Abstract
Eukaryotes have evolved elaborate mechanisms to survive periods of adversity. By manipulating genes that control these mechanisms, researchers have found they can generate more stress resistant, longer-lived organisms. One of these is the PNC1 gene of Saccharomyces cerevisiae, a master “longevity regulatory gene” that translates a variety of environmental stresses into lifespan extension by activating the sirtuin family of longevity deacetylases. Master longevity genes such as PNC1 are highly adaptive because they allow organisms to respond in a concerted way to adversity and to rapidly evolve life strategies to compensate for a changing environment. Hence, they should be well conserved. We propose that there is a functional equivalent of PNC1 in mammals called Nampt (a.k.a. PBEF/Visfatin), a stress-responsive gene that would coordinately regulate metabolism, cell defenses, and resistance to diseases of aging.
Keywords: Caloric restriction, Dietary restriction, Aging, PBEF, Nampt, Lifespan, Sirtuin, Sir2, NAD+ salvage pathway, Nmnat, Longevity, Nicotin-amide, SIRT1, Chromatin, DNA stability, CR, DR
1. Introduction
Caloric restriction (CR), also known as dietary restriction (DR), is the only reproducible way to extend the maximum lifespan of mammals. Although numerous theories have been proposed to explain how CR works, the most favored theory is that it activates a biological defense response that evolved to help organisms survive adversity (Masoro, 2000). Yeast cells, for example, live longer when placed under a variety of mildly stressful conditions including high temperature, high salt, low nutrients, or osmotic stress (Anderson et al., 2003).
A critical question relating to how aging is regulated in eukaryotes is whether each type of biological stress activates a different pathway that extends lifespan or whether there is a master regulatory pathway that responds to a variety of stresses? One could argue that the latter is more likely, based on the realization that organisms might benefit from having master regulatory pathways that respond to a variety of stresses because it would allow them to elicit a broad defense response in response to an insult and because it would allow them to rapidly evolve new strategies in response to a new environment (Kirkwood and Shanley, 2005). A single base change in the promoter of a master regulator could have large effects on energy storage, organismal defenses, reproduction, and somatic maintenance, resulting in perhaps a more fertile yet shorter lived species better adapted to a harsh environment. As we discuss below, Saccharomyces cerevisiae seems to possess just such a regulator and we propose that there is an equivalent gene in more complex organisms that might also regulate survival and lifespan.
2. The sirtuin family of enzymes
Recently, the lifespan-extending effect of CR has been linked to a versatile class of protein deacetylases called “sirtuins”. The founding member of this family is yeast Sir2, which deacetylates histones H3 and H4, catalyzing the formation of silent heterochromatin and stabilizing repetitive DNA (Guarente and Picard, 2005). The longevity function of Sir2 is well conserved because increasing the level or activity of Sir2 also extends lifespan in Caenorhabditis elegans and Drosophila melanogaster (Rogina and Helfand, 2004; Tissenbaum and Guarente, 2001). Even a protozoan parasite, Leishmania, encodes a Sir2 enzyme that promotes its survival (Vergnes et al., 2002). In S. cerevisiae, C. elegans, and Drosophila, nutrient restriction fails to extend lifespan in the absence of Sir2, (Lin et al., 2000; Rogina and Helfand, 2004; Wang and Tissenbaum, 2006). Similarly, a recent study in mice showed that Sir2/SIRT1 knockouts do not show the typical increase in physical activity that occurs with CR (Chen et al., 2005). Together these data indicate that sirtuins underlie certain physiological effects of CR and its effect on lifespan. The focus on Sir2 in the field does not imply that all aspects of CR are mediated via Sir2. In fact, an alternative pathway to yeast Sir2 exists because cells deleted for SIR2 live longer when subjected to intense glucose restriction (0.05% glucose) (Kaeberlein et al., 2004). Our lab has proposed and presented evidence that a backup pathway to SIR2 is requires HST2, a SIR2 homolog (Lamming et al., 2005).
There are five sirtuins in S. cerevisiae: Sir2 and Hst1–4. The precise cellular functions of the Hst proteins are not clear but the analysis of yeast mutants on these genes indicate that they have roles in silencing, mitochondrial metabolism and DNA repair. Hst2 is suspected to be a histone deacetylase because it can substitute for Sir2 (Lamming et al., 2005) and is found associated with chromatin (Halme et al., 2004) (Lamming et al., 2006). In mammals, there are seven sirtuins, SIRT1–7. SIRT1 targets both histone and non-histone proteins involved in such processes as gene regulation, apoptosis, axonal protection, glucose synthesis, and fat mobilization (Araki et al., 2004; Luo et al., 2001; Picard et al., 2004; Pruitt et al., 2006; Vaquero et al., 2004; Vaziri et al., 2001; Yeung et al., 2004). The known targets of SIRT1 are summarized in Table 1.
Table 1.
Known targets and functions of the mammalian Sir2 family of proteins (sirtuins)
| Sirtuin | Cellular location | Protein modifieda | Amino acid modified | Cellular function |
|---|---|---|---|---|
| SIRT1 | Nucleus | Histone H3 | 9, 14 | DNA packing, gene transcription |
| Nucleus | Histone H4 | 16 | DNA packing, gene transcription | |
| Nucleus | Histone H1 | 29 | DNA packing, gene transcription | |
| Nucleus | p53 | 320, 382 | Cell survival | |
| Cytoplasm | Ku70 | 539, 542 | Cell survival | |
| Nucleus | NF-κB(p65) | 310 | Cell survival/anti-inflammatory? | |
| Nucleus | HIV Tat | 50 | HIV replication | |
| Nucleus | p300 | 1020, 1024 | Gene transcription | |
| Nucleus | TAF I 68 | ? | Gene transcription | |
| Nucleus | MyoD | ? | Muscle development | |
| Nucleus | NCOR | ? | Fat metabolism | |
| Nucleus | FOXO1/3/4 | ? | Glucose metabolism/cell defenses | |
| Nucleus | PGC-1α | ? | Glucose metabolism | |
| SIRT2 | Cytoplasm | Tubulin | ? | Cytoskeleton |
| SIRT3 | Mitochondria | ? | ? | Thermogenesis? |
| SIRT4 | Mitochondria | ? | ? | ? |
| SIRT5 | Mitochondria | ? | ? | ? |
| SIRT6 | Nucleus | SIRT6? | ? | Base excision repair |
| SIRT7 | Nucleolus | RNA PolI | rRNA transcription |
SIRT1,2,3,5,7 possess deacetylase activity; SIRT4 and 6 possess ADP-ribosyltransferase activity.
