obese individuals. Each subject participated in four visits, each of which consisted of three parts: 1) 20-min resting period where the subjects remained seated quietly for 10 min and spent 10 min reading aloud; 2) a meal period consisting of unlimited time to eat four food items in a specific order (pizza, yogurt, apple, peanut butter sandwich, and water); 3) a second 20-min resting period with subjects remaining silent for 10 min and reading aloud for 10 min. A total of 66 visits were analyzed in this study. The purpose of reading aloud was to test the ability of the chewing sensor and pattern recognition algorithms to differentiate jaw motion during chewing and talking. The food items consumed during the meal period were selected to represent different physical properties of everyday foods, specifically, different chewing properties such “hardness”, “crunchiness” and “tackiness” [26], [27]. For example, pizza is a typical soft food; yogurt is a semi-solid food that some people chew and some don’t; apple is a hard crunchy food while peanut butter sandwich is soft but very tacky and hard to chew. During each visit, subjects were monitored by a multi-modal sensor system [17] including a jaw motion sensor. By means of custom designed scoring software, the beginning and end of each chewing period were marked labeling each sample of the sensor signal either as ’chewing’ (+1) or ’no chewing’ (−1). Periods containing bites, swallows, speech or inactivity were labeled as ’no chewing’. The number of chews associated with each period was counted. The top plot in Fig. 2 illustrates an example of a signal captured during a 140 s interval by the piezoelectric strain gauge sensor. The bottom plot illustrates the results of the scoring process that marked bites (dashed line) and chewing periods (solid line). Fig. 3 shows the signal collected during a whole visit (more than an hour of data). In both resting periods, the movement of the lower jaw was minimal during subject’s inactivity. Fig. 3 also illustrates that the sensor responds to the jaw motion both during food intake and talking with food intake signal being of higher magnitude, most likely due to higher forces involved in food grinding. The goal of automatic food intake detection is to reject sensor signals not originating from chewing (e.g., talking) and respond only to chewing events. The classifier assigns a class label from the set {’intake’, ’no intake’} to a time window of specific length (epoch) by classifying the state of chewing in the sensor signal as ’chewing’ or ’no chewing’. Before classification, a relevant set of features should be extracted from the sensor signal as described in next section.
Fig. 3.
Example of a chewing signal collected during the three parts of a subject’s visit.
C. Feature Extraction and Selection
The first step in the signal processing stage consisted in normalization with respect to the signal median to compensate for differences in signal amplitude between subjects. The chewing signal was then segmented into non overlapping windows of fixed duration, called decision epochs. This parameter was of special importance since it defined the time resolution of food intake detection. A shorter epoch would give better time resolution and should help to identify shorter periods of food intake, such as snacking. On the other hand, a longer epoch may result in higher accuracy as more of the sensor signal is involved in making the decision. Our earlier study used an epoch size of 30 s to detect food intake events based on the frequency of swallows [18]. In this study, three different epoch sizes were evaluated to determine the most appropriate window size for the detection of food intake based on chewing signal: 15 s, 30 s, and 60 s.
Since the original signal was divided into non overlapping decision epochs which may have parts of the signal labeled as “chewing” and parts of the signal labeled as “no chewing”, a procedure was implemented to derive labels for each epoch from the labels of the original signal. The 50% determination rule was used, which labeled an epoch as ’chewing’ if at least 50% of the labels in the original sensor signal were +1, and it labeled an epoch as ’no chewing’ otherwise.
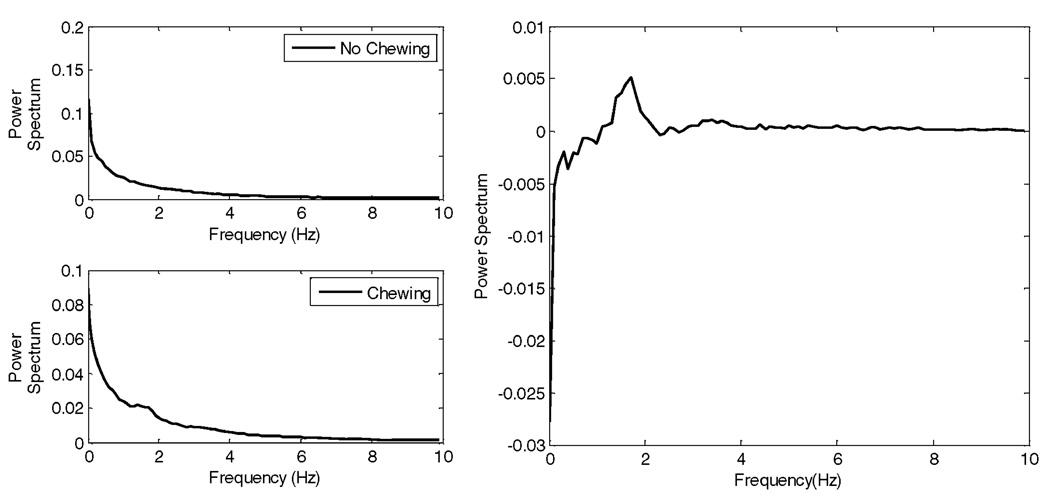
Spectral differences in the sensor signal during chewing and no chewing were utilized for feature computation. During the process of chewing, food is crushed inside the mouth by up-and-down and side-to-side movements of the lower jaw whereas these movements are absent or less pronounced in inactivity and talking periods (’no chewing’). This effect is illustrated in Fig. 4 by calculating the average power spectra for chewing and no chewing epochs (Fig. 4, left) and the difference between them (Fig. 4, right). The frequency interval ranging from 1.0 Hz to 5.0 Hz presented the largest difference between ’chewing’ and ’no chewing’ spectra with the most pronounced difference in the range of 1.25–2.5 Hz. These observations suggested that the characteristic frequencies that distinguish the chewing process fell inside the 1.25–2.5 Hz range, where suitable features could potentially be used to create the classification model. This finding corresponds well with the finding of earlier studies [28], [29] that determined chewing frequency to be in the range 0.7–2 Hz.
