Abstract
North African gundis (Ctenodactylus gundi) were trapped in the Leishmania (L.) tropica focus of cutaneous leishmaniasis, situated in southeast Tunisia and evaluated for Leishmania infection by real-time kinetoplast DNA polymerase chain reaction (PCR). Species identification was performed by internal transcribed spacer one (ITS1)-PCR-restriction fragment length polymorphism (RFLP) and high-resolution melting (HRM) analysis of the 7SL RNA gene. Real-time PCR on blood was positive in 6 of 13 (46.2%) tested gundis. Leishmania tropica was identified in five infected gundis and Leishmania major in one specimen. Alignments of the ITS-1 DNA sequences and 7S-HRM curves analysis indicated that similar genotypes were present in humans, a sandfly, and gundis from the same region suggesting a potential role of this rodent as reservoir host of L. tropica in southeast Tunisia.
Introduction
Leishmania (L.) tropica is widely distributed in north and east Africa, the Middle East, and large parts of Asia. It is a genetically heterogeneous species including multiple genotypes.1 Cutaneous leishmaniasis (CL) caused by L. tropica is usually considered as an anthroponosis and Phlebotomus (P.) sergenti the acting vector.2 However, the occurrence of sporadic human cases in some foci suggests the concomitant presence of zoonotic transmission.3 The rock hyrax is the proven reservoir and its reservoir role was confirmed even experimentally.3–6 This animal species is found throughout sub-Saharan Africa and north-east Africa, being discontinuously distributed from Senegal through southern Algeria, Libya, and Egypt to central and southern Africa. It also extends to the Arabian Peninsula and to the Middle-East, but is not found in Tunisia. Rattus rattus is the suspected rodent reservoir of L. tropica as asymptomatic rats were infectious for P. sergenti many months after L. tropica inoculation.7
The CL caused by L. tropica MON-8 (syn Leishmania killicki) is endemic in the southeast of Tunisia.8 It occurs as scarce scattered cases and prevails in communities living in the rocky mountainside of the Tataouine region.8 Recently, new foci have emerged in the Central part of the country.9 Until now, little data concerning the transmission routes of CL caused by L. tropica in Tunisia have been available. The relative paucity of CL cases and their spatial distribution excludes the anthroponotic character of the disease and suggests that CL caused by L. tropica might be a zoonosis8,9; the putative reservoir host is the North African gundi (Ctenodactylus gundi). In fact, this rodent is extremely abundant in all Tunisian L. tropica foci, where it is found in natural and peri-domestic environments.8–10 On the other hand, natural infection of P. sergenti with Leishmania promastigotes was reported in the CL focus in southeastern Tunisia.11 Likewise, P. sergenti has been recently identified as the L. tropica host in a sylvatic site populated by gundis (Massoutiera mzabi) of a neighboring region, namely Ghardaïa, south Algeria.12
The purpose of this study was to assess the Leishmania infection rate in North African gundis of the Tataouine region, to identify Leishmania species infecting this wild rodent, and to compare the genetic relatedness between parasites characterized in gundis, humans, and sandflies of the same region.
Materials and Methods
Study area.
The study was carried out in the surrounding of the village of Ghomrassen, governorate of Tataouine, southeast Tunisia. This village is situated in a desert region presenting arid rock outcrops. It is built on a rocky mountainside at moderate altitude (300 m a.s.l.) and is a well-known focus of L. tropica CL.8 Ctenodactylus gundi is the most prevalent wild rodent living in the stony mountains around the village.
Sample collection.
Thirteen gundis were live-trapped in October 2009 in natural sites located 6 km from the village, in the rocky mountain at c. 310 m a.s.l. The gundis were manually caught by the local population during the twilight hours near the entrances of caves and crevices inhabited by the rodent and then kept alive in cages. Animals were physically examined. Blood samples were extracted from the 13 gundis into tubes containing EDTA. Biopsies were performed only on three rodents. Nasal biopsies were taken from the three animals, whereas a piece of spleen was taken from only one. Biopsies were stored in a sterile microtube (Deltalab S.L., Spain) at –80°C until DNA extraction. The study was conducted after the authorization of the Ministry of Agriculture and Environment, Forest directory, Tunisia.