3. The sirtuin reaction: implications for regulation
For many years, Sir2 was believed to play a structural role in the formation of silent heterochromatin, but this view began to change in 1998 when the protein was found to have similarity to CobB, an Escherichia coli enzyme that promotes the synthesis of vitamin B12 via a nicotinate mononucleotide phosphoribosyltransferase step (Tsang and Escalante-Semerena, 1998). Roy Frye played a key role by postulating that Sir2 may be an ADP-ribosyltransferase and cloning the entire mammalian Sir2 family (Frye, 1999; Frye, 2000). Danesh Moazed’s group also published that Sir2 has ADP-ribosyltransferase activity and showed that mutation of the catalytic site abolished silencing (Tanny et al., 1999). A more robust histone deacetylase activity was discovered shortly after by the Guarente, Sternglanz, and Smith labs (Imai et al., 2000; Landry et al., 2000b; Smith et al., 2000). Today, over a dozen sirtuins have been characterized from eukaryotes and even the cobB enzyme from E. coli has been shown to possess deacetylase activity and is considered a bacterial sirtuin (Zhao et al., 2004a).
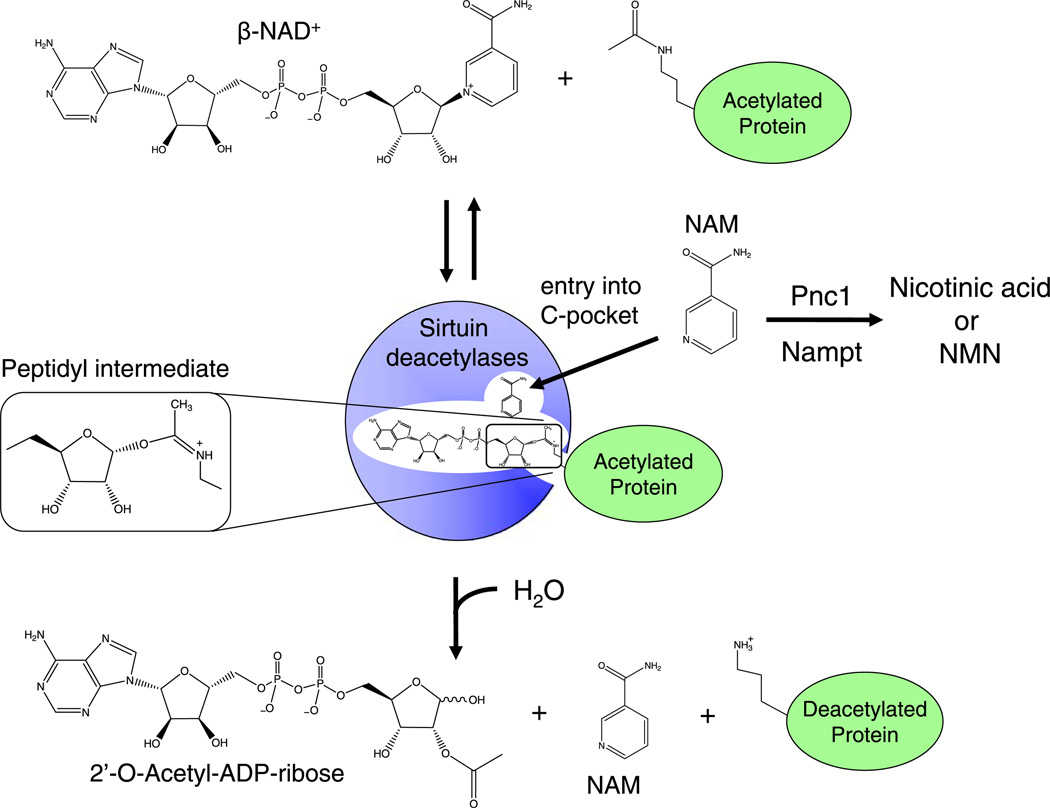
Sirtuins are not typical deacetylases. They utilize a complex two-step reaction to catalyze what should be a simple hydroylsis reaction (Sauve and Schramm, 2003), which is thought to allow for regulation by NAD+ and the reaction products (Lin et al., 2000; Anderson et al., 2003). Each reaction utilizes one molecule of NAD+ and an acetyl-lysine as substrate in a coupled base-exchange then deacetylation step. The first-step of the reaction results in the formation of a stable 1′-O-a-peptidylamidate-ADPR intermediate and nicotinamide (NAM) is released from the active site (Sauve et al., 2001). Step two of the reaction is transfer of acetyl group to the ADP ribose moiety and the release of deacetylated lysine, NAM and the acetyl ester metabolites-2′ and 3′-O-acetyl ADP ribose (Landry et al., 2000a; Tanner et al., 2000) (Fig. 1).
Fig. 1.
The sirtuin/class III HDAC deacetylation reaction and regulation by nicotinamide. Step 1 is a reversible base-exchange reaction in which a peptidyl intermediate is formed between the acetyl group on the lysine of the polypeptide to generate 1′-O-α-peptidylamidate-ADP-ribose, with the release of nicotinamide (NAM). Step two is the deacetylation of the lysine by hydrolytic attack. The peptidyl intermediate is sufficiently stable to permit regeneration of NAD+ in the presence of elevated nicotinamide concentrations. Figure adapted from Bitterman et al. (2002).
Although it will likely be years before we know whether the sirtuins underlie key aspects of CR in mammals, it is clear that in lower organisms Sir2 plays an important role. This raises the important question: How does CR stimulate Sir2? In yeast, CR does not alter NAD+ levels appreciably, ruling out the simplest hypothesis (Araki et al., 2004; Revollo et al., 2004).
