Abstract
Several histone deacetylases (HDACs) are involved in the regulation of forkhead box protein P3 (FOXP3) expression and function by affecting features of FOXP3 protein stability. FOXP3, a forkhead family transcription factor specially expressed in regulatory T (Treg) cells, controls the expression of many key immune-regulatory genes. Treg cells are a population of T lymphocytes that have critical roles in the immune system homeostasis and tolerance to self and foreign antigens, the body’s response to cancer and infectious agents. FOXP3 forms oligomeric complexes with other proteins, the components of which are believed to be regulated dynamically. In addition, HDAC activities influence FOXP3 interactions with other partners to form transcriptional regulatory complexes. By understanding the details of the biochemical and structural basis of the regulation of FOXP3 acetylation, therapeutic strategies for diseases related to Treg cells may emerge.
Keywords: FOXP3, acetylation, HDAC, HAT, Treg cells
Regulatory T cells (Tregs) are a subpopulation of T lymphocytes with suppressive properties for CD4+ effector T cells (Th), CD8+ cytotoxic T cells (Tc), antigen-presenting cells, natural killer cells and B cells.1 The deficiency or dysfunction of Treg cells has been linked to several autoimmune and inflammatory diseases including arthritis, irritable bowel syndrome, atopic dermatitis, psoriasis and the deleterious graft-versus-host disease.2 There are two categories of Treg cells: natural Treg cells that are derived in the thymus and emigrate into the periphery, and adaptive Treg cells that are induced in the periphery by converting CD4+CD25− T cells into CD4+CD25+ Treg cells.
Treg cells use the transcription factor forkhead box protein P3 (FOXP3)3 to mediate suppressive activities. The expression of high levels of FOXP3 is necessary for the suppressive function of Treg. Mutations of the FOXP3 gene lead to the X-linked autoimmunity–allergic dysregulation syndrome4 in human and the lymphoproliferative disease in the Scurfy mouse.5–7 The FOXP3 X-linked recessive mutation results in lethality in hemizygous male mice soon after birth, and is characterized by excessive proliferation of the CD4+ T cells, extensive multi-organ infiltration by leukocytes and systemic elevation of numerous cytokines.8
Histone deacetylases (HDACs) include four families of enzymes that were initially found to remove the acetyl moiety from the e-amine of the lysine residue of histones, including: H2A, H2B, H3 and H4.9 Class I HDAC (HDAC1, HDAC2, HDAC3 and HDAC8) and Class IV (HDAC11) are mainly located in the nucleus and mostly form repression complexes such as mSin3, NuRD10 and CoREST.11
Class II HDAC (HDAC4, HDAC5, HDAC6, HDAC7, HDAC9 and HDAC10) shuttle between the nucleus and the cytoplasm; and class III HDACs (SIRT1-7) are located in various organelles. Class I, II and IV are classical HDACs that are Zn2+-dependent and sensitive to the inhibitor trichostatin A (TSA), whereas class III HDACs are homologs to the yeast sirtuins and can be selectively inhibited by nicotinamide.
Acetylation of histones is reciprocally regulated by another class of enzymes, namely histone acetyltransferases (HATs). Histones are not the only substrates for HDAC and HAT. Many other proteins,12 such as p5313 and c-Myc,14 have been identified to undergo acetylation. One consequence of acetylation is that it affects protein stability by preventing ubiquitination and subsequent proteasomal degradation. Acetylation may modulate DNA binding of transcription factors, as well as protein–protein interactions.15 A growing body of data has shown that FOXP3 is one of the targets for HDAC and HAT. We are concerned with roles of HDAC in the regulation of protein stability and suppressive functions related to Treg cells.
FOXP3 AS A UNIQUE TARGET OF HDACS
HDAC inhibitors (HDACi) are convenient reagents to clarify involvement of HDAC in the transcription of genes and the regulation of protein posttranslational modifications. There are caveats to the use of HDACi, as many of these inhibitors are not specific enough to differentiate individual HDAC members, and what is observed can be either a direct or an indirect outcome of the inhibition of multiple HDACs. Nevertheless, HDACi have been used to study the involvement of HDAC in the development of Treg functions in several animal models.
In one disease model of induced colitis, disease severity was reduced when the mice were treated with the HDACi TSA. TSA was found to boost the number of functional Treg cells by increasing thymic production in normal animals, as well as converting CD4+CD25− cells to CD4+FOXP3+ Tregs cells.16 In a collagen-induced arthritis model for rheumatoid arthritis, treatment with the HDACi valproic acid increased both the number of CD4+CD25+FOXP3+ Tregs in vivo and the suppressive activity of CD4+CD25+ Tregs.17 As a result, disease incidence and severity was significantly reduced by valproic acid. To answer how HDAC is involved, initial studies focused on the regulation of epigenetic status of Foxp3 gene locus and thus, the gene expression. Recently, the focus has shifted to understanding the direct interaction of HDAC with the FOXP3 protein and the acetylation axis of posttranslational modification of FOXP3 that regulates the FOXP3 protein stability and functions.
Regulation of FOXP3 expression by HDAC
Histone acetylation occurs on specific lysine residues and neutralizes the positive charge required for histones to compact chromatin structure.18 With some important exceptions, acetylated histones are generally associated with decondensation of DNA and activation of gene transcription.19 As negative regulators of acetylation, HDACs are potent and able to repress expression of certain genes. HDACs, especially Class I members, can form repression complexes that selectively bind to methylated cytosines in cytosine–phosphate diester–guanine islands, which are frequently observed in the promoter regions of silenced genes. For example, the methyl-cytosine–phosphate diester–guanine-binding domain protein MBD1 forms a complex with HDAC3 before being recruited to its target promoters.20 Della Ragione et al.21 reported that HDACi modulate 23 out of 588 genes studied in a human colon cancer cell line. Many of these genes, such as tob-1, p21 and GATA-2, are involved in the regulation of cell cycle or function as transcription factors.
