Abstract
The crystal structure of Escherichia coli MoaB was determined by multiwavelength anomalous diffraction phasing and refined at 1.6-Å resolution. The molecule displayed a modified Rossman fold. MoaB is assembled into a hexamer composed of two trimers. The monomers have high structural similarity with two proteins, MogA and MoeA, from the molybdenum cofactor synthesis pathway in E. coli, as well as with domains of mammalian gephyrin and plant Cnx1, which are also involved in molybdopterin synthesis. Structural comparison between these proteins and the amino acid conservation patterns revealed a putative active site in MoaB. The structural analysis of this site allowed to advance several hypothesis that can be tested in further studies.
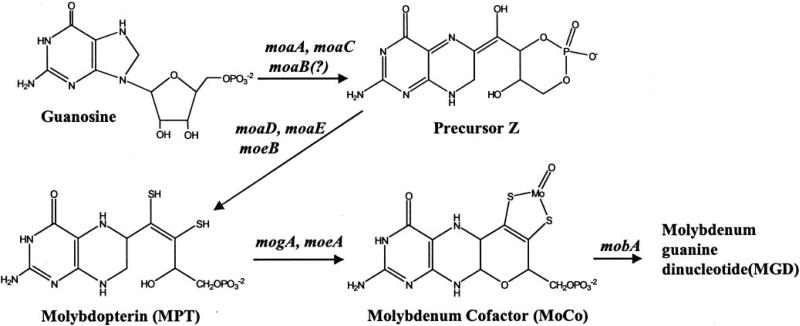
Molybdoenzymes are ubiquitous mediators of many essential redox reactions (1, 2). All molybdoenzymes except nitrogenase contain a molybdenum cofactor (Moco)1 that consists of a mononuclear molybdenum covalently coordinated by molybdopterin (3, 4). In eubacteria, Moco is often modified by various nucleotides. In Escherichia coli, some of the molybdoenzymes contain a modified form of Moco known as molybdopterin guanine dinucleotide (MGD) (5). In some others, the metal ion is often coordinated by two MGDs (6, 7).
The biosynthesis of MGD (see Fig. 1) is catalyzed by the products of the moa, mob, and moe operons, as well as the product of the mogA (8) and mobA (9–11) genes. In eukaryotes, the homologs of MogA and MoeA are fused to form a single multifunctional multidomain protein (12), called gephyrin in mammals (13), cinnamon in Drosophila melanogaster (14), and Cnx1 in Arabidopsis thaliana (15).
Fig. 1. Moco biosynthesis pathway in E. coli.
The four-step synthesis first involves the conversion of a guanosine derivative to an unstable intermediate, precursor Z, through the action of the enzymes MoaC and MoaA. The heterotetrameric enzyme molybdopterin synthase (MoaD2: MoaE2) then incorporates sulfur from the C-terminal thiocarboxylate of MoaE onto precursor Z to form the dithiolene group of molybdopterin (23, 43, 44). The incorporation of molybdenum into molybdopterin to form Moco requires the proteins MogA and MoeA (19). On the last step, a GMP group is transferred to the terminal phosphate of Moco by the product of the gene mobA (9–11) to form MGD.
Mutants in the E. coli MGD biosynthetic pathway are viable under experimental conditions, albeit with compromised phenotypes (16, 17). In humans, mutations in the enzymes of Moco biosynthesis result in a Moco deficiency, leading to several neurological complications and subsequent death at a young age (18).
The structures of members of the E. coli Moco biosynthesis pathway, MoaC (17), MogA (19), MobA (10, 11), MobB (20), ModE (13), MoeA (21, 22), and molybdopterin synthase (MoaD2-MoaE2) (23), have been determined to clarify their roles in the pathway (Fig. 1). However, the exact enzymatic activities of these proteins are not well understood. Similarly, very little is known about the biochemical and physiological roles of the MoaB protein, although it is believed that MoaB may be involved in the synthesis of precursor Z (8). BLAST data base (24) searches also indicate the involvement of MoaB in the biosynthesis of Moco. Here we report the crystal structure of E. coli MoaB at a 1.6-Å resolution. Amino acid sequence and structural similarities with other members of the Moco pathway strengthen the possibility of a role for MoaB.
EXPERIMENTAL PROCEDURES
Cloning and Protein Purification
E. coli MoaB was cloned, expressed, and purified using standard procedures (25). The hexahistidine tag was removed before crystallization. The purified MoaB protein was dialyzed against 10 mm HEPES, pH 7.5, 500 mm NaCl, and concentrated using a BioMax concentrator (Millipore). Before crystallization, any particulate matter was removed from the sample by passage through a 0.2-μm Ultrafree-MC centrifugal filtration device (Millipore).
Crystallization and X-ray Data Collection
Two microliters of the dialyzed MoaB protein were mixed with an equal volume of precipitant containing 2.0 m ammonium sulfate and 20% (v/v) ethylene glycol. Droplets were equilibrated against the precipitant with hanging drop vapor diffusion, and crystals grew after 2–5 days at 21 °C. For diffraction studies, the crystals were flash-frozen in liquid nitrogen with 25% (v/v) ethylene glycol added to the crystallization buffer as the cryoprotectant. The crystals belonged to the hexagonal space group P321, with the cell parameters at 100 K being a = b = 69.11Å, c = 126.22Å, α = β = 90°, γ = 120°, and contained two molecules of protein/asymmetric unit. The Matthews coefficient (26) was 2.33, which corresponds to the solvent content of ~46% in crystals.
A two-wavelength anomalous dispersion experiment with crystals of Se-Met-derivatized protein was carried out on the 19ID beamline of the Structural Biology Center Collaborative Access Team of the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). The absorption peak and the rising inflection point were determined by calculating and plotting f′ and f″ values against energy (27) from the fluorescence spectrum measured for a test crystal. Later, higher resolution data were collected from the unexposed part of the same crystal, which had been stored in liquid nitrogen. All crystallographic data were measured with the custom built 3 × 3 tiled charge-coupled device detector (28) with a 210 × 210-mm2 active area and fast duty cycle (~1.7 s). The experiment, data collection, and visualization were controlled with d*TREK (29), and all data were integrated and scaled with the program package HKL2000 (30). Some of the basic statistics of data collection and processing are provided in Table I.