Another hypothesis is that Sir2 is inhibited in vivo by NADH and that CR reduces NADH levels from 0.85 to 0.39 mM, which relieves inhibition in vitro (Lin et al., 2004). Moreover, genetic manipulations that lower NADH in S. cerevisiae increase Sir2 activity. This model has been challenged by the finding that the IC50 of NADH for Sir2 is 11 mM (Schmidt et al., 2004), considerably higher than some in vivo estimates of the concentration of this molecule (~50–100 µM) (Anderson et al., 2002; Srivastava and Bernhard, 1987). Denu and colleagues found that a 10-fold reduction in the NAD+:NADH ratio changes the rate of deacetylation by only ~0.2% (Schmidt et al., 2004). That said, there remains the very real possibility that there are local high concentrations of NADH in the nucleus or that in vitro conditions fail to recapitulate the nature of the enzyme in vivo.
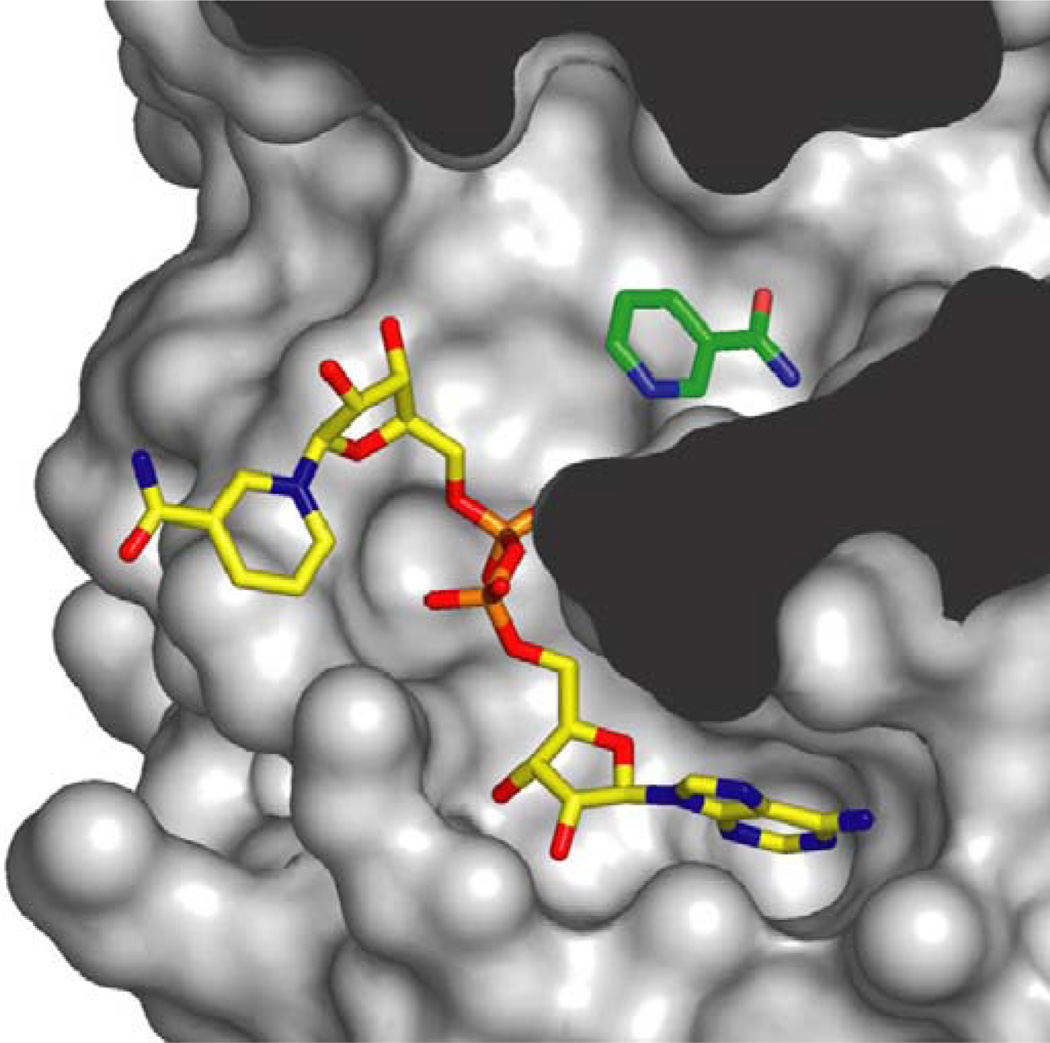
Another hypothesis is that CR works in yeast by depleting NAM, the product and a small molecule inhibitor of sirtuins (Fig. 1). The first clue to the potential importance of NAM in sirtuin biology was the observation that sirtuins are inhibited by NAM in a non-competitive fashion (Bitterman et al., 2002; Landry et al., 2000a), the implication being that NAM does not compete with NAD+ for binding but binds to a distinct site (Avalos et al., 2004; Avalos et al., 2002; Bitterman et al., 2002). The crystal structure of a sirtuin from Archaeoglobis fulgidus, Sir2-Af2, shows that in the absence of substrate NAD+ binds in a ‘non productive’ configuration (Avalos et al., 2002; Min et al., 2001). However, in the presence of peptide substrate, the NAM ring of NAD+ sits in a ‘productive’ conformation in which the NAM moiety sits in the so-called “C-pocket” prior to catalysis (Zhao et al., 2004b). Structures of Sir2-Af2 and Sir2Tm from Salmonella typhimurium co-crystallized with NAM show that the free NAM can also bind the C-pocket, thus allowing it to react with a peptidyl intermediate and drive the reaction in reverse (Fig. 2) (Avalos et al., 2005). Consistent with this, addition of NAM to yeast media completely blocks the ability of Sir2 to function in vivo (Bitterman et al., 2002; Gallo et al., 2004). But is Sir2 inhibition by NAM physiologically relevant? This question was tackled by the Sinclair and Smith laboratories and both labs reached the same conclusion.
Fig. 2.
Space-filling model of Sir2-Af2 and nicotinamide inhibition. Nicotinamide (NAM) in green is shown bound in the “C-pocket” of Sir2-Af2 adjacent to an NAD+ molecule in the active site. Inhibition by NAM is proposed to occur when free NAM binds in the C-pocket and reacts with the relatively long-lived peptidyl intermediate, driving the reaction in reverse to generate NAD+ and acetylated target protein (image provided courtesy of C. Wolberger, Johns Hopkins Medical School).
