Summary
Progress has been made in elucidating the cell surface phenotype of primary adipose progenitors; however, specific functional markers and distinct molecular signatures of fat depot-specific preadipocytes have remained elusive. In this study, we label committed murine adipose progenitors through expression of GFP from the genetic locus for Zfp423, a gene controlling preadipocyte determination. Selection of GFP-expressing fibroblasts from either subcutaneous or visceral adipose-derived stromal vascular cultures isolates stably committed preadipocytes that undergo robust adipogenesis. Immunohistochemistry for Zfp423-driven GFP expression in vivo confirms a perivascular origin of preadipocytes within both white and brown adipose tissues. Interestingly, a small subset of capillary endothelial cells within white and brown fat also express this marker, suggesting a contribution of specialized endothelial cells to the adipose lineage. Zfp423GFP mice represent a simple tool for the specific localization and isolation of molecularly defined preadipocytes from distinct adipose tissue depots.
Keywords: adipose stem cells, adipogenesis, PPARγ, Zfp423, preadipocytes, white adipose tissue, cell fate determination
Introduction
Adipose cells are a key component of mammalian energy homeostasis. Brown fat cells are specialized to dissipate chemical energy in the form of heat, and serve to protect mammals from hypothermia and defend against obesity (Cannon and Nedergaard, 2004). White fat cells represent the main site of energy storage, containing a single large lipid droplet and the enzymatic machinery to both synthesize and hydrolyze triglycerides. Fat cells secrete a large number of polypeptides (adipokines) that affect various aspects of energy homeostasis, including food intake, insulin sensitivity, and blood pressure (Rosen and Spiegelman, 2006). Adipocytes have historically been described as “white” or “brown”; however, more recent molecular studies have revealed that white adipose tissues (WAT) located in different anatomical regions contain adipocytes that exhibit stable and intrinsic differences in gene expression and adipokine secretion (Gesta et al., 2006; Perrini et al., 2008; Vidal, 2001; Vohl et al., 2004). This implies that multiple types of white adipocytes exist in mammals and make different contributions to the maintenance of energy balance. In fact, numerous studies indicate that obese individuals accumulating fat in the visceral (abdominal) compartment are at much greater risk for developing type 2 diabetes and cardiovascular events than BMI-matched individuals whose adiposity is more prevalent in the subcutaneous regions (Kissebah and Krakower, 1994).
The global epidemic of obesity and metabolic syndrome has increased the urgency of understanding more fully how these various types of adipocytes develop. Committed fat cell precursors, or preadipocytes, are fibroblastic cells that are morphologically indistinguishable from non-adipogenic fibroblasts, and can be found among many loose connective tissues. They are particularly abundant within the stromal-vascular (SV) fraction of adult adipose tissue. Light and electron microscopy studies from over 40 years ago first suggested a close association of adipose progenitors to the adipose tissue vasculature, with immature/developing adipocytes residing in a pericyte position along capillary endothelial cells (Barnard, 1969; Cameron and Smith, 1964; Cinti et al., 1984; Flemming, 1871; Hausman et al., 1980; Napolitano, 1963; Poljakoff, 1888; Tedeschi, 1970; Toldt, 1870; Wasserman, 1926, 1929, 1960, 1965). Importantly, subsets of mural cells in the WAT blood vessels express Pparγ, the master regulatory gene of adipocyte differentiation (Tang et al., 2008); in addition, several recent studies have clearly demonstrated that perivascular cells isolated from adipose tissue have the ability to differentiate into adipocytes (Amos et al., 2008; Crisan et al., 2008; Traktuev et al., 2008; Zannettino et al., 2008; Zimmerlin et al., 2010).
A significant barrier to understanding the developmental and functional differences between adipose depots has been the inability to selectively localize and purify comparable populations of stably committed preadipocytes from distinct adipose tissues. Current approaches have utilized cell surface markers to enrich for adipose progenitor populations (Joe et al., 2009; Joe et al., 2010; Rodeheffer et al., 2008; Schulz et al., 2011; Sengenes et al., 2005; Uezumi et al., 2010; Xu et al., 2011; Zimmerlin et al., 2010). Most notably, Friedman and colleagues pioneered an elegant cell sorting strategy to identify and isolate a rare, but highly adipogenic progenitor population capable of differentiating into functional adipocytes both in vitro and in vivo (Rodeheffer et al., 2008). However, the markers utilized (Sca-1, CD24 and CD34) are not individually specific for the adipose lineage, and some are functionally dispensible for the initial formation of adipose progenitors (Staszkiewicz et al., 2011). In addition, the molecular identity and gene expression program of primary committed preadipocytes from different depots remain incompletely understood. Moreover, these markers do not provide a simple method for localizing adipose precursors in vivo.
In other areas of developmental biology, the elucidation of hierarchical relationships among transcription factors that govern the formation of specific cell types has greatly facilitated the localization and isolation of defined progenitors at different stages of differentiation. Defining precursor populations of various developmental stages on the basis of transcription factor gene expression has been particularly useful in the study of myogenesis, hematopoesis, and pancreatic endocrine cell development (David-Fung et al., 2009; Mellitzer et al., 2004; Montarras et al., 2005; White et al., 2008; Wilson et al., 2003). Much of our current understanding of the transcriptional basis of adipogenesis is based on studies using the established 3T3 preadipocyte cell lines first isolated by Green and colleagues (Green and Kehinde, 1975, 1976; Green and Meuth, 1974). Mainly relying on this system, a canonical pathway for the differentiation of fat cells from preadipocytes has been elucidated (Farmer, 2006). Gain and loss of function studies in vitro and in vivo clearly indicate that adipocyte differentiation is driven by the nuclear hormone receptor PPARγ, but also modulated by other key regulators such as the C/EBP and EBF families of transcription factors (Akerblad et al., 2002; Barak et al., 1999; Farmer, 2006; Jimenez et al., 2007; Kubota et al., 1999; Rosen et al., 1999; Tontonoz et al., 1994). Importantly, the functions of many of these regulators identified in the immortalized murine adipose cell systems have been confirmed in vivo, in mouse models or human studies. Most notably, humans with genetically reduced PPARγ function are lipodystrophic and severely insulin-resistant (Barroso et al., 1999).
We recently identified Zfp423, a multi-zinc finger transcriptional regulator, as an essential functional determinant of preadipocyte commitment (Gupta et al., 2010). Unlike many early transcriptional regulators of adipocyte differentiation (e.g. C/EBP family), Zfp423 expression is enriched in numerous immortalized preadipocyte cell lines when compared to numerous fibroblast cell lines with little propensity for adipocyte differentiation. Furthermore, Zfp423 functions within preadipocytes to regulate a large number of preadipocyte genes characteristic of committed murine 3T3 preadipocytes, including the basal preadipocyte expression of Pparγ. Zfp423 is both necessary and sufficient for adipocyte differentiation in culture, and is required for the initial formation of white and brown adipose tissue in vivo. Since Zfp423 acts upstream of Pparγ and other preadipocyte-enriched genes and is expressed abundantly in the SV fraction of brown and white adipose tissues (Gupta et al., 2010), the selective purification of Zfp423-expressing fibroblasts from adipose depots could, in principle, provide a relatively simple, rapid method for localizing and isolating committed and functionally relevant preadipocytes.
