Abstract
We developed a pipeline to integrate the proteomic technologies used from the discovery to the verification stages of plasma biomarker identification and applied it to identify early biomarkers of cardiac injury from the blood of patients undergoing a therapeutic, planned myocardial infarction (PMI) for treatment of hypertrophic cardiomyopathy. Sampling of blood directly from patient hearts before, during and after controlled myocardial injury ensured enrichment for candidate biomarkers and allowed patients to serve as their own biological controls. LC-MS/MS analyses detected 121 highly differentially expressed proteins, including previously credentialed markers of cardiovascular disease and >100 novel candidate biomarkers for myocardial infarction (MI). Accurate inclusion mass screening (AIMS) qualified a subset of the candidates based on highly specific, targeted detection in peripheral plasma, including some markers unlikely to have been identified without this step. Analyses of peripheral plasma from controls and patients with PMI or spontaneous MI by quantitative multiple reaction monitoring mass spectrometry or immunoassays suggest that the candidate biomarkers may be specific to MI. This study demonstrates that modern proteomic technologies, when coherently integrated, can yield novel cardiovascular biomarkers meriting further evaluation in large, heterogeneous cohorts.
Numerous studies have used proteomic strategies to discover candidate protein biomarkers for a range of diseases, including those affecting cardiovascular biology. Yet no protein biomarker identified using proteomics has been introduced into clinical use1–4. To date, no demonstrably successful strategy has emerged to progressively credential (that is, provide additional data to support a candidate’s prioritization for clinical validation) putative protein biomarkers from the discovery phase through to their initial clinical validation. Many groups have used the exceptional sensitivity and dynamic range of modern mass spectrometers for proteomics discovery. However, the available instrumentation has yet to be adapted to specifically address the daunting bottleneck left by findings of unsubstantiated clinical relevance. Comparative discovery proteomics analyses that compare case and control samples generally couple protein and peptide fractionation and enrichment methods with high-performance mass spectrometry (MS) to increase coverage of the proteome, and often generate many hundreds of differentially abundant candidate biomarkers5,6. Discovery proteomics may be most effectively implemented using either tissue or fluids proximal to the site of disease where biomarkers are likely to be enriched. However, clinical tests need to measure biomarkers in patient blood, and there is currently no way to predict which of the candidate proteins identified during the discovery phase are likely to be detectable in plasma, nor which of the hundreds of differentially abundant proteins detected are truly disease-related. Adequate solutions for these serious technological barriers to moving candidate biomarker proteins toward clinical implementation presently do not exist6,7.
Quantitative antibody-based assays are the current method of choice for credentialing candidate biomarkers in patient plasma. Although it is difficult to derive an exact count, it is likely that antibody reagents suitable for configuring sandwich immunoassays currently exist for <2,000 of the >20,000 proteins in the human proteome (Guo-Liang Liu, Epitomics, personal communication). Multiple reaction monitoring MS (MRM-MS, also referred to as selected (S)RM-MS) is a rapidly emerging technology for construction of multiplexed assays for proteins in patient plasma8–10, but generation of quantitative MS-based assays employing stable isotope-labeled peptides is both time consuming and expensive. Generalizable approaches are therefore needed to identify and prioritize the subset of candidate biomarker proteins that are detectable in peripheral blood (a process we refer to as qualification) before investing intensive resources to generate either MS-based assays or immunoassays to quantitatively measure these proteins in additional samples (a process termed verification)6,7. We previously posited a testable discovery-through-verification biomarker pipeline that includes, first, proteomics-based discovery of candidate biomarker proteins in proximal fluid or tissue of patients; second, qualification of discovered candidates in the peripheral blood of additional patient samples using label-free targeted high-performance liquid chromatography (LC)-MS/MS; and third, verification of discovered and qualified candidates in peripheral blood, using targeted, quantitative MS-based assays with isotope-labeled peptide standards6,7,9–11.
Here we present a proof of principle demonstration that this coherent, MS-intensive pipeline, employing high-performance LC-MS/MS, accurate inclusion mass screening11 (AIMS) and stable isotope dilution (SID)-MRM-MS in an integrated fashion for biomarker candidate discovery, analytical qualification and quantitative verification, respectively, yields novel cardiovascular biomarkers that merit further evaluation in large, heterogeneous patient cohorts. We employed a human model of planned MI, septal ablation for hypertrophic cardiomyopathy12,13, to faithfully reproduce clinical aspects of spontaneous MI (Supplementary Results and Discussion). The analytical methods and statistical approaches used should be generalizable to biomarker discovery and verification in any other diseases, particularly in real-world clinical scenarios where individuals serve as their own controls.
RESULTS
Discovery using plasma from the coronary sinuses of PMI patients
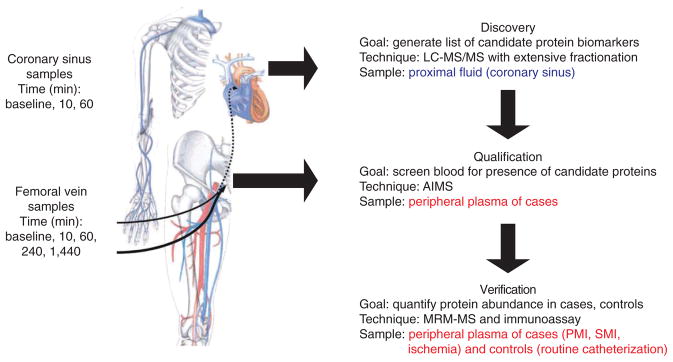
An overview of the proteomics biomarker pipeline and its application to the model of acute myocardial infarction is shown in Figure 1. Clinical characteristics of the patients are detailed in Supplementary Table 1. In the discovery phase, we used blood from the coronary sinuses of three PMI patients sampled at baseline and at 10 and 60 min after PMI (nine samples total) to generate a candidate biomarker list. Plasma was immunoaffinity-depleted of 12 high-abundance proteins, enzymatically digested with LysC followed by trypsin, and extensively fractionated at the peptide level by strong cation exchange chromatography into 80 fractions that were analyzed by nanoflow LC-MS/MS. The MS/MS spectra acquired were searched against the human IPI database using Spectrum Mill Proteomics Workbench.
Figure 1.
A pipeline for biomarker verification and its application to a human model of myocardial injury. Blood samples were collected from the coronary sinus of patients undergoing alcohol septal ablation for hypertrophic cardiomyopathy (planned myocardial infarction or PMI) before ablation (baseline), and at 10 and 60 min after ablation. The coronary sinus samples, comprising a proximal fluid of the heart, were used for discovery proteomics, using abundant protein depletion and extensive fractionation and LC-MS/MS of peptides to generate a prioritized list of biomarker candidates. Peripheral blood was collected from patients undergoing PMI at the same time points and additional time points extending to 24 h after ablation. Blood collected up to 4 h after ablation was used for analytical qualification by AIMS, a process that determines which of the differentially abundant proteins from the discovery experiments are detectable in peripheral blood. Qualified protein biomarker candidates were subsequently quantitatively measured in peripheral blood using immunoassays (when antibodies were available) and SID-MRM-MS when antibody reagents were not available. Figure was adapted from reference 29. Blue type indicates proximal fluid. Red type indicates peripheral blood.
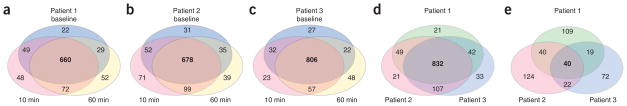
We identified 1,105 unique proteins in the nine coronary sinus plasma samples, with an average of 871 proteins/sample using a minimum of two peptides/protein and a peptide false discovery rate (FDR; Supplementary Methods) ≤1.5% (Fig. 2). A list of all proteins and peptides identified is presented in Supplementary Tables 2 and 3, and the number of distinct proteins identified in each patient and at each time point is shown in Supplementary Figure 1. More than 70% of the proteins identified were observed in all three PMI patients (Fig. 2d).
Figure 2.
Venn diagrams summarizing proteins identified in the coronary sinuses of PMI patients. (a–c) The overlap of proteins identified across all three time points in patients 1, 2 and 3, respectively. Proteins were identified with a minimum of two unique peptides per protein and peptide FDR ≤ 1.5%. (d) We identified 1,105 unique proteins in the nine coronary sinus samples analyzed by LC-MS/MS with >70% of the proteins identified in all three patients. Label-free relative quantification of peptides was done to prioritize candidate proteins for subsequent qualification and verification studies. (e) A minimum of a fivefold change in the MS-derived discovery data between baseline and either the 10-min or 60-min time point was required. 121 proteins met these criteria in all three or any two patients combined.