Table I. Basic data collection and processing statistics for MoaB crystals.
The number of amino acid residues/methionines (including N-terminal Met) was 340/6. The number of molecules in the asymmetric unit was 2. The Rmerge between low- and high-resolution passes was 0.037. Crystal lattice P321 a = b = 69.11, c = 126.22 Å α = β = 90° γ = 120°.
| Inflection point | Peak | Low resolution | High resolution | |
|---|---|---|---|---|
| Energy (keV) | 12.65934 | 12.66075 | 12.000 | 12.000 |
| Wavelength (Å) | 0.97954 | 0.97929 | 1.03321 | 1.03321 |
| Resolution (Å) | 50–1.87 | 50–1.87 | 50–2.7 | 3.0–1.54 |
| No. of observations | 311,387 | 311,440 | 60,562 | 251,553 |
| Uniquea reflections | 55,534 | 55,494 | 8,840 | 45,003 |
| Completeness (%)b | 99.4 (99.0) | 99.2 (98.0) | 86.8 (90.2) | 99.5 (95.8) |
| I/σ(I)b | 26.5 (2.5) | 26.2 (2.3) | 28.9 (14.5) | 22.4 (2.6) |
| Rsymb | 0.057 (0.45) | 0.06 (0.52) | 0.056 (0.137) | 0.067 (0.41) |
Bijvoet pairs for scaling the multiwavelength anomalous dispersion data sets were kept separately.
Statistics in the last resolution shell are in parentheses.
Guanosine Triphosphate Binding
Purified recombinant MoaB protein was subjected to binding assays using reaction conditions consisting of 0.7 mg/ml protein, 50 mm KCl, 100 nCi [α-32P]GTP (3000 Ci/mmol; PerkinElmer Life Sciences), and 50 mm pH buffer (sodium acetate, pH 5.0, MES, pH 6.5, Tris, pH 7.5, Tris, pH 8.0) in a 50-μl reaction volume. Divalent cations were added as MgCl2, CaCl2, or MnCl2 to a concentration of 5 mm. After 15 min at 30 °C, reactions were stopped, and the protein was precipitated by the addition of an equal volume of saturated ammonium sulfate in binding buffer and incubation on ice for 15 min. Pellets were collected by centrifugation, washed twice with saturated ammonium sulfate, and dried. The protein was then resuspended in 10 μl of binding buffer, and the precipitated GTP was monitored by scintillation counting using total input radioactivity as a standard.
Determination of the GTPase activity was essentially as previously described (31). Briefly, pelleted proteins were resuspended in binding buffer and hydrolyzed by adding perchloric acid to 6% concentration for 15 min. The pH was adjusted to 3.5 with potassium phosphate, the reactions were clarified by centrifugation, and aliquots were spotted on polyethyleneimine-cellulose TLC plates (Selecto Scientific Inc.). A GDP standard was enzymatically derived by the incubation of [α-32P]GTP substrate with thermosensitive alkaline phosphatase (Invitrogen) in the manufacturer's supplied reaction buffer. Separation of phosphorylated reaction products was achieved by TLC using 0.65 m KH2PO4 as running buffer. Plates were dried and exposed to Fuji RX film.
RESULTS
Structure Solution and Refinement
Initially, only two Se sites, one from each of the molecules in the asymmetric unit, were found with the CNS (32) program, and phases were calculated to a 2.5-Å resolution. An inspection of the log-likelihood gradient maps revealed the non-isotropic nature of these sites. After phase refinement and accounting for this anisotropy, two other Se sites were also located. However, they were disordered to the extent that the phases did not improve significantly (Table II). Experimental phases were extended from 2.5- to 2.0-Å resolution with density modification, using data collected at the inflection point of the absorption edge.
Table II.
Phasing and refinement statistics for MoaB structure
| Phasing in 50–2.5-Å resolution shell | Inflection point | Peak | FOMa |
|---|---|---|---|
| Number of reflections | 17,750 (1754) | 22,885 (2607) | 23,021 (2641) |
| Phasing power with | |||
| 2 sites | 2.44 (1.71) | 2.42 (1.67) | 0.60 (0.48) |
| 2 sites + disorder | 2.83 (1.93) | 2.74 (1.85) | 0.66 (0.54) |
| 4 sites + disorder | 2.95 (2.18) | 2.90 (2.07) | 0.67 (0.55) |
| Refinement | ||
|---|---|---|
| Resolution (Å) | 62–1.60 | 1.64–1.60 |
| Number of reflections | 42215 | 2867 |
| R-factor % | 18.5 | 20.6 |
| Rfree % | 21.9 | 27.1 |
| Correlation | 96.2 | |
| Correlation free | 95.2 | |
| All nonhydrogen atoms | 2961 | |
| Mean B-factor | 14.77 |
| Deviations from ideal | Refined | Target |
|---|---|---|
| Covalent bonds | 0.020 | 0.021 |
| Bond angles | 1.885 | 1.955 |
| Planarity | 0.007 | 0.02 |
| Chiral centers | 0.107 | 0.20 |
| Torsion angles | 6.285 | 5.0 |
| van der Waals contacts | 0.240 | 0.30 |
Mean FOM after density modification in 50–2.0-Å shell was 0.92.
The initial model for the most of both molecules was built with the program ARP/wARP (33). The remainder of the model was built and all side chains were corrected manually. This model was then refined against the 1.6-Å resolution data with several macrocycles of CNS and subsequently with REFMAC5 (34). Inspections and corrections of the model were made with the programs O (35) and QUANTA (Accelrys, Inc.).
Phasing and refinement parameters are shown in Table II. The structural coordinates have been deposited in the Protein Data Bank (36) with the access code 1MKZ.
Spatial Model of MoaB
The current model of MoaB consists of two crystallographically independent molecules, A and B. Molecule A contains 170 amino acid residues and has Met1 and Ser2 absent. Molecule B has 171 residues, with Met1 absent. Gly171 and Ser172, adducts of cloning, were clearly visible in the electron density maps of both molecules. The Arg22 and Glu100 side chains in molecule A were incomplete because of poor electron densities, and in molecule B the Ser2, Gln3, Arg21, Arg22, Glu83, and Glu112 side chains were incomplete for the same reason.