Results
Derivation of Zfp423GFP Preadipocyte Reporter Mice
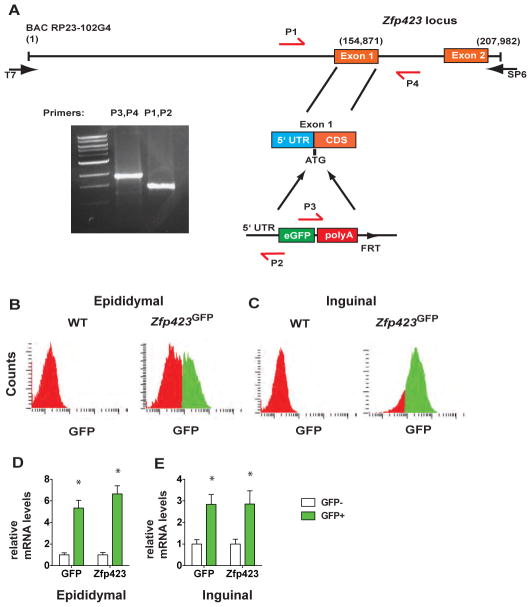
To genetically label committed adipose progenitors, we derived transgenic mice expressing GFP from the Zfp423 genetic locus. Briefly, starting with a 200 kilobase (kb) pair BAC containing 150 kb of sequence upstream of exon 1, the initiation codon of Zfp423 was replaced by the enhanced GFP (eGFP) coding sequence followed by a polyadenylation signal sequence (Figure 1A). Importantly, this modified BAC does not encode any functional domains of Zfp423 or contain full length coding sequence of any other annotated genes. Proper targeting of the engineered BAC was verified by PCR (Figure 1A) and DNA sequencing. We analyzed the RNA expression of GFP in the five founder lines derived and identified two founders in which GFP positive cells were greatly enriched for Zfp423 mRNA levels, compared to the GFP negative cells (see below). In addition, the distribution of GFP mRNA across tissues in adult mice closely followed the pattern of Zfp423 expression (Supplementary Figure 1A). These two founder lines yielded very similar results in the experiments described below, and we therefore have included only data from a single reporter line (Zfp423GFP mice) here.
Figure 1. Derivation and characterization of Zfp423GFP transgenic mice.
(A) Genomic structure of Zfp423GFP transgene modified from bacterial artificial chromosome (BAC) RP23-102G4. Coding sequence of enhanced GFP (eGFP) followed by a polyadenylation signal (polyA) was inserted into the initiation codon of Zfp423 located in exon 1. Proper targeting of the BAC was verified by sequencing and PCR analysis using primers as shown (P1–P4). (B) Quantitation of GFP+ cells in cultures of epididymal SV cells passaged 3–4 times after initial isolation. Cells were defined as GFP+ (shown in green) if fluorescent intensity was clearly greater than background fluorescence levels in wild-type (WT) cultures (shown in red) (See Supplementary Figure 1) ~50% of the cells within the SV cultures from this depot are GFP+. (C) Quantitation of GFP+ cells in cultures of inguinal SV cells passaged 3–4 times after initial isolation. ~85% of the cells within the SV cultures from this depot are GFP+ (D) Relative mRNA levels of GFP and Zfp423 in purified GFP+ and GFP− cells from epididymal SV cultures. (E) Relative mRNA levels of GFP and Zfp423 in purified GFP+ and GFP− cells from inguinal SV cultures. n=3 replicates
GFP expressing cells from both subcutaneous and visceral SV cultures are functional determined preadipocytes
Cultures of SV cells from adult adipose depots contain both stably committed preadipose fibroblasts as well as non-adipogenic fibroblastic cells. Functional markers that define the adipose progenitors within SV cultures of adult adipose tissues are lacking. Moreover, the stable molecular differences between primary adipogenic and non-adipogenic fibroblasts have not been fully elucidated. We first plated cells from the SV fractions of adipose tissues in culture to eliminate non-adherent blood cells, and to enrich for fibroblastic cells, both preadipose and non-preadipose, by allowing them to proliferate for 3–4 days. We then evaluated the abundance of GFP expressing cells in cultures obtained from either epididymal (visceral) or inguinal (subcutaneous) fat depots of 6–8 week-old mice. GFP+ cells can be detected by microscopy in SV cultures from both epididymal and inguinal fat depots (data not shown). The relative abundance of GFP+ cells within SV cultures from each depot was determined by flow cytometry and ~50% and ~85% of the cells are GFP+ within the epididymal and inguinal SV cultures respectively (Figure 1B,C). These numbers generally correlate with the percentage of cells within SV cultures that undergo adipogenesis under potent stimuli in vitro (data not shown; see Methods). Importantly, GFP expression can be detected in nearly all adipocytes derived from differentiated SV cultures, consistent with the continued expression of Zfp423 throughout adipocyte differentiation (Supplementary Figure 1B). To determine if selecting for GFP+ cells enriches for Zfp423 mRNA expression, we sorted GFP− cells and an equal number of GFP+ cells from the undifferentiated SV cultures (Supplementary Figure 2). Both GFP and Zfp423 mRNA levels were enriched in GFP+ cells compared to the GFP− cells, indicating that GFP expression within the adipose lineage of Zfp423GFP mice faithfully mimics Zfp423 expression (Figure 1D and E,).
