Abstract
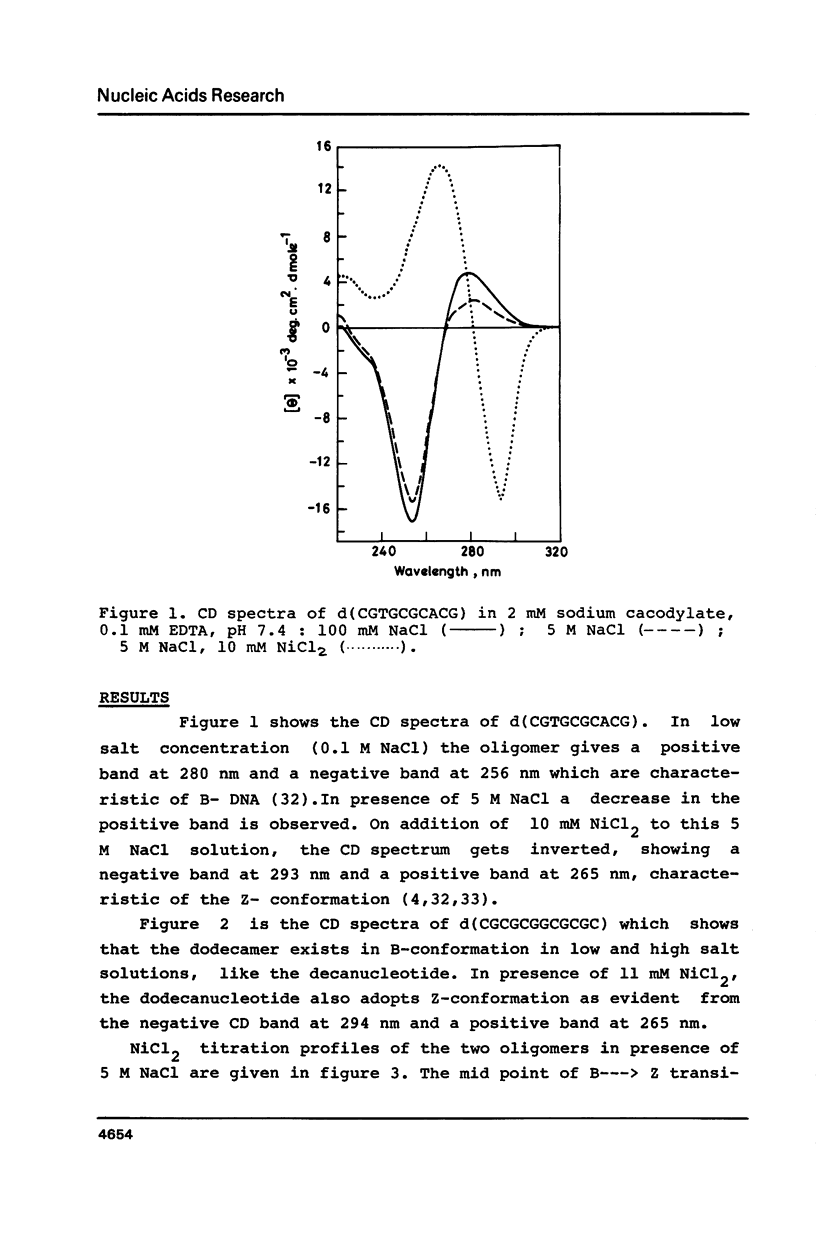
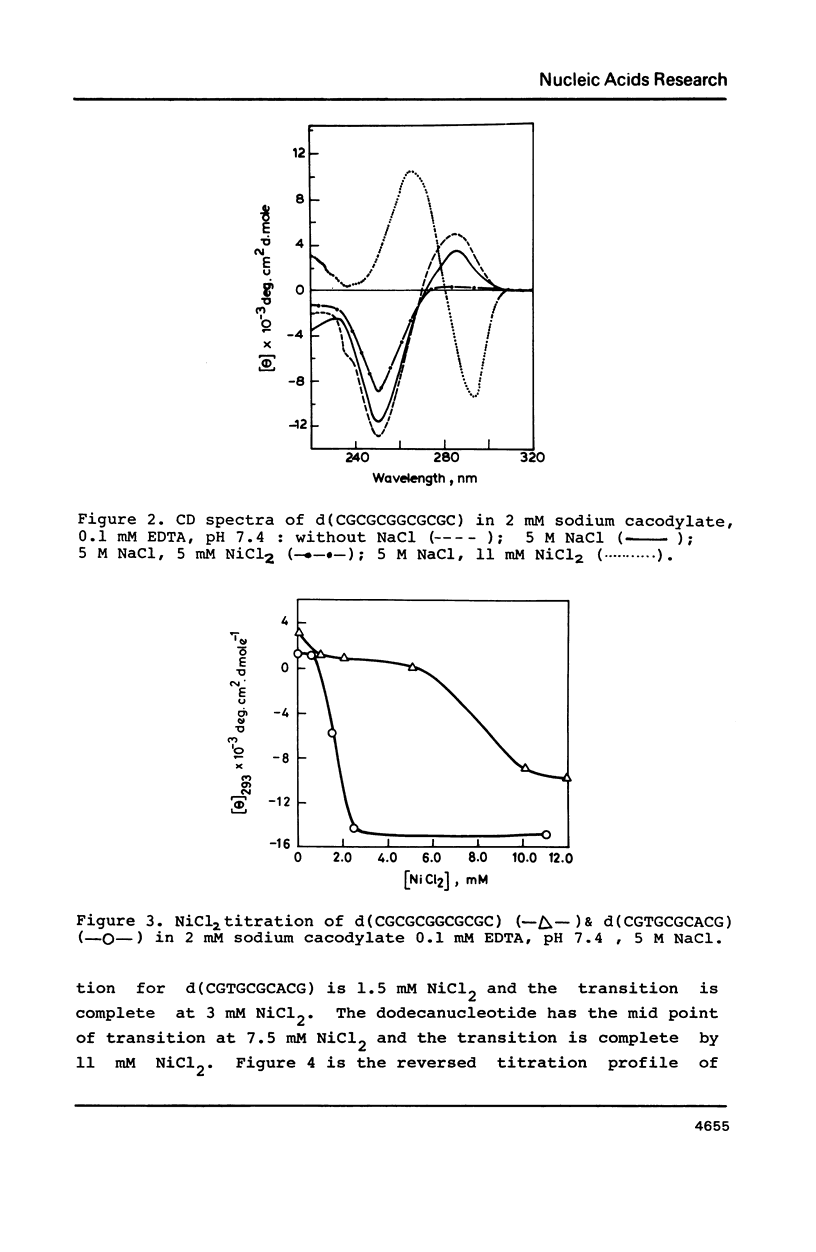
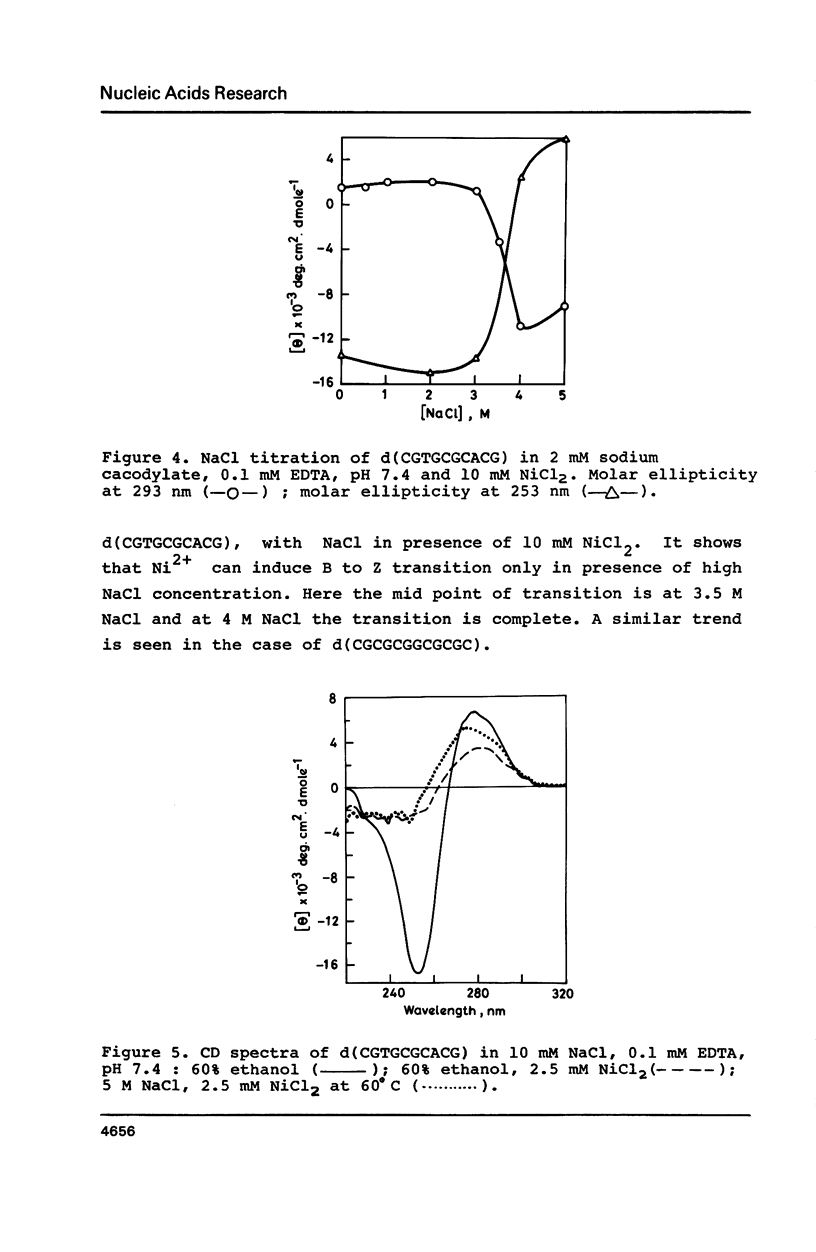
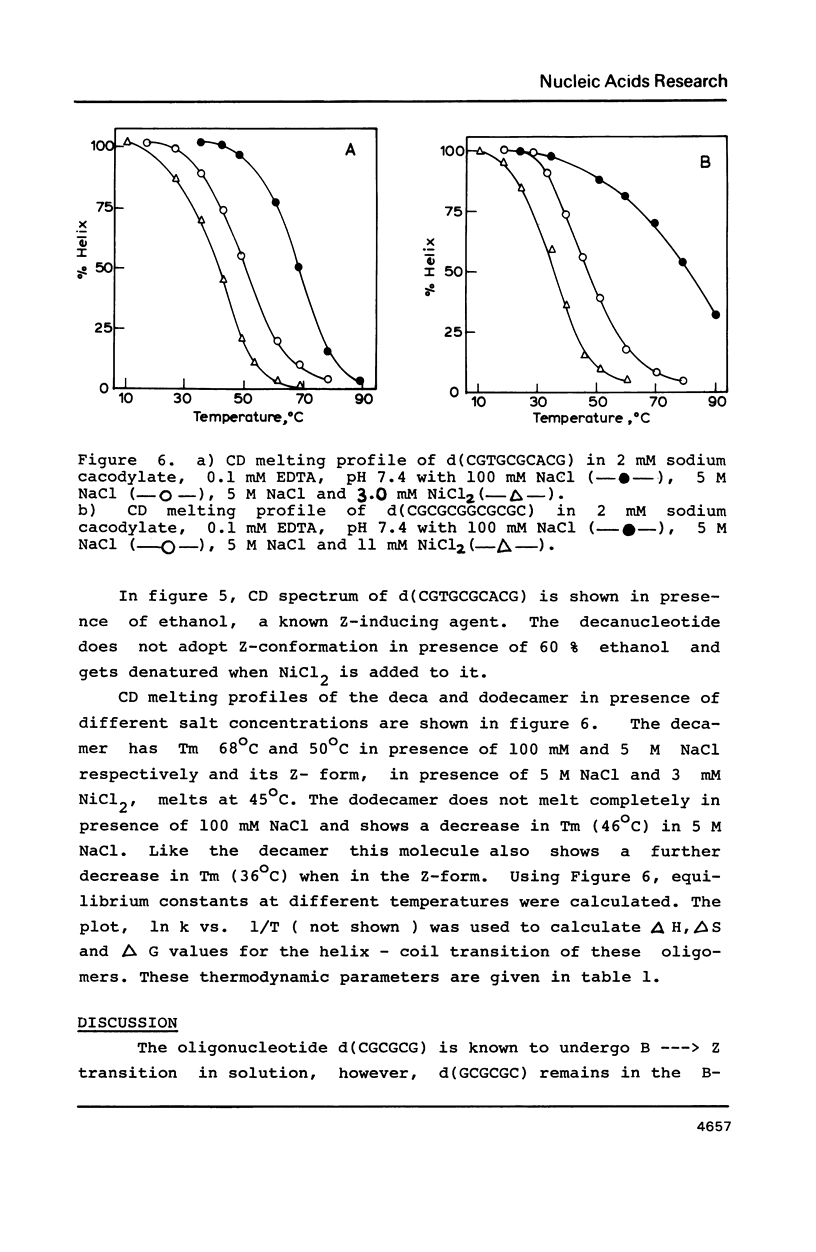
Oligonucleotides containing alternating purines-pyrimidines with AT base pairs have been shown to exist in the Z-form preferably in solid state. We report that oligodeoxyribonucleotides with GG, TG and CA interruptions in their alternating CG sequences can undergo B to Z transition in solution in the absence of any chemical modification or topological constraint. The sequences, d(CGCGCGGCGCGC) and d(CGTGCGCACG) have been synthesised and shown to adopt Z- conformation in presence of millimolar concentrations of Ni2+ under low water activity conditions. Significance of GG, TG and CA interruptions in the B to Z transition is discussed.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albergo D. D., Marky L. A., Breslauer K. J., Turner D. H. Thermodynamics of (dG--dC)3 double-helix formation in water and deuterium oxide. Biochemistry. 1981 Mar 17;20(6):1409–1413. doi: 10.1021/bi00509a001. [DOI] [PubMed] [Google Scholar]
- Brahmachari S. K., Shouche Y. S., Cantor C. R., McClelland M. Sequences that adopt non-B-DNA conformation in form V DNA as probed by enzymic methylation. J Mol Biol. 1987 Jan 5;193(1):201–211. doi: 10.1016/0022-2836(87)90637-1. [DOI] [PubMed] [Google Scholar]
- Butzow J. J., Shin Y. A., Eichhorn G. L. Effect of template conversion from the B to the Z conformation on RNA polymerase activity. Biochemistry. 1984 Oct 9;23(21):4837–4843. doi: 10.1021/bi00316a004. [DOI] [PubMed] [Google Scholar]
- Chatterji D., Wu C. W., Wu F. Y. Nuclear magnetic resonance studies on the role of intrinsic metals in Escherichia coli RNA polymerase. Effect of DNA template on the nucleotide-enzyme interaction. J Biol Chem. 1984 Jan 10;259(1):284–289. [PubMed] [Google Scholar]