Fig. 4.
Left: Power spectrum for “No Chewing” and “Chewing.” Right: Difference between power spectra.
To extract signal features, the sensor signals were first filtered using a bandpass filter with cutoff frequencies of 1.25 Hz and 2.5 Hz. A feature vector fi ∈ ℜd representing each decision epoch (for i = 1, 2, …, N; where N is the total number of epochs) was created by combining a set of 25 scalar features extracted from the filtered and unfiltered signal of each epoch in linear and logarithmic scale. This set of 25 features included time domain and frequency domain features. Time domain features were based on time statistics of: 1) the epoch data, 2) the number of zero crossings inside the epoch, and 3) the number of peaks observed in the epoch. Frequency domain features involved the entropy and standard deviation of the spectrum as well as the peak frequency observed is the epoch data. Table I presents a list with the features used.
TABLE 1.
Scalar Features Used to Extract Information From Chewing Signal.
| Feat # | Description |
|---|---|
| 1 RMS | |
| 2 Entropy (signal randomness) | |
| 3 Base 2 logarithm | |
| 4 Mean | |
| 5 Max | |
| 6 Median | |
| 7 Max to RMS ratio | |
| 8 RMS to Mean ratio | |
| 9 Number of zero crossings | |
| 10Mean time between crossings | |
| 11Max. time between crossings | |
| 12Median time between cross. | |
| 13Minimal time between cross. | |
| 14Std. dev. of time between crossings | |
| 15Entropy of zero crossings | |
| 16Number of peaks | |
| 17Entropy of peaks | |
| 18Mean time between peaks | |
| 19Std. dev. of time between peaks | |
| 20Ratio peaks/zero crossings number | |
| 21Ratio zero crossings/peaks number | |
| 22Entropy of spectrum | |
| 23Std. dev. of spectrum | |
| 24Peak frequency | |
| 25Fractal dimension (uniqueness of the elements inside an epoch) | |
An initial feature vector fi was created when merging several feature subsets that were formed by calculating the 25 scalar features form the filtered and unfiltered epoch and by different feature combinations:
| (1) |
where ffilt and funfilt represent feature subsets extracted from the filtered and unfiltered epochs respectively; ffilt/unfilt and funfilt/filt represent two feature subsets obtained by calculating the ratio between each feature of the ffilt and funfilt subsets and vice versa; and ffilt·unfilt represents another subset of features obtained by calculating the product between each feature of the ffilt and funfilt subsets. These combinations yielded an initial feature vector with 125 dimensions.
A scale equalization was performed to features in the ffilt and funfilt subsets using the natural logarithm. Ratio and product between resulting feature subsets was again calculated to create a log-scaled feature vector with 125 dimensions:
| (2) |
Finally, both the linear and log-scaled feature vectors were concatenated into a single 250-dimension feature vector Fi ∈ ℜ250 that represented each epoch
| (3) |
To account for the time-varying structure of the chewing process, features from neighboring epochs were added to the original epoch feature vector according to the number of lags selected L. Three different lag values were tested: 0, 1, and 2. If this number was greater than zero, then features from L previous and L subsequent epochs were included in the final feature vector τi:
| (4) |
Some of the 250 features extracted may not be as important as others to discriminate between chewing and no chewing epochs. A forward selection procedure [30] was used to find a subset with the most relevant features based on 20-fold cross-validation classification accuracy. In the first step, for a total of D = 250 features, D models were created containing only one feature each. The most relevant feature was given by the model that presented the highest classification accuracy. In the second step, the selected feature was added to the subset and D − 1 new subsets were created by pairing the chosen feature with the remaining D − 1 features, one at a time. D − 1 models were created using those subsets and the pair of features that presented the higher classification accuracy was selected. In the jth step, D−j models were created using D−j subsets that combined the j previously selected features with the each one of the remaining D − j features. The model that yielded the highest classification accuracy was used to selected the (j + 1)th most relevant feature. The algorithm stopped when the addition of new features did not increase the classification accuracy. The resulting subset contained the most relevant set of features that presented the highest classification accuracy.