Mantel et al.22 reported hyperacetylation of histone H4 in the promoter and the surrounding chromatin of actively transcribed Foxp3 genes. Cavassani et al.23 demonstrated that acetylation of histone H3 at the K9 site, but not the K12 site on histone H4, was associated with increased FOXP3 expression. These studies suggest that higher HAT activity and reduced HDAC activity is preferred for FOXP3 expression and Treg development. As chromatin condensation is changed after histone acetylation, the Foxp3 promoter may become accessible to certain transcription factors that thereafter regulate FOXP3 expression. In human CD4+CD25− T cells, Fayyad-Kazan et al.24 showed that Ets-1 and Ets-2 bind to Foxp3 promoter in the presence of HDACi and induce FOXP3 expression.
Tao et al.16 studied the effect of HDACi on the Foxp3 gene expression by analyzing both non-Treg (CD4+CD25−) and Treg (CD4+CD25+) cells from mice treated with HDACi TSA (1mg kg−1 per day; 7 days). TSA treatment significantly increased Foxp3 expression in Treg cells, but not in non-Treg cells.16 Other genes upregulated by TSA included CTLA4, PD-1, GITR and IL-10.
In normal mice, TSA increases the number of FOXP3+ Treg cells by increasing thymic production, but in Treg-depleted hosts, TSA appears to promote the conversion of CD4+CD25− cells to CD4+FOXP3+ Treg cells.16 When human CD4+CD25− T cells were treated with HDACi MS-275 and SAHA, Foxp3 expression was induced.25 Increased Foxp3 expression was linked to Treg conversion promoted by the activity of splenic dendritic cells. As a result of the increased Treg activity, mice display an increase in tumor growth compared with sham-treated mice with a syngeneic tumor.23 HDACs can clearly affect Foxp3 expression and have an important role in the development of both natural and adaptive Treg cells.
Despite many studies in this field, there is little consensus on which HDACs are specifically involved in the expression of Foxp3. When Treg functions and interleukin (IL)-10 production was induced by the green tea polyphenol (−)-Epigallocatechin-3-Gallate (EGCG), increased HDAC2 expression was noted in Treg cells from human subjects.26 Using quantitative PCR, Tao et al.16 surveyed HDAC expression in Treg and non-Treg cells, and found similar expression for Class I HDACs and only subtle differences for Class II, except that HDAC9 expression was higher in Treg. HDAC9 may have a complex role in the upregulation of FOXP3 expression in Treg cells, as HDAC9 appears to physically interact with FOXP3. In addition, HDAC9−/− cells demonstrate higher FOXP3 expression and Treg activity.16 Recently, Treg cells in HDAC6−/− mice were found to express more FOXP3 than wild-type mice.27 It is likely that multiple HDACs work together to regulate FOXP3 expression. Due to the broad involvement of HDACs in gene expression,21 HDACi are unlikely to be used in a clinical setting as a specific regulator of FOXP3 expression.
Acetylation enhances the stability of FOXP3 proteins
Li et al.28 found that anti-FOXP3 antibody immunoprecipitates acetylated FOXP3 proteins from nuclear extracts, suggesting that endogenous FOXP3 protein in primary human CD4+CD25+ Treg cells is acetylated. Acetylation may regulate the function of certain protein by altering the protein stability. For some proteins, such as cyclin A, acetylation leads to protein degradation.29 In some other proteins (e.g., p53), acetylation inhibits ubiquitination and increases protein stability.30 Five lysine residues at the C-terminus of p53 are subjected to either acetylation or ubiquitination modification. Acetylation of the lysine residue prevents p53 from being ubiquitinated on these sites. In addition, acetylation, as demonstrated by the use of the K→Q mutant that mimics the effect of acetylation on lysine, also appears to affect ubiquitination on other sites. Acetylation changes p53 conformation and render those lysine residues inaccessible to ubiquitination.30
FOXP3 protein also becomes more stable when it is acetylated. van Loosdregt et al.31 reported that hyperacetylation of FOXP3 prevented polyubiquitination and subsequent proteasomal degradation. Ectopically expressed HAT p300, as well as treatment of HDACi (TSA and nicotinamide), led to higher FOXP3 protein levels. HDACi clearly prevented FOXP3 degradation, which was examined with the help of cycloheximide to inhibit synthesis of new proteins. Stable FOXP3 protein levels yield phenotypic change, both in vitro and in vivo. Treatment of HDACi significantly increases the number and functions of Treg cells in mouse splenocytes, human peripheral blood mononuclear cells and skin-derived T cells.
However, the exact acetylation sites in FOXP3 that are involved in ubiquitination and protein stability are still unknown. A FOXP3 mutant (K22XR) was used to obtain stable ubiquitination-free FOXP3, but all 22 lysine sites were removed in this mutant.31 Although the K22XR mutant is stable, it is unlikely that it will still have normal FOXP3 activity, as acetylation also affects other FOXP3 functions. We have observed reduced FOXP3 activity with the removal of even one potential acetylation site (data not shown). Our initial study indicates that K250 and K252 in human FOXP3 protein are critically involved in the p300-mediated acetylation and some aspects of protein stability,32 but may not be relevant to other functionalities.
Involvement of HDACs in FOXP3 ensembles
FOXP3 functions as a transcriptional factor to regulate the expression of target genes, mostly as a repressor for some genes, but also as enhancer for others. Stable expression of FOXP3 is sufficient to control Treg cell development.33,34 In non-Treg cells, induced expression of FOXP3 can also convert the cell to a Treg phenotype. For example, ectopic expression of FOXP3 in non-Treg leads to repression of the IL-2 and interferon γ (IFNγ) genes, gain of suppressor function and induction of genes such as CD25, GITR and CTLA-4.35 Furthermore, FOXP3 binding to promoter/regulatory elements correlates with histone deacetylation in repressed genes (IL-2 and IFNγ), but with acetylation in induced genes (GITR, CD25 and CTLA-4).35 FOXP3 also associates with a number of other transcription factors including nuclear factor-κB,36 nuclear factor of activated T cells (NFAT),37 AML1/Runx-138 and retinoic-acid-related orphan receptor α (RORα)39 to function as a negative regulator of gene transcription. So far, three HDAC proteins have been reported to associate with FOXP3: HDAC7, HDAC9 and Sirt1.