4. Nicotinamide: a physiological regulator of sirtuins
The yeast PNC1 gene encodes a nicotinamidase that depletes nicotinamide from the cell by converting NAM to nicotinic acid (vitamin B3) as part of the NAD salvage pathway. Overexpression of PNC1 leads to increases in Sir2-mediated silencing and recombination and a ~50% increase in replicative lifespan (Anderson et al., 2003; Gallo et al., 2004). This effect does not require increased NAD+ levels because deletion of NPT1, a gene in the NAD salvage pathway does not block the ability of PNC1 to increase Sir2 activity (Anderson et al., 2003). As further evidence that lower NAM concentrations are key, expression of a human nicotinamide methyltransferase gene (NNMT) or its putative yeast homolog also activate Sir2. Together, these results strongly indicate that physiological levels of NAM regulate Sir2.
The NAM regulation hypothesis has gained considerable support from the work of Suave and colleagues who took a biochemical approach to the problem. Iso-NAM is an analog of nicotinamide that increases the Vmax of Sir2 in vitro, most likely by preventing NAM from entering the C-pocket, and can enhance Sir2 activity in vivo (Sauve and Schramm, 2003). Concentrations of NAM in a yeast cell are between 10–150 µM, depending on whether you assume a concentration gradient across the cell or whether NAM is even distributed. The IC50 for NAM is ~50 µM, consistent with NAM being a physiological inhibitor of Sir2.
Given that additional copies of PNC1 activate Sir2 and extend lifespan, our best hypothesis was that CR works in yeast is by increasing PNC1 expression. Consistent with this, PNC1 is highly upregulated in response to glucose restriction, the yeast form of CR. In fact, a decrease in glucose concentration, from 2% to 0.5% is sufficient to boost PNC1 levels by ~4-fold. On the restricted medium, cells still grow at a normal pace, showing it is not a severe stress response or related to the pace of the cell cycle. Interestingly, PNC1 levels are also boosted more than 4-fold in response to other stimuli that extend yeast lifespan, such as low amino acids, heat stress, and osmotic stress (Anderson et al., 2003). These findings support the Hormesis Hypothesis of CR: that the lifespan extension the consequence of an active cellular response to sub-optimal conditions. What is particularly new about the finding is that longevity stimuli act through a master gene, PNC1. It is worth noting that these data do not imply that PNC1 is the only way Sir2 can be regulated or pathway by which CR can extend lifespan. In fact, the Guarente lab showed that if NAM levels are kept low in a pnc1 Δ strain by over-expressing a putative yeast nicotinamide methyltransferase (NNMT), then glucose restriction could still extend lifespan, demonstrating there are probably alternate mechanisms (Lin et al., 2004).
5. Is there a functional equivalent of PNC1 in mammals?
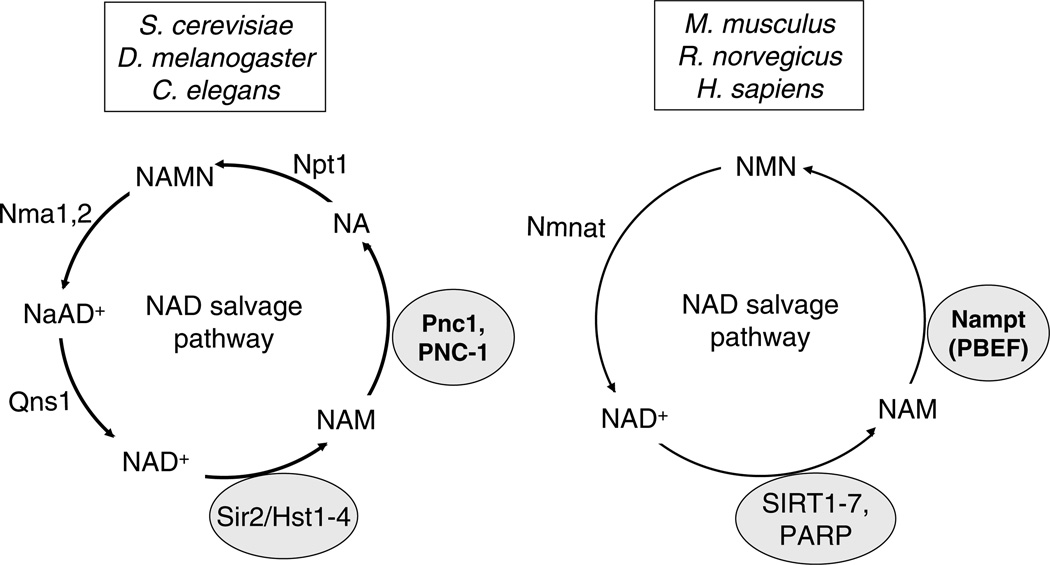
The key role of PNC1 in yeast longevity and environmental sensing led us to hypothesize that a similar master regulator of aging may exists in higher organisms including mammals (Anderson et al., 2003). Clear PNC1 homologs exist in C. elegans and Drosophila but in mammals the pathway for NAD salvage has diverged allowing NAD to be recycled in two steps instead of four (Fig. 3).
Fig. 3.
The two known pathways for the salvage of NAD+ from nicotinamide. Mammals recycle NAD from nicotinamide (NAM) in two steps rather than four, bypassing the production of nicotinic acid (NA). Pnc1, NAM deamidase; Nampt, NAM phosphoribosyltransferase; Nmnat, nicotinamide mononucleotide adenylyltransferase.
In mammals, instead of being converted to nicotinic acid (NA), NAM is converted to nicotinamide mononucleotide (NMN), followed by an adenylyltransferase step to regenerate NAD+. Although there is no PNC1 homolog in mammals, we propose that there is a functional homolog: the a nicotinamide phosphoribosyl transferase that carries out the first step of the NAD+ salvage pathway, known by three different names, Nampt, PBEF, and Visfatin. For simplicity and consistency, we will refer to the gene as Nampt.