The following side chains were modeled in alternative conformations, Molecule A, Thr6, Glu24, Glu25, Thr28, Gln38, Glu83, Arg97, Ser110, Glu113, Thr118, and Ile133 and molecule B, Thr6, Thr28, Leu92, Arg97, Glu100, Se-Met108, Thr118, Ile133, and Arg160. Phe134 in the hydrophobic core of the molecule was replaced by leucine, probably as a mutation introduced by a polymerase chain reaction. Seven sulfate ions, three acetate molecules, and 238 water molecules were also revealed in the model. Four of the sulfates are bound to molecule A and three to molecule B.
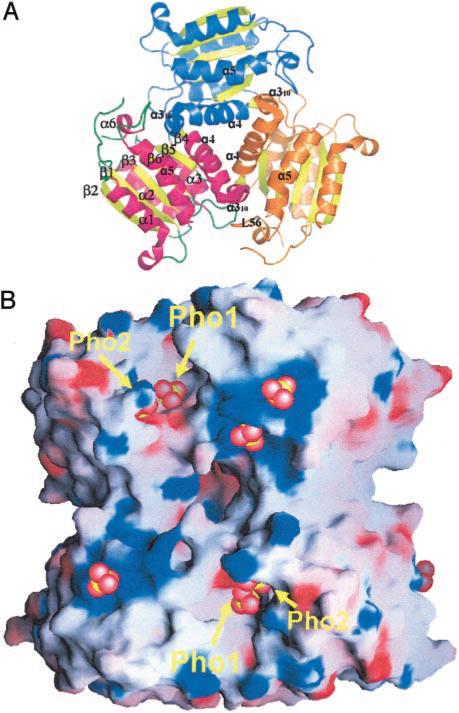
The MoaB monomer is ~32 × 35 × 50 Å3 and consists of a six-strand β sheet, six α helices, and one 310 helix packed in a modified Rossman fold (Fig 2A). The twisted β sheet with a β2 ↑ β11 ↑ β3 ↑ β6 ↑ β5 ↓ β4 ↑ topology is formed by five parallel β strands, β2 (residues 43–50), β1 (12–18), β3 (72–76), β6 (131–136), β4 (97–98), and one antiparallel β5 (124–128) inserted between β6 and a very short β4. The β sheet is sandwiched between four α helices, a1 (residues 23–40) and α5 (139–150) on one side and α2 (53–67) and α3 (86–92) on the other. The three remaining helices are α4 (99–115), a one-turn α6 (164–168), and a 310 helix (116–121). Helices α1 and α5 are distorted to various degrees.
Fig. 2. Oligomerization of MoaB.
A, view of a trimer along the crystallographic 3-fold axis. Six strands of the β sheet (yellow), along with α and 310 helices (pink), are labeled in one monomer. The 310 helix (perpendicular to the plane of the paper) of one monomer forms rich hydrophobic interactions in the cleft created by helices α4, α5, and the loop between α5 and α6 (labeled L56) of the other. B, electrostatic representation of the surface of MoaB hexamer. Red and blue correspond to the negatively and positively charged areas, respectively, whereas white represents neutral regions. The hexamer can be approximated as a cylinder 62-Å high with a radius of 35 Å. Longitudinal and equatorial grooves are also visible intersecting at the central hole created between the four monomers. Bound sulfate ions are represented with red (oxygens) and yellow (sulfur) spheres. Two sulfate ions, which could mimic the phosphate groups in the binding site, are labeled Pho1 and Pho2. The binding of remaining sulfates is probably opportunistic. Fig. 2B was prepared with GRASP (42).
Each of the molecules in the asymmetric unit is incorporated in a compact trimer generated by the 3-fold crystallographic axis. In this formation, each monomer interacts with two other monomers (Fig. 2A). The buried surface area within each pair of monomers in the trimer is ~1700 Å2, whereas the total surface area of a monomer is ~8400 Å2. The major contributor to trimer stability is the insertion of the 310 helix of one monomer into the hydrophobic groove of another, which is formed between helices α4, α5, and the loop connecting α5 and α6. There is also a network of hydrogen bonds and salt bridges between each pair of monomers (Table III).
Table III. Polar interactions in the interface within the trimer.
Some of the residues are highly conserved in MoaB sequences across 22 organisms. For example, Glu89 is invariant, whereas residues 97, 104, 107, and 122 have conserved charges.
| Atom in monomer 1 | Atom in monomer 2 | Distance |
|---|---|---|
| (Å) | ||
| O Glu83 | NH-2, Arg97 | 3.17 |
| OE-2 Glu89 | NH-2 Arg97 | 3.29 |
| OE-2 Glu89 | NZ Arg97 | 2.78 |
| OE-1 Glu89 | NH-1 Arg122 | 3.30 |
| NH-2 Arg107 | OE-2 Glu100 | 3.32 |
| NH-2 Arg107 | OE-2 Glu104 | 2.98 |
| NZ Arg107 | OE-1 Glu104 | 2.62 |
| OG Ser117 | N Cys162 | 3.21 |
| OG Ser117 | O Cys162 | 2.65 |
| OG-1 Thr116 | NE-2 Gln154 | 2.89 |
| NE-2 Gln120 | OE-1 Gln154 | 2.85 |
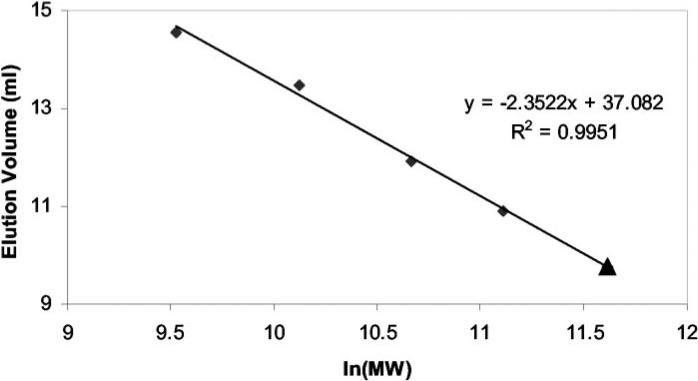
Two trimers form a hexamer, mostly through hydrophobic interactions at their interface. Size-exclusion chromatography showed that MoaB is a hexamer in solution as well (Fig. 3). In crystals, hexamerization buries a total of 2680 Å2 (32%) of the surface of each monomer. Strong interactions in the observed hexamer, together with the conservation of some residues involved in its formation (Table III), lead us to believe that the oligomeric state of MoaB in crystals is the same as in solution and is biologically relevant.