Histone deacetylase 7
Li et al.28 reported that FOXP3 recruits HAT TIP60 (Tat-interactive protein, 60 kDa) and class II deacetylases, HDAC7 and HDAC9. In later work, HDAC1 was also detected in the same large transcriptional complex. The N-terminal proline-rich domain (amino acids 106–190) of FOXP3, which is critical for transcriptional repression, appears to be involved in the interaction with HDAC7 and TIP60. HDAC7 is by itself able to interact with TIP60 and enhance its co-repressor function.40 Maximal inhibition of IL-2 production by FOXP3 was observed when both TIP60 and HDAC7 were expressed,28 indicating that all three proteins (FOXP3, HDAC7 and TIP60) are needed for the FOXP3 ensemble. Association between FOXP3 and HDAC7 under physiological conditions can also be detected using nuclear extracts from primary CD4+CD25+ Treg cells, and interaction with HDAC7 is not dependent on FOXP3’s capability to form dimers.28
HDAC7 is related to HDAC4 and HDAC5 in Class II sub-family. It has the nuclear localization signal in the N-terminus and the catalytic domain (Figure 1) in the C-terminal half of the protein. Like HDAC4 and HDAC5, HDAC7 has binding domains for C-terminal-binding protein (CtBP), myocyte enhancer factor 2 (MEF2) and 14-3-3 in the N-terminus.41 As a DNA-binding transcription factor, MEF2 has an essential role in muscle differentiation. By binding to MEF2, HDAC7 represses the function of MEF2 and blocks muscle cell differentiation.42 Splicing forms of HDAC7 are also involved in the differentiation of embryonic stem cells into smooth muscle cells.43
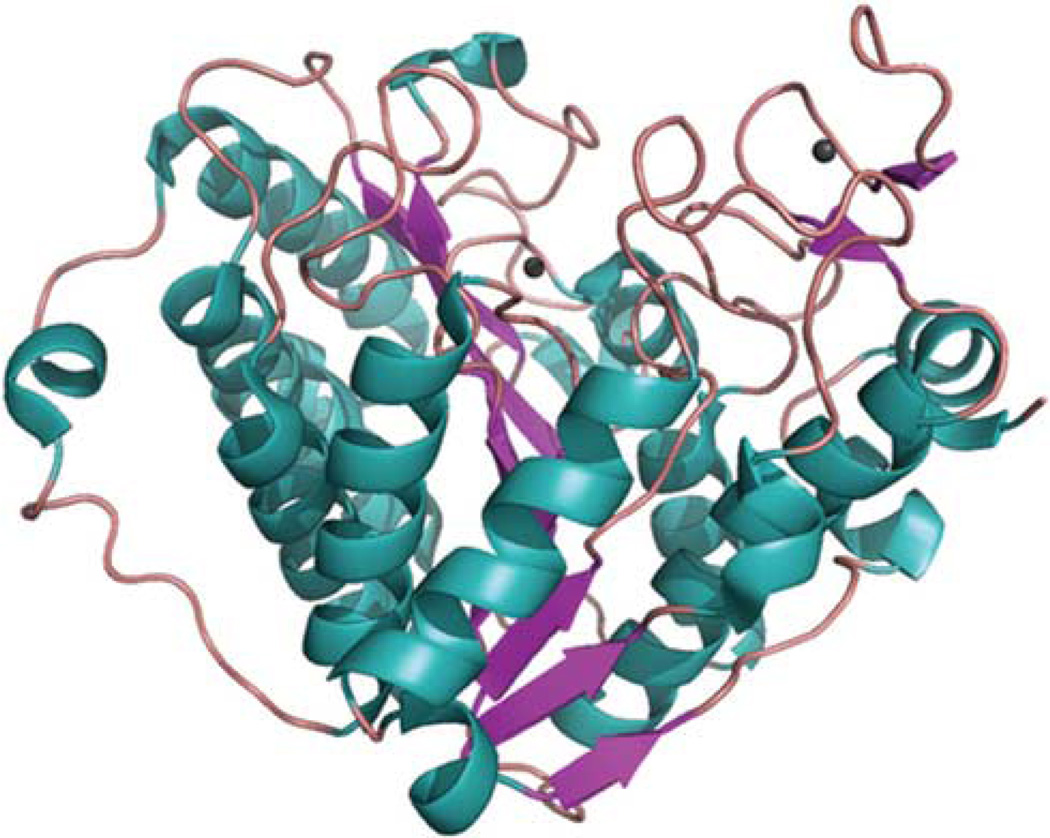
Figure 1.
Structure of the HDAC7 catalytic domain. The crystal structure of the HDAC catalytic domain solved by Schuetz et al.60 (PDB code: 3c0y) is shown as a structural cartoon. Different secondary structures (alpha helix, beta strand and loop) are colored teal, magentas and salmon, respectively. The two coordinated zinc ions are colored grey.