D. Signal Classifier
Support Vector Machines (SVM) is a supervised machine learning technique that have been used for classification of wide variety of data sets showing outstanding performance in most cases [31]. Robustness and high generalization are two powerful properties that place SVM over other binary classification methods. These properties become beneficial when training group models, which would classify data sets with high variability between subjects. Consequently, SVM was selected in this study as the classification algorithm for chewing detection. SVM models were trained using the LibSVM software package [32]. A linear kernel function k(x, xi) = x·xi was used. Since the features may not be linearly separable in the new feature space, the parameter C was introduced to penalize the training points lying in the wrong side of the decision margin. The optimal value for the parameter C = 10n was chosen by a search procedure using four values of n ∈ (−2, −1, 0, 1).
During the training stage, a subject-independent group model was created by incorporating chewing information from all of the subjects into the training dataset to account for the inter-subject variability. The group model allows detection of food intake without a need for individual calibration. Each point in the training set was a pair of the form {τi, ti}, where τi was the feature vector representing epoch ei and ti ∈ {−1, 1} was the class label that indicated if the feature vector represented a ’chewing’ (ti = 1) or a ’no chewing’ (ti = −1) epoch. The resulting group model classified an epoch’s feature vector belonging to the validation set as ’chewing’ or ’no chewing’ which represented an ’intake’ or ’no intake’ epoch, respectively. A 20-fold cross-validation procedure was used to train and validate the model. The selection of 20 folds allowed the training of the model with features from 19 of the 20 subjects recruited and the validation using independent features from the remaining subject. Thus, each subject was used once for model validation and the results were averaged across all the validation results.
To evaluate the performance of the model, the class labels assigned to each epoch in the validation set were compared to manual scores performed by experienced human raters [17]. The accuracy value defined as the average between precision and recall was the parameter used for comparison:
| (5) |
| (6) |