Fig. 3. Size-exclusion chromatography of E. coli MoaB.
Size-exclusion fast protein liquid chromatography was performed using a Superdex-75 column (Amersham Biosciences). The purified MoaB protein (▴) eluted from the column with an apparent molecular mass of ~111.2 kDa. Standard markers (◆), albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa), and ribonuclease A (13.7 kDa) (Amersham Biosciences), were used to calibrate the column. The best-fit line is shown with the R2 value of the fit indicated.
The shape of the hexamer can be roughly described as an ~62-Å-high cylinder with a radius of ~35 Å. It has an equatorial groove formed between the trimers and three vertical clefts each formed by four molecules of the hexamer (Fig. 2B). The vertical clefts of the hexamer are lined with polar and mostly positively charged residues. The sides of the hexamer between each pair of vertical clefts are mostly neutral but with some negatively charged and a considerable number of hydrophobic residues. Three holes, ~8 × 11 Å, on the sides of the hexamer lead to a large cave inside of it. The bases of the cylinder are rather flat, formed by three pairs of helices α1 and α5 from each monomer of the trimer. Judging by the area of buried surface and the number and character of interactions, the trimer appears to be a more stable formation than the hexamer.
Analysis of Structural Similarities and the Putative Active Site
A structural similarity search using the DALI algorithm (37) indicated that MoaB shares a common fold with several proteins also involved in Moco synthesis. MoaB had highly significant similarity with the G domains of the multifunctional mammalian protein gephyrin (12, 38), the plant protein Cnx1 (38), with E. coli MogA (19), and with the largest domain of MoeA (21, 22). Z-scores were 22.0, 21.9, 19.5, and 10.9, respectively. The corresponding root mean square deviations with these homologous structures were 1.38 Å for the 150 aligned Cα atoms, 1.47 Å for 147 Cα atoms, 1.73 Å for 147 Cα atoms, and 1.88 Å for 126 Cα atoms.
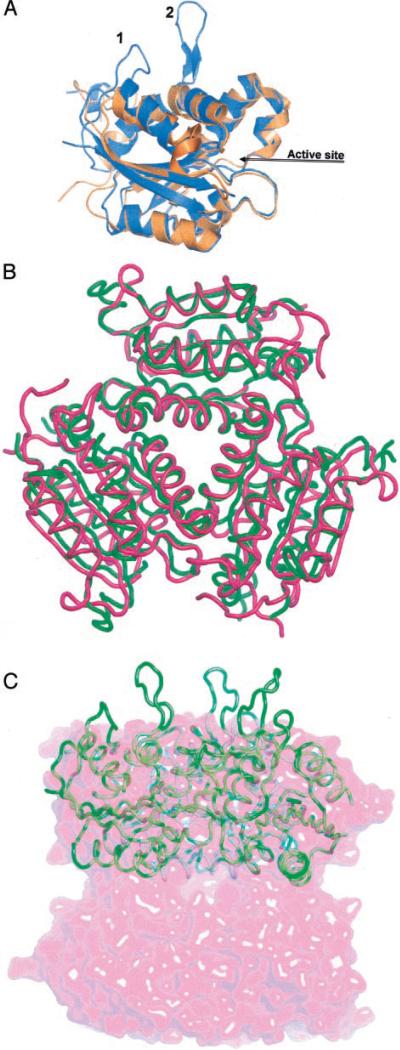
In addition to the structural conservation of several proteins along the Moco pathway, oligomerization of these proteins is also remarkably well preserved (Fig. 4). Thus, the functional trimer of MogA (19) is also seen in gephyrin and Cnx1 oligomers, although the latter two are multidomain proteins. The MoaB hexamer comprises two such trimers (Fig. 4C).
Fig. 4. Structural similarity of MoaB with MogA and gephyrin.
A, superposition of the MoaB (orange) and MogA (marine) monomers shows very good alignment of secondary structure elements and many of the loops. The only significant difference is caused by two insertions in the MogA amino acid sequence, corresponding to the protruding β turn (labeled 1) and β hairpin (labeled 2). B, superposition of Cα traces of MoaB (pink) and gephyrin G domain (green) trimers. The latter is more similar to MoaB than MogA, because it does not have protrusions, which are characteristic for MogA and shown in A. C, the MoaB hexamer is shown with a pink semitransparent surface. A wire model of the MogA trimer fits in half of the hexamer very well, except for the protrusions shown in A.
The extent of structural similarity between MoaB and the gephyrin and Cnx1 proteins is not reflected in amino acid sequence similarity. The sequence identities of MoaB with the G domains of Cnx1 and gephyrin, with MogA, and with the largest domain of MoeA are 33, 32, 27, and 21.5%, respectively. The structural similarity between MoaB and two other proteins from the Moco synthesis pathway, MoaC and MobA, is significantly lower, with respective root mean square deviations of 3.22 Å for 57 Cα atoms and 3.38 Å for 72 Cα atoms.
A number of residues are highly conserved among MoaB orthologs. Asp27, Gly30, Asp85, Glu89, Gly126, Gly138, Ser139, and the motif Gly77-Gly78-Thr79-Gly80 are invariant in MoaB proteins from 22 genomes. Pro137, which is conserved in all but two of these sequences forms another conserved motif Pro137-Gly138-Ser139. Sixteen other residues are similar, indicating conservation of mostly hydrophobic and a few polar interactions. These residues are Ile/Leu/Val15, Leu/Ile/Val16, Ala/Thr87, Leu/Ile/Val94, Phe/Leu/Ile95, Arg/Lys97, Phe/Ile102, Phe/Ile106, Arg/His107, Ser/Thr121, Arg/Lys-122, residues 132–134, Met/Leu136, and Ile/Leu/Val150.