HDAC7 is not primarily an element of the SMRT/N-CoR repressor. However, Class I members such as HDAC3 are required to associate with HDAC4, HDAC5 or HDAC7, to establish a functional SMRT/N-CoR complex, indicating an adaptor role for HDAC7 to link DNA-binding recruiters and repressor HDACs.44 HDAC7 is also able to interact with co-repressors BCoR (Bcl-6-interacting co-repressor) and BcoR-L1.45
HDAC7 has a nuclear export sequence at the C-terminus. The same sequence has also been identified in HDAC4 and HDAC5. McKinsey et al.46 have shown that in HDAC5, the nuclear export sequence is activated once the chaperone protein 14-3-3 is attached. Phosphorylated HDAC5 is then exported from the nucleus to the cytoplasm. Ca2+/calmodulin-dependent protein kinase (CaMK) I appears to be important for the phosphorylation of the 14-3-3 docking site, as an HDAC7 mutant that lacks the putative CaMK I sites (S178A/S344A/S479A) loses the capability to bind to 14-3-3 and localizes exclusively in the nucleus.47 However, the regulation of HDAC7 phosphorylation is even more complex. Ectopically expressed CaMK I selectively phosphorylates S178 and only has modest effects on S344 and S479. Studies with phospho-specific HDAC7 antibody indicate that HDAC7 with S344 or S479 phosphorylation is retained in the nucleus, whereas S178-phosphorylated species reside in both the nucleus and the cytoplasm.48 Studies by Gao et al.48 also reveal that CRM1 can provide an alternative nuclear exporting pathway for HDAC7, as overexpression of CRM1 rescues the nuclear export of the HDAC7 S178A/S344A/S479A mutant.
Histone deacetylase 9
HDAC9 can also complex with FOXP3 in T cells, but only in the absence of T-cell stimulation.28 T cell receptor (TCR), plus CD28 stimulation, sufficiently disrupts the association between HDAC9 and FOXP3. However, in the presence of HDACi TSA, FOXP3 and HDAC9 form much more stable complex even in the presence of T-cell stimulation.28
HDAC9 is a unique Class II member as it carries the catalytic domain at the N-terminus, just like Class I HDACs. HDAC9 has several isoforms.49 One splice form, termed HDAC9c, lacks the catalytic domain, but is able to recruit HDAC3 as a catalytic domain to function. Harrison et al.50 reported that protein kinase C-related kinase 2 strongly phosphorylated S253 within the nuclear localization signal of HDAC9. An HDAC9 splice variant, MEF2 interacting transcription repressor (MITR), which lacks nuclear export sequence, is thus primarily localized to the nuclei. Upon phosphorylation of two 14-3-3-binding sites (Ser218 and Ser448) by CaMK I,51 MITR changed from discrete nuclear structures to diffusely distributed staining within the nucleus. A MITR mutant S253D, which has Ser253 substituted by aspartic acid (to mimic phosphorylation), was still localized to discrete foci in the nucleus. However, in the presence of activated CaMK I, MITR (S253D) dramatically migrated to the cytoplasm.50 This CaMK I-mediated redistribution was not observed with the MITR S253A mutant, or with MITR (S253D/S448A) that lacks a 14-3-3-binding site.
HDAC9 appears to be a negative regulator of FOXP3 and Treg functions. Higher local expression of HDAC9 is found in mice with colitis, and HDAC9−/− mice are resistant to the development of colitis.52 HDAC9 levels in Treg cells may be influenced by Hsp70, which was also found to interact with FOXP3.52,53 In comparison with wild-type mice, HDAC9−/− mice have more FOXP3+CD4+ T cells, but not CD4+CD25+ T cells in the spleen.16 In addition, Treg cells from HDAC9−/− are much more active. Nevertheless, HDAC9 expression is increased in Treg cells and TCR activation further increases HDAC9 expression.16 It is likely that HDAC9 interacts and represses FOXP3 function, but after TCR activation, the HDAC9 disassociates and releases FOXP3, resulting in higher Treg activity.
Sirt1
van loosdregt et al.31 noted a higher level of FOXP3 when both TSA and nicotinamide were used. Nicotinamide targets class III HDACs (i.e., Sirt deacetylases). Sirt1 of this class interacts with FOXP3, when both proteins were overexpressed in 293T cells.31 Sirt1 is known to be negatively associated with T-cell activation. It has been shown that loss of Sirt1 function results in abnormally increased T-cell activation and a breakdown of CD4+ T-cell tolerance, whereas upregulation of Sirt1 expression leads to T-cell anergy.54 Experimental allergic encephalomyelitis, as well as spontaneous autoimmunity, has been noted in Sirt1-deficient mice. Zhang et al.54 then demonstrated that the role of Sirt1 in T-cell activation is related to the acetylation and inactivation of AP-1. Currently, it is uncertain whether the effect of Sirt1 on AP-1 is cell type-specific and whether Sirt1 can also interact with endogenous FOXP3 in Treg cells.
CURRENT STRUCTURAL BASIS OF FOXP3 ENSEMBLE
Although previous biochemical studies by our laboratory have suggested that the zinc finger leucine zipper region of FOXP3 is necessary and sufficient to mediate both homo-association and hetero-association with FOXP1,55–57 it is not clear how hetero- and homo-dimers are formed and how posttranslational modifications such acetylation affect the ensemble.
We have recently solved the crystal structure of mouse FOXP3 oligomerization domain containing the zinc finger and leucine zipper motifs (G196-K276). As anticipated, the leucine zipper (D224-K262) region, but not the zinc finger motif (V197-L223), is directly involved in dimerization via an anti-parallel coiled-coil structure. K251, a lysine residue that was found acetylated in mass spectrometric studies, appears to mediate dimerization by promoting the inter-molecule hydrogen bond between E248 and Q234 (Song et al., submitted). In the human sequence, the corresponding site is K252. Substitution of K252 with glutamine (K252Q) mimics the effect of acetylation on this residue and reduces human FOXP3 homodimer and FOXP3-FOXP1 heterodimer formation. However, the K252Q mutation does not obliterate dimer formation, suggesting that acetylation at this site is able to modulate the suppressive behavior of Treg cells. Our structural study established a mechanistic model of the coiled-coil motif as a regulatory unit for the FOXP3 complex ensemble. These data also identify the complexity of acetylation and its ability to regulate the structural conformation and assembly of the FOXP3 complex. As acetylation has been shown to interfere with FOXP3 ubiquitination and degradation, it is possible that FOXP3 undergoes deacetylation at K252 to become active for a short term and then proceed to degradation as a way to prevent the Treg cells from being overactive.