Polymerase chain reaction (PCR) assays.
The DNA was extracted from 100 μL of blood using the QIAamp DNA mini kit (Qiagen, Hilden, Germany) as recommended by the manufacturer. The DNA extraction from tissue homogenates was performed using the DNeasy Tissue Kit (Qiagen) according to the manufacturer's instructions. All samples were analyzed by kinetoplast DNA (kDNA) real-time PCR13 and internal transcribed spacer 1 (ITS1) PCR,14 and if positive, species were identified by ITS1-restriction fragment length polymorphism (RFLP),14 ITS 1 sequencing, and high-resolution melting (HRM) analysis of the 7SL RNA gene.15
Briefly, kDNA real-time PCR was conducted in a final volume of 25 μL by using a TaqMan universal master mixture (Roche, Palo Alto, CA) containing 100 μM of direct primer (5′-CTTTTCTGGTCCTCCGGGTAGG-3′), 100 μM of reverse primer (5′-CCACCCGGCCCTATTTTACACCAA-3′), 50 μM of probe (FAM-5′-TTTTCGCAGAACGCCCCTACCCGC-3′-TAMRA), and 1 μL of DNA extract.13 The DNA was amplified in an ABI PRISM 7700 sequence detector (Applied Biosystems, Foster City, CA) for 50 cycles at 95 and 60°C. Each sample was tested in duplicate, and each run included both positive and negative controls. Samples were checked for inhibition by being retested at a 1:10 dilution and by spiking these samples with 1 μL of the positive control. A standard curve, plotted from a dilution series of Leishmania DNA (extracted from 106 Leishmania infantum promastigotes), allowed parasite quantification by PCR. The volume or the weight of collected samples and the dilution rates applied during DNA extraction and amplification were taken into account to estimate the parasite load in each collected specimen.
Concerning ITS1-PCR, LITSR and L5.8S primers were used to amplify the genes encoding the small subunit and 5.8S rRNA.14 The positive ITS1-PCR products were digested by the HaeIII restriction enzyme following the protocol described earlier.14 The ITS1 products were also purified and sequenced in both directions with the forward and reverse primers on an ABI Prism 377 DNA sequencer (Applied Biosystems) according to the manufacturer's instructions. The consensus sequences were deposited in GenBank and compared with nucleotide sequences of 12 L. tropica complex strains and 5 Leishmania major strains. These 17 reference strains were isolated in the study site (three isolates from humans and one from P. sergenti) and in other geographical regions. A phylogenic tree was generated using MEGA5 software. Relationships were inferred based on genetic distances by the neighbor-joining option. The evolutionary distances were computed using the Kimura 2-parameter method with L. mexicana as an out-group and 2,000 bootstraps.
The 7SL-HRM PCR assay using the primers CJ7SLF (5′-ACG TGG ACC AGC GAG GGT-3′) and QRT7SLR (5′-CGG TTC CCT CGC TTC AAC-3′) was performed in 20 μL, final volume of the PCR mix, containing 2× SsoFast EvaGreen supermix (Bio-Rad), 0.5 μM of each primer, and 2 μL of DNA extract.15 Amplification conditions were as follows: 15 min of denaturation at 95°C, followed by 40 cycles of denaturation for 5 s at 95°C; annealing for 30 s at 63°C; and a final extension for 2 min at 72°C. The HRM ramping was carried out at 0.2°C/s from 65 to 95°C. The HRM PCR and analysis were performed by a CFX96 real-time PCR detection system (Bio-Rad) and the Precision Melt Analysis Software (Bio-Rad). Positive and negative control reactions were included in each experiment. The PCR efficiency was evaluated using the threshold cycle (Cq), and a normalized melt window, 88 to 90, allowed the analysis of HRM curves. The sensitivity, specificity, and discrimination between L. major and L. tropica species were analyzed according to strains promastigotes, isolated from human CL cases living in the same region, and typed by iso-enzyme analysis. Sensitivity was determined using 10-fold dilutions of each DNA (data not shown).