Label-free, relative quantification of peptides was used to identify proteins changing in abundance in the discovery data and to generate a list of candidate biomarker proteins of PMI for subsequent qualification and verification (Fig. 1). For a protein to be nominated as a candidate, we required a minimum fivefold change in the MS-derived precursorion abundance for a minimum of two unique peptides/protein between baseline and either the 10-min or 60-min samples (Online Methods, calculations which demonstrate the ability to detect fivefold changes in biomarkers for various values of coefficient of variation (CV) and number of sample pairs). A subset of the proteins that met these criteria is presented in Table 1 with the full list of proteins presented in Supplementary Table 4. Levels of 40 proteins were increased more than fivefold as compared to baseline in all three patients at either or both the 10-min or 60-min time points, whereas levels of 81 additional proteins were increased more than fivefold after injury in at least two patients (Fig. 2e).
Table 1.
Summary of data related to 52 protein biomarker candidates that were detected in coronary sinus plasma of PMI patients by discovery proteomics, qualified as being detectable in peripheral plasma of PMI patients and prioritized for quantitative assay development by MRM-MS
| No. | Candidate biomarker protein | AIMS
|
Correlation
|
Clusterc | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Normalized total intensity (×106)
|
Relative ratio
|
|||||||||
| Baseline (BL) | 10 min | 60 min | 10:BL | 60:BL | 60:10 | AIMSa | DDAb (pool) | |||
| 1 | GPI | 76 | 82 | 162 | 1.1 | 2.1 | 2.0 | 1.0 | 0.9 | 5 |
| 2 | CYCS | 14 | 38 | 146 | 2.7 | 10.4 | 3.9 | 1.0 | 1.0 | 5 |
| 3 | LOC100133739 | 2,440 | 4,930 | 1,990 | 2.0 | 0.8 | 0.4 | 1.0 | 1.0 | 3 |
| 4 | MPO | 520 | 2,920 | 2,230 | 5.6 | 4.3 | 0.8 | 1.0 | 1.0 | 1 |
| 5 | CKM | 818 | 1,180 | 3,150 | 1.4 | 3.9 | 2.7 | 1.0 | 1.0 | 5 |
| 6 | LIPC | 51 | 106 | 70 | 2.1 | 1.4 | 0.7 | 1.0 | n.a. | 3 |
| 7 | MDH1 | 49 | 180 | 883 | 3.7 | 18.1 | 4.9 | 1.0 | 1.0 | 5 |
| 8 | CKB | 0 | 0 | 71 | 0.0 | >20 | >20 | 1.0 | n.a. | 5 |
| 9 | SCUBE1 | 0 | 176 | 60 | >20 | >20 | 0.3 | 1.0 | 1.0 | 3 |
| 10 | ADH1C | 0 | 0 | 150 | 0.0 | >20 | >20 | 1.0 | 0.6 | 5 |
| 11 | CSRP3 | 0 | 0 | 78 | 0.0 | >20 | >20 | 1.0 | 1.0 | 5 |
| 12 | SMOC1 | 0 | 393 | 96 | >20 | >20 | 0.2 | 1.0 | 1.0 | 3 |
| 13 | FABP3 | 0 | 41 | 144 | >20 | >20 | 3.5 | 1.0 | 1.0 | 5 |
| 14 | FSTL1 | 230 | 302 | 307 | 1.3 | 1.3 | 1.0 | 1.0 | 1.0 | 1 |
| 15 | ENSP00000348237 | 226 | 307 | 840 | 1.4 | 3.7 | 2.7 | 1.0 | 1.0 | 5 |
| 16 | GPD1 | 0 | 0 | 2 | 0.0 | >20 | >20 | 1.0 | n.a. | 5 |
| 17 | PGAM2 | 95 | 171 | 418 | 1.8 | 4.4 | 2.4 | 1.0 | 1.0 | 5 |
| 18 | CRELD1 | 11 | 34 | 26 | 3.2 | 2.4 | 0.7 | 1.0 | −0.8 | 1 |
| 19 | ACLP1 | 48 | 130 | 37 | 2.7 | 0.8 | 0.3 | 1.0 | 1.0 | 3 |
| 20 | MB | 333 | 1,670 | 3,160 | 5.0 | 9.5 | 1.9 | 1.0 | 1.0 | 5 |
| 21 | SERPINA11 | 592 | 1,010 | 702 | 1.7 | 1.2 | 0.7 | 1.0 | 0.9 | 3 |
| 22 | SOD1 | 268 | 660 | 654 | 2.5 | 2.4 | 1.0 | 1.0 | 1.0 | 1 |
| 23 | PSMA7 | 64 | 85 | 80 | 1.3 | 1.3 | 0.9 | 1.0 | n.a. | 1 |
| 24 | AK1 | 43 | 62 | 95 | 1.4 | 2.2 | 1.5 | 1.0 | −0.1 | 5 |
| 25 | CLSTN1 | 73 | 213 | 93 | 2.9 | 1.3 | 0.4 | 1.0 | 0.8 | 3 |
| 26 | LOC654188 | 171 | 302 | 341 | 1.8 | 2.0 | 1.1 | 1.0 | −1.0 | 1 |
| 27 | SPON1 | 223 | 492 | 253 | 2.2 | 1.1 | 0.5 | 0.9 | 1.0 | 3 |
| 28 | TPI1 | 190 | 370 | 864 | 1.9 | 4.5 | 2.3 | 0.9 | 0.9 | 5 |
| 29 | PEBP1 | 41 | 27 | 156 | 0.7 | 3.8 | 5.9 | 0.9 | 0.9 | 5 |
| 30 | TFPI | 309 | 628 | 240 | 2.0 | 0.8 | 0.4 | 0.9 | 1.0 | 3 |
| 31 | MMRN2 | 29 | 114 | 98 | 3.9 | 3.3 | 0.9 | 0.9 | −0.6 | 1 |
| 32 | LTF cDNA FLJ78440 | 918 | 3,130 | 2,420 | 3.4 | 2.6 | 0.8 | 0.9 | 0.9 | 1 |
| 33 | YWHAQ | 178 | 148 | 211 | 0.8 | 1.2 | 1.4 | 0.9 | 1.0 | 2 |
| 34 | TPM1 | 265 | 350 | 428 | 1.3 | 1.6 | 1.2 | 0.8 | 1.0 | 5 |
| 35 | GAPDH | 934 | 1,780 | 2,390 | 1.9 | 2.6 | 1.3 | 0.8 | 0.8 | 1 |
| 36 | YWHAH | 186 | 145 | 223 | 0.8 | 1.2 | 1.5 | 0.8 | −0.8 | 2 |
| 37 | CAST | 6 | 25 | 27 | 4.4 | 4.7 | 1.1 | 0.8 | n.a. | 1 |
| 38 | CTSF | 172 | 234 | 135 | 1.4 | 0.8 | 0.6 | 0.8 | 0.6 | 3 |
| 39 | PPBP | 10,600 | 13,300 | 9,160 | 1.3 | 0.9 | 0.7 | 0.8 | 1.0 | 3 |
| 40 | AKAP12 | 0 | 12 | 9 | >20 | >20 | 0.8 | 0.7 | n.a. | 1 |
| 41 | PECAM1 | 0 | 0 | 24 | 0.0 | >20 | >20 | 0.7 | n.a. | 5 |
| 42 | MDH2 | 0 | 26 | 278 | >20 | >20 | 10.7 | 0.7 | 0.7 | 5 |
| 43 | PGM1 cDNA FLJ92549 | 18 | 22 | 69 | 1.2 | 3.9 | 3.2 | 0.7 | n.a. | 5 |
| 44 | GOT1 | 686 | 433 | 992 | 0.6 | 1.4 | 2.3 | 0.6 | 0.9 | 2 |
| 45 | CRYZ | 0 | 0 | 6 | 0.0 | >20 | >20 | 0.6 | n.a. | 5 |
| 46 | NME1 | 0 | 18 | 0 | >20 | 0.0 | 0.0 | 0.6 | −0.1 | 3 |
| 47 | GP5 | 319 | 368 | 165 | 1.2 | 0.5 | 0.4 | 0.5 | 1.0 | 4 |
| 48 | MYL3 | 0 | 18 | 129 | >20 | >20 | 7.1 | 0.5 | 0.4 | 5 |
| 49 | DLD | 0 | 285 | 144 | >20 | >20 | 0.5 | 0.5 | 0.6 | 1 |
| 50 | PF4V1 | 1,580 | 2,700 | 1,320 | 1.7 | 0.8 | 0.5 | 0.5 | 1.0 | 3 |
| 51 | FHL1 | 21 | 28 | 114 | 1.4 | 5.6 | 4.0 | 0.5 | 0.4 | 5 |
| 52 | FLNC | 0 | 61 | 0 | >20 | 0.0 | 0.0 | 0.5 | 1.0 | 3 |
121 proteins were analyzed by AIMS for analytical qualification using an independent pool of peripheral plasma collected from 10 PMI patients at baseline and 10 min and 60 min after ablation. For each of the 121 proteins in the inclusion list, its temporal trend in AIMS (Correlation – AIMS) across the 3 time points was correlated with averaged temporal profiles in the discovery proteomics of individual patients. 52 proteins have a moderate or better Pearson correlation score (correlation coefficient ≥0.4) and are prioritized for quantitative assay development by MRM-MS. Proteins only detected by AIMS versus data-dependent analysis of pooled peripheral plasma are in boldface type. See Supplementary Table 5 for the complete set of AIMS results.