- Delseny M., Laroche M., Penon P. Detection of sequences with Z-DNA forming potential in higher plants. Biochem Biophys Res Commun. 1983 Oct 14;116(1):113–120. doi: 10.1016/0006-291x(83)90388-1. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Conformation and dynamics in a Z-DNA tetramer. J Mol Biol. 1981 Nov 15;152(4):723–736. doi: 10.1016/0022-2836(81)90124-8. [DOI] [PubMed] [Google Scholar]
- Drew H., Takano T., Tanaka S., Itakura K., Dickerson R. E. High-salt d(CpGpCpG), a left-handed Z' DNA double helix. Nature. 1980 Aug 7;286(5773):567–573. doi: 10.1038/286567a0. [DOI] [PubMed] [Google Scholar]
- Fujii S., Wang A. H., Quigley G. J., Westerink H., Van der Marel G., Van Boom J. H., Rich A. The octamers d(CGCGCGCG) and d(CGCATGCG) both crystallize as Z-DNA in the same hexagonal lattice. Biopolymers. 1985 Jan;24(1):243–250. doi: 10.1002/bip.360240118. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987 Jul 20;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Garner M. M., Felsenfeld G. Effect of Z-DNA on nucleosome placement. J Mol Biol. 1987 Aug 5;196(3):581–590. doi: 10.1016/0022-2836(87)90034-9. [DOI] [PubMed] [Google Scholar]
- Giedroc D. P., Coleman J. E. Structural and functional differences between the two intrinsic zinc ions of Escherichia coli RNA polymerase. Biochemistry. 1986 Aug 26;25(17):4969–4978. doi: 10.1021/bi00365a037. [DOI] [PubMed] [Google Scholar]