In theory, Nampt could co-regulate the sirtuins and other NAD+-dependent processes by (i) lowering the NAM concentration and (ii) boosting NAD+ levels in the cytoplasm, nucleus and, perhaps even in mitochondria where three of the sirtuins are found. In 1994, Nampt was first cloned from a human peripheral blood lymphocyte cDNA library and named “PBEF”, for pre-B-cell colony enhancing factor, for its ability to enhance the effect of interleukin-7 and Sertoli cell factor (SCF) on pre-B-cell colony formation (Ognjanovic et al., 2001; Samal et al., 1994). Even though the Nampt polypeptide lacks any apparent signal sequence for secretion, it was found in conditioned media of activated lymphocytes and HeLa cells. The protein is also found in the medium of Nampt-expressing COS-7 cells, PA317 cells, CHO cells and even baculovirus cells, and was classified as a secreted cytokine (Samal et al., 1994). Interestingly, the finding that Nampt is a secreted protein would be rediscovered over 10 years later by Fukuhara and colleagues in their study of an adipokine that binds the insulin receptor (Fukuhara et al., 2004).
Nampt has been extensively studied by Bryant-Greenwood and colleagues, who characterized Nampt (PBEF) as a mechanically-induced gene that is up regulated when pre-term and full-term thickness fetal membranes are distended in vitro (Nemeth et al., 2000; Ognjanovic et al., 2001). They postulate that Nampt has a central role in the mechanism of infection-induced premature births (Ognjanovic et al., 2001). Nampt is also upregulated at the mRNA level by TALL-1 stimulation in B lymphoma cells and primary B lymphocytes, making it a possible target for intervention of lupus-like autoimmune diseases (Xu et al., 2002). Clearly, Nampt is involved in a complex regulatory loop involving cytokine signaling but exactly in what capacity is not yet known.
6. Nampt is an NAD+ biosynthetic enzyme and adipokine
For almost 8 years following the initial discovery of Nampt, it was considered a secreted cytokine whose levels increase during infection. No one suspected it had a critical function in the cell, recycling NAD+ and regulating enzymes that promote survival. Based on the homology between Nampt and the nadV gene of Haemophilus ducreyi Rongvaux et al. proposed that the protein is a nicotinamide phosphoribosyltransferase (NaMPRTase) (Rongvaux et al., 2002). The nadV gene had been cloned a couple of years earlier by virtue of its requirement for growth on complex media lacking NAD and the ability of bacteria to convert nicotinamide to NAD (Martin et al., 2001). Rongvaux et al. confirmed that Nampt is in fact an NAD biosynthetic enzyme by complementing the bacterial NaMPRTase-deficient strain with the cDNA for Nampt (Rongvaux et al., 2002). Unlike previous reports, Nampt was detected in the cytoplasm and was not secreted. Release into the medium was occurred only when cells underwent apoptosis.
Then in 2004, an adipokine called Visfatin was shown to be identical to the Nampt/PBEF protein (Fukuhara et al., 2004). Fukuhara et al. used a differential display technique to identify 31 new genes that are expressed only in visceral fat compared to subcutaneous fat (Fukuhara et al., 2004). One of the genes corresponded to Nampt (PBEF). The protein was found not only in serum but in the medium of cultured COS-1 and 3T3-L1 adipocytes (Nemeth et al., 2000; Ognjanovic et al., 2001). Plasma Nampt levels correlated with the amount of visceral fat in human subjects and in diabetes-prone KKAy mice (Fukuhara et al., 2004). Intravenous injection of recombinant murine Nampt into mice resulted in decreased plasma glucose levels within 30 min along with increased expression of genes involved in adipogenesis, increased phosphorylation of insulin receptor and the receptor substrates IRS-1 and -2 in liver, and activation of the insulin signaling pathway. Nampt may be secreted from adipocytes into the bloodstream, where it binds to and activates insulin receptors of peripheral tissues, thus lowering glucose. Biochemical studies show that Nampt binds to the insulin receptor tightly, with a dissociation constant of 4.4 nM, at a site that is different from the insulin binding site. Serum levels of Nampt are only 3–10% that of insulin, suggesting it plays a minor role in glucose control (Fukuhara et al., 2004). There remains some question in the field as to whether Nampt/Visfastin is actually secreted or whether it simply leaks out of dying or dead adipocytes but the fact that the same protein has previously been identified as a secreted factor and other proteins like IL-1β are secreted without an obvious signal sequence, argues that Nampt is likely to be a secreted by at least some cell types.
7. Compartmentalization of NAD biosynthesis?
Nampt has been detected in the nucleus and cytoplasm of cells, and this distribution changes in 3T3 cells following treatment with NGF or when cells reach confluency (Kitani et al., 2003). Our lab has detected NAD biosynthetic enzymes, including Pnc1 and Nma1/2 in the nucleus, cytoplasm and Pnc1 in peroxisomes (Anderson et al., 2002; Anderson et al., 2003). In mammals, the Nmnat enzyme has three isoforms: Nmnat1, -2, -3, which are found in the nucleus, Golgi apparatus and mitochondria, respectively, and each possesses different kinetic properties (Berger et al., 2005).