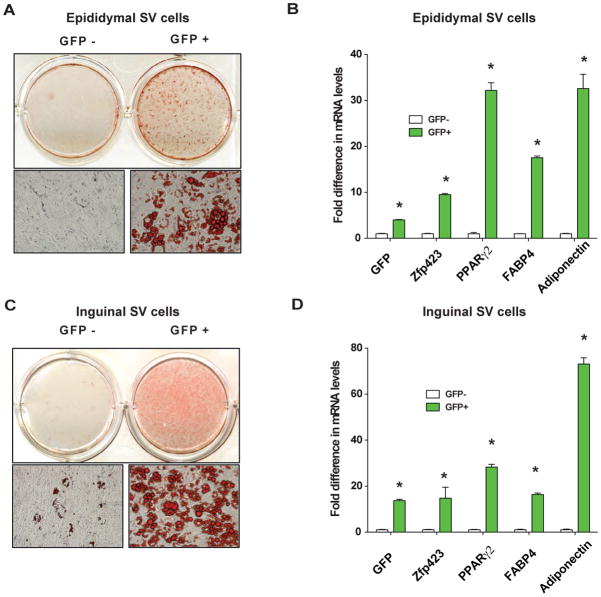
GFP+ and GFP− cells grew at similar rates in culture and had similar morphologies; we then tested the functional capacity of these cells to undergo adipocyte differentiation. Cultures from epididymal GFP+ cells differentiated robustly into lipid-containing adipocytes (Figure 2A), expressing genes characteristic of fully differentiated fat cells such as Fabp4 (aP2) and adiponectin (Figure 2B). On the other hand, even under these strongly pro-adipogenic conditions (See Methods), the GFP− cells from this depot failed to undergo adipocyte differentiation, as evidenced by absence of lipid-containing adipocytes or adipocyte gene expression in differentiated cultures (Figure 2A,B). Similarly, GFP+ cells isolated from inguinal fat SV cultures differentiated robustly into lipid-containing adipocytes (Figure 2C), abundantly expressing Fabp4 (aP2) and adiponectin (Figure 2D). The GFP− SV cells derived from the inguinal depot undergo adipocyte differentiation to a far less extent, with only few lipid-containing adipocytes and low levels of adipocyte gene expression present in differentiated cultures (Figure 2C,D). Taken together, these data indicate that GFP driven by the Zfp423 locus allows selection of functional preadipocyte populations from both the subcutaneous and epididymal SV cultures.
Figure 2. GFP-expressing cells from both subcutaneous and visceral SV cultures are functional preadipocytes.
(A) Oil red-O staining of purified GFP− and GFP+ cells from epididymal SV cultures 6 days following the induction of adipocyte differentiation. (B) Expression of adipocyte selective genes in the differentiated cultures shown in (A). (C) Oil red-O staining of purified GFP− and GFP+ cells from inguinal SV cultures 6 days following the induction of adipocyte differentiation. (D) Expression of adipocyte selective genes in the differentiated cultures shown in (C). n=3 replicates.
Isolated preadipocytes from subcutaneous and visceral SV cultures exhibit distinct cell surface phenotypes and gene expression profiles
We next compared cultured GFP+ preadipocytes from both the subcutaneous and visceral WAT depots. The cell surface expression of proteins previously regarded as WAT preadipocyte markers in other studies was examined first using flow cytometry. Two adipogenic populations from the non-hematopoetic fraction from white adipose tissue depots have recently been described (Rodeheffer et al 2008); one population is CD29+; CD34+; Sca1+; CD24− while the other is CD29+; CD34+; Sca1+; CD24+. Greater than 95% of cultured SV cells from either depot were CD45− (data not shown). Thus, the plating of cells provides a simple selection against most hematopoetic cells. GFP+ preadipocytes from both inguinal and epididymal SV cultures were CD45− and CD29+ (Supplementary Figure 3A). CD34 expression was not detected in any of the cells of SV culture; however, it is known that CD34 expression is often lost in many cultured adipose-derived stromal cells (Suga et al., 2009). Interestingly, the cell surface expression of Sca1 in GFP+ preadipocytes was depot-dependent; inguinal preadipocytes were Sca1+ while epididymal preadipocytes were largely Sca1− (Supplementary Figure 3A). This is consistent with a recent study demonstrating that Sca1+ cells from inguinal, but not epididymal, SVF differentiate into fat cells (Schulz et al., 2011). The cell surface expression of CD24 in GFP+ preadipocytes was also depot-dependent. Most preadipocytes from inguinal SV cultures lacked CD24 expression. Notably, two distinct populations of GFP+ cells were observed in epididymal cultures; ~60% of the precursors were CD24− and 40% were CD24+. Together, these data suggest that the cell surface phenotype of depot-specific preadipocytes is distinct.
We next examined the mRNA levels of genes previously described as preadipocyte-enriched., Pparγ, a gene enriched in preadipose fibroblasts and a key transcriptional target of Zfp423 in preadipocytes (Gupta et al., 2010), was enriched in both epididymal and inguinal GFP+ cells compared to the GFP− cells (Supplementary Figure 3B). Primary preadipocytes from both WAT depots showed enriched levels of Pparγ1 while the Pparγ2 isoform was not detected. This is in striking contrast to 3T3 embryonic preadipocyte cell lines, where levels of the Pparγ2 isoform, but not the Pparγ1 isoform, correlate with adipogenic capacity of the cells (Gupta et al., 2010). Some of the mRNAs recently described by Tang et al as being enriched in Pparγ-defined primary preadipocytes were elevated in GFP+ cells from either the inguinal (Supplementary Figure 3C) or epididymal depot (Supplementary Figure 3D), while three of the six correlated with GFP expression in both fat depots (Tang et al., 2008). We also directly examined the expression of Dlk-1 (Pref-1), a gene commonly referred to as preadipocyte marker (Sul, 2009). Dlk-1 mRNA was not elevated in GFP+ SV cells. In fact, Dlk-1 gene expression was rather enriched in GFP− epididymal SV cells as compared to GFP+ cells from the same depot (Supplementary Figure 3B).
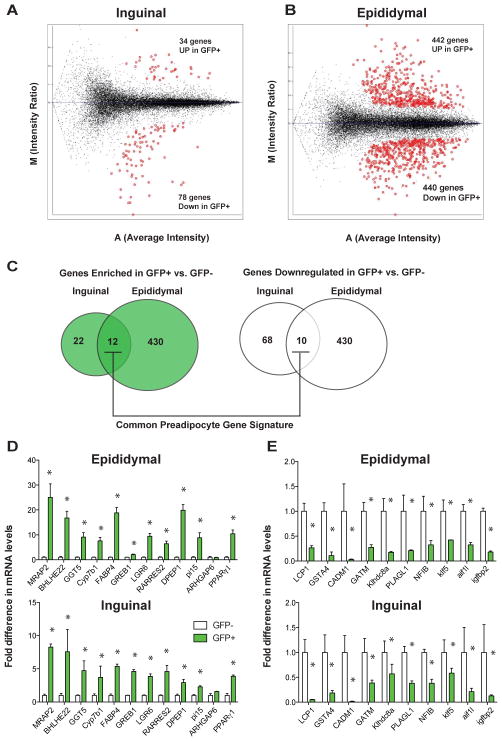
Many of previously suggested preadipocyte markers were identified through analyses of isolated precursors from pooled visceral and subcutaneous WAT depots (Rodeheffer et al., 2008; Tang et al., 2008). The data above indicate that preadipocytes from distinct WAT depots possess distinct gene expression profiles. Thus, to define the molecular signatures of WAT depot-specific preadipocytes, and elucidate a preadipocyte signature common to both white preadipocyte sub-types, we determined the global gene expression profiles of isolated GFP+ and GFP− SV cells from both the inguinal and epididymal fat depots. When comparing inguinal GFP+ SV cells to inguinal GFP− SV cells we found only 112 unique genes significantly differentially expressed with a magnitude of difference greater than 2-fold (Figure 3A, Supplementary Table 1,2). In contrast, far more genes (882 unique genes) were differentially expressed between GFP+ SV cells and GFP- SV cells from epididymal cultures (Figure 3B, Supplementary Table 3,4). This may be explained by the differences in heterogeneity between GFP− cells from these depots. Of the 34 genes whose expression is enriched in inguinal preadipocytes, 12 of these genes were also enriched in epididymal preadipocytes (Figure 3C,D Supplementary Table 5). Of the 78 genes whose expression is relatively repressed in inguinal GFP+ SV cells, 10 of these where also relatively reduced in epididymal preadipocytes (Figure 3C,E, Supplementary Table 5). Thus, these 22 genes define a core molecular signature common to both primary visceral and subcutaneous WAT preadipocytes. Most notable from the genes in this signature is the enrichment in Pparγ1 and the well-characterized Pparγ target gene Fabp4. These data indicate active PPARγ signaling even in the preadipocyte stage, consistent with the long-standing observations that treatment of 3T3 preadipocytes with PPARγ agonists alone is sufficient to initiate adipocyte differentiation (Lehmann et al., 1995; Tontonoz and Spiegelman, 2008).