- Gilmour R. S., Spandidos D. A., Vass J. K., Gow J. W., Paul J. A negative regulatory sequence near the mouse beta-maj globin gene associated with a region of potential Z-DNA. EMBO J. 1984 Jun;3(6):1263–1272. doi: 10.1002/j.1460-2075.1984.tb01961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Kakunaga T. Potential Z-DNA forming sequences are highly dispersed in the human genome. Nature. 1982 Jul 22;298(5872):396–398. doi: 10.1038/298396a0. [DOI] [PubMed] [Google Scholar]
- Hayes T. E., Dixon J. E. Z-DNA in the rat somatostatin gene. J Biol Chem. 1985 Jul 5;260(13):8145–8156. [PubMed] [Google Scholar]
- Ho P. S., Frederick C. A., Quigley G. J., van der Marel G. A., van Boom J. H., Wang A. H., Rich A. G.T wobble base-pairing in Z-DNA at 1.0 A atomic resolution: the crystal structure of d(CGCGTG). EMBO J. 1985 Dec 16;4(13A):3617–3623. doi: 10.1002/j.1460-2075.1985.tb04125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski A., Hsieh W. T., Blaho J. A., Larson J. E., Wells R. D. Left-handed DNA in vivo. Science. 1987 Nov 6;238(4828):773–777. doi: 10.1126/science.3313728. [DOI] [PubMed] [Google Scholar]
- Klysik J., Stirdivant S. M., Singleton C. K., Zacharias W., Wells R. D. Effects of 5 cytosine methylation on the B-Z transition in DNA restriction fragments and recombinant plasmids. J Mol Biol. 1983 Jul 25;168(1):51–71. doi: 10.1016/s0022-2836(83)80322-2. [DOI] [PubMed] [Google Scholar]
- Novotný J., Auffray C. A program for prediction of protein secondary structure from nucleotide sequence data: application to histocompatibility antigens. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):243–255. doi: 10.1093/nar/12.1part1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J., Nordheim A., Rich A., Wang J. C. Flipping of cloned d(pCpG)n.d(pCpG)n DNA sequences from right- to left-handed helical structure by salt, Co(III), or negative supercoiling. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4560–4564. doi: 10.1073/pnas.79.