Abstract
Mitochondrial structure and distribution are regulated by division and fusion events. Mitochondrial division is regulated by Dnm1/Drp1, a dynamin-related protein that forms helices around mitochondria to mediate fission. Little is known about what determines sites of mitochondrial fission within the mitochondrial network. The ER and mitochondria exhibit tightly coupled dynamics and have extensive contacts. We tested whether ER plays a role in mitochondrial division. We found that mitochondrial division occurred at positions where ER tubules contacted mitochondria and mediated constriction prior to Drp1 recruitment. Thus, ER tubules may play an active role in defining the position of mitochondrial division sites.
Regulation of mitochondrial division is critical to normal cellular function; excess division is linked to numerous diseases, including neurodegeneration and diabetes (1, 2). The central player in mitochondrial division is the highly conserved dynamin-related protein (Drp1 in mammals, Dnm1 in yeast), which belongs to a family of large GTPases that self-assemble to regulate membrane structure (3). Division dynamins are likely to work by oligomerizing in a GTP-dependent manner into helices that wrap around mitochondria; locally controlled assembly-stimulated GTP hydrolysis is thought to provide the mechanochemical force that completes fission of the outer and inner membranes (4). There are additional proteins required for mitochondrial division, such as the outer membrane protein Mff (mitochondrial fission factor), which is present only in mammals (5). Although general mechanisms exist for recruiting Dnm1/Drp1 to mitochondria, it is not known whether there are specific sites on mitochondria that are marked for division (6). Furthermore, both Dnm1 and Drp1 oligomerize into helices that are much smaller than the diameter of mitochondria (Dnm1 helices have reported mean diameters of 109 nm in yeast and 129 nm in vitro), suggesting that Dnm1/Drp1-independent mitochondrial constriction may be needed to facilitate mitochondrial division (4, 6–9).
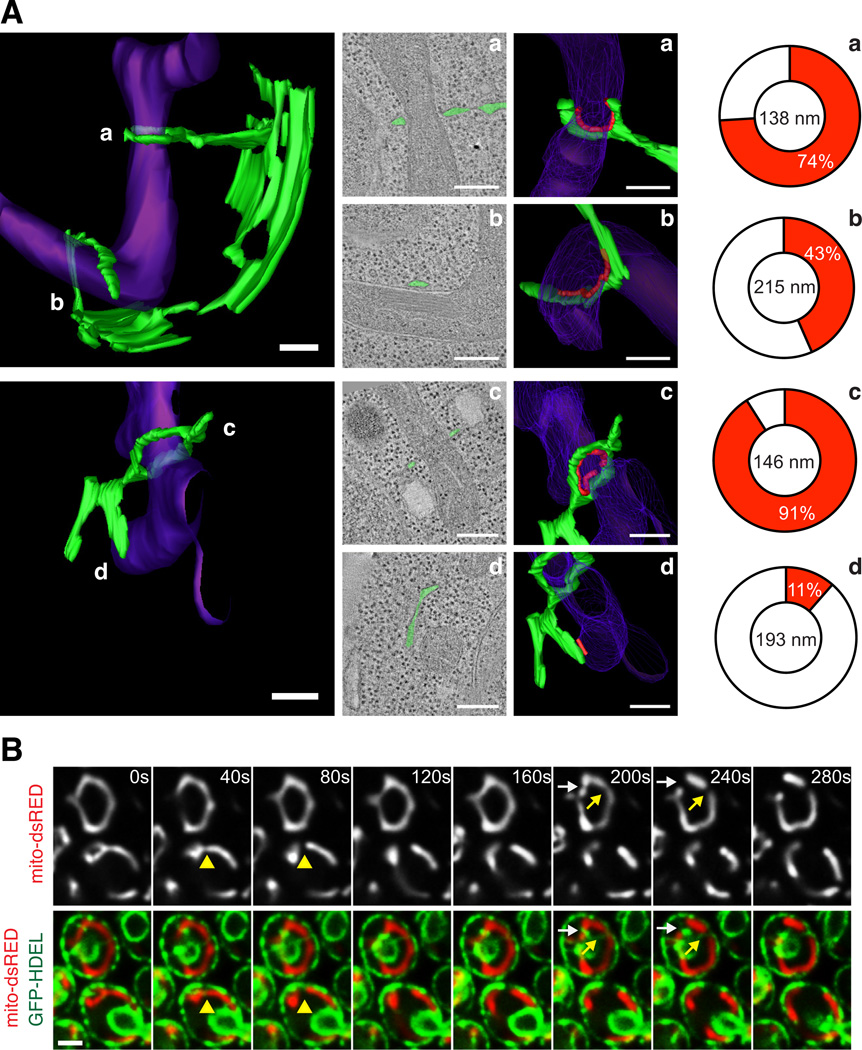
Contact sites exist between mitochondria and the endoplasmic reticulum (ER) and are important for phospholipid synthesis and calcium signaling (for review, see (10)). Based on recent data, there are likely several types of molecular bridges that mediate these contacts, such as the ERMES complex identified in yeast and the mitochondrial fusion protein mitofusin 2 (Mfn2) in mammalian cells (11, 12). These physical contacts are persistent and maintained under dynamic conditions (13), suggesting that the ER-mitochondrial