Results
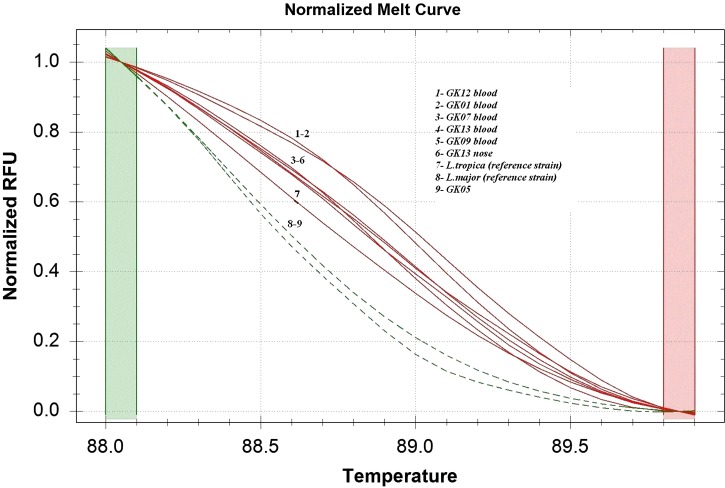
All trapped gundis have no lesions. Using kDNA real-time PCR, 6 of the 13 gundis had Leishmania DNA in their blood (46.2%). Among these six positive rodents, spleen and/or nose biopsies, taken from two specimens, were positive (Table 1). Samples quantification yielded low parasite loads, ranging from 0.5 to 5.6 parasites per 100 μL of blood samples and 6.7 to 9.4 parasites per 100 mg of nose and spleen tissues. All these samples were also found positive using 7SL-HRM, allowing the species identification (Table 1). The average melting temperatures (Tm) ± SD for 40 7SL-HRM runs were as follows: L. major, 88 ± 0.04; L. tropica, 89 ± 0.02. The HRM analysis typed correctly eight samples as L. tropica, and one as L. major compared with reference strains (Figure 1, Table 1).
Table 1.
Diagnosis of Leishmania infection in gundis by different PCR assays
| Specimen | Sample | Parasite load* | ITS PCR | ITS PCR RFLP | Sequencing | 7SL HRM PCR |
|---|---|---|---|---|---|---|
| GK 01 | Blood | 0.72 | + | L. tropica | JF719996 | L. tropica |
| Nose | 6.8 | + | NO | L. tropica | ||
| GK 05 | Blood | 5.6 | + | L. major | JN242001 | L. major |
| GK 07 | Blood | 0.65 | + | NO | L. tropica | |
| GK 09 | Blood | 0.66 | + | NO | L. tropica | |
| GK 12 | Blood | 0.5 | − | NO | L. tropica | |
| GK 13 | Blood | 0.9 | + | L. tropica | L. tropica | |
| Nose | 6.78 | + | L. tropica | JF719995 | L. tropica | |
| Spleen | 9.4 | + | NO | L. tropica |
Number of parasites per 100 μL of blood or per 100 mg of tissue (nose and spleen).
PCR = polymerase chain reaction; ITS = internal transcribed spacer; HRM = high-resolution melting; NO = non observed.
Figure 1.
Comparison of normalized high-resolution melting (HRM) curves of the 7SL polymerase chain reaction (PCR) amplicon obtained for Tunisian Leishmania references species (Leishmania tropica MON8 and Leishmania major) and seven gundi samples (GK01, GK05, GK07, GK09, GK12, GK13 blood and GK13 nose).