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qasba P. K., Safaya S. K. Similarity of the nucleotide sequences of rat alpha-lactalbumin and chicken lysozyme genes. Nature. 1984 Mar 22;308(5957):377–380. doi: 10.1038/308377a0. [DOI] [PubMed] [Google Scholar]
- Quadrifoglio F., Manzini G., Dinkelspiel K., Crea R. Simultaneous stability of short alternating Z and B helices in synthetic DNA concatamers. Nucleic Acids Res. 1982 Jun 25;10(12):3759–3768. doi: 10.1093/nar/10.12.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrifoglio F., Manzini G., Vasser M., Dinkelspiel K., Crea R. Conformational stability of alternating d (CG) oligomers in high salt solution. Nucleic Acids Res. 1981 May 11;9(9):2195–2206. doi: 10.1093/nar/9.9.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrifoglio F., Manzini G., Yathindra N. Short oligodeoxynucleotides with d(G-C)n sequence do not assume left-handed conformation in high salt conditions. J Mol Biol. 1984 May 25;175(3):419–423. doi: 10.1016/0022-2836(84)90358-9. [DOI] [PubMed] [Google Scholar]
- Ramesh N., Shouche Y. S., Brahmachari S. K. Recognition of B and Z forms of DNA by Escherichia coli DNA polymerase I. J Mol Biol. 1986 Aug 20;190(4):635–638. doi: 10.1016/0022-2836(86)90248-2. [DOI] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Saffer J. D., Lerman M. I. Unusual class of Alu sequences containing a potential Z-DNA segment. Mol Cell Biol. 1983 May;3(5):960–964. doi: 10.1128/mcb.3.5.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K., Klysik J., Stirdivant S. M., Wells R. D. Left-handed Z-DNA is induced by supercoiling in physiological ionic conditions. Nature. 1982 Sep 23;299(5881):312–316. doi: 10.1038/299312a0. [DOI] [PubMed] [Google Scholar]