The true positive (T+) was the number of correctly classified ’chewing’ epochs, the false negative (F−) was the number of times that the model failed to classify a ’chewing’ epoch and the false positive (F+) was the number of times the model incorrectly classified an epoch as ’chewing’.
III. Results
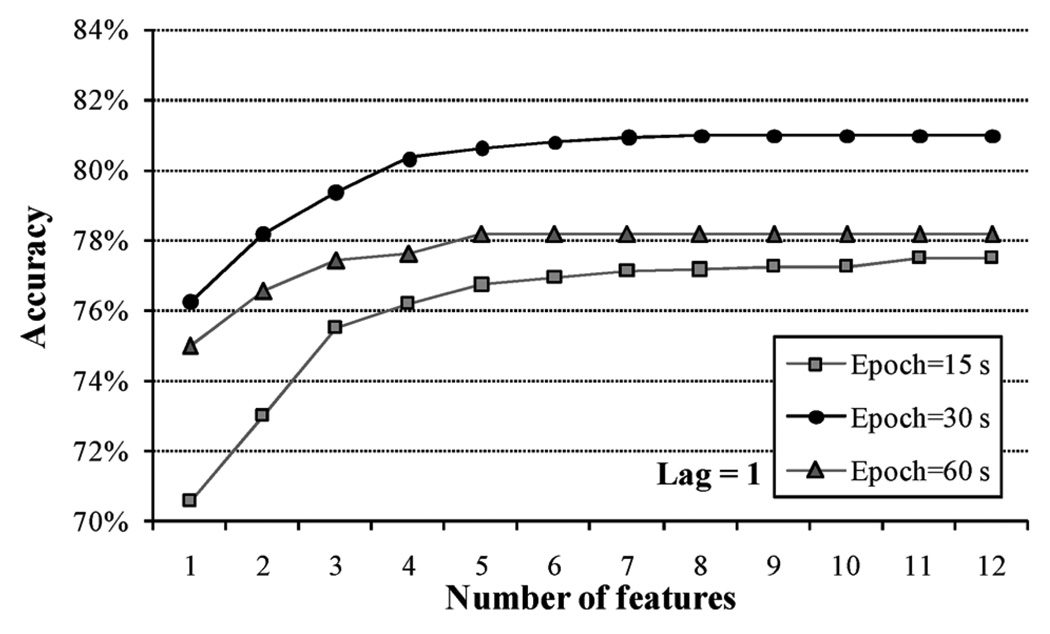
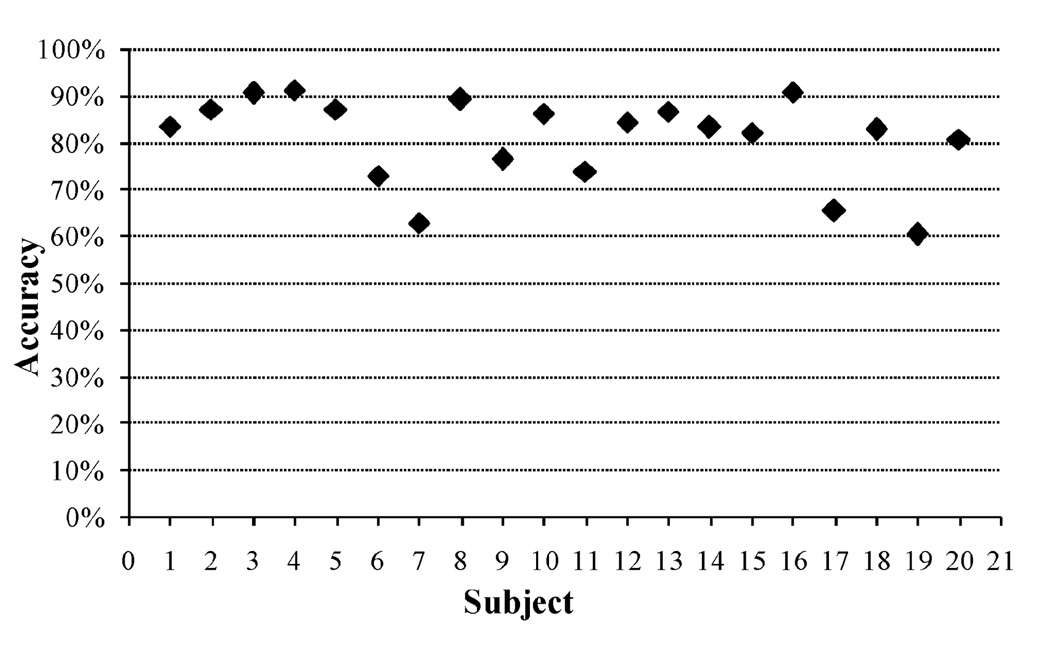
The size of the epoch that best represented the chewing signals was selected from three different values. Results of 20-fold cross-validation indicated that a 30 s epoch size presented higher accuracy than 15 s and 60 s epoch sizes. In all cases, the classifiers were trained with features from the current epoch plus features from one adjacent epoch data (lag L = 1) to account for the time variability of the chewing signal. Fig. 5 illustrates an improvement in the classification accuracy for the three epoch sizes as more features were added to the training vector. No further improvement was observed after the training vector comprised a certain number of features. For an epoch size of 30 s, only 8 features were needed to reach the maximum accuracy value of 80.98% whereas for an epoch size of 15 s the accuracy increment stopped after 11 features were added. When the epoch size increased up to 60 s, the classifier discriminated ’chewing’ and ’no chewing’ epochs with an accuracy of 78.18% using only 5 features although this value did not increase when more features were included in the training vector. Using 30 s epoch (L = 1), the group model trained with 8 features presented per-subject classification accuracies that ranged from 60.5% to 90.8% (Fig. 6).
Fig. 5.
Classification accuracy for three epoch sizes as more features are included in the feature vector. Epochs of 30 s presented the higher accuracy.
Fig. 6.
Classification accuracy for each subject for a time resolution of 30 s.
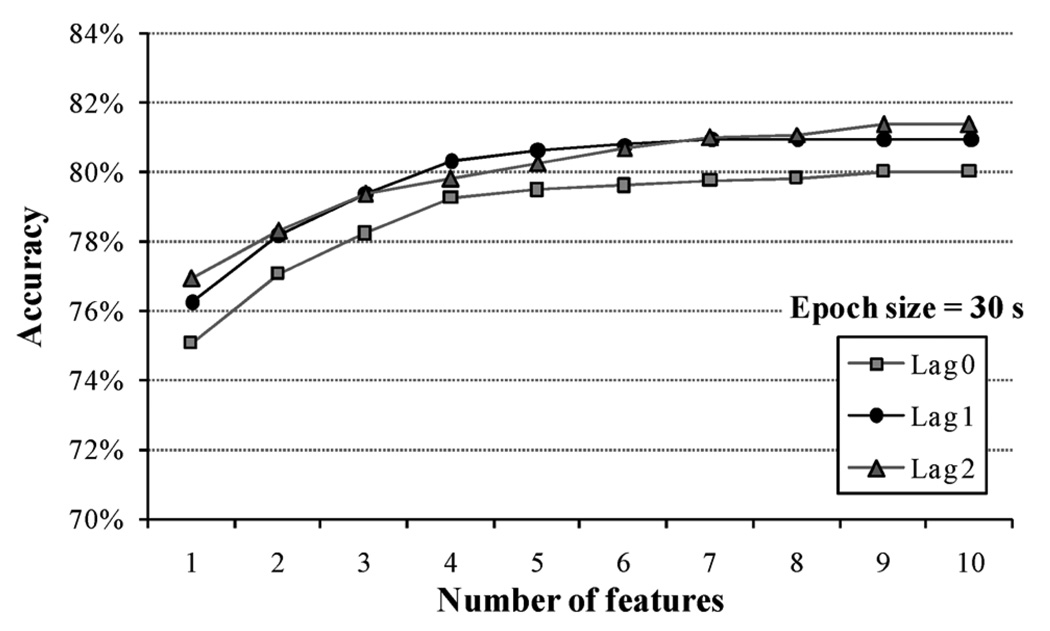
Fig. 7 shows the results using an epoch size of 30 s for three different lag values. The best performance was found to be 81.39% for a feature vector that grouped features from the current epoch and from L = 2 adjacent epochs on each side. This result was very close to the 80.98% accuracy value obtained with L = 1. Statistical analysis showed that the group models for lagged features presented significant difference in the classification accuracy than the group model for non lagged features (two-tailed t-test, p < 0.05), which may indicate the need of including adjacent epoch data to the current epoch.