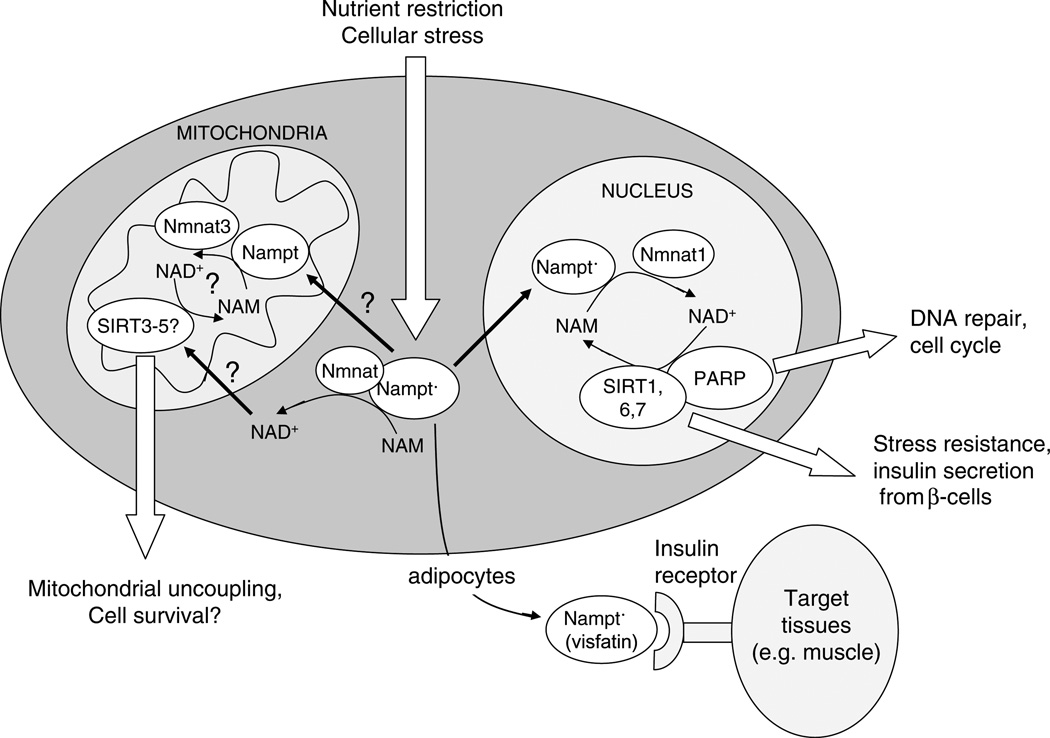
From these observations we can make the following predictions: (i) NAD biosynthesis in eukaryotes occurs within distinct intracellular compartments, (ii) Nampt will likely be found in the Golgi apparatus and mitochondria, completing the NAD+ salvage pathway in those organelles, and (iii) NAD+ levels in those organelles are likely regulated differentially, such that NAD+ synthesis in one compartment might be kept considerably higher than another, say, during times when DNA is damaged and depleted of NAD+, allowing NAD+-dependent enzymes in those regions to continue functioning (Fig. 4).
Fig. 4.
Nampt (PBEF/Visfatin) as a regulator of cell survival, metabolism and longevity. In this model, Nampt is upregulated by nutrient deprivation, caloric restriction (CR) and other forms of mild stress, and localizes with Nmnat to various organelles to catalyze NAM depletion and NAD+ production. This stimulates sirtuins, PARP and other NAD+ dependent enzymes that boost cell survival and alter metabolism to promote the survival of mammals during adversity. Nampt is also secreted from adipocytes as Visfatin and it bind to the insulin receptor to trigger glucose uptake by peripheral tissues. Prolonged upregulation of Nampt might mimic aspects of CR and extend mammalian lifespan, as PNC1 does in S. cerevisiae.
8. Is Nampt a master regulator of aging?
For Nampt to be considered the functional equivalent of yeast Pnc1 it needs to satisfy four criteria. It should: (i) be induced by biological stress and/or nutrient deprivation, (ii) catalyze a rate-limiting step in the removal of NAM and regeneration of NAD, (iii) control the activity of sirtuins, and (iv) increase cellular resistance to damage and alter metabolism consistent with lifespan extension. Over the past two years, many of these criteria have been met. With regards the first criterion, our laboratory has found that Nampt is upregulated following treatment of cells with DNA damaging agents and in rats subjected to fasting (Hongying Yang, and D. Sinclair, unpublished results). Satisfying the second criterion, Imai and colleagues showed that overexpression of Nampt causes a 55% increase in total NAD+, demonstrating that Nampt catalyzes a ratelimiting step in the NAD salvage pathway (Revollo et al., 2004). Overexpression of the other NAD+ salvage pathway gene, NMNAT, does not increase NAD levels in animals or cells (Araki et al., 2004; Revollo et al., 2004) unless the cells are severely depleted for NAD+ (Pillai et al., 2005).
It is not yet known whether Nampt reduces NAM levels in mammals. Measuring NAM levels in cell culture could be misleading because NAM is a small molecule that can diffuse across the plasma membrane. Measuring NAM in tissues using mass spectrometry or a similarly quantitative method would make for a convincing case but no one has yet attempted this. The Km for nicotinamide is calculated to be 0.92 µM (Revollo et al., 2004), which is low relative to the estimated concentration of NAM in eukaryotes (~100 µM), indicating that Nampt levels dictate NAM clearance. The same study also satisfied the third criterion by showing that Nampt boosts SIRT1 activity. Increased dosage of Nampt in mammalian cells repressed a SIRT1-regulated lacZ reporter and gene expression changes due to overexpression of Nampt or SIRT1 had significant overlap (Revollo et al., 2004).
There is also evidence for the fourth and final criterion: that Nampt modulates cell survival in the same way CR does. Neutrophils are part of innate immune response to injury or infection, and dictate the inflammatory response at an injury site. Multiple lines of evidence implicate activated neutrophils in the pathogenesis of sepsis, inflammatory bowel disease, arthritis, and Alzheimer disease. Neutrophils are constitutively apoptotic, but they also have the ability to bypass cell death in response to stimuli from the microenvironment. Prolonged neutrophil survival during inflammation results from active inhibition of apoptosis, a process that requires new gene expression and the synthesis of survival factors, including IL-1. It is well-known that the survival of human neutrophils declines with age and this is dependent on modulation of p42/p44 MAPK and Bax/Bcl-xL pathways (Fulop et al., 2004; Larbi et al., 2005), although there is no information on the impact of CR on neutrophil survival. Jia and colleagues detected increased synthesis of Nampt mRNA in stimulated neutrophils and a corresponding decrease when treated with an agonistic anti-CD95 antibody (Jia et al., 2004). Overexpression of Nampt in neutrophils or addition of recombinant Nampt to the culture medium inhibited apoptosis, while antisense oligonucleotides against Nampt completely blocked the anti-apoptotic activity of LPS and a variety of host-derived inflammatory cytokines, including IL-1β, TNF-αGM-CSF, and IL-8. Together, the work shows that Nampt is a suppressor of neutrophil apoptosis, a phenomenon that increases with age in humans.