Using ITS1 PCR, 5 out of 13 gundis had Leishmania DNA in blood (38%). Among these five positive rodents, spleen and/or nose biopsies, taken from two specimens, were positive. Among the five positive blood samples, only three had successful species identification by sequencing and/or RFLP analysis. However, among the three tissue specimens only one had successful species identification (Table 1).
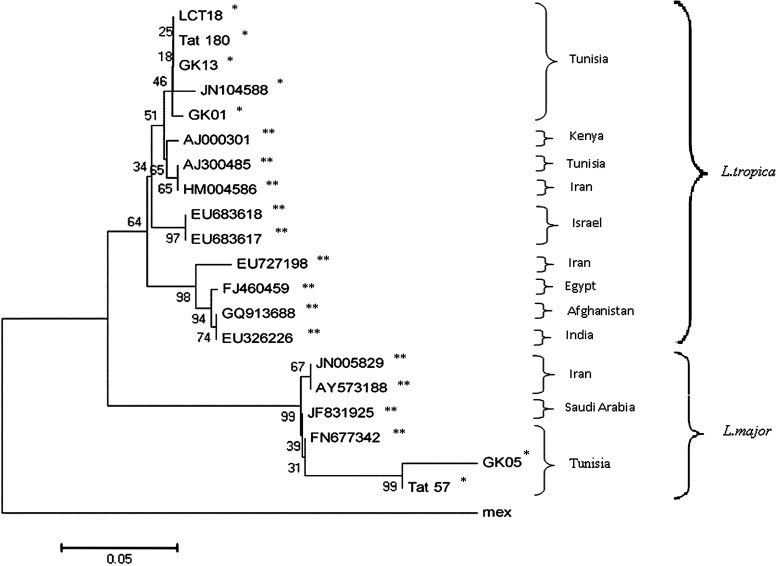
Species characterization of all samples correlated 100% using the three assays (Table 1). Three samples infected by L. tropica or L. major parasites from three different infected gundis were analyzed by the ITS1 sequencing. Phylogenic analysis showed that ITS1 DNA sequences of L. tropica parasites isolated from gundis were highly similar to those isolated from human CL cases and the incriminated peri-domestic vector P. sergenti of the same region (Figure 2). On the other hand, sequences of L. major parasites isolated from one gundi were also highly similar to those isolated from human CL cases of the Tataouine region (Figure 2).
Figure 2.
Phylogenic analysis of internal transcribed spacer one (ITS1) sequences from human, gundi, and Phlebotomus sergenti (Tree obtained by the method of neighbor-joining, Leishmania mexicana as an out-group, bootstrap values based on 2,000 replicates). *Our samples: GK13, GK01, and GK05 were from gundis, LCT18, Tat180, and Tat57 were from humans and JN104588 was from Phlebotomus sergenti. **Nucleotide sequences from GenBank.
Discussion
Ctenodactylus gundi ranges from eastern Morocco, through Algeria and Tunisia, to western Libya. It is found in deserts with arid outcrops where it shelters in fissures and rock crevices, and under rock formations. In Tunisia, North African gundis are extremely common in the L. tropica foci in the Tataouine region, where they inhabit crevices within the rocky mountainside surrounding the villages. It was shown in previous studies that this habitat affords suitable breeding sites for sandflies.10 Thus, as gregarious diurnal mammals, sleeping gundis are an easy blood source available for night-questing phlebotomine females. Furthermore, gundis live for 5–6 years in the wild and may survive through any non-transmission season. Accordingly, they have all the behavioral and ecological requirements of good reservoir hosts for L. tropica CL in southeast Tunisia.16
The detection of Leishmania in naturally infected rodents in endemic areas is of utmost importance to involve these rodents as reservoir hosts.16 To our knowledge, this is the first study that implements quantitative PCR for detection of Leishmania infection in North African gundis. Our results revealed a high rate of L. tropica infection in asymptomatic animals (38.5%). Interestingly, the transmission of L. tropica from black rats without any visible cutaneous changes was previously reported and suggested that an asymptomatic host would be of great epidemiological significance in field conditions.7 On the other hand, the low parasitic load observed in all specimens (a mean of 0.69 parasites per 100 μL of blood) was already reported in hyraxes.4 This may explain the difficulty of parasite isolation by culture (data not shown).