Fig. 7.
Classification accuracy for an epoch size of 30 s and for three values of the lag parameter L. Lagged features overcame non-lagged features.
Results of the forward selection procedure for 30 s epochs are presented in Table II. Only the results for the lagged feature sets are presented due to they showed better performance than non-lagged features. The feature set combined linear and log scaled features from filtered and unfiltered epochs. A common feature observed in both sets was the entropy of the filtered epoch, which in both cases was the first feature selected. More than 70% of accuracy was obtained by using entropy as the only one feature meaning that the measure of the randomness of the filtered signal represents an acceptable predictor of chewing epochs. For L = 1, most of the class discriminatory information was given by the mean and standard deviation of the time between peaks of the filtered and unfiltered epochs as well as by the fractal dimension. For L = 2, results indicated that the most relevant features were mostly log scaled magnitudes from filtered and unfiltered epochs. Mean and standard deviation of the time between peaks was also chosen.
TABLE II.
Features Selected After a Forward Selection Procedure.
| Epoch size = 30s - Lag = 1 | Epoch size = 30s - Lag = 2 | ||
|---|---|---|---|
| # | Features Selected* | # | Features Selected* |
| 2 | Entropy - (F) | 2 | Entropy - (F) |
| 193 | Mean time between peaks - (logF/logNF) | 168 | Mean time Between peaks - (logNF) |
| 129 | Mean - (logF) | 219 | Std. dev. time between peaks - (logNF/logF) |
| 244 | Std. dev. time between peaks (logF*logNF) | 3 | Base 2 log-(F) |
| 50 | Fractal dimension - (NF) | 57 | Max to RMS ratio - (F/NF) |
| 19 | Std dev time between peaks -(F) | 183 | RMS to mean ratio - (logF/logNF) |
| 94 | Std dev time between peaks -(NF/F) | 233 | RMS to mean ratio - (logF*logNF) |
| 25 | Fractal dimension -(F) | 130 | Max - (logF) |
| 224 | peak frequency - (logNF/logF) | ||
(F) filtered epochs, (NF) unfiltered epochs, (logF) log scaled filtered epochs, (logNF) log-scaled unfiltered epochs.
IV. Discussion
The presented food intake sensor is based on a very simple principle of capturing skin strain during chewing events rather than chewing sound [16]. Reliance on low-frequency chewing motion allows for use of inexpensive and low-power piezoelectric film sensor as the sensing element that can be unobtrusively worn below the outer ear. Use of low-frequency signals also reduces the energy consumption for digital signal processing and feature extraction which is important for the envisioned wearable sensor system. Further miniaturization of the sensor and integration of on-board signal processing is possible, potentially enabling creation of a small wireless “band-aid” wearable sensor system for objective monitoring of ingestive behavior.
Signals captured during periods of resting, talking and food ingestion were used to create a group model that discriminated between ’chewing’ and ’no chewing’ epochs with an averaged accuracy of 80.98% and a time resolution of 30 s. This group model was developed on approximately 72 hours of sensor data collected from 20 subjects. The reason behind creating a population-based model was to incorporate the inter-individual variability of the chewing signals in the training stage so a robust classification model requiring no individual calibration could be achieved.
The training dataset grouped the most relevant features that were selected by the forward selection procedure. Through this procedure, features were chosen based on the classification accuracy of SVM models created using individual and combined features. This fact does not necessarily imply that the selected features are not correlated with each other. However, the projection of these features using the linear kernel function provided the largest separation between classes (largest margin) in the new feature space which was essential to discriminate between ’chewing’ and ’no chewing’ epochs. Feature selection results showed that no more than 11 features were required to achieve the highest classification accuracy for any epoch size or number of lags used. This number represented only 4.4% of the original set containing 250 time domain and frequency domain features and therefore can be calculated on limited computing resources of a wearable device. The graphs in Figs. 5 and 7 showed classification accuracies growing asymptotically until they reached a plateau meaning that the addition of more features to the training vector did not significantly increase the classification accuracy. The feature sets giving the highest classification accuracies (Table II) contained features extracted both from the filtered and unfiltered epoch. These results not only highlighted the importance of combining filtered and unfiltered features but also suggested that the frequency band selected for filtering (1.25–2.5 Hz) indeed encompassed discriminatory information to distinguish chewing and no chewing epochs.