When the MoaB sequence is aligned with the sequences of four other Moco synthesis proteins (referred to here as the Moco synthesis group), E. coli MogA, mammalian gephyrin, insect cinnamon, and plant Cnx1, 19 residues are invariant and a number of others show strong conservation (Fig. 5). Most of the invariant residues in the MoaB orthologs and the Moco synthesis group overlap, and only a few belong to either group exclusively. For example, the amino acids in positions (according to the MoaB sequence) 57, 90, 98, and 119 are invariant only in the Moco synthesis group, whereas the residues Gly30, Gly126, and Ser139 are highly conserved in MoaB orthologs only.
Fig. 5. Sequence alignment of E. coli MoaB, MogA, Drosophila cinnamon, Arabidopsis Cnx1, and human gephyrin performed with ClustalW (45).
For the last three proteins, only portions homologous to MoaB and MogA are presented. Numbers after the protein names indicate the starting amino acid number of a given sequence. asterisk, depicts invariant residues; colon, corresponds to conservation of a residue type (e.g. hydrophobic, positively charged, carrying hydroxyl group); dot, indicates that only one residue interrupts an otherwise conserved type. Residues invariant in 22 MoaB orthologs are shown in bold red. Pro137, although not identified here as invariant in MoaB orthologs, changes to Ala in only 2 of 22 sequences.
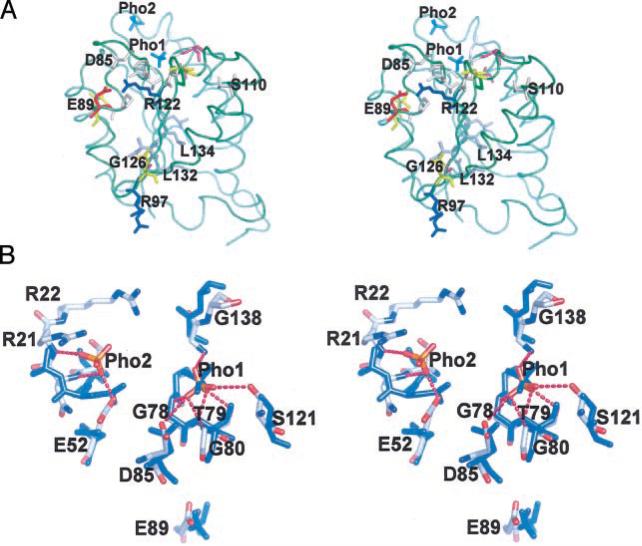
All of the invariant residues throughout the MoaB orthologs and the Moco synthesis group are in or near the depression on the surface of MoaB between two monomers of the MoaB trimer (Fig. 2B) where two of the bound sulfates are found (Figs. 2B and 6). A very similar depression is found in the MogA, gephyrin, and Cnx1 structures.
Fig. 6. Conserved residues and the binding site of MoaB.
A, the overall fold of MoaB is shown in green with two sulfate ions in cyan. Light beige residues are conserved in all 22 orthologs as well as in the other Moco proteins listed (not labeled). Blue residues (Arg97 and Arg122 in MoaB) vary only to lysine, thus conserving a positive charge. Glu89, which is invariant in MoaB orthologs, is aspartic acid in other Moco proteins. Gray represents the residues conserving the hydrophobicity in MoaB orthologs and in other four Moco proteins. Three residues shown in magenta, Glu30, Gly125, and Ser139, are invariant in the 22 MoaB sequences but not in gephyrin, MogA, Cnx1, or cinnamon. Conversely, the yellow residues in positions 57, 90, 98, and 137 (MoaB nomenclature) are invariant in the latter group but vary in MoaB sequences. The Arg97 and Ser110 side chains are shown in two alternative conformations. B, ion binding details in MoaB and MogA. The MoaB residues, interacting with bound sulfates are shown with gray carbons, blue nitrogens, and red oxygens. Corresponding residues from MogA are light blue. The region around Pho2 is flexible with higher B factors. Although it is shown in full, only parts of the Arg22 side chain could be modeled because of its disorder. This segment is more disordered and is missing in MogA structure (19). Both sulfate groups are primarily bound by their interactions (magenta dashed lines) with the backbone nitrogens. Motif Gly77-Gly78-Thr79-Gly80, a major contributor to Pho1 binding, is particularly well conserved structurally. The putative catalytic Asp85 also interacts with this Pho1.
The polypeptide segments in the area of the MoaB surface depression are structurally the most preserved. These segments are formed by two motifs, Gly77-Gly78-Thr79-Gly80 and Pro137-Gly138-Ser139. In both the MoaB and MogA structures, these motifs are responsible for binding one of the sulfates (Fig. 6B). The Ser121 side chain, using the MoaB nomenclature, also contributes to binding. This residue is Ser or Thr in all MoaB orthologs conserving the interacting hydroxyl group.
It is likely that these structurally conserved sites are occupied by phosphate groups of physiological compounds, a conjecture that is supported by the electron density map. As can be seen on Fig. 7, the electron density in this region cannot be entirely accounted for by Pho1. Remarkably, a very similar feature can be found in the structure of MogA. It has been suggested (19) that this could be a remnant of some phosphate-containing compound that was bound to the protein strongly enough so as not to be completely dissociated during purification and crystallization. Another sulfate in our structure is bound to a much less conserved region of the same depression (see Figs. 6 and 7). This region is generally quite flexible as indicated by high B factors. Part of this binding site is missing in the MogA model due to disorder as is the second bound ion. Two more sulfate ions in molecule A and one in molecule B are bound at the C terminus. It is difficult to assess the physiological importance of these ions, because two of the residues at this end are adducts of cloning.
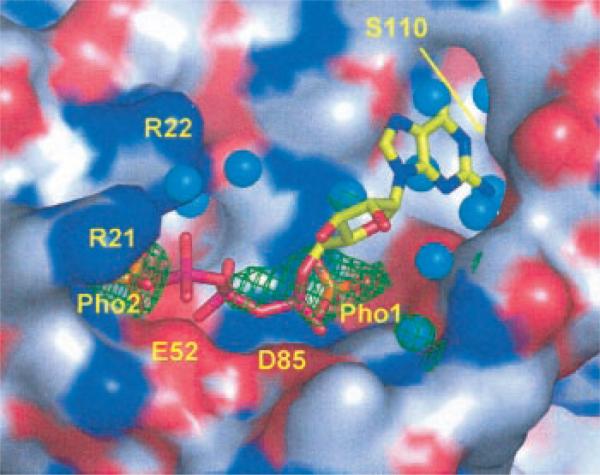
Fig. 7. Modeled GTP in the binding site of MoaB.