Correlation of AIMS and
correlation of DDA in pooled plasma samples to original discovery done in individual patient samples.
Trends in temporal abundance response for the qualified candidate biomarkers across the time points were clustered into five distinct groups (Online Methods and Supplementary Fig. 3).
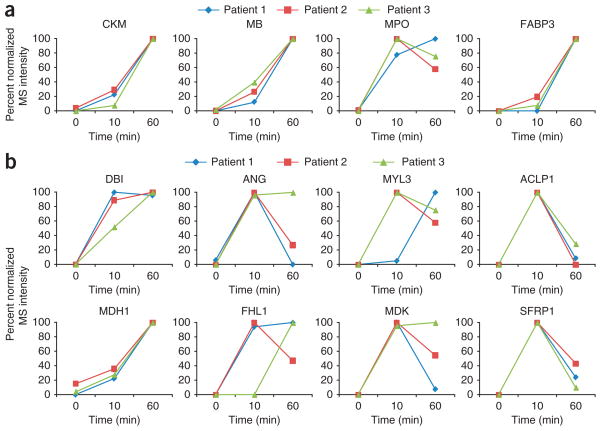
The list of differentially regulated proteins detected in the coronary sinus plasma samples from multiple PMI patients contains many known markers of myocardial injury including myoglobin (MB), myeloperoxidase (MPO), creatine kinase-myocardial isoform B (CKB), creatine kinase-myocardial isoform M (CKM) and fatty-acid binding protein (FABP)14,15. Cardiac troponin T (cTnT) was also observed in the discovery data from two patients, although only a single high-scoring peptide of this low abundance protein was detected. The list also contains many potentially novel biomarkers of cardiovascular disease, including aortic carboxypeptidase-like protein (ACLP1), a transcriptional repressor implicated in cardiovascular wound healing16; four-and-a-half LIM domain protein 1 (FHL1), a cardiomyocyte protein that mediates a hypertrophic biomechanical stress response17; angiogenin (ANG), a potent mediator of new blood vessel formation18; and MYL3, the regulatory light chain of myosin that may serve as a target for caspase-3 in dying cardiomyocytes19. Kinetic analyses of the discovery MS data for the known (Fig. 3a) and putative biomarkers (Fig. 3b) revealed that these proteins were at very low to undetectable levels in the coronary sinus at baseline, then increased by more than fivefold at 10 and 60 min after PMI in each of the three patients. Almost all of the MS changes documented at 10 min were also observed at 60 min, underscoring the consistency of our findings.
Figure 3.
Kinetic analyses of known and putative biomarkers for acute myocardial infarction in PMI patients from discovery proteomics. (a) Known markers, such as CKM, MB, MPO and FABP showed little to no detection at baseline in coronary sinus plasma followed by a >5-fold increase at 10 min and 60 min after ablation in three PMI patients. (b) Eight potentially novel candidate biomarkers from discovery proteomics. These proteins showed no to little detection at baseline in coronary sinus plasma, then increased in MS abundance by at least fivefold at 10 min or 60 min after ablation in all three PMI patients. MRM-MS assays were configured for ACLP1, myosin light chain 3 and four-and-a-half LIM domain protein 1 to quantify these candidates in peripheral plasma of four PMI patients. Antibodies available for ACBP, ANG, MDK, malate dehydrogenase and ACLP1 were used either in ELISAs or western blot analyses to verify these candidates in additional patients (Supplementary Methods).
Qualification in peripheral plasma of PMI patients by AIMS
We incorporated AIMS technology11 into our pipeline to ascertain which of the proteins discovered in proximal fluid (e.g., coronary sinus plasma) could also be detected in peripheral blood samples from a distinct set of subjects in the ‘qualification’ step (Fig. 1)6. AIMS is a targeted, label-free MS approach for relative quantification on the Orbitrap or similar hybrid high-performance MS systems in which MS/MS spectra are triggered and acquired only when accurate mass and charge pairs on the inclusion list are detected. Here we used AIMS to help prioritize the list of candidate biomarkers obtained using discovery proteomics and to identify specific peptides from these candidates that are likely to be well-suited for developing quantitative SID-MRM-MS assays11, thereby facilitating this resource-intensive activity. Plasma processing for analysis by AIMS differed in three important ways from that used in the discovery phase (Supplementary Methods). First, we used peripheral patient plasma rather than coronary sinus plasma; second, we used a pooling strategy to dampen differences due to interindividual variability, strengthen signal from coherently modulated proteins and reduce the total number of samples requiring analysis (the rationale for the pooling strategy used is described in detail in Supplementary Results and Discussion); and third, the number of strong cation exchange peptide fractions we analyzed was cut in half relative to data-dependent LC-MS/MS, taking advantage of the increased sensitivity of AIMS to increase throughput.
Peptides from the 121 candidate biomarker proteins identified in the coronary sinus by discovery proteomics were targeted for analysis by AIMS in three discrete pools of peripheral plasma from ten new PMI patients, with one pool for each of the baseline, 10 min and 60 min post-ablation samples (Table 1 and Supplementary Table 5). The AIMS inclusion list consisted of 1,904 mass and charge pairs for peptides derived from the 121 proteins that were upregulated by at least fivefold between baseline and either of the 10-min and 60-min time points in the discovery proteomics experiments performed using individual patient samples. The inclusion list included peptides derived from proteins that were observed in the discovery proteomics data as well as additional peptides that were computationally predicted to be among the highest responding peptides for each candidate protein using the program ESP20 (Supplementary Table 6). A protein was considered to have been qualified by AIMS if two or more peptides derived from that protein passed the autovalidation criteria in any of the three time-point pools (Supplementary Methods). Peptides uniquely derived from 83 of the 121 candidate biomarker proteins from discovery proteomics experiments were detected and sequenced by AIMS (Table 1, Supplementary Table 5 and Supplementary Fig. 2).
The list of qualified proteins (Table 1 and Supplementary Table 5) contains all of the proteins found in discovery proteomics of individual patients that are known to be associated with myocardial injury, as well as many of the potentially novel biomarkers of cardiovascular injury (that is, those proteins not previously identified in the published literature as being associated with cardiovascular disease, but that both were upregulated fivefold and showed clear temporal trends within each patient).
To further prioritize the list of 83 qualified candidates for MRM assay development, we analyzed the correlation in temporal abundance for candidate biomarker proteins from the discovery proteomics data with the AIMS results at the three time points. For each of the 121 unique proteins in the AIMS inclusion list, we determined the Pearson correlation of its temporal trend across the three time points in the AIMS experiments with the average temporal profile of each protein in the discovery proteomics experiments for each individual patient (Online Methods). Of the 83 proteins detected and qualified by AIMS, 52 proteins (63%) have a moderate or better Pearson correlation score (correlation coefficient ≥0.4) to the corresponding data for individual patients obtained using discovery proteomics (Table 1). AIMS, together with the new correlation analysis, reduced the list of 121 protein candidates from discovery by 57% to a prioritized list of 52 candidates for quantitative assay development by MRM-MS. The abundance profile across time points for the 83 qualified candidate biomarkers clustered into five subgroups (Supplementary Fig. 3 summarizes the temporal trends that the markers follow). Although these clusters were not used to prioritize candidates, it is important to note that a majority of the 52 qualified candidates fall into clusters 1, 3 and 5. Clusters 1 and 5 are profiles that potentially could satisfy clinical utility for a candidate or a panel of candidates based on their early increase and sustained high levels after PMI.
To determine whether qualification by AIMS provides more information than performing additional discovery proteomics experiments using data-dependent acquisition (DDA) of sequencing data, we carried out additional discovery proteomics experiments on the same pooled, peripheral plasma samples in parallel with the targeted, AIMS experiments described above. Data from both sets of experiments were analyzed by Spectrum Mill using the same extraction, search and autovalidation criteria, and the results were compared for the candidate proteins on the inclusion list. Significantly more of the candidate biomarker proteins (17/83 proteins, or 20%) from the inclusion list were detected by AIMS than by discovery proteomics using DDA. Of the 17 proteins not detected at all by the DDA analysis of the pooled samples, about 47% (8/17 proteins) had a very strong correlation (≥0.7 or higher) between AIMS and the original discovery proteomics data for the individual patients.