Along similar lines, another study (Pillai et al., 2005) showed that in cardiac myocytes, Nampt mimics the antiapoptotic effects of CR (Murakami et al., 2003; Sinclair, 2005). CR is known to increase cellular resistance to stress and attenuate the loss of muscle mass during aging. CR also provides protection from cell death and dysfunction following cardiac ischemia (Abete et al., 2002), which is achieved, in large part, by decreasing the susceptibility of myocytes to apoptosis. The death of cardiac myocytes in cardiac failure can be mimicked by an aortic banding procedure, which leads to cell death from oxidant-induced DNA damage and the resulting over-stimulation of the NAD+-dependent enzyme poly-ADP-ribose polymerase (PARP), which depletes NAD+. Overexpression of Nampt protects primary cardiac myocytes from PARP-induced death by maintaining NAD+ levels and stimulating SIRT1 activity (Pillai et al., 2005). The same SIRT1-dependent protective effect is observed for resveratrol, a SIRT1 activating molecule (Howitz et al., 2003).
Rodents on a CR diet are relatively resistant to neurodegeneration as a result of aging, brain injury, and Alzheimer disease (AD) (Hiona and Leeuwenburgh, 2004; Wang et al., 2005). Jeffrey Milbrandt and colleagues showed that the amplification of the NMNAT gene in the WldS mutant mouse is highly protective against axonal degeneration (a.k.a. Wallerian degeneration) that follows the transection or crushing of a nerve (Araki et al., 2004). NMNAT encodes the enzyme that lies immediately downstream of Nampt (see Fig. 3), and most likely protects because it increases NAD+ production and boosts SIRT1 activity (Araki et al., 2004). In fact, NAD+ itself is highly protective of neurons. No change in steady state NAD levels were associated with additional Nampt, with the researchers arguing, as our lab did for the yeast enzymes, that the effect is due to increased metabolic flux through the NAD pathway or highly local increases in NAD+ concentration (Anderson et al., 2002). Although it has not yet been tested, the work implies that overexpression of Nampt should also have a strong neuroprotective effect.
9. Perspective
We have seen that yeast cells utilize Pnc1, a regulator of NAD+ biosynthesis, to control lifespan in response to environmental conditions. We have also discussed some tantalizing clues that there may be a similar mechanism in mammals that works through a very similar mechanism. The clearest test of this hypothesis will be to test the health and lifespan of a mouse that overexpresses Nampt, with the prediction that the animal will be longer-lived and resistant to diseases of aging.
With regards the predicted relationship between Nampt and cancer in vivo, is not yet clear whether Nampt will be an oncogene, a tumor suppressor or neither. One could argue that its high level of expression in cancer cells and its ability to promote cell survival suggests that it will promote cancer. Blocking Nampt activity with a small molecule FK866 effectively induces cell death by apoptosis in HepG2 human liver carcinoma cells with an IC50 of approximately 1 nM (Drevs et al., 2003), but whether overexpression will cause cancer is a very different issue. The systems that promote cell survival and cell death are highly complex and context dependent, so with regards Nampt cancer, the jury is still out.
The correlation between Nampt expression levels and obesity is seemingly inconsistent with it being involved in CR. Nevertheless, we have observed increases in the intracellular form of Nampt during fasting, arguing that the intracellular and extracellular forms of Nampt can be regulated differently. Yet, the ability of Nampt to bind to the insulin receptor and promote glucose uptake is consistent with the low serum glucose levels observed in CR animals. It is not known whether the WldS mutant mouse, with its amplification of the NMNAT gene, lives longer. But one might not expect them to, given that additional NMNAT does not deplete nicotinamide nor do Wld mice have increased NAD+ levels. Whether or not Nampt-over-expressing mice will have the physiology of CR animals and live longer will not be known for years but, if so, it will be certainly worth the wait.
Acknowledgements
We thank C. Wolberger, B. North and J. Baur for advice and assistance with this manuscript. The Sinclair lab is supported by RO1 grants from NIH and NIA, and the Glenn Laboratories for the Molecular Biology of Aging. H. Yang was supported by a Harvard/Hartford Advanced Research Award. David A. Sinclair is a co-founder and advisor to Sirtris Pharmaceuticals, Cambridge, MA.
References
- Abete P, Testa G, Ferrara N, De Santis D, Capaccio P, Viati L, Calabrese C, Cacciatore F, Longobardi G, Condorelli M, et al. Cardioprotective effect of ischemic preconditioning is preserved in food-restricted senescent rats. Am. J. Physiol. Heart Circ. Physiol. 2002;282:H1978–H1987. doi: 10.1152/ajpheart.00929.2001. [DOI] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, Lin SS, Manchester JK, Gordon JI, Sinclair DA. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 2002;277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and Pnc1 govern lifespan extension by calorie restriction in S cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- Avalos JL, Celic I, Muhammad S, Cosgrove MS, Boeke JD, Wolberger C. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol. Cell. 2002;10:523–535. doi: 10.1016/s1097-2765(02)00628-7. [DOI] [PubMed] [Google Scholar]
- Avalos JL, Boeke JD, Wolberger C. Structural basis for the mechanism and regulation of Sir2 enzymes. Mol. Cell. 2004;13:639–648. doi: 10.1016/s1097-2765(04)00082-6. [DOI] [PubMed] [Google Scholar]
- Avalos JL, Bever KM, Wolberger C. Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol. Cell. 2005;17:855–868. doi: 10.1016/j.molcel.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Berger F, Lau C, Dahlmann M, Ziegler M. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J. Biol. Chem. 2005;280:36334–36341. doi: 10.1074/jbc.M508660200. [DOI] [PubMed] [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- Drevs J, Loser R, Rattel B, Esser N. Antiangiogenic potency of FK866/K22.175, a new inhibitor of intracellular NAD biosynthesis, in murine renal cell carcinoma. Anticancer Res. 2003;23:4853–4858. [PubMed] [Google Scholar]
- Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- Fukuhara A, Matsuda M, Nishizawa M, Segawa K, Tanaka M, Kishimoto K, Matsuki Y, Murakami M, Ichisaka T, Murakami H, et al. Visfatin: a protein secreted by visceral fat that mimics the effects of insulin. Science. 2004 doi: 10.1126/science.1097243. [DOI] [PubMed] [Google Scholar]