interface is vital for function. We have used EM and tomography to analyze the 3-dimensional (3-D) structure of contacts between the ER and mitochondria in the yeast Saccharomyces cerevisiae. We observed the high-resolution (~4 nm) structure and 3-D models of four ER-mitochondrial contacts taken from two cells (Fig. 1A). In these examples, the ER was wrapped around mitochondria to varying degrees. In two of the four examples, the ER almost completely circumscribed the mitochondrial outer membrane and mitochondria were constricted at the point of contact (mitochondrial diameter 138 nm and 146 nm circumscribed versus 215 nm and 193 nm uncircumscribed at ER contact; Figs. 1A and S1A–B and Movies S1–S2). These data suggest that ER tubules associate with and may mediate mitochondrial constriction sites.
Figure 1.
Mitochondrial constriction and division occurs at ER-mitochondrial contacts in yeast. (A) 3-D models (left panels) of ER (green) and mitochondria (purple) at contact domains were imaged by EM and tomography of high pressure frozen yeast cells. Middle two panels are 2-D tomographs of contact sites (second panels, ER drawn in green) and the corresponding 3-D models of each (third panels). Contact is defined as regions where the ER membrane comes within 30 nm of the mitochondrial membrane and ribosomes are excluded, and is outlined in red (third panel). Right panel is a schematic demonstrating the percentage of the mitochondrial circumference that makes contact with the ER membrane (red is contact, white is not; (19)). The diameter of each mitochondrion at positions of ER contact is shown. Note that regions where the mitochondria are constricted (models a and c) have a high percent of ER wrapping. Additional EM tomographs and analysis of constrictions are shown in Fig. S1A–B. (B) Time-lapse images of yeast cells expressing mito-dsRED and GFP-HDEL (ER). A single focal plane is shown. Arrows indicate sites of mitochondrial division. Corresponding z-series is shown in Fig. S1C. Scale bars: (A) 200 nm; (B) 2 µm.
We thus examined the role of ER in mitochondrial division using fluorescence microscopy in live yeast cells transformed with GFP-HDEL (ER) and mito-dsRED to image the behavior of ER and mitochondria simultaneously over time. The vast majority of mitochondrial division events were spatially linked to sites of ER-mitochondrial contact (87%, n=112 from 281 cells; Fig. 1B). ER tubules crossed over (Fig. 1B, yellow arrows) and wrapped around mitochondria (Figs. 1B, white arrows and S1C). At ER-mitochondrial contact sites, mitochondrial constriction followed by mitochondrial division was observed (Fig. 1B).