The predilection sites of Leishmania may differ according to reservoir host.6 Indeed, in hyraxes, L. tropica is usually found in the skin above the nose where sandfly vectors prefer to feed.3,6 The presence of L. tropica DNA in nose biopsies of infected gundis suggests that the nose may correspond to a potential feeding site for local sandfly vectors. On the other hand and as reported for hyraxes, the presence of L. tropica DNA in gundi blood demonstrated in the current study could be caused by either the circulation of live amastigotes present in phagocytes or to degraded parasites.4 The Leishmania DNA positivity observed in one spleen sample suggests that L. tropica infection may visceralize. However, the possibility of infection involving visceral organs and circulatory blood dissemination in gundis requires substantiation in further studies.
On the other hand, sequence analysis of ITS1 fragments showed perfect correlation with L. tropica strains isolated in human CL cases and in P. sergenti of the same region. Therefore, Ctenodactylus gundi could be considered, as a potential reservoir of L. tropica in this region. However, further studies must be conducted to show that gundis are experimentally susceptible to L. tropica. Moreover, infected gundis should be confirmed as an infective feeding for potential vectors, namely Phlebotomus riouxi and P. sergenti.10,11
Recently, zoonotic CL caused by Leishmania (L.) major has emerged likewise in the Tataouine region. A previous study showed that cases of L. major are clustered at the margin of some villages where the gerbil reservoir hosts (Meriones species) are widespread.8 The presence of L. major infection, with relatively high parasitic load, in one infected gundi stresses the need of further experimental investigation in this wild rodent. However, it is highly probable that these animals are not reservoir hosts, and they might be just considered as incidental hosts because the infection is scarce and the relative abundance of the proven vector of L. major and Phlebotomus papatasi in gundis' habitats is low.10
ACKNOWLEDGMENTS
We are grateful to Mohamed Raouane and his staff of the Regional Directory of Public Health of Tataouine for their contribution to the achievement of this work. We also thank Souha Benaberrazak and Mehdi Driss from Pasteur Institute of Tunis for their help in phylogenic analysis.
Footnotes
Financial support: This study was supported by the network of Pasteur Institutes (Actions Concertées Inter-Pasteuriennes, ACIP A 04 2007).
Authors' addresses: Nadia Bousslimi, Soumaya Ben-Ayed, Imène Ben-Abda, Karim Aoun, and Aïda Bouratbine, Laboratoire de Parasitologie, Institut Pasteur de Tunis, Tunis Belvédère, Tunisia, E-mails: nguetari@gmail.com, Soumayatba@yahoo.fr, benabda.imen@gmail.com, karim.aoun@pasteur.rns.tn, and aida.bouratbine@pasteur.rns.tn.