Verification in peripheral plasma by SID-MRM-MS
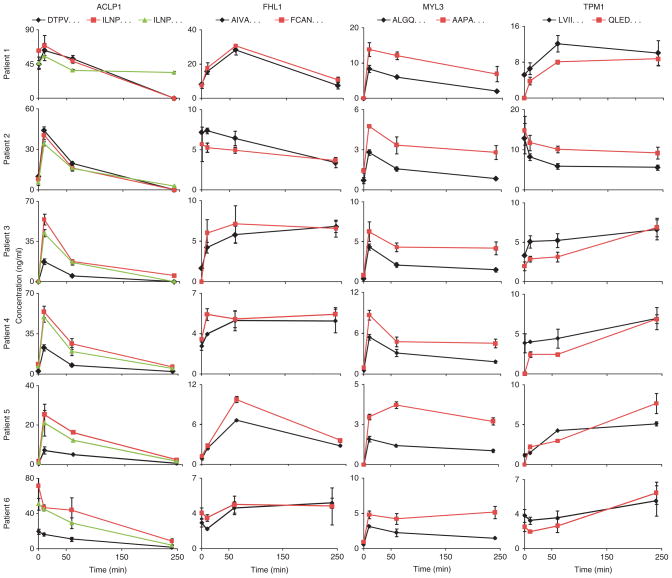
Quantitative verification of candidate biomarkers was conducted in the peripheral plasma of PMI patients using available antibodies (see below) as well as by SID-MRM-MS, a targeted, quantitative MS approach (Fig. 1). SID-MRM-MS proved to be essential, as antibody reagents suitable for construction of enzyme-linked immunosorbent assays (ELISAs) (that is, two per protein) were available for only 3 of the 52 protein biomarker candidates detected by AIMS (Supplementary Table 5). As a demonstration that SID-MRM-MS can be used to assay novel proteins in the absence of antibodies for quantitative immunoassay construction, we applied our SID-MRM-MS strategy (Supplementary Fig. 4) to develop quantitative assays to measure four of the novel biomarkers that were discovered and subsequently qualified by AIMS (ACLP1, FHL1, MYL3 and tropomyosin 1 (TPM1)). These four proteins were selected because their expression is enriched in the heart16,17,21. In addition, these proteins exhibited temporal trends representing two of the five temporal classes observed in the AIMS results up to 60 min after ablation (Supplementary Fig. 3), and their behavior at the 240-min time point in peripheral plasma of patients was of interest. Quantitative assays were successfully configured for ACLP1, FHL1, MYL3 and TPM1 using at least two and no more than five tryptic peptides per protein, which were observed in the discovery MS and/or the AIMS data (Supplementary Table 7 and Supplementary Fig. 5). These four novel proteins, together with several known markers of myocardial injury, including C-reactive protein (CRP), MPO and cTnT were measured at four time points in process triplicate (that is, new aliquots of patient plasma from the same time point were depleted, reduced and alkylated, trypsin digested, fractionated by SCX and analyzed by SID-MRM-MS) in six additional PMI patient samples using a 20-plex SID-MRM-MS assay we constructed.
ACLP1, FHL1, MYL3, TPM1, MPO and cTnT were readily quantified at multiple time points in the patient samples, with measured values ranging from ~1 ng/ml to 50 ng/ml, whereas measured levels of CRP ranged from 160 ng/ml to 5.5 μg/ml across all patients and time points (Fig. 4 and Supplementary Table 8). The temporal intensity profiles of three of the four novel proteins (ACLP1, FHL1 and TPM1) detected in AIMS experiments on pooled PMI patient samples are highly correlated to the temporal intensity profiles obtained by SID-MRM-MS in individual patients samples, with Pearson correlation values >0.85, and MYL3 with a moderate correlation of 0.36 (Supplementary Results and Discussion).
Figure 4.
Verification of novel candidate biomarkers in peripheral blood of PMI patients by targeted, quantitative MS. Multiplexed SID-MRM-MS–based assays were configured for six candidate proteins to precisely quantify their changes in peripheral blood from PMI patients at 10 min, 60 min and 240 min after ablation. Multiple signature peptides derived from each protein were used to quantify protein levels (Supplementary Tables 7 and 8). Measured concentrations for the four novel proteins ranged from 1 ng/ml to ~50 ng/ml across all patients and time points. Error bars indicate s.e.m. concentration measured at each time point (n = 3). Signature peptides are represented by the first four residues. ACLP1, aortic carboxypeptidase-like protein 1; FHL1, four-and-a-half LIM domain protein 1; MYL3, myosin light chain 3; TPM1, tropomyosin 1. Three known markers of cardiovascular disease were also monitored (Supplementary Table 8b).
Verification of protein changes by western blot analysis and ELISA
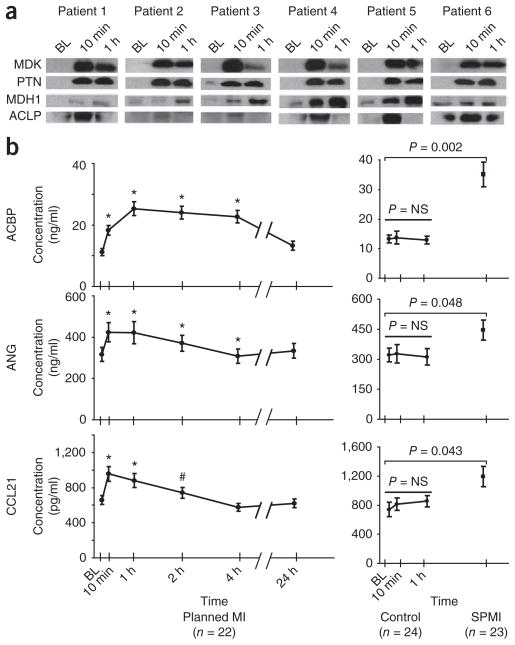
Antibody reagents for western blot analysis and ELISAs were available for the ten candidate proteins listed in Supplementary Table 5. Single antibody reagents were available for 7 of the 121 prioritized candidate biomarkers (Supplementary Table 5). We attempted to use these reagents for western blot analyses on coronary sinus samples from six additional subjects who underwent the PMI procedure. Only four of the ten antibodies detected recombinant protein spiked into normal plasma from a healthy female donor by western blot analysis. Findings for midkine (MDK), pleiotrophin (PTN), malate dehydrogenase 1 (MDH1) and ACLP1 were consistent with the discovery MS data (Fig. 5a). By contrast, the antibodies for MYL3, FHL1 and TPM1 did not detect endogenous protein in the PMI samples. Antibodies specific for MYL3, FHL1 and TPM1 detected recombinant protein at 10 ng/ml in buffer, but did not detect these proteins when spiked into human plasma, suggesting interference by other proteins or possible proteolysis of the spiked proteins by active proteases in the plasma matrix (data not shown).
Figure 5.
Verification of candidate biomarkers by western blot analysis and ELISA. (a) Single antibody reagents suitable for western blot analysis were available for MDK, PTN, malate dehydrogenase 1 (MDH1) and aortic carboxypeptidase-like protein 1 (ACLP1). Kinetic analysis of coronary sinus samples from six patients show consistency in the protein changes between the western blot results shown here and the MS-derived temporal trends shown in Figure 3 for the identical proteins. (b) For ANG, ACBP, and C-C motif chemokine 21 (CCL21), sandwiched immunoassays were either constructed (ANG) or commercially available (ACBP and CCL21), and were used to verify protein changes in peripheral plasma from a larger set of PMI patient samples, control samples and spontaneous MI cohorts. In the PMI cohort (b, left) ELISA results confirm significant changes in these candidate biomarkers as early as 10 min after the onset of myocardial injury. In patients with spontaneous MI (b, right) presenting for acute coronary angiography and intervention, significantly higher levels of these proteins were observed as compared to levels in patients who presented to the cardiac catheterization suite with non-acute coronary artery disease (controls, b, center). BL, baseline; NS, not significant; #, P < 0.05; *, P< 0.01.
For ANG, MDK, decorin (DCN) and secreted frizzled-related protein, two different commercially available antibodies recognized distinct regions of each of these proteins, enabling construction of ELISAs. In addition, ELISA kits were commercially available to detect C-C motif chemokine 21 (CCL21) and acyl CoA binding protein (ACBP). ELISAs for these six candidate proteins were constructed or purchased, and used for initial candidate verification and to conduct more extensive kinetic analyses using peripheral blood samples from an additional 22 individuals undergoing the ablation procedure (Fig. 5b, left). These studies confirmed highly significant changes in ANG, CCL21 and ACBP as early as 10 min after the onset of myocardial injury, with continued elevation of the proteins 2–4 h after injury.
Clinical validation of potential, novel biomarkers
Using available immunoassays, we explored the specificity of the findings observed in the PMI cohort by examining blood samples from patients undergoing routine cardiac catheterization, without the induction of myocardial infarction that occurs in the unique ablation injury model. Levels of ACBP, ANG and CCL21 were unchanged up to 60 min after routine catheterization in patients who had nonacute coronary artery disease (Fig. 5b (right panel, control)) and were similar to preinjury levels of PMI subjects (Fig. 5b, left panel).