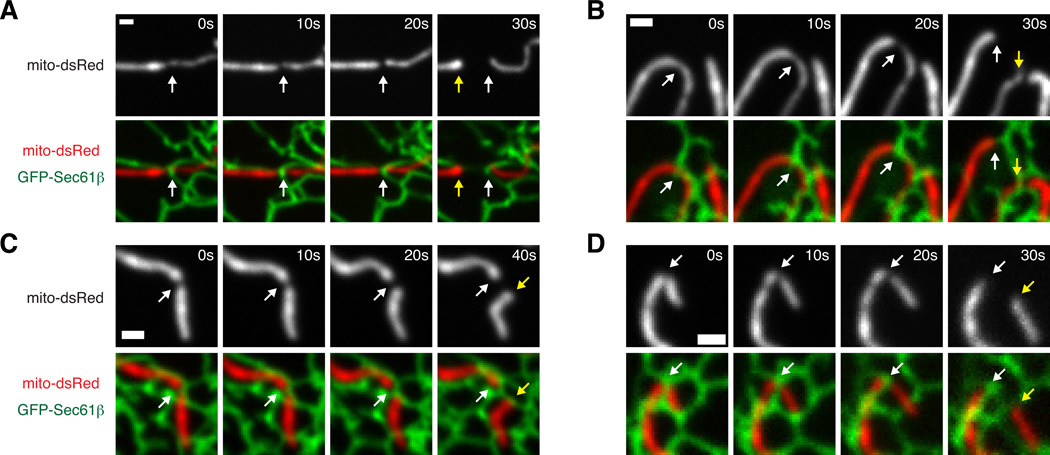
We next tested whether ER plays a similar role in mammalian mitochondrial division using fluorescence microscopy of live Cos-7 cells transiently transfected with fluorescent markers for ER (GFP-Sec61β) and mitochondria (mito-dsRed). We imaged regions of the cell periphery where contacts between the mitochondria and ER were well-resolved, and observed that mitochondrial division events predominantly occurred at sites of contact between ER and mitochondria (94%, n=32 from 23 cells; Figs. 2 and S2A and Movies S3–S4). Furthermore, the majority of events (88%) were sites of ER tubules crossing over the mitochondria, suggesting that the structural context of the interaction is important. The frequency of ER-associated mitochondrial division is much higher than would be predicted based on the area of mitochondria covered by crossing ER tubules as determined by co-localization of mitochondrial and ER markers (Fig. S2B).
Figure 2.
Mitochondrial division occurs at ER-mitochondrial contact sites in mammalian cells. (A–D) Four examples of mitochondrial division over timecourses shown in Cos-7 cells expressing GFP-Sec61β (ER) and mito-dsRed. The site of mitochondrial division (white arrows) and the position of the newly formed mitochondrial ends (yellow arrows) are shown. Additional examples are included in Fig. S2A. Scale bars: 1 µm.
Thus, in both yeast and mammalian cells, ER tubules are at mitochondrial division sites and may be involved in mitochondrial constriction during this process. Next, we asked whether mitochondrial division occurs in yeast cells that have significantly reduced levels of tubules due to the absence of the membrane shaping proteins Rtns/Yop1 (14, 15). Using both EM and fluorescence microscopic analyses, we observed that in regions of mutant cells where ER tubules were dramatically reduced, short ER tubules were observed to extend out of the massive ER cisternae and associate with mitochondrial constrictions and division events (Fig. S3). Thus, ER tubules are a consistent feature of ER contact at mitochondrial constrictions even under conditions where most tubules are depleted. Furthermore, Rtns/Yop1 are dispensable for the biogenesis of the ER tubules that associate with mitochondrial division events.
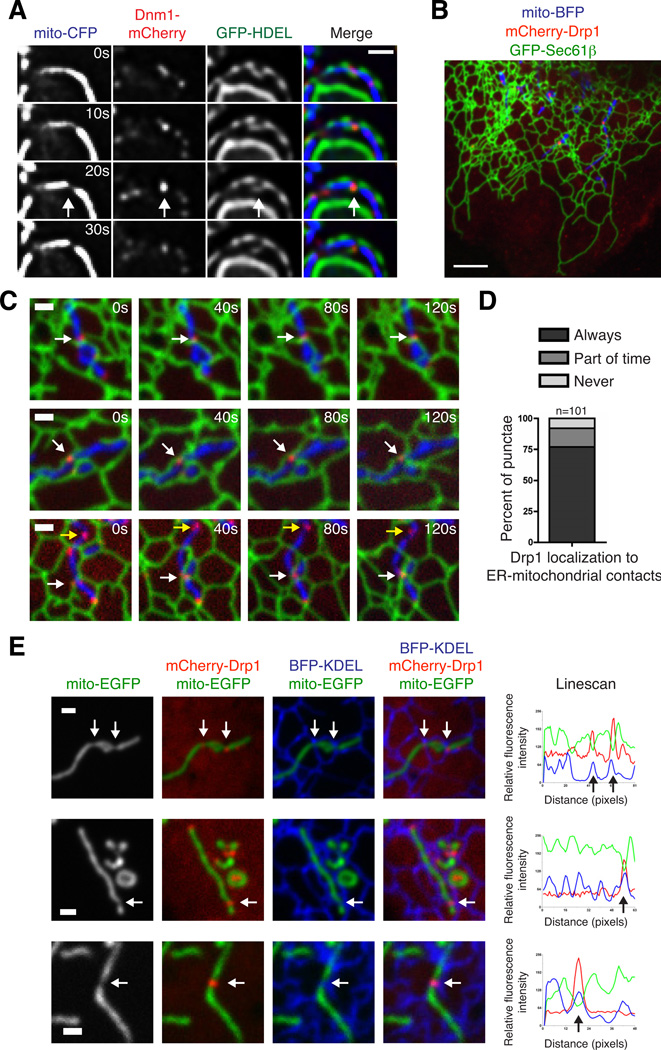