- Fulop T, Larbi A, Douziech N, Fortin C, Guerard KP, Lesur O, Khalil A, Dupuis G. Signal transduction and functional changes in neutrophils with aging. Aging Cell. 2004;3:217–226. doi: 10.1111/j.1474-9728.2004.00110.x. [DOI] [PubMed] [Google Scholar]
- Gallo CM, Smith DL, Jr, Smith JS. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol. Cell Biol. 2004;24:1301–1312. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L, Picard F. Calorie restriction – the SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Halme A, Bumgarner S, Styles C, Fink GR. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116:405–415. doi: 10.1016/s0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]
- Hiona A, Leeuwenburgh C. Effects of age and caloric restriction on brain neuronal cell death/survival. Ann. NY Acad. Sci. 2004;1019:96–105. doi: 10.1196/annals.1297.018. [DOI] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Jia SH, Li Y, Parodo J, Kapus A, Fan L, Rotstein OD, Marshall JC. Pre-B cell colony-enhancing factor inhibits neutrophil apoptosis in experimental inflammation and clinical sepsis. J. Clin. Invest. 2004;113:1318–1327. doi: 10.1172/JCI19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood TB, Shanley DP. Food restriction, evolution and ageing. Mech. Ageing Dev. 2005;126:1011–1016. doi: 10.1016/j.mad.2005.03.021. [DOI] [PubMed] [Google Scholar]
- Kitani T, Okuno S, Fujisawa H. Growth phase-dependent changes in the subcellular localization of pre-B-cell colony-enhancing factor. FEBS Lett. 2003;544:74–78. doi: 10.1016/s0014-5793(03)00476-9. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2006;312:1312. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- Landry J, Slama JT, Sternglanz R. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem. Biophys. Res. Commun. 2000a;278:685–690. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA. 2000b;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larbi A, Douziech N, Fortin C, Linteau A, Dupuis G, Fulop T., Jr The role of the MAPK pathway alterations in GM-CSF modulated human neutrophil apoptosis with aging. Immun. Ageing. 2005;2:6. doi: 10.1186/1742-4933-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Martin PR, Shea RJ, Mulks MH. Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence. J. Bacteriol. 2001;183:1168–1174. doi: 10.1128/JB.183.4.1168-1174.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoro EJ. Caloric restriction and aging: an update. Exp. Gerontol. 2000;35:299–305. doi: 10.1016/s0531-5565(00)00084-x. [DOI] [PubMed] [Google Scholar]
- Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- Murakami S, Salmon A, Miller RA. Multiplex stress resistance in cells from long-lived dwarf mice. FASEB. J. 2003;17:1565–1566. doi: 10.1096/fj.02-1092fje. [DOI] [PubMed] [Google Scholar]
- Nemeth E, Tashima LS, Yu Z, Bryant-Greenwood GD. Fetal membrane distention: I, Differentially expressed genes regulated by acute distention in amniotic epithelial (WISH) cells. Am. J. Obstet. Gynecol. 2000;182:50–59. doi: 10.1016/s0002-9378(00)70490-x. [DOI] [PubMed] [Google Scholar]
- Ognjanovic S, Bao S, Yamamoto SY, Garibay-Tupas J, Samal B, Bryant-Greenwood GD. Genomic organization of the gene coding for human pre-B-cell colony enhancing factor and expression in human fetal membranes. J. Mol. Endocrinol. 2001;26:107–117. doi: 10.1677/jme.0.0260107. [DOI] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2α deacetylase activity. J. Biol. Chem. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad Sci. USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A, Shea RJ, Mulks MH, Gigot D, Urbain J, Leo O, Andris F. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur. J. Immunol. 2002;32:3225–3234. doi: 10.1002/1521-4141(200211)32:11<3225::AID-IMMU3225>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Samal B, Sun Y, Stearns G, Xie C, Suggs S, McNiece I. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol. Cell Biol. 1994;14:1431–1437. doi: 10.1128/mcb.14.2.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve AA, Schramm VL. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry. 2003;42:9249–9256. doi: 10.1021/bi034959l. [DOI] [PubMed] [Google Scholar]
- Sauve AA, Celic I, Avalos J, Deng H, Boeke JD, Schramm VL. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry. 2001;40:15456–15463. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- Schmidt MT, Smith BC, Jackson MD, Denu JM. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J. Biol. Chem. 2004;279:40122–40129. doi: 10.1074/jbc.M407484200. [DOI] [PubMed] [Google Scholar]
- Sinclair DA. Toward a unified theory of caloric restriction and longevity regulation. Mech. Ageing Dev. 2005 doi: 10.1016/j.mad.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava DK, Bernhard SA. Biophysical chemistry of metabolic reaction sequences in concentrated enzyme solution and in the cell. Annu. Rev. Biophys. Biophys. Chem. 1987;16:175–204. doi: 10.1146/annurev.bb.16.060187.001135. [DOI] [PubMed] [Google Scholar]
- Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product-1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Tsang AW, Escalante-Semerena JC. CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J. Biol. Chem. 1998;273:31788–31794. doi: 10.1074/jbc.273.48.31788. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Eaton EN, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Vergnes B, Sereno D, Madjidian-Sereno N, Lemesre JL, Ouaissi A. Cytoplasmic SIR2 homologue overexpression promotes survival of Leishmania parasites by preventing programmed cell death. Gene. 2002;296:139–150. doi: 10.1016/s0378-1119(02)00842-9. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech. Ageing Dev. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Wang J, Ho L, Qin W, Rocher AB, Seror I, Humala N, Maniar K, Dolios G, Wang R, Hof PR, Pasinetti GM. Caloric restriction attenuates beta-amyloid neuropathology in a mouse model of Alzheimer’s disease. FASEB J. 2005 doi: 10.1096/fj.04-3182fje. [DOI] [PubMed] [Google Scholar]
- Xu LG, Wu M, Hu J, Zhai Z, Shu HB. Identification of downstream genes up-regulated by the tumor necrosis factor family member TALL-1. J. Leukoc. Biol. 2002;72:410–416. [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Chai X, Marmorstein R. Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Escherichia coli. J. Mol. Biol. 2004a;337:731–741. doi: 10.1016/j.jmb.2004.01.060. [DOI] [PubMed] [Google Scholar]
- Zhao K, Harshaw R, Chai X, Marmorstein R. Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases. Proc. Natl. Acad. Sci. USA. 2004b;101:8563–8568. doi: 10.1073/pnas.0401057101. [DOI] [PMC free article] [PubMed] [Google Scholar]