The group model created was based on a per-epoch classification instead of per-meal classification, which would allow the detection of short events of food ingestion such as snacking. The original sensor signal was divided into non-overlapping epochs of fixed size that were labeled as ’chewing’ or ’no chewing’ epochs. Three different epoch sizes were investigated and the optimal size was selected after comparing the classification accuracy of the models created. Results in Fig. 5 showed that a 30 s epoch presented the highest classification accuracy, meaning that the group model was able to detect food intake with 30 s time resolution. The amount of information used for epoch classification increased when features from neighbor epochs were added to the current epoch features. The lag parameter L indicated the total number of adjacent epochs used to train the model. Fig. 7 shows that lagged features (L = 1 and L = 2) improved the classification accuracy observed when features were not lagged (L = 0), which suggested the need of including features form adjacent epochs to account for the time variability of food ingestion. Reported results indicated that the classification accuracy increased when more features were lagged. For an epoch size of 30 s, L = 1 and L = 2 produced statistically similar accuracy values (two-tailed t-test, p > 0.05). This result added to the fact that feature vectors with L = 1 require less computing time than with L = 2 suggested that features from only one adjacent epoch may be added to the current epoch features without jeopardizing the performance of the system.
Classification accuracy may potentially be improved by creating individual models instead of the group model. The implementation of individual models based on subject-specific data would have to provide for means of individual calibration which could potentially adapt to subject’s individual pattern of chewing and therefore increase classification accuracy.
Ingestion of liquids may not be accurately detected by the proposed model although further studies are needed to confirm this statement. Typically, the chewing process is absent during liquid consumption. However, the sensor signals captured during the sip and swallowing process may present typical features that can be sufficient to detect liquids consumption. Another interesting point of conflict may arise during chewing of food surrogates such as gum. This event is not associated with food intake and therefore the model may accurately detect chewing but mistakenly predicting food intake. However, further tests are needed to determine how the methodology behaves on food surrogates as their chewing pattern may be different from that of food.
Results of a previous study estimated that an accuracy of 95% can be achieved in the detection of food ingestion when measuring chews and swallows [18]. Another study used only swallowing sounds captured with a miniature microphone located over the laryngopharynx to create individual models that detected periods of food intake with an average accuracy of 83.2% [19]. The results reported here showed that comparable classification accuracy but without the need of individual calibration can be achieved by monitoring of jaw motion. The simple structure of the chewing strain sensor may result in a less intrusive and simpler way to detect food intake. Two more advantages support that statement. First, the strain sensor is simply attached to the skin by using medical tape and may last for a 24-hour period with a low risk of sensor detach. Second, the strain sensor is easily available in the market at a low cost. On the other hand, changes in temperature as well as vibrations caused by daily living activities (e.g., walking, talking, sitting, etc) may affect the performance of the sensor. New studies are being performed in our laboratory to include such artifacts into the training data for long-term applications under free-living conditions. The simple implementation of the chewing sensor presented in this paper plus the automatic classification of food intake using only chewing events constitutes an important step towards the development of a wearable device for monitoring of the ingestive behavior of individuals under free-living conditions.
V. Conclusion
This paper presented a sensor and signal processing and pattern recognition methodology to detect periods of food intake by monitoring characteristic jaw motion during food consumption. The most relevant features representing the sensor signals were chosen by a forward selection procedure. The population-based model created was able to classify chewing epochs with an averaged accuracy of 80.98% and a time resolution of 30 s. This epoch-based classification approach would allow the monitoring of short events of food consumption, such as snacking. Practical implementation of this methodology in a wearable device is highly possible due to the use of a simple and inexpensive strain gauge sensor to monitor chews plus a simple classification algorithm that can be easily implemented in a microcontroller. The development of such wearable device would allow the real-time monitoring of ingestive behavior under free-living conditions.
Acknowledgments
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases under Grant R21DK085462. The associate editor coordinating the review of this manuscript and approving it for publication was Prof. Okyay Kaynak.
Biographies

Edward S. Sazonov (M’02) received the Diploma of Systems Engineer from Khabarovsk State University of Technology, Russia, in 1993, and the Ph.D. degree in computer engineering from West Virginia University, Morgantown, in 2002.