Figure 3. Elucidation of a core preadipocyte gene program.
(A, B) M vs. A plot of gene expression data obtained from microarray analysis of GFP+ and GFP− SV cells from inguinal or epididymal depots. “M” represents intensity ratio [log2 GFP+ − log2 GFP−] and “A” represents average intensity value of the gene across all samples [1/2 × (log2 GFP+ + log2 GFP−)]. Red spots represents genes significantly differentially expressed greater than 2-fold (p< 0.05). (C) Venn diagram illustrating overlap between gene signatures derived from expression analysis of epididymal and inguinal GFP+ and GFP− cultures. (D) Real-time PCR confirmation of the 12 genes depicted in (B) as being preadipocyte enriched genes. (E) Real-time PCR confirmation of the 10 genes depicted in (B) as being enriched in non-adipogenic GFP− SV cells. n=3 replicates.
GFP expression localizes to a subset of pericytes and endothelial cells of the adipose tissue blood vessels
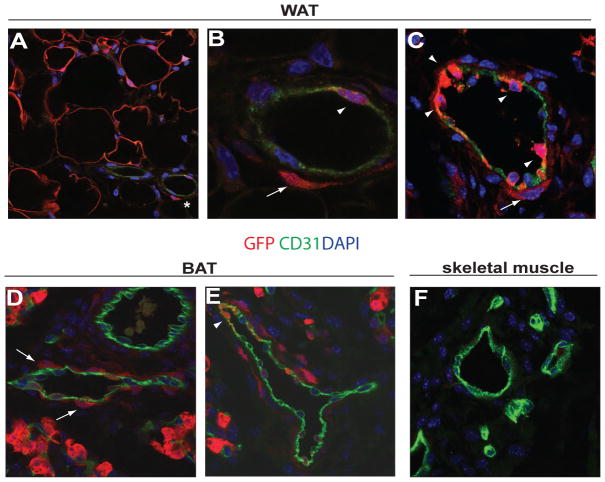
The identification of Zfp423 as a functional marker of committed adipocyte progenitors affords the opportunity to localize preadipocytes within the developing adipose tissue in vivo. As described above, adipose progenitors have long been thought to reside in or near the vasculature of adipose tissue and have been often referred to as specialized pericytes (Barnard, 1969; Cinti et al., 1984; Cinti et al., 1985; Hausman et al., 1980; Napolitano, 1963). To determine the localization of Zfp423-expressing progenitors in white adipose tissue, we first performed indirect immunofluorescence on sections of inguinal WAT from adult mice with antibodies specific to GFP. As expected, GFP expression was found in lipid-containing adipocytes (Figure 4A). In addition, GFP+ cells were located in or near some blood vessels within the developing white adipose tissue (Figure 4A). A closer inspection of the adipose vasculature reveals that GFP expression can be found in some mural cells lining the capillary endothelium. Notably, we also found that a very small subset of CD31+ capillary endothelial cells exhibited clear GFP expression (Figure 4B). This observation was also clearly evident in the developing inguinal WAT of newborn mice (postnatal day 4), prior to appearance of fully mature adipocytes (Figure 4C). This stands in contrast to Pparγ, whose expression in the adipose vasculature is confined to mural cells (Tang et al., 2008).
Figure 4. GFP+ preadipocytes reside in the adipose vasculature as a subset of both pericytes and capillary endothelial cells.
(A,B) Confocal images of adult inguinal WAT stained with antibodies recognizing GFP (red) and the endothelial cell protein CD31 (green), with nuclei counterstained with DAPI. In (A) note the expression of GFP in mature adipocytes and in some blood vessels (*). In (B) note the expression of GFP in a subset of perivascular cells (arrow) and in a subset of endothelial cells (arrowhead) of the blood vessel highlighted in (A). (C) Confocal image of developing inguinal WAT from postnatal day 4 mice stained with antibodies recognizing GFP (red) and the endothelial cell protein CD31 (green). Note the expression of GFP in a subset of perivascular cells (arrow) and in a strong subset of endothelial cells (arrowhead) even before the full development of mature adipocytes at this stage. (D,E) Confocal images of embryonic day 18.5 interscapular BAT stained with the same antibodies shown in (A–C). In (D) note the expression of GFP in mature adipocytes and in numerous perivascular cells (arrows). In (E) note the expression of GFP in a subset of endothelial cells (arrowhead). (F) Confocal image of skeletal muscle directly adjacent to the interscapular BAT shown in (D–E). Note the absence of GFP+ cells in the vasculature of this tissue.
Since Zfp423 is also abundantly expressed in brown adipose tissue and is a critical regulator of brown preadipocyte Pparγ expression and brown adipose tissue development (Gupta et al., 2010), we also examined GFP expression in the interscapular BAT region of embryonic day 18.5 Zfp423GFP embryos (Supplementary Figure 4A). Likewise, GFP expression is found in mature adipocytes as well as in some, but not all, blood vessels within the interscapular BAT (Figure 4D). At this embryonic stage, numerous GFP+ cells in brown adipose tissue can be observed lining the capillaries and appear to be endothelium (Figure 4E). Confocal analysis of sections incubated with antibodies recognizing GFP and the pericyte marker, PDGFRβ, clearly demonstrated that GFP expression is found in some, but not all, PDGFRβ+ pericytes along these blood vessels (Supplementary Figure 4B), providing additional evidence to the hypothesis that preadipocytes are specialized pericytes. In addition, a small subset of CD31+ capillary endothelial cells in this depot also exhibited clear GFP expression (Figure 4E). Importantly, GFP+ endothelial cells were only found in the same adipose capillaries containing GFP+ pericytes and neither GFP+ pericytes nor GFP+ endothelial cells were detected in the vasculature of the developing skeletal muscle of these same embryos (Figure 4F, Supplementary Figure 4A).