The FOXP3 ensemble attaches to DNA through the forkhead domain of the FOXP3 protein. When three lysine residues in this forkhead region, K383, K393 and K415, were replaced by histidine residues and the FOXP3 mutants were introduced into CD4+CD25− cells, the Treg-like suppressive ability of transduced cells is greatly enhanced.16 This contrived study suggests that acetylation might be involved in the attachment of FOXP3 ensemble to target DNA, as these lysines are potential acetylation sites. It has yet to be determined whether acetylation in fact occurs on these sites in vivo and which HDAC/HAT is involved in the maintenance of acetylation on these sites.
Acetylation as a mechanism in the regulation of FOXP3 ensemble in response to extracellular stimuli
Recent studies suggest that the FOXP3 ensemble is dynamically regulated.2 Nuclear transport of FOXP3 relies on three distinct domains, which can form complexes with other proteins containing nuclear localization signal.58 Activation stimuli induce the translocation of FOXP3 from a primarily cytoplasmic/perinuclear residence to a dense nuclear subcellular localization, where FOXP3 and the ensemble can bind to the regulatory elements of target genes such as cytokine (e.g., IL-2 and IFNγ). Transforming growth factor-β, which is known to induce murine FOXP3(+) Treg cells, also increases FOXP3 binding to chromatin in human T-cell line SZ-4.59
Acetylation has an important role in the binding of FOXP3 ensemble to chromatin, as the HDACi sodium butyrate also increases FOXP3 binding to chromatin in cells. There may be more binding sites after chromatin decondensation that are associated with acetylated histones. However, sodium butyrate also inhibits effects of transforming growth factor-β and IL-6 on FOXP3 binding to chromatin, indicating a more complicated regulation of the FOXP3 ensembles that might be related to the acetylation status of certain components of the ensemble.59 It is argued that binding of FOXP3 ensembles to repressed and inducible genes should be studied separately.
In addition to the histone axis of acetylation that affects the chromatin binding of FOXP3 ensemble, acetylation may directly affect FOXP3 or other proteins in the ensemble and change the activity of FOXP3. We have shown that the acetylation of K252 in FOXP3 can lead to formation of less FOXP3–FOXP3 homodimers and FOXP3–FOXP1 heterodimers (Song, et al., submitted). We have also shown that FOXP3–HDAC9 association is interrupted by T-cell receptor stimulation.28 These studies highlight the dynamic nature of the FOXP3 ensemble in terms of its components and location due to the changing cellular stimuli. A more extensive study of each component in response to stimulus signals is currently under way to dissect the complex regulation of FOXP3 functions.
HAT FOR FOXP3
The action of HAT enzymes on protein substrates is opposite to that of HDAC. The current knowledge of HAT proteins that are involved in the regulation of FOXP3 is reviewed elsewhere.32 There is evidence that TIP60 (Figure 2) interacts with FOXP3. In human CD4+CD25+ Treg cells, endogenous TIP60 co-localizes with FOXP3 in the nucleus. In 293T cells co-transfected with HA–FOXP3 and FLAG–TIP60, FOXP3 can be co-immunoprecipitated with TIP60.28 The N-terminal proline-rich region of FOXP3 was identified to be responsible for binding to TIP60. TIP60 appears to be responsible for FOXP3 acetylation and subsequent changes of function. Overexpression of TIP60 leads to increase of acetylated FOXP3 proteins in the 293T cells. In addition, FOXP3-mediated transcriptional repression can be enhanced by the overexpression of wild-type TIP60, but not the HAT activity-deficient TIP60 mutant, and dramatically reduced by the knockdown of endogenous TIP60 with shRNA.28
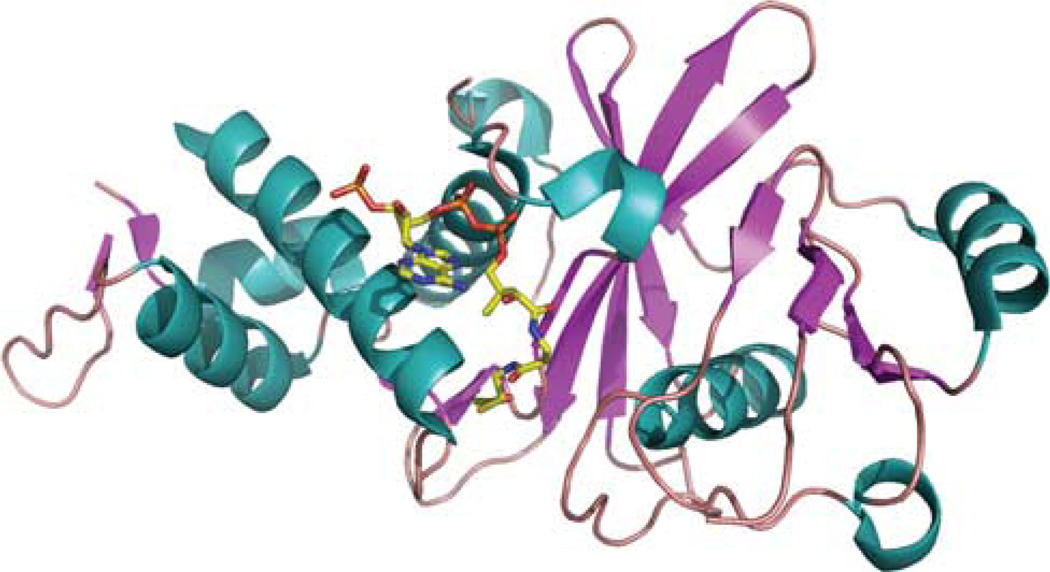
Figure 2.