We next examined whether our findings were applicable to a cohort of patients with spontaneous MI (SMI) presenting for acute coronary angiography and intervention. The onset of SMIs relative to sample collection was heterogeneous (162 ± 102 min), as was the extent of myocardial injury. The baseline characteristics for these patients are listed in Supplementary Table 1. We observed significantly higher levels of ACBP, ANG and CCL21 in the SMI patients, as compared to levels in those who presented to the cardiac catheterization suite with nonacute coronary artery disease (control) (Fig. 5b, right panel). SMI levels were similar to peak levels seen in PMI. Of note, cardiac catheterization alone was associated with changes in the levels of MDK, PTN, DCN and secreted frizzled-related protein, as observed with in-house constructed ELISAs (data not shown). Proteins with changes that were not specific to myocardial injury and that may instead reflect procedural events such as arteriotomy, catheter manipulation or drug therapy were eliminated for further evaluation using the appropriate patient controls.
Finally, as proteins were released early after the onset of the planned myocardial infarction, we examined whether levels were also higher in the setting of reversible myocardial ischemia. A total of 52 individuals undergoing exercise stress testing with myocardial perfusion imaging served as the study population: 26 with no evidence of ischemia (controls) and 26 with evidence of inducible ischemia (cases). The baseline characteristics and stress test performance parameters for these individuals are listed in Supplementary Table 9. The mean ages of the two groups were comparable, though as expected, patients with inducible ischemia had slightly more cardiac risk factors (3.0 ± 0.9 versus 2.1 ± 0.9) and were more likely to have a documented history of coronary disease.
The exercise stress test results of cases and controls are shown in Supplementary Figure 6. By design, all 26 cases had reversible perfusion defects, with the mean percentage of myocardium with a reversible perfusion defect being 17 ± 8%, whereas no controls had any degree of a reversible perfusion defect. We were interested to find that for two of the proteins, ACBP and ANG, baseline levels were higher in the ischemic as compared to the at-risk control patients. Furthermore, for ACBP, we also documented a modest augmentation in protein levels in the setting of myocardial ischemia that was not observed in the control subjects.
DISCUSSION
Emerging proteomics profiling technologies hold enormous promise for illuminating new biomarkers. However, successful applications to human disease are still lacking. This is due, in large part, to the lack of a coherent, demonstrably successful pipeline enabling systematic building of credentialing information around biomarker candidate proteins emerging from discovery proteomics experiments. Here we developed an MS-intensive pipeline that coherently integrates high-performance LC-MS/MS, AIMS and SID-MRM-MS for biomarker candidate discovery, analytical qualification and quantitative verification, respectively, and applied the pipeline in the context of cardiovascular disease to yield novel cardiovascular biomarkers meriting further evaluation in large, heterogeneous patient cohorts. The discovery, qualification and verification steps systematically informed the next stage of the pipeline and the analyses took specific advantage of key attributes of the MS-based technology platforms used at each stage. An essential feature of this pipeline is transitioning from the analysis of proximal fluid (or tissue) for biomarker candidate discovery to peripheral blood for qualification and verification of candidates.
We applied our pipeline approach, beginning with discovery proteomics, to a unique clinical model of MI that allowed for precise kinetic analysis in patients who serve as their own biological controls. Coronary sinus catheterization provided the opportunity to sample directly from the organ of interest. This approach enabled the use of a proximal fluid of the heart for discovery of candidate biomarker proteins, rather than peripheral plasma where proteins arising from the myocardium would have been more diluted. The consistent temporal changes in the levels of candidate biomarkers within individual patients and in comparisons between patients (Fig. 3) underscore the biological plausibility of the observed association between proteomic changes and MI. This study emphasizes the important point that small numbers of samples may be used for discovery if the effect size is large and if these initial findings can be followed up with methods (specifically AIMS and SID-MRM-MS) that enable large numbers of candidates to be further credentialed or discarded by analysis of additional patient samples. In the current study, we began with samples from three time points in three patients undergoing PMI, and focused on changes of at least fivefold in protein abundance before identifying a protein as a candidate. This experimental design enhanced our power to identify statistically meaningful changes. It is important to emphasize that the MS tools, data acquisition and analysis methods as well as the statistical tests used are not specific to this human model, but are broadly applicable to the analysis of any perturbational experiment, including the more common biomarker discovery paradigm in which cases and controls come from different patients.
Using untargeted, data-dependent LC-MS/MS–based proteomics for discovery, we identified 1,105 unique total proteins with two or more peptides and FDR ≤ 1.5% in the plasma from the coronary sinus, or 999 proteins after excluding immunoglobulins and common contaminants such as keratins. The identified proteins spanned ~6 or 7 orders of magnitude of abundance, based on detection of peptides from REG3, IGFBP4 and LCN2, all of which are known to be present at 1–130 ng/ml levels in the plasma of healthy people22. Consistent with prior studies23,24, our pipeline underscores the need for abundant protein depletion combined with extensive peptide- or protein-level fractionation before LC-MS/MS for identification of proteins present at low ng/ml range in plasma. In the present study, the nine discovery samples yielded >700 sample subfractions, necessitating ~2,800 h of instrument time on the Orbitrap for LC-MS/MS analyses. The resulting list of proteins detected with high confidence in plasma also adds to the list of high-quality studies of the human plasma proteome23,24.
Qualification by AIMS is an essential element of our pipeline for biomarker prioritization (Fig. 1), providing a reliable and relatively high-throughput method to prioritize lengthy lists of biomarker candidate methods discovered in proximal fluids or tissues. AIMS, a targeted, label-free, relative quantification method, can be thought of as the MS equivalent of a highly multiplexed western blot. Using AIMS, we effectively configured 121 MS-based western blots in a single series of analyses without the need for antibody reagents. AIMS analyses in peripheral plasma, together with the temporal correlation analysis, qualified 52 of the 121 candidate proteins (43%) derived from discovery proteomics in the coronary sinus, thereby prioritizing the candidate list to focus critical resources for quantitative assay development by MRM-MS on those qualified protein biomarker candidates with a high likelihood for success in being detected and quantified in peripheral patient plasma.
The inability to rediscover 17 out of 83 AIMS-qualified candidate biomarker proteins by discovery proteomics is important, and underscores that DDA is not as efficient as AIMS for candidate qualification. The high degree of correlation (>0.85 for three out of the four proteins verified by SID-MRM-MS) between the temporal behavior in protein abundance observed by AIMS and the more quantitative approach of SID-MRM-MS further demonstrates the utility of AIMS as a method to prioritize candidates for the resource-intensive SID-MRM-MS assay development. Together, these results suggest that AIMS is an essential technology in a functioning biomarker-discovery-through-verification pipeline, and that AIMS provides increased sensitivity for candidate biomarker qualification compared to data-dependent methods (Supplementary Results and Discussion). Although, in principle, it is possible to proceed directly from discovery proteomics to MRM-MS-based assay configuration, doing so is prohibitive with respect to the cost and time involved. Use of AIMS to prioritize assay development results in considerable time and cost savings (Supplementary Results and Discussion and Supplementary Methods).
The third step of our pipeline is verification6 using SID-MRM-MS, or ELISA for the minority of cases where antibodies are available (Supplementary Table 5). Antibodies suitable for construction of ELISAs were available for only four of the novel candidate biomarker proteins that emerged from discovery and single-antibody reagents or commercial ELISAs were available for seven other candidate proteins. In principle, antibody-based measurements could be used at all steps in the validation process. However, few immunoassay-grade antibodies of sufficient quality and number (two per protein candidate) are available. Moreover, because developing a new, clinically deployable immunoassay is both expensive and time consuming, such development is normally restricted to but a short list of already highly credentialed candidates. The need for alternate methods to rapidly configure quantitative assays to credential novel protein candidate biomarkers is highlighted by a recent study of pancreatic cancer22. Over 600 proteins were quantified in plasma of which 165 (~27%) were found to change in abundance with development of pancreatic cancer. In their verification studies, antibody reagents for only 11 of these proteins were available, including an antibody specific for CA-19-9, a marker of pancreatic cancer already in clinical use. Owing to the lack of antibody reagents, no follow-up studies were done for the remaining proteins of interest.
As a proof of principle, we developed quantitative SID-MRM-MS assays for four of the novel, heart-specific proteins discovered, together with additional cardiovascular-related proteins already in clinical use or of growing interest10. Highly consistent temporal trends were observed when we measured two or three peptides for each of the novel candidate proteins in four patients. Additionally, there was a high degree of correlation between AIMS and SID-MRM-MS results for the novel candidates, further supporting our findings that AIMS is a useful initial method for label-free quantification. Levels of MYL3, TPM1 and FHL1 all remained sufficiently high at 240 min after ablation to warrant further investigation in larger clinical cohorts. Inaccurate quantification can occur in SID-MRM-MS due to problems in MRM-MS data acquisition and analysis8,25, but potential problems can be circumvented (Supplementary Results and Discussion).
Our unbiased analysis also rediscovered many of the known cardiovascular biomarkers, including creatine kinase, MB, FABP and MPO and extended prior work by identifying many new proteins not previously associated with acute myocardial injury in humans. Supplementary Results and Discussion details a number of candidate biomarkers with published reports of proteins potentially associated with cardiovascular disease.