Discussion
The ability to localize and purify functional progenitor cells is essential to fully understand how any tissue or organ is normally formed during development. Given the current epidemic of obesity and related disorders, this need is particularly acute for adipose tissues. The stromal vascular cells of adipose tissue have long been considered a rich source of adipose progenitors (Hauner et al., 1989; Reyne et al., 1989; Van et al., 1976). Primary cultures of SV cells obtained from adult adipose tissue contain fibroblasts with varying propensities for adipocyte differentiation; however, functional markers that define the preadipose fibroblast have been lacking. The identification of Zfp423 as a preadipocyte determination factor afforded an excellent opportunity to isolate committed and functionally relevant adipose progenitors. Here we demonstrate through the use of Zfp423GFP transgenic mice that Zfp423/GFP expression defines the preadipocyte population among fibroblastic cells of adult adipose depots.
Current cell sorting strategies to enrich for adipose progenitors rely on combinations of antibodies positively selecting for cell surface proteins commonly expressed in progenitor populations and negatively selecting against well-defined non-adipose cells (e.g. immune cells). These methods have been successful in collecting highly adipogenic and proliferative populations that differentiate in vitro and can reconstitute adipose tissue in lipodystrophic mice in vivo (Rodeheffer et al., 2008). Cultured GFP+ preadipocytes from Zfp423GFP transgenic mice express many of the cell surface proteins present in the populations described by Rodeheffer et al; however, the precise expression pattern of these proteins appears fat-depot dependent. It is worth noting that in the sorting experiments described here, both GFP+ and GFP− cells were collected from cultures of primary SV cells. We cannot exclude the possibility that this approach is associated with potential artifacts due to the initial expansion of SV cells in culture; however, our experience is that all cells directly sorted from freshly isolated SVF differentiate poorly and their numbers are, of course, much more limited. It is also important to note that in this study we evaluate Zfp423 as a functional marker of preadipose fibroblasts residing within the stromal fraction of adipose tissue and within the adipose tissue vasculature in vivo. These are the most likely locations of the functional adipose progenitors that contribute to the development or expansion of adipose tissues. Adipose progenitors residing outside the adipose tissue have been suggested (Crossno et al., 2006; Majka et al., 2011; Majka et al., 2010); whether Zfp423 is expressed in these cell populations is not known.
We previously demonstrated that Zfp423 and Pparγ are enriched in preadipose 3T3 fibroblasts and that Zfp423 functions to regulate Pparγ expression in these cells (Gupta et al., 2010). Consistent with this model, Pparγ expression is enriched in primary Zfp423-expressing SV cells. Tang et al genetically labeled functional adipose progenitors through expression of GFP from the Pparγ locus (Tang et al., 2008). It is likely that GFP+ preadipocytes isolated from our model overlap with these Pparγ-defined adipose progenitors. In fact, many of these genes described by Tang et al as preadipocyte enriched are also enriched in GFP+ preadipocytes from Zfp423GFP transgenic mice. Unlike Zfp423, however, Pparγ expression is robustly and quickly elevated during the initial stages of the adipocyte differentiation process (Chawla et al., 1994; Tontonoz et al., 1994). Hence, selection for the highest expressing Pparγ+ cells from the SV fraction of adipose tissue may bias the purification towards cells further along the adipose lineage.
Within the adipose tissue we localize GFP+ preadipocytes to a subset of pericytes in some, but not all, adipose blood vessels. We do not detect the presence of GFP+ cells in the vasculature of skeletal muscle and other tissues surrounding the developing interscapular fat in embryos. However, it is not possible to exclude that GFP+ pericytes, similar to Pparγ+ pericytes, arise in the vasculature of other tissues in older animals (Tang et al., 2008). Interestingly, GFP+ preadipocytes in both BAT and WAT depots reside in the vasculature. This data suggest that despite the distinct origin of WAT and classical BAT depots during early embryogenesis (Seale et al., 2008), committed brown and white adipocytes may independently pass through a pericyte lineage.
Importantly, Zfp423 appears to be actively expressed in a crucial subset of endothelial cells in the developing and adult adipose tissues. The presence of this preadipocyte determination factor in a subset of adipose endothelial cells, but not in endothelial cells of other embryonic tissues examined, suggests that Zfp423+ endothelial cells may contribute to the adipose lineage. In fact, such cells may represent the “birth place” of the first cells committed to this lineage. Of note, isolated adipocytes can dedifferentiate in culture into functional endothelial cells, strongly suggesting a developmental relationship between these two lineages (Planat-Benard et al., 2004). Notably, Medici et al demonstrated that mesenchymal cells derived from a BMP-dependent endothelial-mesenchymal transition have adipogenic capacity (Medici et al., 2010). We have previously shown that Zfp423, a known co-activator of SMAD transcription factors, amplifies the pro-adipogenic and Pparγ-inducing actions of BMP signals (Gupta et al., 2010; Hata et al., 2000) Thus, Zfp423+ endothelial cells may undergo an endothelial-mesenchymal transition into Zfp423+; Pparγ+ pericytes that serve as a pool of committed preadipocytes. Consistent with this model, electron microscopy analyses demonstrated that a small number of endothelial cells of the adipose vasculature exhibited morphological markers characteristic of developing adipose cells, such as large mitochondria packed with long cristae and small lipid droplets which were not observed in most other endothelial cells (Cinti, 1995).
The Zfp423 mice described here allow for the use of a single marker to separate preadipocytes from non-adipogenic cells within stromal-vascular cultures.. Zfp423GFP mice, which will be made widely available, (See Methods) provide a tool for the study of adipogenesis. The gene signatures distinguishing depot-specific preadipocytes may represent a starting point for determining the gene programs that drive the formation depot-specific adipocytes. Comparing preadipocytes to non-adipogenic fibroblasts may shed light on the mechanisms controlling preadipocyte commitment and Zfp423 function. In addition, visualization of GFP expression within the adipose vasculature of Zfp423GFP mice provides a simple method to localize committed adipose progenitors in vivo under different physiological conditions. The ability to localize and purify preadipocytes should aid in discovering the mechanisms that drive the development and physiological regulation of adipose tissue.