Structure of the TIP60 acetyltransferase domain. The crystal structure of human TIP60 isoform 3 (PDB code: 2ou2) in a complex with acetyl coenzyme A is shown as a structural cartoon (Wu et al., to be published). Different secondary structures (alpha helix, beta strand and loop) are colored teal, magentas and salmon, respectively. Acetyl coenzyme A is shown as sticks and colored by atomic type (carbon: yellow; nitrogen: blue; oxygen: red; phosphorus: orange).
It is clear that other HATs are also involved in the acetylation of FOXP3. van Loosdregt et al.31 and our laboratory demonstrated that p300 (Figure 3) is capable to bind and acetylate FOXP3 when both molecules are overexpressed in HEK 293 cells. The p300 appears to stabilize FOXP3 protein levels by preventing ubiquitination. In addition, we have also found that p300/CBP-associated factor (pCAF) also acetylates FOXP3 on sites distinct from those of p300 (Xiao and Greene; unpublished). Studies are underway to investigate the exact role of p300 and other additional HATs in the regulation of FOXP3 function.
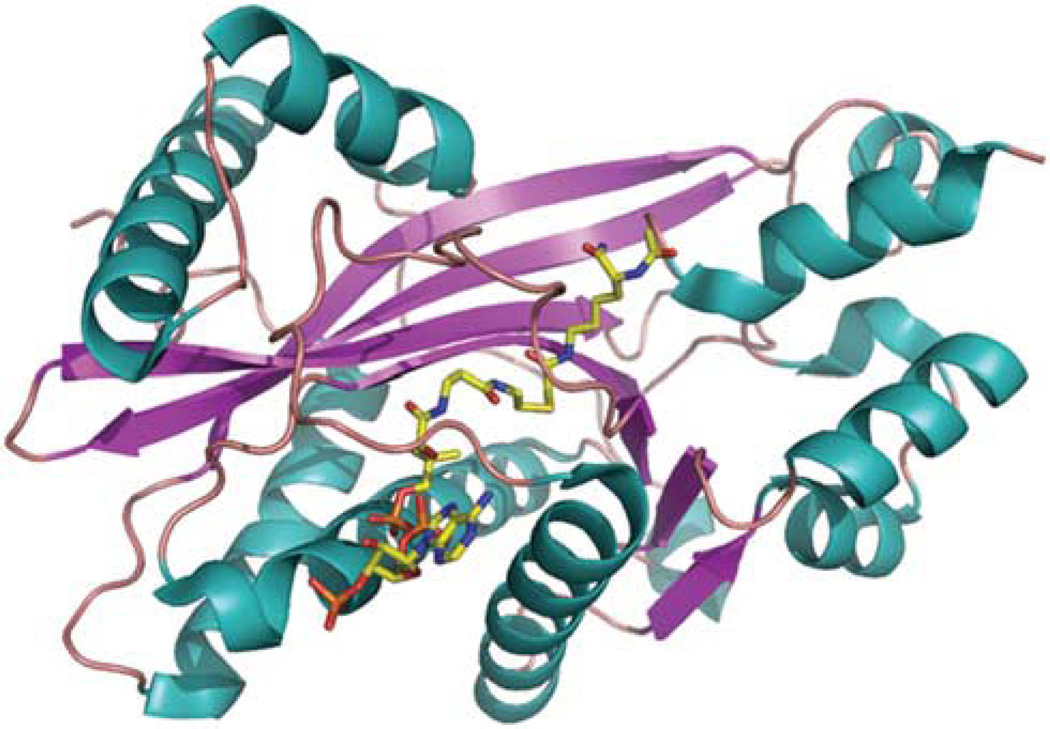
Figure 3.
Structure of p300 HAT domain. Crystal structure of p300 HAT domain (PDB code: 3biy) was solved in complex with a bisubstrate inhibitor, Lys-CoA.61 Two HAT subdomains, amino acids 1287–1522 and amino acids 1569–1666, were co-purified and used for crystallization. The structure is showed as cartoon and colored by secondary structure (alpha helix: teal, beta strand: magentas; loop: salmon). The inhibitor in the structure is presented as sticks and colored by atomic type (carbon: yellow; nitrogen: blue; oxygen: red; phosphorus: orange).
CONCLUSION
Acetylation has evolved to be a critical factor in the regulation of functions of Treg cells by manipulating the expression of the Foxp3 gene and the activity of the FOXP3 protein. However, an orchestra of multiple HDACs and HATs might be required for the regulation of FOXP3 expression. There is a lack of evidence to which HDAC is particularly involved in the regulation of FOXP3 at the transcriptional level. The current focus of research in our laboratory is to develop small molecular therapies targeting distinct HDACs and HATs, in particular by manipulating their stability, complex assembly, or functions, to modify Treg cell activity. Our rational design approach is facilitated by the knowledge of solved crystal structures of HDACs and HATs (Figures 1–3). A clear understanding of the involvement of HDACs in the FOXP3 complex assembly will lead to insights to develop new strategies to modulate Treg functions for human disease.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (to MIG 5P01AI073489-04) and the Abramson Family Cancer Research Institute (to MIG). We thank Gabriela Canales for her help with the manuscript.