- Stirdivant S. M., Kłysik J., Wells R. D. Energetic and structural inter-relationship between DNA supercoiling and the right- to left-handed Z helix transitions in recombinant plasmids. J Biol Chem. 1982 Sep 10;257(17):10159–10165. [PubMed] [Google Scholar]
- Taboury J. A., Adam S., Taillandier E., Neumann J. M., Tran-Dinh S., Huynh-Dinh T., Langlois d'Estaintot B., Conti M., Igolen J. The B----Z transition in two synthetic oligonucleotides: d(C-2-amino-ACGTG) and d(m5CGCAm5CGTGCG) studied by IR, NMR and CD spectroscopies. Nucleic Acids Res. 1984 Aug 10;12(15):6291–6305. doi: 10.1093/nar/12.15.6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taboury J. A., Bourtayre P., Liquier J., Taillandier E. Interaction of Z form poly(dG-dC).poly(dG-dC) with divalent metal ions: localization of the binding sites by I.R. spectroscopy. Nucleic Acids Res. 1984 May 25;12(10):4247–4258. doi: 10.1093/nar/12.10.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taboury J. A., Taillandier E. Right-handed and left-handed helices of poly(dA-dC) X (dG-dT). Nucleic Acids Res. 1985 Jun 25;13(12):4469–4483. doi: 10.1093/nar/13.12.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillandier E., Taboury J. A., Adam S., Liquier J. Left-handed helical structure of poly[d(A-C)].poly[d(G-T)] studied by infrared spectroscopy. Biochemistry. 1984 Nov 20;23(24):5703–5706. doi: 10.1021/bi00319a007. [DOI] [PubMed] [Google Scholar]
- Travers A. A. Structure and function of E. coli promoter DNA. CRC Crit Rev Biochem. 1987;22(3):181–219. doi: 10.3109/10409238709101483. [DOI] [PubMed] [Google Scholar]