Experimental Procedures
Generation of Zfp423GFP mice
The BAC clone RP23-102G4, containing ~150 kb and ~50 kb of DNA flanking the 5′ and 3′ ends of exon 1 of Zfp423, respectively, was obtained from the BAC resource at Children’s Hospital Oakland Research Institute. The eGFP cassette was inserted at the initiation codon of the Zfp423 coding sequence, which is located within exon 1, by the use of standard BAC recombineering techniques (See Supplementary Experimental Procedures) (Warming et al., 2006). Purified Zfp423GFP BAC DNA was microinjected into fertilized embryos by the Beth Israel Deaconess Medical Center Transgenic Core Facility using standard pronuclear injection techniques. Founder mice carrying the BAC transgene and subsequent offspring were identified by PCR with forward primer 5′-aagggcatcgacttcaaggag-3′ and reverse primer 5′-gatgccgttcttctgcttgtc-3′ yielding a 108-bp product. Two faithfully expressing founder lines were produced and all offspring appeared grossly normal.
All animal experiments were performed according to procedures approved by the Dana-Farber Cancer Institute’s and Beth Israel Deaconess Medical Center’s Institutional Animal Care and Use Committee. Mice were maintained on a standard rodent chow diet with 12 hr light and dark cycles. For timed matings, the first appearance of vaginal plugs was designated as E0.5. Zfp423GFP mice will be deposited to Jackson laboratories.
FACS analysis
FACS experiments were carried out on a FACSAria™ flow cytometer (Dana-Farber Cancer Institute Flow Cytometry Core Facility). Briefly, subconfluent SV cultures passaged 3–4 times following the initial fractionation were trypsinized, centrifuged, resuspended in 2% FBS/PBS at a concentration of 106 cells/mL, and then filtered through a 40 μm strainer. For sorting, cells were initially selected by size, on the basis of forward scatter (FSC) and side scatter (SSC). Dead or dying cells were excluded on the basis of uptake of propidium iodide. Live cells were then gated on both SSC and FSC singlets, ensuring that individual cells were analyzed. SV cultures from wild-type mice were used to determine background fluorescence levels. Cells were sorted into PBS containing 2% FBS and antibiotics and then returned to culture in complete SV culture medium. Antibodies used for flow cytometry analysis can be found in Supplementary Experimental Procedures.
Gene expression profiling
Total RNA extraction, cDNA synthesis, and real-time PCR analysis were performed as previously described (Gupta et al., 2010). Relative expression of mRNAs was determined after normalization to Rps18 levels using the ΔΔ-Ct method. Student’s t test was used to evaluate statistical significance. All primer sequences are available upon request. For microarray analysis total RNA was isolated from indicated cells in triplicate. Array hybridization and scanning were performed by the Dana-Farber Cancer Institute Core Facility using Affymetrix GeneChip Mouse Genome 430A 2.0 arrays according to established methods (Lockhart et al., 1996). The array data were normalized and analyzed using the DNA-Chip Analyzer (dChip) software (Li and Wong, 2001). Complete array data has been deposited to Gene Expression Omnibus (GEO) (Accession # GSE34775).
SV culture and adipocyte differentiation assays
Inguinal or epididymal SV fractions were obtained from 6–8 week-old male mice following standard procedures (See Supplementary Experimental Procedures). For adipocyte differentiation assays, SV cells were plated onto collagen-coated dishes and grown to confluence in SV culture medium (DMEM/F12 (1:1 Invitrogen) plus Glutamax, Pen/Strep, and 10% FBS (Omega Lot #152071). At confluence, cells were exposed to the adipogenic cocktail containing dexamethasone (1 μM), insulin (5 μg/ml), isobutylmethylxanthine (0.5 mM) (DMI) and rosiglitazone (1 μM) in SV culture medium. 48 hours after induction, cells were maintained in SV culture medium containing insulin (5 μg/ml) and rosiglitazone (1 μM) until harvest. Oil-red-O staining was performed as previously described (Jimenez et al., 2007). Lipid staining of adipocyte cultures was performed using LipidTox™ Red neutral lipid stain (Invitrogen) according to manufacturer’s instructions.
Histological analysis of adipose tissues
Histological analysis of embryonic or newborn adipose tissue was performed on decapitated embryos or neonates which were fixed in 4% paraformaldehyde for 24 hours, equilibrated in 30% sucrose for 24 hours, then embedded in OCT freezing medium. Cryosections of the interscapular region of E18.5 embryos were used for indirect immunofluorescence staining of embryonic BAT (See Supplement to Figure 4). Cryosections of the lower abdominal region of 4 day-old mice were used for indirect immunofluorescence staining of the developing inguinal WAT. For histological analysis of adult adipose tissue, adult mice (4–5 week-old) were first perfused with 4% paraformaldehyde and then dissected inguinal WAT was further fixed in 4% formaldehyde for an additional 24 hours. Following fixation, tissues were equilibrated in 30% sucrose for 24 hours, embedded in OCT freezing medium, and then cryosectioned. A detailed protocol for indirect immunofluorescence assays can be found in Supplementary Experimental Procedures.
Statistical analysis
In all figures, bars represent mean ± standard deviation from the mean. * denotes p< 0.05 from student’s t-test.
Supplementary Material
Research Highlights.
Adipose progenitors are labeled through expression of GFP from the locus for Zfp423.
GFP+ fibroblasts from adipose stromal vascular cultures undergo adipogenesis.
Zfp423-driven GFP expression localizes to perivascular cells of WAT and BAT.
A subset of capillary endothelial cells within WAT and BAT also express this marker.
Acknowledgments
The authors are grateful to J. Lawitts (Beth Israel Deaconess Medical Center Transgenic Core) for derivation of BAC transgenic mice, S. White (Beth Israel Deaconess Medical Center Histology Core) for cryosectioning and histology services, E. Fox (Dana-Farber Cancer Institute Microarray Core) for Affymetrix gene chip analysis, M. Curry (Dana-Farber Cancer Institute Flow cytometry Core) for assistance with FACS analysis, and L. Cameron (Dana-Farber Cancer Institute Microscopy Core) for assistance with confocal microscopy. R.K.G is supported by NIH K01 DK090120-02, J.C.L. and P.C. are supported by NIH T32 HL007604, and the research described in this study was supported by the NIH grant DK31405 to B.M.S. and by funding from Universita Politecnica delle Marche to S.C.