References
- 1.Rudensky AY, Campbell DJ. In vivo sites and cellular mechanisms of T reg cell-mediated suppression. J Exp Med. 2006;203:489–492. doi: 10.1084/jem.20060214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li B, Greene MI. Special regulatory T-cell review: FOXP3 biochemistry in regulatory T cells—how diverse signals regulate suppression. Immunology. 2008;123:17–19. doi: 10.1111/j.1365-2567.2007.02774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakaguchi S, Ono M, Setoguchi R, Yagi H, Hori S, Fehervari Z, et al. Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol Rev. 2006;212:8–27. doi: 10.1111/j.0105-2896.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- 4.Chatila TA, Blaeser F, Ho N, Lederman HM, Voulgaropoulos C, Helms C, et al. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest. 2000;106:R75–R81. doi: 10.1172/JCI11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 6.Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 7.Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 8.Godfrey VL, Wilkinson JE, Russell LB. X-linked lymphoreticular disease in the scurfy (sf) mutant mouse. Am J Pathol. 1991;138:1379–1387. [PMC free article] [PubMed] [Google Scholar]
- 9.Stimson L, Wood V, Khan O, Fotheringham S, La Thangue NB. HDAC inhibitor-based therapies and haematological malignancy. Ann Oncol. 2009;20:1293–1302. doi: 10.1093/annonc/mdn792. [DOI] [PubMed] [Google Scholar]
- 10.Feng Q, Zhang Y. The NuRD complex: linking histone modification to nucleosome remodeling. Curr Top Microbiol Immunol. 2003;274:269–290. doi: 10.1007/978-3-642-55747-7_10. [DOI] [PubMed] [Google Scholar]
- 11.Lakowski B, Roelens I, Jacob S. CoREST-like complexes regulate chromatin modification and neuronal gene expression. J Mol Neurosci. 2006;29:227–239. doi: 10.1385/JMN:29:3:227. [DOI] [PubMed] [Google Scholar]
- 12.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 13.Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ, et al. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol. 2004;24:10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 17.Saouaf SJ, Li B, Zhang G, Shen Y, Furuuchi N, Hancock WW, et al. Deacetylase inhibition increases regulatory T cell function and decreases incidence and severity of collagen-induced arthritis. Exp Mol Pathol. 2009;87:99–104. doi: 10.1016/j.yexmp.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villa R, Morey L, Raker VA, Buschbeck M, Gutierrez A, De Santis F, et al. The methyl-CpG binding protein MBD1 is required for PML-RARalpha function. Proc Natl Acad Sci USA. 2006;103:1400–1405. doi: 10.1073/pnas.0509343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Della Ragione F, Criniti V, Della Pietra V, Borriello A, Oliva A, Indaco S, et al. Genes modulated by histone acetylation as new effectors of butyrate activity. FEBS Lett. 2001;499:199–204. doi: 10.1016/s0014-5793(01)02539-x. [DOI] [PubMed] [Google Scholar]
- 22.Mantel PY, Ouaked N, Ruckert B, Karagiannidis C, Welz R, Blaser K, et al. Molecular mechanisms underlying FOXP3 induction in human T cells. J Immunol. 2006;176:3593–3602. doi: 10.4049/jimmunol.176.6.3593. [DOI] [PubMed] [Google Scholar]
- 23.Cavassani KA, Carson WF, Moreira AP, Wen H, Schaller MA, Ishii M, et al. The post sepsis-induced expansion and enhanced function of regulatory T cells create an environment to potentiate tumor growth. Blood. 2010;115:4403–4411. doi: 10.1182/blood-2009-09-241083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fayyad-Kazan H, Rouas R, Merimi M, El Zein N, Lewalle P, Jebbawi F, et al. Valproate treatment of human cord blood CD4-positive effector T cells confers on them the molecular profile (microRNA signature and FOXP3 expression) of natural regulatory CD4-positive cells through inhibition of histone deacetylase. J Biol Chem. 2010;285:20481–20491. doi: 10.1074/jbc.M110.119628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lucas JL, Mirshahpanah P, Haas-Stapleton E, Asadullah K, Zollner TM, Numerof RP. Induction of Foxp3+ regulatory T cells with histone deacetylase inhibitors. Cell Immunol. 2009;257:97–104. doi: 10.1016/j.cellimm.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Yun JM, Jialal I, Devaraj S. Effects of epigallocatechin gallate on regulatory T cell number and function in obese v. lean volunteers. Br J Nutr. 2010;103:1771–1777. doi: 10.1017/S000711451000005X. [DOI] [PubMed] [Google Scholar]
- 27.de Zoeten EF, Wang L, Butler K, Beier UH, Akimova T, Sai H, et al. Histone deacetylase 6 heat shock protein 90 control the functions of Foxp3(+) T-regulatory cells. Mol Cell Biol. 2011;31:2066–2078. doi: 10.1128/MCB.05155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B, Samanta A, Song X, Iacono KT, Bembas K, Tao R, et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci USA. 2007;104:4571–4576. doi: 10.1073/pnas.0700298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mateo F, Vidal-Laliena M, Canela N, Busino L, Martinez-Balbas MA, Pagano M, et al. Degradation of cyclin A is regulated by acetylation. Oncogene. 2009;28:2654–2666. doi: 10.1038/onc.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem. 2002;277:50607–50611. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- 31.van Loosdregt J, Vercoulen Y, Guichelaar T, Gent YY, Beekman JM, van Beekum O, et al. Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood. 2010;115:965–974. doi: 10.1182/blood-2009-02-207118. [DOI] [PubMed] [Google Scholar]
- 32.Xiao Y, Li B, Zhou Z, Hancock WW, Zhang H, Greene MI. Histone acetyltransferase mediated regulation of FOXP3 acetylation and Treg function. Curr Opin Immunol. 2010;22:583–591. doi: 10.1016/j.coi.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3.[comment] Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 34.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 35.Chen C, Rowell EA, Thomas RM, Hancock WW, Wells AD. Transcriptional regulation by Foxp3 is associated with direct promoter occupancy and modulation of histone acetylation. J Biol Chem. 2006;281:36828–36834. doi: 10.1074/jbc.M608848200. [DOI] [PubMed] [Google Scholar]