- Vardimon L., Rich A. In Z-DNA the sequence G-C-G-C is neither methylated by Hha I methyltransferase nor cleaved by Hha I restriction endonuclease. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3268–3272. doi: 10.1073/pnas.81.11.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visentin D., Harley C. B. Generation and statistical mechanical modeling of Z-DNA in the mouse metallothionein I promoter. Biochemistry. 1987 Oct 20;26(21):6578–6583. doi: 10.1021/bi00395a002. [DOI] [PubMed] [Google Scholar]
- Walmsley R. M., Szostak J. W., Petes T. D. Is there left-handed DNA at the ends of yeast chromosomes? Nature. 1983 Mar 3;302(5903):84–86. doi: 10.1038/302084a0. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Hakoshima T., van der Marel G., van Boom J. H., Rich A. AT base pairs are less stable than GC base pairs in Z-DNA: the crystal structure of d(m5CGTAm5CG). Cell. 1984 May;37(1):321–331. doi: 10.1016/0092-8674(84)90328-3. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wang Y., Thomas G. A., Peticolas W. L. Sequence dependence of the B to Z transition in crystals and aqueous NaCl solutions for deoxyoligonucleotides containing all four canonical DNA bases. Biochemistry. 1987 Aug 11;26(16):5178–5186. doi: 10.1021/bi00390a042. [DOI] [PubMed] [Google Scholar]
- Zacharias W., Larson J. E., Kilpatrick M. W., Wells R. D. HhaI methylase and restriction endonuclease as probes for B to Z DNA conformational changes in d(GCGC) sequences. Nucleic Acids Res. 1984 Oct 25;12(20):7677–7692. doi: 10.1093/nar/12.20.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Sande J. H., McIntosh L. P., Jovin T. M. Mn2+ and other transition metals at low concentration induce the right-to-left helical transformation of poly[d(G-C)]. EMBO J. 1982;1(7):777–782. doi: 10.1002/j.1460-2075.1982.tb01247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