The surface of the protein containing the binding site is color-coded with red oxygens, blue nitrogens, and gray carbons. Experimentally observed sulfate ions (orange S and red O atoms) are shown with their corresponding 2Fo – Fc electron densities contoured at a 1σ level (green mesh). Water molecules are shown as cyan spheres. Sulfates are labeled Pho1 and Pho2 because they likely are phosphate groups in vivo. Associated with Pho1, electron density has an additional feature, which cannot be explained with the sulfate ion and which extends toward Pho2. It is likely the remnant of a phosphate-containing compound from the physiological complex. Arg21 and Arg22 are also labeled. The guanidinium group of the Arg22 side chain is not entirely visible in our structure but is added in this figure to show its proximal position and to avoid the creation of an apparent extended surface depression that could lead to erroneous interpretations. The ridge, separating two deeper cavities on either side of Pho1, is formed by two conserved motifs, Gly77-Gly78-Thr79-Gly80 and Pro137-Gly138-Ser139. Modeled GTP is shown with stick model color coded as yellow carbons, blue nitrogens, red oxygens, and magenta phosphors. The locations of the Glu52 and Asp85 carboxyl groups are indicated with labels, and the location of Ser110 is shown with an arrow.
Labeled GTP Binding in Solution
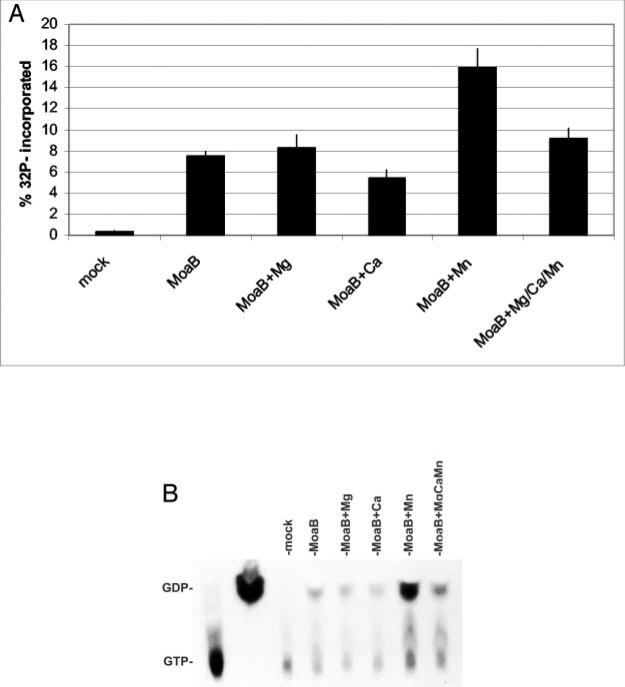
Our modeling studies indicate that GTP may bind in the putative active site of MoaB. To establish reaction conditions for potential GTP binding by the MoaB protein, pH was modulated from 5.0 to 8.0, and [α-32P]GTP binding was monitored by protein precipitation followed by scintillation counting. GTP binding was most efficient at pH 7.5; therefore, reactions were performed at pH 7.5. Between 5 and 20% of input radioactivity was recovered, compared with less than 1% in mock reactions containing no protein, indicating the binding of GTP. Various divalent cations were added to determine their effect on MoaB GTP binding. The results showed that Mg2+ had no impact on binding, Ca2+ was inhibitory, and Mn2+ was stimulatory (Fig. 8A). GTPase activity was assessed by monitoring reaction products with thin-layer chromatography (Fig. 8B). Ca2+ again appeared to decrease overall activity, whereas Mn2+ acted both to increase overall binding and to modulate the ratio of GDP to GTP bound to the MoaB protein. In the presence of all three cations (Mg2+, Ca2+, and Mn2+) simultaneously, the stimulatory and modulatory effects of Mn2+ were abated.
Fig. 8. GTP binding and GTPase activity in MoaB.
A, incorporation of radio-labeled GTP. Binding reactions were assembled using [α-32P]GTP as a tracer and either no protein (mock), MoaB protein alone (MoaB), or MoaB protein with the listed divalent cations (+Mg/Ca/Mn). Protein precipitations were performed as described under “Experimental Procedures.” The amount of bound [α-32P]GTP was determined by scintillation counting and is expressed as a percentage of the total input [α-32P]GTP. B, GTPase activity. Binding reactions were assembled as in A. Protein was precipitated, acid-hydrolyzed, and analyzed by thin-layer chromatography as described under “Experimental Procedures.” Input [α-32P]GTP (lane 1) and enzymatically derived [α-32P]GDP (lane 2) were used as running standards.
DISCUSSION
The exact role of MoaB in Moco biosynthesis is unknown. Strong structural similarity of MoaB to MogA and to G domains of mammalian gephyrin and plant Cnx1 (Fig. 4), which are also involved in the Moco biosynthesis, together with the sequence (Fig. 5) and spatial (Fig. 6) conservation of several residues, suggests a conserved function among these proteins. Although the bound molybdopterin has not been observed crystallographically, MogA, mammalian gephyrin, and plant Cnx1 bind to it with high affinity (12, 19, 39). The high degree of sequence and structural conservation in the surface depression suggests that this cavity may be a consensus binding site for molybdopterin.
Liu et al. (19) mutated several highly conserved residues in MogA to Ala to study their possible role in function. The MogA residues studied were Ser12, Asp49, Thr76, Arg81, Asp82, Ser107, and Ser117; the corresponding residues in MoaB are Ser19, Glu52, Thr79, Arg84, Asp85, Ser110, and Ser121, many of which are also well conserved (Figs. 4–6). The only mutations that had a measurable effect on MogA activity were D49A and D82A. They were assumed to disrupt catalysis, because molybdopterin binding in the two mutant proteins was shown to be improved (19). The corresponding residues in MoaB, Glu52 and Asp85, are highly conserved both in primary and in tertiary structures and could play a similar role. In MogA, the proposed catalytic site contained electron density that was modeled as a sulfate ion but was proposed to correspond to the remnants of molybdopterin (19). In the MoaB structure, we observed very similar electron density (Fig. 7). Thus, the overall conservation of the binding site and of the interaction with the bound anion suggests that MoaB might bind MTP similarly to MogA, gephyrin, and Cnx1.