- 36.Bettelli E, Dastrange M, Oukka M. Foxp3 interacts with nuclear factor of activated T cells and NF-kappa B to repress cytokine gene expression and effector functions of T helper cells. Proc Natl Acad Sci USA. 2005;102:5138–5143. doi: 10.1073/pnas.0501675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 38.Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T, et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- 39.Du J, Huang C, Zhou B, Ziegler SF. Isoform-specific inhibition of ROR alpha-mediated transcriptional activation by human FOXP3. J Immunol. 2008;180:4785–4792. doi: 10.4049/jimmunol.180.7.4785. [DOI] [PubMed] [Google Scholar]
- 40.Xiao H, Chung J, Kao HY, Yang YC. Tip60 is a co-repressor for STAT3. J Biol Chem. 2003;278:11197–11204. doi: 10.1074/jbc.M210816200. [DOI] [PubMed] [Google Scholar]
- 41.Bertos NR, Wang AH, Yang XJ. Class II histone deacetylases: structure, function, and regulation. Biochem Cell Biol. 2001;79:243–252. [PubMed] [Google Scholar]
- 42.McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 43.Margariti A, Xiao Q, Zampetaki A, Zhang Z, Li H, Martin D, et al. Splicing of HDAC7 modulates the SRF-myocardin complex during stem-cell differentiation towards smooth muscle cells. J Cell Sci. 2009;122:460–470. doi: 10.1242/jcs.034850. [DOI] [PubMed] [Google Scholar]
- 44.Fischle W, Dequiedt F, Fillion M, Hendzel MJ, Voelter W, Verdin E. Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. J Biol Chem. 2001;276:35826–35835. doi: 10.1074/jbc.M104935200. [DOI] [PubMed] [Google Scholar]
- 45.Pagan JK, Arnold J, Hanchard KJ, Kumar R, Bruno T, Jones MJ, et al. A novel corepressor, BCoR-L1, represses transcription through an interaction with CtBP. J Biol Chem. 2007;282:15248–15257. doi: 10.1074/jbc.M700246200. [DOI] [PubMed] [Google Scholar]
- 46.McKinsey TA, Zhang CL, Olson EN. Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol Cell Biol. 2001;21:6312–6321. doi: 10.1128/MCB.21.18.6312-6321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kao HY, Verdel A, Tsai CC, Simon C, Juguilon H, Khochbin S. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J Biol Chem. 2001;276:47496–47507. doi: 10.1074/jbc.M107631200. [DOI] [PubMed] [Google Scholar]
- 48.Gao C, Li X, Lam M, Liu Y, Chakraborty S, Kao HY. CRM1 mediates nuclear export of HDAC7 independently of HDAC7 phosphorylation and association with 14-3-3s. FEBS Lett. 2006;580:5096–5104. doi: 10.1016/j.febslet.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 49.Zhou X, Marks PA, Rifkind RA, Richon VM. Cloning and characterization of a histone deacetylase, HDAC9. Proc Natl Acad Sci USA. 2001;98:10572–10577. doi: 10.1073/pnas.191375098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harrison BC, Huynh K, Lundgaard GL, Helmke SM, Perryman MB, McKinsey TA. Protein kinase C-related kinase targets nuclear localization signals in a subset of class IIa histone deacetylases. FEBS Lett. 2010;584:1103–1110. doi: 10.1016/j.febslet.2010.02.057. [DOI] [PubMed] [Google Scholar]
- 51.Zhang CL, McKinsey TA, Olson EN. The transcriptional corepressor MITR is a signal-responsive inhibitor of myogenesis. Proc Natl Acad Sci USA. 2001;98:7354–7359. doi: 10.1073/pnas.131198498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Zoeten EF, Wang L, Sai H, Dillmann WH, Hancock WW. Inhibition of HDAC9 increases T regulatory cell function and prevents colitis in mice. Gastroenterology. 2010;138:583–594. doi: 10.1053/j.gastro.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li B, Saouaf SJ, Samanta A, Shen Y, Hancock WW, Greene MI. Biochemistry and therapeutic implications of mechanisms involved in FOXP3 activity in immune suppression. Curr Opin Immunol. 2007;19:583–588. doi: 10.1016/j.coi.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 54.Zhang J, Lee SM, Shannon S, Gao B, Chen W, Chen A, et al. The type III histone deacetylase Sirt1 is essential for maintenance of T cell tolerance in mice. J Clin Invest. 2009;119:3048–3058. doi: 10.1172/JCI38902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chae WJ, Henegariu O, Lee SK, Bothwell AL. The mutant leucine-zipper domain impairs both dimerization and suppressive function of Foxp3 in T cells. Proc Natl Acad Sci USA. 2006;103:9631–9636. doi: 10.1073/pnas.0600225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li B, Samanta A, Song X, Iacono KT, Brennan P, Chatila TA, et al. FOXP3 is a homooligomer and a component of a supramolecular regulatory complex disabled in the human XLAAD/IPEX autoimmune disease. Int Immunol. 2007;19:825–835. doi: 10.1093/intimm/dxm043. [DOI] [PubMed] [Google Scholar]
- 57.Lopes JE, Torgerson TR, Schubert LA, Anover SD, Ocheltree EL, Ochs HD, et al. Analysis of FOXP3 reveals multiple domains required for its function as a transcriptional repressor. J Immunol. 2006;177:3133–3142. doi: 10.4049/jimmunol.177.5.3133. [DOI] [PubMed] [Google Scholar]
- 58.Hancock WW, Ozkaynak E. Three distinct domains contribute to nuclear transport of murine Foxp3. PLoS One. 2009;4:e7890. doi: 10.1371/journal.pone.0007890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samanta A, Li B, Song X, Bembas K, Zhang G, Katsumata M, et al. TGF-beta and IL-6 signals modulate chromatin binding and promoter occupancy by acetylated FOXP3. Proc Natl Acad Sci USA. 2008;105:14023–14027. doi: 10.1073/pnas.0806726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schuetz A, Min J, Allali-Hassani A, Schapira M, Shuen M, Loppnau P, et al. Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity. J Biol Chem. 2008;283:11355–11363. doi: 10.1074/jbc.M707362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, et al. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. 2008;451:846–850. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]