Our approach to enhanced biomarker discovery emphasized the in-depth analysis of a small, extensively phenotyped patient cohort. Promising proteins were then validated in additional, more heterogeneous cohorts. Some limitations are implicit in this approach. First, although serial sampling within patients constrains interindividual variability and improves signal-to-noise ratios, the small discovery population means that changes in proteins that failed to reach nominal significance in our study may still be scientifically important and warrant further investigation. Second, the marked cardiac perturbation that characterizes the PMI model may have influenced the type and magnitude of protein alterations and hence the ultimate clinical utility of our markers. Notably, however, the finding that several of the biomarkers appear elevated in subjects with spontaneous MI and reversible myocardial ischemia supports the clinical relevance of the model. Finally, although our proteomics markers had excellent discriminatory power in subjects with spontaneous ischemic disease and myocardial injury, these findings must be further evaluated in larger populations, improving estimates of predictive value, permitting comparison to and adjustment for traditional cardiovascular risk factors, and allowing evaluation within subgroups of interest including those defined by gender, race and comorbidities.
In summary, we have developed a generalizable, proteomics-based, discovery-through-verification pipeline and demonstrated its value by identifying novel protein biomarkers of myocardial injury. We have demonstrated that this pipeline can successfully credential candidate biomarkers using MS-based targeted assays and immunoassays when the appropriate reagents exist. We have developed and deployed assays for targets enriched in myocardium, and are applying our methods to interrogate the remaining candidates from our discovery proteomics studies. In addition to markers of infarction, our candidates include several proteins that may serve as markers of reversible myocardial ischemia, a condition for which there are no circulating biomarkers. Markers emerging from these studies can be integrated with established biomarkers to create multimarker risk scores, providing additional information to help guide cardiovascular disease management. We anticipate that our strategy could be used in many other clinically relevant scenarios where planned perturbational experiments are performed to elicit pathological phenotypes. These treatments might include drug administration, oral glucose challenge for diabetes26, exercise testing for cardiovascular disorders27 or dialysis for kidney disorders28.
ONLINE METHODS
Patients with hypertrophic cardiomyopathy undergoing septal ablation
Twenty-two patients undergoing PMI using alcohol septal ablation for the treatment of symptomatic hypertrophic cardiomyopathy were included in this study. Inclusion criteria for this cohort were: (i) primary hypertrophic cardiomyopathy; (ii) septal thickness of 16 mm or greater; (iii) resting outflow tract gradient of >30 mm Hg, or an inducible outflow tract gradient of ≥50 mm Hg; (iv) symptoms refractory to optimal medical therapy; and (v) appropriate coronary anatomy. The most proximal accessible septal branch was accessed using standard angioplasty guiding catheters and guidewires and 1.5 or 2.0 mm × 9 mm Maverick balloon catheters. Radiographic and echocardiographic contrast injections confirmed proper selection of the septal branch and balloon catheter position. Ethanol was infused through the balloon catheter at 1 ml/min. Additional injections in the same or other septal branches were administered as needed, causing cessation of blood flow to the isolated myocardium, and to reduce the gradient to <20 mm Hg30. Blood was drawn at baseline, just before the onset of ablation, and at 10, 60, 120, 240 and 1,450 min after injury (Fig. 1). Of the 22 patients, 11 consented to the placement of a catheter to the coronary sinus during the ablation, allowing for the simultaneous sampling of blood from the coronary sinus and femoral catheters at baseline, 10 min and 60 min. The coronary sinus catheter was subsequently removed before the patient left the catheterization suite.
Patients undergoing elective cardiac catheterization
A cohort of 24 patients undergoing elective, diagnostic cardiac catheterization for cardiovascular disease, but not acute myocardial ischemia, was used as controls for the PMI and spontaneous MI patients. Blood was drawn before the onset of cardiac catheterization and at 10 min and 1 h after the procedure was begun.
Patients with spontaneous acute coronary syndromes
We enrolled a cohort of 23 patients undergoing emergent cardiac catheterization for acute ST-segment elevation, spontaneous MI within 8 h of symptom onset. For this cohort, blood samples were obtained in the coronary catheterization suite upon initial presentation.
Patients undergoing cardiac stress testing
The exercise tolerance test cohort consisted of patients who underwent stress testing with myocardial perfusion imaging at Massachusetts General Hospital (MGH) and who were enrolled in a prospective biomarker cohort study. The Human Research Committee approved the study protocol and all patients provided written informed consent. All patients who were referred for stress testing for the evaluation of possible myocardial ischemia were eligible for participation. Patients who underwent pharmacologic testing were excluded. Data were obtained on each patient’s age, sex, race, weight, cardiac risk factors (including hypertension, diabetes mellitus, smoking and hyperlipidemia), prior cardiac disease (including angina, MI, congestive heart failure, angiographically confirmed significant coronary artery disease, percutaneous coronary intervention and coronary artery bypass grafting) and cardiac medications.
Patients underwent exercise testing using the standard Bruce protocol30. Symptoms, heart rate, blood pressure and a 12-lead electrocardiogram (ECG) were recorded before the test, midway through each stage and during recovery. The stress test was terminated if there was physical exhaustion, severe angina, >2 mm horizontal or downsloping ST-segment depression, ≥20 mm Hg fall in systolic blood pressure or sustained ventricular arrhythmia. Duration of the stress test, metabolic equivalents achieved, peak heart rate and peak blood pressure were recorded. If the patient developed angina during the test, the timing, quality (typical versus atypical) and effect on the test (limiting or nonlimiting) were noted. The maximal horizontal or downsloping ST segment changes were recorded in each ECG lead.
A stress-rest imaging protocol was used. Tetrofosmin labeled with technetium (99mTc) was administered at peak stress and imaging was performed soon thereafter. Four hours later, a second injection was administered and repeat imaging was performed. Quantitative analysis of perfusion was performed using the CEqual method to calculate the percent reversible and fixed perfusion defects. Patients with >5% reversible perfusion defect were selected as cases (26 patients) and those without any perfusion defect were selected as controls (26 patients). Left ventricular ejection fraction was calculated using commercially available software31. Blood samples were obtained just before the test (baseline), at peak exercise (peak) and 1 h after cessation of exercise (post).
Blood collection
All protocols for blood collection, including the coronary sinus sampling protocol, were approved by the MGH Institutional Review Board, and all subjects gave written informed consent. All blood samples were collected in K2EDTA-treated tubes (Becton Dickinson) and were centrifuged at 2000g for 10 min to pellet cellular elements. The supernatant plasma was then aliquoted and immediately frozen at −80 °C. Additional blood samples were sent to the clinical chemistry laboratory for evaluation of the standard cardiac markers creatine kinase, CKM, CKB and Troponin T (Roche Diagnostics). Detailed protocols for plasma processing, sample preparation, mass spectrometry and data analysis for discovery proteomics, AIMS and SID-MRM can be found in Supplementary Methods.
Statistical analyses for discovery proteomics
For label-free, relative quantification, the sum of the precursor-ion signal intensities of all peptides derived from each protein was used as an approximation of that protein’s expression level across time points. The peak area for the extracted ion chromatogram of each precursor ion in the intervening high-resolution MS1 scans of the data-dependent LC-MS/MS runs was calculated automatically by the Spectrum Mill software using narrow windows around each individual member of the isotope cluster. Peak widths in both the time and m/z domains were dynamically determined based on MS scan resolution, precursor charge and m/z subject to quality metrics on the relative distribution of the peaks in the isotope cluster versus theoretical. Total intensity information for all proteins across patients and time points was exported to Excel where ratios of the total intensities at 10 min and 60 min were calculated with respect to baseline. Because equivalent amounts of plasma protein were processed for each patient and time point, and all samples were subsequently treated equivalently, no further normalization was done when calculating relative protein abundance ratios between samples.
We employed a minimum detectable fivefold change between baseline and either 10-min or 60-min samples. This was based on preliminary calculations summarized in the table in Online Methods, which shows the power to detect threefold and fivefold changes in biomarkers for various values of CV and number of sample pairs. Derivation of this table is based on the t-test, and assumes that the measurements are normally distributed (or can achieve a normal distribution after log transformation), with the CV fixed irrespective of the magnitude of the measurement (that is, a very conservative CV is used). Furthermore, the significance level is an indicator of the probability that a specific biomarker is a false positive when it has a fold-change larger than the minimum noted in the table. This is, therefore, a nominal P-value, and does not correct for multiple testing to account for the many hundreds of markers that will be evaluated. We used the table generated to provide ballpark estimates for minimum detectable fold-change, and statistical power attainable for a chosen fold-change level (specifically threefold and fivefold). These power calculations suggest that we would have ~60–80% power to detect changes of fivefold or greater, based on having 6–8 sample pairs (respectively), a nominal significance level of 0.05, and a conservative CV of 50% for discovery proteomic findings. We effectively have six sample pairs when selecting protein candidates that have a fivefold average change over the combined 10- and 60-min samples, compared to baseline. For independently detecting changes in the 10- or 60-min samples, this power will be rapidly attained as more samples are analyzed. We then used a staged approach as described in the main text to credential markers, and assess for any false positives that may be introduced by the process as detailed in the Results.