To ask whether ER-associated division events are spatially linked to the mitochondrial division machinery, we determined the relationship of ER-mitochondrial contacts to the division dynamin, Dnm1/Drp1. Dnm1 and Drp1 assemble into punctate structures at steady state and a subset of these structures are found on mitochondria and at mitochondrial division sites (6, 16, 17). We imaged live yeast transformed with Dnm1-mCherry, mito-CFP and GFP-HDEL (ER) and observed that a large percentage of Dnm1 punctae were at sites of mitochondrial-ER contact (46%, n=225). These Dnm1 punctae could be observed at sites where ER tubule crossover and mitochondrial division occurred (Fig. 3A). In Cos-7 cells transiently transfected with GFP-Sec61β (ER), mito-BFP, and mCherry-Drp1, we observed that the majority of Drp1 punctae stably associated with mitochondria and localized to ER-mitochondrial contacts over time (Fig. 3B–D and Movie S5). Furthermore, a subset of Drp1 at these contacts was associated with a mitochondrial constriction site (78%, excluding punctae localized to mitochondrial tips, n=50). The mitochondrial constrictions marked by Drp1 punctae were always at either ER tubule crossovers (81%) or adjacent to them (19%; Figs. 3E and S4). Together, the localization of the mitochondrial division dynamins in yeast and mammalian cells to regions of ER-mitochondrial contacts and the observations that these regions are associated with constricted mitochondria and subsequent division indicate a direct role of the ER in the process of mitochondrial division.
Figure 3.
Dnm1/Drp1-mediated mitochondrial division occurs at ER contact sites. (A) Time-lapse images of wild type yeast cells expressing mito-CFP, GFP-HDEL (ER) and Dnm1-mCherry. A single focal plane is shown. Arrows indicate the site of mitochondrial division, which is notably marked by both ER-mitochondria contact and Dnm1. (B) Merged image of a live Cos-7 cell expressing GFP-Sec61β (ER), mito-BFP, and mCherry-Drp1. (C) Examples of cells as in (B) that show that Drp1 punctae maintain co-localization with positions of ER-mitochondrial contact over time. White arrows indicate Drp1 punctae that maintain contact with both the ER and mitochondria. Yellow arrows indicate a rare example of Drp1 that does not contact the ER. (D) Graph indicating the percentage of mitochondrial Drp1 punctae that co-localize with the ER membrane over a two-minute timecourse. (E) Examples of mitochondrial constrictions at ER contact sites marked by Drp1. Left panels show Cos-7 cells expressing mito-EGFP, BFP-KDEL (ER), and mCherry-Drp1, merged as indicated. Right panels are Linescans drawn through the mitochondria and show the relative fluorescence intensity of mitochondria (green), ER (blue) and Drp1 (red) along its length. White arrows at constrictions on images correspond to black arrows shown on the Linescan. Additional examples are shown in Fig. S4. Scale bars: (A, C, E) 1 µm; (B) 5 µm.