Currently, he is an Associate Professor with the Department of Electrical and Computer Engineering, University of Alabama, Tuscaloosa, and the head of the Laboratory of Ambient and Wearable Systems. His research interests span bioengineering, computational intelligence, wireless, ambient and wearable devices. Current research projects include development of methods and wearable sensors for non-invasive monitoring of ingestion; methods and devices for monitoring of physical activity and energy expenditure; wearable platforms for rehabilitation of stroke patients and monitoring of the risk of falling in elderly; and other methods for noninvasive monitoring of human behavior. His work has been supported by national (National Science Foundation, National Institutes of Health, National Academies of Science) and state agencies, and private industry.

Juan M. Fontana received the B.Sc. degree in electrical engineering from the National University of Cordoba, Cordoba, Argentina, in 2005, and the Ph.D. degree in biomedical engineering from Louisiana Tech University, Ruston, in 2010.
He is currently a Post-Doctoral Fellow with the Computer Laboratory of Ambient and Wearable Devices, University of Alabama, Tuscaloosa. His research interests include biosignal processing and pattern recognition for non-invasive physiological monitoring applications.
Contributor Information
Edward S. Sazonov, Email: esazonov@eng.ua.edu.
Juan M. Fontana, Email: jmfontana@bama.ua.edu, juanmfontana@gmail.com.
References
- 1.World Health Organization | Obesity and Overweight. 2011 Mar. [Online]. Available: http://www.who.int/mediacentre/factsheets/fs311/en/
- 2.Bray GA. How do we get fat? An epidemiologic and metabolic approach. Clin. Dermatol. 2004 Jul.vol. 22(no. 4):281–288. doi: 10.1016/j.clindermatol.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Kral TVE, Rolls BJ. Energy density and portion size: Their independent and combined effects on energy intake. Physiol. Behavior. 2004 Aug.vol. 82(no. 1):131–138. doi: 10.1016/j.physbeh.2004.04.063. [DOI] [PubMed] [Google Scholar]
- 4.Prentice AM, Black AE, Murgatroyd PR, Goldberg GR, Coward WA. Metabolism or appetite: Questions of energy balance with particular reference to obesity. J. Human Nutrition Dietetics. 1989 Apr.vol. 2(no. 2):95–104. [Google Scholar]
- 5.Champagne CM, et al. Energy intake and energy expenditure: A controlled study comparing dietitians and non-dietitians. J. Amer. Dietetic Assoc. 2002 Oct.vol. 102(no. 10):1428–1432. doi: 10.1016/s0002-8223(02)90316-0. [DOI] [PubMed] [Google Scholar]
- 6.De Castro JM. Methodology, correlational analysis, and interpretation of diet diary records of the food and fluid intake of free-living humans. Appetite. 1994 Oct.vol. 23(no. 2):179–192. doi: 10.1006/appe.1994.1045. [DOI] [PubMed] [Google Scholar]
- 7.Day N, McKeown N, Wong M, Welch A, Bingham S. Epidemiological assessment of diet: A comparison of a 7-day diary with a food frequency questionnaire using urinary markers of nitrogen, potassium and sodium. Int. J. Epidemiol. 2001 Apr.vol. 30(no. 2):309–317. doi: 10.1093/ije/30.2.309. [DOI] [PubMed] [Google Scholar]
- 8.Kaczkowski CH, Jones PJH, Feng J, Bayley HS. Four-day multimedia diet records underestimate energy needs in middle-aged and elderly women as determined by doubly-labeled water. J. Nutrition. 2000 Apr.vol. 130(no. 4):802–805. doi: 10.1093/jn/130.4.802. [DOI] [PubMed] [Google Scholar]
- 9.Black AE, Goldberg GR, Jebb SA, Livingstone MB, Cole TJ, Prentice AM. Critical evaluation of energy intake data using fundamental principles of energy physiology: 2. Evaluating the results of published surveys. Eur. J. Clin. Nutrition. 1991 Dec.vol. 45(no. 12):583–599. [PubMed] [Google Scholar]
- 10.Livingstone MBE, Black AE. Markers of the validity of reported energy intake. J. Nutrition. 2003 Mar.vol. 133(Suppl 3):895S–920S. doi: 10.1093/jn/133.3.895S. no. [DOI] [PubMed] [Google Scholar]