Preliminary calculations, which show the power to detect threefold and fivefold changes in biomarkers for various values of CV and number of sample pairs.
| Significance level (P-value) | CV of assay | Number of sample pairs | Minimum detectable fold change | Power
|
||
|---|---|---|---|---|---|---|
| Min. detectable fold change | Threefold change | Five fold change | ||||
| 0.05 | 0.2 | 6 | 1.35 | 0.45 | 0.99 | 1.00 |
| 7 | 1.30 | 0.45 | 1.00 | 1.00 | ||
| 8 | 1.27 | 0.45 | 1.00 | 1.00 | ||
| 10 | 1.23 | 0.45 | 1.00 | 1.00 | ||
| 0.3 | 6 | 1.59 | 0.41 | 0.86 | 0.95 | |
| 7 | 1.50 | 0.42 | 0.93 | 0.98 | ||
| 8 | 1.44 | 0.42 | 0.97 | 0.99 | ||
| 10 | 1.36 | 0.43 | 0.99 | 1.00 | ||
| 0.5 | 6 | 2.33 | 0.36 | 0.46 | 0.61 | |
| 7 | 2.06 | 0.37 | 0.55 | 0.70 | ||
| 8 | 1.90 | 0.37 | 0.63 | 0.78 | ||
| 10 | 1.71 | 0.38 | 0.76 | 0.89 | ||
Correlation analyses for AIMS and pooled DDA experiments
The correlation between AIMS and discovery proteomics performed on individual patients was calculated for each protein using Pearson correlation based on the intensity values measured in AIMS at all time points (baseline, 10 min and 60 min) and the average intensity values (calculated from the three replicates) of the individual patients from discovery proteomics at all three time points. A similar method was used to calculate the correlation between the discovery proteomics of individual patients and the data collected by data-dependent acquisition on pooled samples.
Correlation over the three time points (n = 3) is underpowered to show statistical significance. We nonetheless use the magnitude of the correlation coefficient to assess the strength of the correlation (quantitative significance, 32). Based on established cut points in biostatistical literature32,33, a correlation of 0.4–0.7 reflects moderate correlation, and correlation ≥0.7 is indicative of high correlation.
Cluster analysis for AIMS data
The AIMS data for each protein were standardized (that is, normalized to have a mean of 0 and a s.d. of 1), and clustered using fuzzy c-means clustering34–36 implemented in the mfuzz R package36 (http://www.r-project.org/). The final number of clusters, five, was decided based on inspection of the coherence of the profiles contained in each cluster. The clustering assigns, to each protein, a membership value between 0 and 1 for each cluster. The cluster numbers shown in Table 1 and Supplementary Table 5 are the most probable cluster memberships for the respective proteins. Supplementary Figure 3 shows the corresponding profiles for this analysis.
Western blot analysis
The following commercial antibodies were purchased for western blot analysis of depleted, peripheral plasma from PMI patients: goat anti-human PTN (Abcam), rabbit anti-human MDK (Antigenix), mouse anti-human MDH1 (Novus Biological), rabbit anti-human MYL3 (Proteintech group), mouse anti-human FHL1 (Abnova), mouse anti-human TPM1 (Novus Biological) and rabbit anti-human ACLP1 (Affinity BioReagents). Depleted peripheral plasma protein was mixed with 6× protein loading buffer and boiled to denature proteins completely, then loaded onto 10% SDS–PAGE gels. SDS gels were then placed into transfer buffer (25 mM Tris, 192 mM glycine, 20% vol/vol methanol, pH 8.3) for 5 min and the separated proteins were transferred onto nitrocellulose filters. The filter was blocked with 5% nonfat milk powder in TBST (0.05% Tween-20) for 1 h, probed with goat anti-human PTN (0.1 μg/ml), rabbit anti-human MDK (0.2 μg/ml), mouse anti-human MDH1 (1:500 dilution), rabbit anti-human MYL3 (1:1,000 dilution), mouse anti-human FHL1 (1:1,000 dilution), mouse anti human TPM1 (1:500 dilution) or rabbit anti-human ACLP1 (0.2 μg/ml) respectively at 4 °C overnight and incubated with secondary antibody horseradish peroxidase (HRP) labeled anti-rabbit (1:3,000), anti-goat (1:5,000) or anti-mouse (1:3,000) respectively for 1 h. The signal was detected by enhanced chemiluminescence detection reagents (Amersham, Life Science). In initial verification studies, western blot analysis failed to detect changes noted by MS in three cases. Ongoing studies are presently examining the cause of the discrepancies.
ELISA detection
Peripheral plasma concentrations of CCL21 (human CCL21/6CKine immunoassay, R&D Systems), and ACBP (human diazepam binding inhibitor ELISA kit, Young In Frontier) were measured with commercially available kits according to manufacturer’s instructions.
Lower and upper limits of quantification (LLOQ and ULOQ, respectively) supplied with the kits were reported as follows:
CCL21. LLOQ, 78.1 pg/ml; ULOQ, 5,000 pg/ml; intra CV < 4%, inter CV < 9.2%.
ACBP. LLOQ, 25 pg/ml; ULOQ, 1,600 pg/ml; intra CV < 5.6%, inter CV < 8.9%.
For in-house ELISA development, the clear high-binding, flat-bottom poly-styrene microplates (R&D Systems) was coated with 100 μl of 1 μg/ml of mouse anti-human MDK (Antigenix), 0.5 μg/ml of mouse anti-human ANG (R&D Systems), 2 μg/ml of mouse anti-human DCN (R&D Systems) or 0.5 μg/ml of rabbit anti-human sFRP1 in PBS at 4 °C overnight, respectively, decant all the capture solution the next day and wash the plate with PBST four times after the blockage of the nonbinding area with 200 μl of 5% BSA in PBS at 25 °C for 1 h. After removal of 5% BSA in PBS, 100 μl of EDTA-plasma and serial diluted standard of human MDK (Antigenix), ANG (R&D Systems), DCN (R&D Systems) or sFRP1 (R&D Systems) was added to each well respectively and incubated at 25 °C for 2 h. The plate was washed again with PBST four times and 100 μl of 0.2 μg/ml of biotin-conjugated rabbit anti-human MDK (Antigenix), 0.4 μg/ml of biotin-conjugated goat anti-human ANG (R&D Systems), 0.25 μg/ml of biotin-conjugated mouse anti-human DCN (R&D Systems) or 0.2 μg/ml of goat anti-human sFRP1 (R&D Systems) in 0.1% BSA in PBS was added to each well, respectively, and incubated at 25 °C. The solution was removed 1h later by washing with PBST four times. For MDK, ANG and DCN detection, 100 μl of 1:200 diluted streptavidin-HRP (R&D Systems) with 0.1% BSA in PBS was added in each well and incubated at 25 °C for 30 min in the dark. For sFRP1 detection, 100 μl of 1:2,000 diluted HRP-conjugated rabbit anti-goat immunoglobulin (DAKO North America) with 0.1% BSA in PBS was added in each well and incubated at 25 °C for 1h in the dark. The plate was washed four times with PBST and 100 μl of substrate reagent (R&D Systems) was added in each well and incubated at 25 °C in the dark for 10 min, then terminated the reaction by adding 100 μl of 2 M sulfuric acid. Determine the optical density (OD450) of each well immediately by using the microplate reader set to 450 nm. The limits of quantification were determined as the following:
MDK. LLOQ, 0.156 pg/ml; ULOQ, 20 ng/ml; intra CV<6.2%, inter CV < 9.2%.
ANG. LLOQ, 78.1 pg/ml; ULOQ, 5,000 pg/ml; intra CV<5%, inter CV < 9%.
DCN. LLOQ, 31.25 pg/ml; ULOQ, 2,000 pg/ml; intra CV <4.3%; inter CV < 7.8%.
sFRP1. LLOQ, 0.195 ng/ml; ULOQ, 25 ng/ml; intra CV <4.8, inter CV < 8.8%.
Statistical analyses for clinical data and ELISA findings
For clinical characteristics, values for continuous variables are presented as mean ± s.d., and comparisons between groups were performed using two-sample t-tests. Association between categorical variables was assessed using the Fisher’s Exact Test. To evaluate whether metabolic changes observed in the PMI patients were generalizable to spontaneous MI, we studied proteins for which ELISAs were available that displayed significant changes from baseline at 1, 2 and 4 h in the derivation and validation planned MI cohorts (P < 0.05 at all three time points). A Wilcoxon Rank-Sum test was used to examine levels of these individual proteins in the patients who had spontaneous MI as compared to control patients who were admitted to the cardiac catheterization suite with nonacute cardiovascular disease. The ELISA results of ACBP, ANG and CCL21 in the patients are presented as mean ± s.e.m., the paired student’s t-test are used to display the significant changes compared to the baseline in PMI patients and the patients for elective cardiac catheterization. The random Student’s t-test is used to display the significant changes between the spontaneous MI and the control patients.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge support from the US National Institutes of Health (NIH) National Heart, Lung, and Blood Institute U01HL083141 and R01HL096738-02 to R.E.G., S.A.C. and M.S.S., the Donald W. Reynolds Foundation (to R.E.G. and M.S.S.) and Foundation Leducq (to R.E.G.). S.A.C. also acknowledges support from the NIH 1U24 CA126476 as part of the National Cancer Institute (NCI)’s Clinical Proteomic Technologies Assessment in Cancer Program, from the Women’s Cancer Research Fund of the Entertainment Industry Foundation and to D.R.M. from the NCI Clinical Proteomic Technologies Initiative (R01 CA126219). We also appreciate the excellent technical assistance of C. Bodycombe.