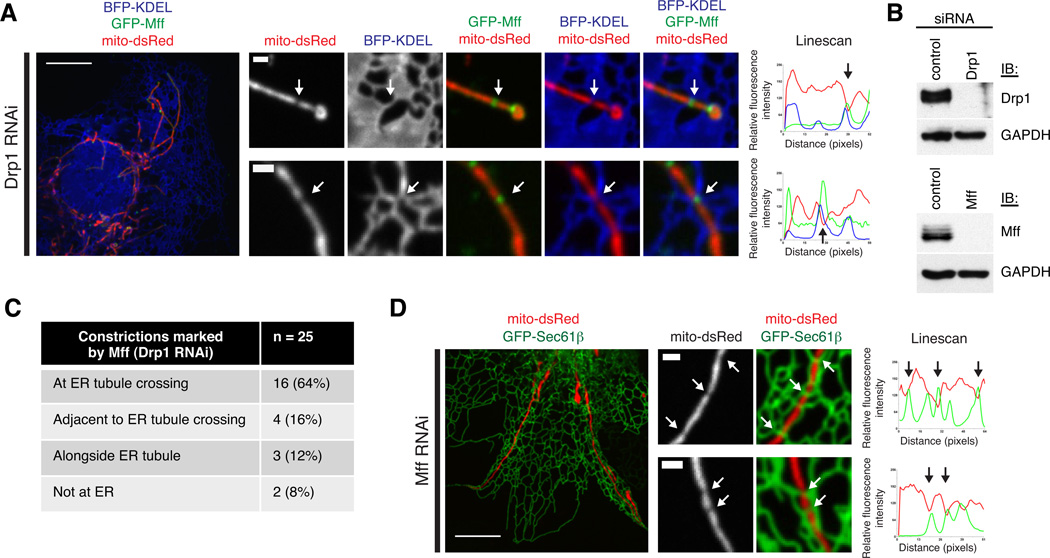
Mff is a mammalian-specific mitochondrial outer membrane protein required for mitochondrial localization of Drp1 and division (5, 18). Drp1 and Mff co-localize in punctate structures on mitochondria and Mff punctae persist in cells where Drp1 expression is reduced by RNAi (18). Thus, Mff punctae may mark the future sites of mitochondrial division prior to Drp1 recruitment (18). In Cos-7 cells transiently transfected with GFP-Mff, mCherry-Drp1 and mito-BFP, we observed that Mff circumscribed and localized to punctae on mitochondria, the majority of which co-localized with Drp1 (Fig. S5A–C). To test whether Mff punctae localize to ER contacts independently of Drp1, we depleted Drp1 from Cos-7 cells with siRNA, and co-transfected these cells with GFP-Mff, mito-dsRed, and BFP-KDEL (ER). Immuno-Drp1 was significantly depleted in Drp1 RNAi cells in comparison to the control cells (Fig. 4B). Selective depletion of Drp1 was further supported by the aberrant and elongated mitochondrial morphology in Drp1 RNAi cells (Figs. 4A and S5D). As expected (18), in Drp1 depleted cells, Mff punctae localized to mitochondria (Fig. 4A). We asked whether mitochondria were constricted at Mff punctae in the absence of Drp1, and if so, whether these sites localized to ER contacts. Of the 25 constrictions we resolved, 16 were at an ER crossover (64%), and another 4 were adjacent to an ER tubule crossing (16%; Figs. 4A, 4C and S6). Thus, Mff localizes in a Drp1-independent manner to mitochondrial constrictions at sites of ER contact. We next asked whether the ER localizes to regions of mitochondrial constriction in the absence of Mff. Cos-7 cells were depleted of Mff by siRNA and co-transfected with GFP-Sec61β (ER) and mito-dsRed. As expected, mitochondrial morphology was elongated in these cells (Figs. 4B, 4D, and S5E). In cells depleted of Mff, we observed mitochondrial constriction at sites of ER contact, indicating that ER-mitochondrial contacts form and mark positions of mitochondrial constriction independently of both Mff and Drp1 recruitment (Fig. 4D).
Figure 4.
The ER localizes to mitochondrial constrictions prior to Drp1 and Mff recruitment. (A) Examples of mitochondrial constrictions at ER contacts marked by Mff in Cos-7 cells depleted of Drp1. Left and center panels show images of these cells expressing mito-dsRed, BFP-KDEL (ER), and GFP-Mff, merged as indicated. Right panels are Linescans drawn through the mitochondria and show the relative fluorescence intensity of mitochondria (red), ER (blue) and Mff (green) along their length. White arrow position at constrictions corresponds to black arrows on the Linescan. Additional examples are shown in Fig. S6. (B) Western blots with antibody against Drp1 (top panel) or Mff (bottom panel) and GAPDH demonstrate depletion of Drp1 in lysates from cells transfected with siRNA against Drp1 (as in Fig. 4A) or Mff (as in Fig. 4D) compared to control RNAi cells. (C) Table indicating the number of Mff-localized mitochondrial constrictions in Drp1-depleted cells that co-localize with ER tubules, from 23 cells. (D) As in (A), for cells depleted of Mff and expressing GFP-Sec61β (ER; green on Linescan) and mito-dsRed (red on Linescan). Scale bars: (A and C, left panels) 5 µm; (A and C, right panels) 1 µm.
Here, we have shown that ER-mitochondrial contacts are a conserved feature of mitochondrial division. We envision two ways that ER contact might directly regulate mitochondrial division: (1) ER proteins intimately participate in division and/or (2) ER tubules physically wrap around and constrict mitochondria to a diameter comparable to Dnm1/Drp1 helices to facilitate their recruitment and assembly to complete fission (Fig. S9). The latter is attractive given that the diameter of Dnm1 helices (~110–130 nm) is significantly narrower than that of mitochondria and is quite similar to the diameter of constricted mitochondria at ER tubule contacts (138 nm and 146 nm) (4, 6–9).