The two presumed phosphate ions in MoaB could be consistent with a binding of GTP, which was easily modeled in the putative binding site of MoaB (Fig. 7). In this model, the α and γ phosphate groups of GTP are very close to the positions of the ions bound in the crystal structure. The putative catalytic Asp85 and Glu52 interact directly with these phosphates and are ideally poised to play a role, perhaps in the transformation of GTP into precursor Z. The base of the modeled GTP fits very well in the deep cavity, which is occupied by several water molecules in our crystal structure. The invariant Ser110 (Figs. 5 and 6) is located in a deep cleft where the base of the modeled GTP is bound (Fig. 7) and could interact with the N-1 and/or N-2 atoms of the latter. Our results show that MoaB indeed binds GTP (Fig. 8A), but with low affinity. The protein also possessed a low GTPase activity (Fig. 8B). Both binding and enzymatic potential were affected by some divalent ions, especially by Mn2+. Coincidentally, when some divalent ions were added in crystallization trials of another Moco synthesis protein, MoeA, Mn2+ appeared to have more drastic effects than the others.2
Based on the possibility of binding various compounds of Moco pathway by MoaB and on its structural similarity with MogA and MoeA, we suggest that MoaB might play a substrate-shuttling role in the biosynthetic process. The intermediates of Moco synthesis and the cofactor are unstable, suggesting that the reactions, including the transfer of Moco to apomolybdoenzymes, occur while the corresponding compounds are still bound to the enzymes or to transporter/storage proteins, avoiding their dissociation into the external solvent.
The protection of the substrates against solvent might occur through specific protein-protein interactions among the enzymes. It is becoming apparent that protein-protein interactions play crucial role in Moco biosynthesis. Magalon et al. (40) reported that numerous protein pairs from the Moco pathway do form complexes in vivo, including MobB-MoeA, MobB-MogA, MoeA-MogA, MoeA-MobA, and MobB-MobA and that some compounds of this pathway influence these interactions. Divalent ions, as shown here, can also influence binding of some compounds in the Moco pathway, although the mechanism of such influence is not yet clear.
Possible involvement of MoaB in such protein-protein interactions is supported indirectly by several observations. First, in Synechococcus sp., the MoaC ortholog is fused to the homolog of either MoaB or MogA (41). Second, MogA and MoeA are combined to form the multifunctional protein gephyrin in mammals (13), Cnx1 in plants (15), and cinnamon in insects (14). All of the protein-protein complexes observed in vivo (40, except for MobB-MobA, contain either MogA or MoeA, the structural homologs of MoaB. If MoaB can indeed bind GTP, precursor Z, or molybdopterin as discussed above and if it can form complexes with several other proteins of Moco synthesis pathway, it is tempting to suggest that MoaB plays a transporting and/or storage role.
The presence of six independent binding sites in the MoaB hexamer would enhance the capacity of the protein in such a role. As reported by Schwarz et al. (39), the G domain of Cnx1 can be present in both hexameric and trimeric forms. Because the trimer of Cnx1 G domain is very similar to the trimer of MoaB, and because the trimer of MoaB seems to be much more stable than its hexamer, it is possible that MoaB is also present in these two states of oligomerization. In our experiments with size-exclusion chromatography, MoaB was not found in a trimeric state. However, equilibrium between trimeric and hexameric states could be modulated by particular compounds, such as reaction substrates and products, ions, or other proteins of the pathway, all of which were absent in our experiments. In the trimeric state MoaB could bind MobB and MoeA much the same way as does MogA (40). On the other hand, MogA could replace one-half of the MoaB hexamer, as shown on Fig. 4C. The largest structural differences between MoaB and MogA are at the bases of the MoaB hexamer, away from the trimer-trimer interface. Formation of such heterooligomers could be an effective way of substrate/product exchange along the Moco pathway without their dissociation into the external solvent. An inferred promiscuity of MoaB in protein-protein complex formation would be an asset for a transporter/storage protein.
Summary
The crystal structure of E. coli MoaB was determined at 1.6-Å resolution. The strong structural similarity of MoaB with other Moco synthesis proteins, such as MogA, gephyrin, and Cnx1, along with the amino acid conservation patterns, indicated that MoaB might bind some of the same compounds, namely molybdopterin. The presence of a sulfate ion in the putative binding site, the experimental verification of MoaB/GTP binding, and modeling studies suggest that GTP and possibly precursor Z could bind in the same binding site.
MoaB displayed very strong structural similarity to MogA and MoeA, two proteins that form numerous complexes with other proteins within the Moco synthesis pathway in E. coli, suggesting that MoaB might be part of such complexes and may play a role in transport or storage of several compounds in the Moco synthesis pathway.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The atomic coordinates and structure factors (code 1MKZ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The abbreviations used are: Moco, molybdenum cofactor; GTP, guanosine triphosphate; GDP, guanosine diphosphate; MGD, molybdopterin guanine dinucleotide; MES, 4-morpholineethanesulfonic acid.