Footnotes
Accession codes. All mass spectrometric data files associated with this manuscript may be downloaded from the ProteomeCommons.org Tranche network using the following hashes:
bquYtp//Q8fY6EdnRMoUEe5Jme2WHwYKLj50kWO/JvsNdVn QV6UCgTixEFP9uQiBYJo9emQVc2lRzbDQLKURot4mEXEAAAAAAAGNYQ.==
WNWMJg68D/CjrNnR26osMAfjmCNGbxEMweFdZJTps7AUv8Aw9pIhsfbMehM4lGYJSIKVx6CbZS/pBCftDf8XvDuKidMAAAAAAABCMg==
Accessible data include all LC-MS/MS files for discovery proteomics, AIMS experiments collected on an LTQ-Orbitrap and SID-MRM-MS files collected on a 5500 hybrid triple quadrupole/linear ion trap mass spectrometer.
Note: Supplementary information is available on the Nature Biotechnology website.
AUTHOR CONTRIBUTIONS
T.A.A., S.A.C. and R.E.G. wrote the manuscript. S.A.C. and M.A.G. conceived of the biomarker pipeline used here. R.E.G. conceived of the PMI as a model for proteomic discovery, and along with M.A.F., M.S.S., G.D.L. and L.A.F. developed the human studies protocols included in the manuscript, and performed the phenotyping of the patient populations. H.K., X.S. and T.A.A. carried out all of the sample preparation, conducted the MS-based proteomics experiments for discovery and AIMS and interpreted the results. H.K. and M.B. conducted all of the MRM-MS experiments for assaying proteins by M.S.S. X.S. tested, developed and applied all antibody-based measurements, with contributions from D.S. M.A.G. was responsible for design of the AIMS experiments. K.R.C. designed and adapted the Spectrum Mill software for peptide and protein identification, label-free quantification and calculation of peptide-level FDR and participated in data analysis. D.R.M., M.S.S. and K.R.C. were responsible for statistical design and analysis.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Edwards AVG, White MY, Cordwell SJ. The role of proteomics in clinical cardiovascular biomarker discovery. Mol Cell Proteomics. 2008;7:1824–1837. doi: 10.1074/mcp.R800007-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Jacquet S, et al. Identification of cardiac myosin-binding protein C as a candidate biomarker of myocardial Infarction by proteomics analysis. Mol Cell Proteomics. 2009;8:2687–2699. doi: 10.1074/mcp.M900176-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu Q, Van Eyk JE. Proteomics and heart disease: identifying biomarkers of clinical utility. Expert Rev Proteomics. 2006;3:237–249. doi: 10.1586/14789450.3.2.237. [DOI] [PubMed] [Google Scholar]
- 4.Anderson NL. The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clin Chem. 2010;56:177–185. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- 5.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 6.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 7.Paulovich AG, Whiteaker JR, Hoofnagle AN, Wang P. The interface between biomarker discovery and clinical validation: the tar pit of the protein biomarker pipeline. Proteomics Clin Appl. 2008;2:1386–1402. doi: 10.1002/prca.200780174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Addona TA, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keshishian H, et al. Quantification of cardiovascular biomarkers in patient plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2009;8:2339–2349. doi: 10.1074/mcp.M900140-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaffe JD, et al. Accurate inclusion mass screening: a bridge from unbiased discovery to targeted assay development for biomarker verification. Mol Cell Proteomics. 2008;7:1952–1962. doi: 10.1074/mcp.M800218-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sigwart U. Non-surgical myocardial reduction for hypertrophic obstructive cardiomyopathy. Lancet. 1995;346:211–214. doi: 10.1016/s0140-6736(95)91267-3. [DOI] [PubMed] [Google Scholar]
- 13.Knight C, et al. Nonsurgical septal reduction for hypertrophic obstructive cardiomyopathy: outcome in the first series of patients. Circulation. 1997;95:2075–2081. doi: 10.1161/01.cir.95.8.2075. [DOI] [PubMed] [Google Scholar]
- 14.de Lemos JA, et al. The prognostic value of serum myoglobin in patients with non–ST-segment elevation acute coronary syndromes: results from the TIMI 11B and TACTICS-TIMI 18 studies. J Am Coll Cardiol. 2002;40:238–244. doi: 10.1016/s0735-1097(02)01948-4. [DOI] [PubMed] [Google Scholar]
- 15.O’Donoghue M, et al. Prognostic utility of heart-type fatty acid binding protein in patients with acute coronary syndromes. Circulation. 2006;114:550–557. doi: 10.1161/CIRCULATIONAHA.106.641936. [DOI] [PubMed] [Google Scholar]
- 16.Layne MD, et al. Impaired abdominal wall development and deficient wound healing in mice lacking aortic carboxypeptidase-like protein. Mol Cell Biol. 2001;21:5256–5261. doi: 10.1128/MCB.21.15.5256-5261.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheikh F, et al. An FHL1-containing complex within the cardiomyocyte sarcomere mediates hypertrophic biomechanical stress responses in mice. J Clin Invest. 2008;118:3870–3880. doi: 10.1172/JCI34472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kishimoto K, Liu S, Tsuji T, Olson KA, Hu GF. Endogenous angiogenin in endothelial cells is a general requirement for cell proliferation and angiogenesis. Oncogene. 2005;24:445–456. doi: 10.1038/sj.onc.1208223. [DOI] [PubMed] [Google Scholar]
- 19.Moretti A, et al. Essential myosin light chain as a target for caspase-3 in failing myocardium. Proc Natl Acad Sci USA. 2002;99:11860–11865. doi: 10.1073/pnas.182373099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fusaro VA, Mani DR, Mesirov JP, Carr SA. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat Biotechnol. 2009;27:190–198. doi: 10.1038/nbt.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richard P, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 22.Faca VM, et al. A mouse to human search for plasma proteome changes associated with pancreatic tumor development. PLoS Med. 2008;5:e123. doi: 10.1371/journal.pmed.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.States DJ, et al. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat Biotechnol. 2006;24:333–338. doi: 10.1038/nbt1183. [DOI] [PubMed] [Google Scholar]
- 24.Schenk S, Schoenhals GJ, de Souza G, Mann M. A high confidence, manually validated human blood plasma protein reference set. BMC Med Genomics. 2008;1:41–68. doi: 10.1186/1755-8794-1-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbatiello SE, Mani DR, Keshishian H, Carr SA. Automated detection of inaccurate and imprecise transitions in quantitative assays of peptides by multiple monitoring mass spectrometry. Clin Chem. 2010;56:291–305. doi: 10.1373/clinchem.2009.138420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaham O, et al. Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol Syst Biol. 2008;4:214. doi: 10.1038/msb.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis GD, et al. Metabolic signatures of exercise in human plasma. Sci Transl Med. 2010;2:33ra37. doi: 10.1126/scitranslmed.3001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhee EP, et al. Metabolite profiling identifies markers of uremia. J Am Soc Nephrol. 2010;21:1041–1051. doi: 10.1681/ASN.2009111132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Hall G, et al. Leg and arm lactate and substrate kinetics during exercise. Am J Physiol Endocrinol Metab. 2003;284:E193–E205. doi: 10.1152/ajpendo.00273.2002. [DOI] [PubMed] [Google Scholar]
- 30.Baggish AL, et al. Pathological effects of alcohol septal ablation for hypertrophic obstructive cardiomyopathy. Heart. 2006;92:1773–1778. doi: 10.1136/hrt.2006.092007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horiba M, et al. Midkine plays a protective role against cardiac ischemia/reperfusion injury through a reduction of apoptotic reaction. Circulation. 2006;114:1713–1720. doi: 10.1161/CIRCULATIONAHA.106.632273. [DOI] [PubMed] [Google Scholar]
- 32.Feinstein AR. Principles of Medical Statistics. Chapman & Hall/CRC; 2002. [Google Scholar]
- 33.Guilford JP. Fundamental Statistics in Psychology and Education. McGraw Hill; 1956. [Google Scholar]
- 34.Futschik ME, Carlisle B. Noise-robust soft clustering of gene expression time-course data. J Bioinform Comput Biol. 2005;3:965–988. doi: 10.1142/s0219720005001375. [DOI] [PubMed] [Google Scholar]
- 35.Kumar L, Futschik ME. Mfuzz: A software package for soft clustering of microarray data. Bioinformation. 2007;2:5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.