- 11.Thompson FE, Subar AF, Loria CM, Reedy JL, Baranowski T. Need for technological innovation in dietary assessment. J. Amer. Dietetic Assoc. 2010 Jan.vol. 110(no. 1):48–51. doi: 10.1016/j.jada.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiss R, Stumbo PJ, Divakaran A. Automatic food documentation and volume computation using digital imaging and electronic transmission. J. Amer. Dietetic Assoc. 2010 Jan.vol. 110(no. 1):42–44. doi: 10.1016/j.jada.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Six BL, et al. Evidence-based development of a mobile telephone food record. J. Amer. Dietetic Assoc. 2010 Jan.vol. 110(no. 1):74–79. doi: 10.1016/j.jada.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun M, et al. A wearable electronic system for objective dietary assessment. J. Amer. Dietetic Assoc. 2010 Jan.vol. 110(no. 1):45. doi: 10.1016/j.jada.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amft O, Tröster G. Recognition of dietary activity events using on-body sensors. Artif. Intell. Medicine. 2008 Feb.vol. 42:121–136. doi: 10.1016/j.artmed.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Amft O. A wearable earpad sensor for chewing monitoring. Proc. IEEE Sensors. 2010:222–227. [Google Scholar]
- 17.Sazonov E, et al. Non-invasive monitoring of chewing and swallowing for objective quantification of ingestive behavior. Physiolog. Meas. 2008;vol. 29(no. 5):525–541. doi: 10.1088/0967-3334/29/5/001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sazonov ES, et al. Toward objective monitoring of ingestive behavior in free-living population. Obesity. 2009;vol. 17(no. 10):1971–1975. doi: 10.1038/oby.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Meyer P, Makeyev O, Schuckers S, Melanson E, Neuman M, Sazonov E. Detection of food intake from swallowing sequences by supervised and unsupervised methods. Ann. Biomed. Eng. 2010;vol. 38(no. 8):2766–2774. doi: 10.1007/s10439-010-0019-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sazonov E, Makeyev O, Lopez-Meyer P, Schuckers S, Melanson E, Neuman M. Automatic detection of swallowing events by acoustical means for applications of monitoring of ingestive behavior. IEEE Trans. Biomed. Eng. 2010 Mar.vol. 57(no. 3):626–633. doi: 10.1109/TBME.2009.2033037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peyron M-A, Blanc O, Lund JP, Woda A. Influence of age on adaptability of human mastication. J. Neurophysiol. 2004 Aug.vol. 92(no. 2):773–779. doi: 10.1152/jn.01122.2003. [DOI] [PubMed] [Google Scholar]
- 22.Fueki K, Sugiura T, Yoshida E, Igarashi Y. Association between food mixing ability and electromyographic activity of jaw-closing muscles during chewing of a wax cube. J. Oral Rehab. 2008 May;vol. 35(no. 5):345–352. doi: 10.1111/j.1365-2842.2008.01849.x. [DOI] [PubMed] [Google Scholar]
- 23.Kohyama K, Hatakeyama E, Sasaki T, Azuma T, Karita K. Effect of sample thickness on bite force studied with a multiple-point sheet sensor. J. Oral Rehab. 2004 Apr.vol. 31(no. 4):327–334. doi: 10.1046/j.1365-2842.2003.01248.x. [DOI] [PubMed] [Google Scholar]
- 24.Bousdras VA, et al. A novel approach to bite force measurements in a porcine model in vivo. Int. J. Oral Maxillofacial Surgery. 2006 Jul.vol. 35(no. 7):663–667. doi: 10.1016/j.ijom.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 25.Anthon G. The Use of Thin-Film Corrugated Strain Gauge Biosensors for the Detection and Monitoring of Human Chewing Frequency. Michigan Technolog. Univ.; 2008. [Google Scholar]
- 26.Foster KD, Woda A, Peyron MA. Effect of texture of plastic and elastic model foods on the parameters of mastication. J. Neurophysiol. 2006 Jun.vol. 95(no. 6):3469–3479. doi: 10.1152/jn.01003.2005. [DOI] [PubMed] [Google Scholar]
- 27.Kohyama K, Hatakeyama E, Sasaki T, Dan H, Azuma T, Karita K. Effects of sample hardness on human chewing force: A model study using silicone rubber. Archives Oral Biol. 2004 Oct.vol. 49(no. 10):805–816. doi: 10.1016/j.archoralbio.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 28.Veyrune J-L, Miller CC, Czernichow S, Ciangura CA, Nicolas E, Hennequin M. Impact of morbid obesity on chewing ability. Obesity Surgery. 2008 Nov.vol. 18(no. 11):1467–1472. doi: 10.1007/s11695-008-9443-9. [DOI] [PubMed] [Google Scholar]
- 29.Woda A, Mishellany A, Peyron M-A. The regulation of masticatory function and food bolus formation. J. Oral Rehab. 2006 Nov.vol. 33(no. 11):840–849. doi: 10.1111/j.1365-2842.2006.01626.x. [DOI] [PubMed] [Google Scholar]
- 30.Kohavi R, John GH. Wrappers for feature subset selection. Artif. Intell. 1997 Dec.vol. 97(no. 1–2):273–324. [Google Scholar]
- 31.Meyer D, Leisch F, Hornik K. The support vector machine under test. Neurocomputing. 2003 Sep.vol. 55(no. 1–2):169–186. [Google Scholar]
- 32.Chih-Chung C, Chih-Jen L. LIBSVM: A Library for Support Vector Machines. [Online]. Available: http://www.csie.ntu.edu.tw/~cjlin/libsvm. [Google Scholar]