Footnotes
Conflicting interests statement. The authors declare that they have no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akerblad P, Lind U, Liberg D, Bamberg K, Sigvardsson M. Early B-cell factor (O/E-1) is a promoter of adipogenesis and involved in control of genes important for terminal adipocyte differentiation. Mol Cell Biol. 2002;22:8015–8025. doi: 10.1128/MCB.22.22.8015-8025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos PJ, Shang H, Bailey AM, Taylor A, Katz AJ, Peirce SM. IFATS collection: The role of human adipose-derived stromal cells in inflammatory microvascular remodeling and evidence of a perivascular phenotype. Stem Cells. 2008;26:2682–2690. doi: 10.1634/stemcells.2008-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak Y, Nelson MC, Ong ES, Jones YZ, Ruiz-Lozano P, Chien KR, Koder A, Evans RM. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- Barnard T. The ultrastructural differentiation of brown adipose tissue in the rat. J Ultrastruct Res. 1969;29:311–322. doi: 10.1016/s0022-5320(69)90109-9. [DOI] [PubMed] [Google Scholar]
- Barroso I, Gurnell M, Crowley VE, Agostini M, Schwabe JW, Soos MA, Maslen GL, Williams TD, Lewis H, Schafer AJ, et al. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402:880–883. doi: 10.1038/47254. [DOI] [PubMed] [Google Scholar]
- Cameron IL, Smith RE. Cytological Responses of Brown Fat Tissue in Cold-Exposed Rats. J Cell Biol. 1964;23:89–100. doi: 10.1083/jcb.23.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- Chawla A, Schwarz EJ, Dimaculangan DD, Lazar MA. Peroxisome proliferator-activated receptor (PPAR) gamma: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology. 1994;135:798–800. doi: 10.1210/endo.135.2.8033830. [DOI] [PubMed] [Google Scholar]
- Cinti S, Cigolini M, Bosello O, Bjorntorp P. A morphological study of the adipocyte precursor. J Submicrosc Cytol. 1984;16:243–251. [PubMed] [Google Scholar]
- Cinti S, Cigolini M, Gazzanelli G, Bosello O. An ultrastructural study of adipocyte precursors from epididymal fat pads of adult rats in culture. J Submicrosc Cytol. 1985;17:631–636. [PubMed] [Google Scholar]
- Cinti SaMM. Brown adipocyte precursor cells: a morphological study. Ital J Anat Embryol. 1995 [PubMed] [Google Scholar]
- Crisan M, Yap S, Casteilla L, Chen CW, Corselli M, Park TS, Andriolo G, Sun B, Zheng B, Zhang L, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Crossno JT, Jr, Majka SM, Grazia T, Gill RG, Klemm DJ. Rosiglitazone promotes development of a novel adipocyte population from bone marrow-derived circulating progenitor cells. J Clin Invest. 2006;116:3220–3228. doi: 10.1172/JCI28510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Fung ES, Butler R, Buzi G, Yui MA, Diamond RA, Anderson MK, Rowen L, Rothenberg EV. Transcription factor expression dynamics of early T-lymphocyte specification and commitment. Dev Biol. 2009;325:444–467. doi: 10.1016/j.ydbio.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming W. On the formation and regression of fat cells in connective tissue with comment on the structure of the latter. Arch Mikrosk Anat. 1871;7:32. [Google Scholar]
- Gesta S, Bluher M, Yamamoto Y, Norris AW, Berndt J, Kralisch S, Boucher J, Lewis C, Kahn CR. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc Natl Acad Sci U S A. 2006;103:6676–6681. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H, Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- Green H, Kehinde O. Spontaneous heritable changes leading to increased adipose conversion in 3T3 cells. Cell. 1976;7:105–113. doi: 10.1016/0092-8674(76)90260-9. [DOI] [PubMed] [Google Scholar]
- Green H, Meuth M. An established pre-adipose cell line and its differentiation in culture. Cell. 1974;3:127–133. doi: 10.1016/0092-8674(74)90116-0. [DOI] [PubMed] [Google Scholar]
- Gupta RK, Arany Z, Seale P, Mepani RJ, Ye L, Conroe HM, Roby YA, Kulaga H, Reed RR, Spiegelman BM. Transcriptional control of preadipocyte determination by Zfp423. Nature. 2010;464:619–623. doi: 10.1038/nature08816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A, Seoane J, Lagna G, Montalvo E, Hemmati-Brivanlou A, Massague J. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell. 2000;100:229–240. doi: 10.1016/s0092-8674(00)81561-5. [DOI] [PubMed] [Google Scholar]
- Hauner H, Entenmann G, Wabitsch M, Gaillard D, Ailhaud G, Negrel R, Pfeiffer EF. Promoting effect of glucocorticoids on the differentiation of human adipocyte precursor cells cultured in a chemically defined medium. J Clin Invest. 1989;84:1663–1670. doi: 10.1172/JCI114345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausman GJ, Campion DR, Martin RJ. Search for the adipocyte precursor cell and factors that promote its differentiation. J Lipid Res. 1980;21:657–670. [PubMed] [Google Scholar]
- Jimenez MA, Akerblad P, Sigvardsson M, Rosen ED. Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol Cell Biol. 2007;27:743–757. doi: 10.1128/MCB.01557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe AW, Yi L, Even Y, Vogl AW, Rossi FM. Depot-specific differences in adipogenic progenitor abundance and proliferative response to high-fat diet. Stem Cells. 2009;27:2563–2570. doi: 10.1002/stem.190. [DOI] [PubMed] [Google Scholar]
- Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FM. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissebah AH, Krakower GR. Regional adiposity and morbidity. Physiol Rev. 1994;74:761–811. doi: 10.1152/physrev.1994.74.4.761. [DOI] [PubMed] [Google Scholar]
- Kubota N, Terauchi Y, Miki H, Tamemoto H, Yamauchi T, Komeda K, Satoh S, Nakano R, Ishii C, Sugiyama T, et al. PPAR gamma mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- Majka SM, Barak Y, Klemm DJ. Concise review: adipocyte origins: weighing the possibilities. Stem Cells. 2011;29:1034–1040. doi: 10.1002/stem.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka SM, Fox KE, Psilas JC, Helm KM, Childs CR, Acosta AS, Janssen RC, Friedman JE, Woessner BT, Shade TR, et al. De novo generation of white adipocytes from the myeloid lineage via mesenchymal intermediates is age, adipose depot, and gender specific. Proc Natl Acad Sci U S A. 2010;107:14781–14786. doi: 10.1073/pnas.1003512107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici D, Shore EM, Lounev VY, Kaplan FS, Kalluri R, Olsen BR. Conversion of vascular endothelial cells into multipotent stem-like cells. Nat Med. 2010;16:1400–1406. doi: 10.1038/nm.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellitzer G, Martin M, Sidhoum-Jenny M, Orvain C, Barths J, Seymour PA, Sander M, Gradwohl G. Pancreatic islet progenitor cells in neurogenin 3-yellow fluorescent protein knock-add-on mice. Mol Endocrinol. 2004;18:2765–2776. doi: 10.1210/me.2004-0243. [DOI] [PubMed] [Google Scholar]
- Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, Buckingham M. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- Napolitano L. The Differentiation of White Adipose Cells. An Electron Microscope Study. J Cell Biol. 1963;18:663–679. doi: 10.1083/jcb.18.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrini S, Laviola L, Cignarelli A, Melchiorre M, De Stefano F, Caccioppoli C, Natalicchio A, Orlando MR, Garruti G, De Fazio M, et al. Fat depot-related differences in gene expression, adiponectin secretion, and insulin action and signalling in human adipocytes differentiated in vitro from precursor stromal cells. Diabetologia. 2008;51:155–164. doi: 10.1007/s00125-007-0841-7. [DOI] [PubMed] [Google Scholar]
- Planat-Benard V, Silvestre JS, Cousin B, Andre M, Nibbelink M, Tamarat R, Clergue M, Manneville C, Saillan-Barreau C, Duriez M, et al. Plasticity of human adipose lineage cells toward endothelial cells: physiological and therapeutic perspectives. Circulation. 2004;109:656–663. doi: 10.1161/01.CIR.0000114522.38265.61. [DOI] [PubMed] [Google Scholar]
- Poljakoff P. A new type of fat-forming organ in loose connective tissue. Arch Mikrosk Anat. 1888;32:123–136. [Google Scholar]
- Reyne Y, Nougues J, Dulor JP. Differentiation of rabbit adipocyte precursor cells in a serum-free medium. In Vitro Cell Dev Biol. 1989;25:747–752. doi: 10.1007/BF02623728. [DOI] [PubMed] [Google Scholar]
- Rodeheffer MS, Birsoy K, Friedman JM. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135:240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TJ, Huang TL, Tran TT, Zhang H, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, et al. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci U S A. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengenes C, Lolmede K, Zakaroff-Girard A, Busse R, Bouloumie A. Preadipocytes in the human subcutaneous adipose tissue display distinct features from the adult mesenchymal and hematopoietic stem cells. J Cell Physiol. 2005;205:114–122. doi: 10.1002/jcp.20381. [DOI] [PubMed] [Google Scholar]
- Staszkiewicz J, Gimble JM, Dietrich MA, Gawronska-Kozak B. Diet-Induced Obesity in Stem Cell Antigen-1 KO Mice. Stem Cells Dev. 2011 doi: 10.1089/scd.2010.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, Matsumoto D, Eto H, Inoue K, Aoi N, Kato H, Araki J, Yoshimura K. Functional implications of CD34 expression in human adipose-derived stem/progenitor cells. Stem Cells Dev. 2009;18:1201–1210. doi: 10.1089/scd.2009.0003. [DOI] [PubMed] [Google Scholar]
- Sul HS. Minireview: Pref-1: role in adipogenesis and mesenchymal cell fate. Mol Endocrinol. 2009;23:1717–1725. doi: 10.1210/me.2009-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, Graff JM. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi CG. The fat cell origin and structure. Conn Med. 1970;24:33–40. [PubMed] [Google Scholar]
- Toldt C. Contribution to the histology and physiology of adipose tissue. Sitzungsber Akad Wiss Wien Math Naturwiss Kl. 1870;62:445. [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid- activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- Traktuev DO, Merfeld-Clauss S, Li J, Kolonin M, Arap W, Pasqualini R, Johnstone BH, March KL. A population of multipotent CD34-positive adipose stromal cells share pericyte and mesenchymal surface markers, reside in a periendothelial location, and stabilize endothelial networks. Circ Res. 2008;102:77–85. doi: 10.1161/CIRCRESAHA.107.159475. [DOI] [PubMed] [Google Scholar]
- Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat Cell Biol. 2010;12:143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- Van RL, Bayliss CE, Roncari DA. Cytological and enzymological characterization of adult human adipocyte precursors in culture. J Clin Invest. 1976;58:699–704. doi: 10.1172/JCI108516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal H. Gene expression in visceral and subcutaneous adipose tissues. Ann Med. 2001;33:547–555. doi: 10.3109/07853890108995965. [DOI] [PubMed] [Google Scholar]
- Vohl MC, Sladek R, Robitaille J, Gurd S, Marceau P, Richard D, Hudson TJ, Tchernof A. A survey of genes differentially expressed in subcutaneous and visceral adipose tissue in men. Obes Res. 2004;12:1217–1222. doi: 10.1038/oby.2004.153. [DOI] [PubMed] [Google Scholar]
- Warming S, Rachel RA, Jenkins NA, Copeland NG. Zfp423 is required for normal cerebellar development. Mol Cell Biol. 2006;26:6913–6922. doi: 10.1128/MCB.02255-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman F. The fat organs of man: development, structure and systematic place of the so-called adipose tissue. A Zellforsch Microskop Anat Abt Histochem. 1926;3:325–329. [Google Scholar]
- Wasserman F. On storage, mobilization, and renewed storage in fat organs. Ergeb Anat Anz. 1929;67:181–194. [Google Scholar]
- Wasserman F. Electron microscopic investigation of the surface structures of the fat cell and of their changes during depletion of cell. Z Zellforsch Microskop Anat Abt Histochem. 1960;52:778–787. doi: 10.1007/BF00336627. [DOI] [PubMed] [Google Scholar]
- Wasserman F. The development of adipose tissue. In: AERaGFC Jr, editor. Handbook of Physiology. Washington, DC: American Physiological Society; 1965. pp. 87–100. [Google Scholar]
- White P, May CL, Lamounier RN, Brestelli JE, Kaestner KH. Defining pancreatic endocrine precursors and their descendants. Diabetes. 2008;57:654–668. doi: 10.2337/db07-1362. [DOI] [PubMed] [Google Scholar]
- Wilson ME, Scheel D, German MS. Gene expression cascades in pancreatic development. Mech Dev. 2003;120:65–80. doi: 10.1016/s0925-4773(02)00333-7. [DOI] [PubMed] [Google Scholar]
- Xu X, Ying Z, Cai M, Xu Z, Li Y, Jiang SY, Tzan K, Wang A, Parthasarathy S, He G, et al. Exercise ameliorates high-fat diet-induced metabolic and vascular dysfunction, and increases adipocyte progenitor cell population in brown adipose tissue. Am J Physiol Regul Integr Comp Physiol. 2011;300:R1115–1125. doi: 10.1152/ajpregu.00806.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannettino AC, Paton S, Arthur A, Khor F, Itescu S, Gimble JM, Gronthos S. Multipotential human adipose-derived stromal stem cells exhibit a perivascular phenotype in vitro and in vivo. J Cell Physiol. 2008;214:413–421. doi: 10.1002/jcp.21210. [DOI] [PubMed] [Google Scholar]
- Zimmerlin L, Donnenberg VS, Pfeifer ME, Meyer EM, Peault B, Rubin JP, Donnenberg AD. Stromal vascular progenitors in adult human adipose tissue. Cytometry A. 2010;77:22–30. doi: 10.1002/cyto.a.20813. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.