References
- 1.Schwenkenbecher JM, Wirth T, Schnur LF, Jaffe CL, Schallig H, Al-Jawabreh A, Hamarsheh O, Azmi K, Pratlong F, Schönian G. Microsatellite analysis reveals genetic structure of Leishmania tropica. Int J Parasitol. 2006;36:237–246. doi: 10.1016/j.ijpara.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 2.WHO The leishmaniasis. Report of a WHO Expert Committee. World Health Organ Tech Rep Ser. 1984;701:1–140. [PubMed] [Google Scholar]
- 3.Svobodova M, Votypka J, Peckova J, Dvorak V, Nasereddin A, Baneth G, Sztern J, Kravchenko V, Orr A, Meir D, Schnur LF, Volf P, Warburg A. Distinct transmission cycles of Leishmania tropica in 2 adjacent foci, northern Israel. Emerg Infect Dis. 2006;12:1860–1868. doi: 10.3201/eid1212.060497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talmi-Frank D, Jaffe CL, Nasereddin A, Warburg A, King R, Svobodova M, Peleg O, Baneth G. Leishmania tropica in rock hyraxes (Procavia capensis) in a focus of human cutaneous leishmaniasis. Am J Trop Med Hyg. 2010;82:814–818. doi: 10.4269/ajtmh.2010.09-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sang DK, Njeru WK, Ashford RW. A zoonotic focus of cutaneous leishmaniasis due to Leishmania tropica at Utut, Fift Valley Province, Kenya. Trans R Soc Trop Med Hyg. 1994;88:35–37. doi: 10.1016/0035-9203(94)90486-3. [DOI] [PubMed] [Google Scholar]
- 6.Svobodová M, Volf P, Votýpka J. Experimental transmission of Leishmania tropica to hyraxes (Procavia capensis) by the bite of Phlebotomus arabicus. Microbes Infect. 2006;8:1691–1694. doi: 10.1016/j.micinf.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 7.Svobodová M, Votýpka J, Nicolas L, Volf P. Leishmania tropica in the black rat (Rattus rattus): persistence and transmission from asymptomatic host to sand fly vector Phlebotomus sergenti. Microbes Infect. 2003;5:361–364. doi: 10.1016/s1286-4579(03)00046-7. [DOI] [PubMed] [Google Scholar]
- 8.Bousslimi N, Aoun K, Ben-Abda I, Ben-Alaya-Bouafif N, Raouane M, Bouratbine A. Epidemiologic and clinical features of cutaneous leishmaniasis in southeastern Tunisia. Am J Trop Med Hyg. 2010;83:1034–1039. doi: 10.4269/ajtmh.2010.10-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouratbine A, Aoun K, Ghrab J, Harrat Z, Ezzedini MS, Etlijani S. Spread of Leishmania killicki to central and south-west Tunisia. Parasite. 2005;12:59–63. doi: 10.1051/parasite/2005121059. [DOI] [PubMed] [Google Scholar]
- 10.Tabbabi A, Ghrab J, Aoun K, Ready PD, Bouratbine A. Habitats of the sandfly vectors of Leishmania tropica and L. major in a mixed focus of cutaneous leishmaniasis in southeast Tunisia. Acta Trop. 2011;119:131–137. doi: 10.1016/j.actatropica.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Tabbabi A, Bousslimi N, Rhim A, Aoun K, Bouratbine A. First report on natural infection of Phlebotomus sergenti with Leishmania promastigotes in the cutaneous leishmaniasis focus in southeastern Tunisia. Am J Trop Med Hyg. 2011;85:646–647. doi: 10.4269/ajtmh.2011.10-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boubidi SC, Benallal K, Boudrissa A, Bouiba L, Bouchareb B, Garni R, Bouratbine A, Ravel C, Dvorak V, Votypka J, Volf P, Harrat Z. Phlebotomus sergenti (Parrot, 1917) identified as Leishmania killicki host in Ghardaïa, south Algeria. Microbes Infect. 2011;13:691–696. doi: 10.1016/j.micinf.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Mary C, Faraut F, Lascombe L, Dumon H. Quantification of Leishmania infantum DNA by a real-time PCR assay with high sensitivity. J Clin Microbiol. 2004;42:5249–5255. doi: 10.1128/JCM.42.11.5249-5255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schonian G, Nasereddin A, Dinse N, Schweynoch C, Schallig HD, Presber W, Jaffe CL. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis. 2003;47:349–358. doi: 10.1016/s0732-8893(03)00093-2. [DOI] [PubMed] [Google Scholar]
- 15.Nasereddin A, Jaffe CL. Rapid diagnosis of Old World leishmaniasis by high-resolution melting analysis of the 7SL RNA gene. J Clin Microbiol. 2010;48:2240–2242. doi: 10.1128/JCM.00553-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashford RW. Leishmaniasis reservoirs and their significance in control. Clin Dermatol. 1996;14:523–532. doi: 10.1016/0738-081x(96)00041-7. [DOI] [PubMed] [Google Scholar]