Regardless of the exact mechanism, the ER appears to mark the division site and is likely to be an active participant in this process because it remains in contact with the mitochondria through the entire fission event. Many human diseases are associated with excessive mitochondrial division, raising the intriguing possibility that these diseases could involve an alteration of ER-mitochondrial contacts.
Supplementary Material
Acknowledgments
This work is supported by NIH grant R01 GM083977 and a Searle Scholar award (to G.K. Voeltz), NIH training grant GM08759 (to J.R. Friedman), NIH grant R01 GM062942 and an American Heart Innovative Research Grant (to J. Nunnari), grants from the Biological Sciences Initiative (BURST grant) and the Undergraduate Research Opportunity Program at the University of Colorado (to J.R. DiBenedetto). We thank the Boulder 3D Electron Microscopy facility for shared equipment and helpful suggestions.
Footnotes
Supporting Online Material
Materials and Methods
SOM Text
Figs. S1–S9
References (20–29)
Movies S1–S5
References and Notes
- 1.Cho DH, Nakamura T, Lipton SA. Mitochondrial dynamics in cell death and neurodegeneration. Cell. Mol. Life Sci. 2010;67:3435. doi: 10.1007/s00018-010-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoon Y, Galloway CA, Jhun BS, Yu T. Mitochondrial dynamics in diabetes. Antioxid. Redox Signal. 2011;14:439. doi: 10.1089/ars.2010.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lackner LL, Nunnari JM. The molecular mechanism and cellular functions of mitochondrial division. Biochim. Biophys. Acta. 2009;1792:1138. doi: 10.1016/j.bbadis.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingerman E, et al. Dnml forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 2005;170:1021. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 2008;19:2402. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Legesse-Miller A, Massol RH, Kirchhausen T. Constriction and Dnmlp recruitment are distinct processes in mitochondrial fission. Mol. Biol. Cell. 2003;14:1953. doi: 10.1091/mbc.E02-10-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 1999;4:815. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 8.Yoon Y, Pitts KR, McNiven MA. Mammalian dynamin-like protein DLP1 tubulates membranes. Mol. Biol. Cell. 2001;12:2894. doi: 10.1091/mbc.12.9.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mears JA, et al. Conformational changes in Dnml support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 2011;18:20. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Brito OM, Scorrano L. An intimate liaison: spatial organization of the endoplasmic reticulum-mitochondria relationship. EMBO J. 2010;29:2715. doi: 10.1038/emboj.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 12.Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman JR, Webster BM, Mastronarde DN, Verhey KJ, Voeltz GK. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J. Cell Biol. 2010;190:363. doi: 10.1083/jcb.200911024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West M, Zurek N, Hoenger A, Voeltz GK. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 2011;193:333. doi: 10.1083/jcb.201011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 16.Sesaki H, Jensen RE. Division versus fusion: Dnmlp and Fzolp antagonistically regulate mitochondrial shape. J. Cell Biol. 1999;147:699. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drpl is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 2001;12:2245. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otera H, et al. Mff is an essential factor for mitochondrial recruitment of Drpl during mitochondrial fission in mammalian cells. J. Cell Biol. 2010;191:1141. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Materials and methods are available as supporting material on Science Online.
- 20.Naylor K, et al. Mdvl interacts with assembled dnml to promote mitochondrial division. J. Biol. Chem. 2006;281:2177. doi: 10.1074/jbc.M507943200. [DOI] [PubMed] [Google Scholar]
- 21.Lackner LL, Horner JS, Nunnari J. Mechanistic analysis of a dynamin effector. Science. 2009;325:874. doi: 10.1126/science.1176921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossanese OW, et al. A role for actin, Cdclp, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J. Cell Biol. 2001;153:47. doi: 10.1083/jcb.153.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prinz WA, et al. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 2000;150:461. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westermann B, Neupert W. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast. 2000;16:1421. doi: 10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 25.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 26.Song Z, Ghochani M, McCaffery JM, Frey TG, Chan DC. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol. Biol. Cell. 2009;20:3525. doi: 10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata Y, et al. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem. 2008;283:18892. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zurek N, Sparks L, Voeltz G. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic. 2011;12:28. doi: 10.1111/j.1600-0854.2010.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yasukawa K, et al. Mitofusin 2 inhibits mitochondrial antiviral signaling. Set Signal. 2009;2:ra47. doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.