R. Sanishvili, unpublished results.
REFERENCES
- 1.Hille R. Essays Biochem. 1999;34:125–137. doi: 10.1042/bse0340125. [DOI] [PubMed] [Google Scholar]
- 2.Romao MJ, Knablein J, Huber R, Moura JJ. Prog. Biophys. Mol. Biol. 1997;68:121–144. doi: 10.1016/s0079-6107(97)00022-9. [DOI] [PubMed] [Google Scholar]
- 3.Rajagopalan KV, Johnson JL. J. Biol. Chem. 1992;267:10199–10202. [PubMed] [Google Scholar]
- 4.Rajagopalan KV. Biochem. Soc. Trans. 1997;25:757–761. doi: 10.1042/bst0250757. [DOI] [PubMed] [Google Scholar]
- 5.Rajagopalan KV. Adv. Enzymol Relat. Areas Mol. Biol. 1991;64:215–290. doi: 10.1002/9780470123102.ch5. [DOI] [PubMed] [Google Scholar]
- 6.Rothery RA, Blasco F, Magalon A, Weiner JH. J. Mol. Microbiol. Biotechnol. 2001;3:273–283. [PubMed] [Google Scholar]
- 7.Boyington JC, Gladyshev VN, Khangulov SV, Stadtman TC, Sun PD. Science. 1997;275:1305–1308. doi: 10.1126/science.275.5304.1305. [DOI] [PubMed] [Google Scholar]
- 8.Rajagopalan KV. In: Escherichia coli and Salmonella. Neidhardt FC, editor. ASM Press; Washington D.C.: 1996. pp. 674–679. [Google Scholar]
- 9.Palmer T, Vasishta A, Whitty PW, Boxer DH. Eur. J. Biochem. 1994;222:687–692. doi: 10.1111/j.1432-1033.1994.tb18913.x. [DOI] [PubMed] [Google Scholar]
- 10.Stevenson CE, Sargent F, Buchanan G, Palmer T, Lawson DM. Structure Fold. Des. 2000;8:1115–1125. doi: 10.1016/s0969-2126(00)00518-9. [DOI] [PubMed] [Google Scholar]
- 11.Lake MW, Temple CA, Rajagopalan KV, Schindelin H. J. Biol. Chem. 2000;275:40211–40217. doi: 10.1074/jbc.M007406200. [DOI] [PubMed] [Google Scholar]
- 12.Sola M, Kneussel M, Heck IS, Betz H, Weissenhorn W. J. Biol. Chem. 2001;276:25294–25301. doi: 10.1074/jbc.M101923200. [DOI] [PubMed] [Google Scholar]
- 13.Ramming M, Kins S, Werner N, Hermann A, Betz H, Kirsch J. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10266–10271. doi: 10.1073/pnas.97.18.10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamdar KP, Primus JP, Shelton ME, Archangeli LL, Wittle AE, Finnerty V. Biochem. Soc. Trans. 1997;25:778–783. doi: 10.1042/bst0250778. [DOI] [PubMed] [Google Scholar]
- 15.Stallmeyer B, Nerlich A, Schiemann J, Brinkmann H, Mendel RR. Plant J. 1995;8:751–762. doi: 10.1046/j.1365-313x.1995.08050751.x. [DOI] [PubMed] [Google Scholar]
- 16.Kozmin SG, Pavlov YI, Dunn RL, Schaaper RM. J. Bacteriol. 2000;182:3361–3367. doi: 10.1128/jb.182.12.3361-3367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wuebbens MM, Liu MT, Rajagopalan K, Schindelin H. Structure Fold. Des. 2000;8:709–718. doi: 10.1016/s0969-2126(00)00157-x. [DOI] [PubMed] [Google Scholar]
- 18.Reiss J. Hum. Genet. 2000;106:157–163. doi: 10.1007/s004390051023. [DOI] [PubMed] [Google Scholar]
- 19.Liu MT, Wuebbens MM, Rajagopalan KV, Schindelin H. J. Biol. Chem. 2000;275:1814–1822. doi: 10.1074/jbc.275.3.1814. [DOI] [PubMed] [Google Scholar]
- 20.McLuskey K, Harrison JA, Schuettelkopf AW, Boxer DH, Hunter WN. J. Biol. Chem. 2003;278:23706–23713. doi: 10.1074/jbc.M301485200. [DOI] [PubMed] [Google Scholar]
- 21.Xiang S, Nichols J, Rajagopalan KV, Schindelin H. Structure (Lond.) 2001;9:299–310. doi: 10.1016/s0969-2126(01)00588-3. [DOI] [PubMed] [Google Scholar]
- 22.Schrag JD, Huang W, Sivaraman J, Smith C, Plamondon J, Larocque R, Matte A, Cygler M. J. Mol. Biol. 2001;310:419–431. doi: 10.1006/jmbi.2001.4771. [DOI] [PubMed] [Google Scholar]
- 23.Rudolph MJ, Wuebbens MM, Rajagopalan KV, Schindelin H. Nat. Struct. Biol. 2001;8:42–46. doi: 10.1038/83034. [DOI] [PubMed] [Google Scholar]
- 24.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang RG, Skarina T, Katz JE, Beasley S, Khachatryan A, Vyas S, Arrowsmith CH, Clarke S, Edwards A, Joachimiak A, Savchenko A. Structure (Lond.) 2001;9:1095–1106. doi: 10.1016/s0969-2126(01)00675-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matthews BW. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 27.Evans G, Pettifer RF. J. Appl. Crystallogr. 2001;34:8286. [Google Scholar]
- 28.Westbrook EM, Naday I. Methods Enzymol. 1997;276:244–268. [PubMed] [Google Scholar]
- 29.Pflugrath JW. Acta Crystallogr. D. Biol. Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Scheffers DJ, Driessen AJ. Mol. Microbiol. 2002;43:1517–1521. doi: 10.1046/j.1365-2958.2002.02828.x. [DOI] [PubMed] [Google Scholar]
- 32.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Perrakis A, Morris R, Lamzin VS. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. D. Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr. Sect. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 36.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holm L, Sander C. Proteins. 1994;19:165–173. doi: 10.1002/prot.340190302. [DOI] [PubMed] [Google Scholar]
- 38.Schwarz G, Schrader N, Mendel RR, Hecht HJ, Schindelin H. J. Mol. Biol. 2001;312:405–418. doi: 10.1006/jmbi.2001.4952. [DOI] [PubMed] [Google Scholar]
- 39.Schwarz G, Boxer DH, Mendel RR. J. Biol. Chem. 1997;272:26811–26814. doi: 10.1074/jbc.272.43.26811. [DOI] [PubMed] [Google Scholar]
- 40.Magalon A, Frixon C, Pommier J, Giordano G, Blasco F. J. Biol. Chem. 2002;277:48199–48204. doi: 10.1074/jbc.M205806200. [DOI] [PubMed] [Google Scholar]
- 41.Rubio LM, Flores E, Herrero A. J. Bacteriol. 1998;180:1200–1206. doi: 10.1128/jb.180.5.1200-1206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicholls A, Sharp KA, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 43.Pitterle DM, Rajagopalan KV. J. Biol. Chem. 1993;268:13499–13505. [PubMed] [Google Scholar]
- 44.Leimkuhler S, Rajagopalan KV. J. Biol. Chem. 2001;4:4. doi: 10.1074/jbc.M102072200. [DOI] [PubMed] [Google Scholar]
- 45.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]