Summary
Rescuing the function of mutant p53 protein is an attractive cancer therapeutic strategy. Using the NCI anticancer drug screen data, we identified two compounds from the thiosemicarbazone family that manifest increased growth inhibitory activity in mutant p53 cells, particularly for the p53R175 mutant. Mechanistic studies reveal that NSC319726 restores WT structure and function to the p53R175 mutant. This compound kills p53R172H knock-in mice with extensive apoptosis and inhibits xenograft tumor growth in a 175-allele specific mutant p53 dependent manner. This activity depends upon the zinc ion chelating properties of the compound as well as redox changes. These data identify NSC319726 as a p53R175 mutant reactivator and as a lead compound for p53 targeted drug development.
INTRODUCTION
TP53 is the most frequently mutated gene in human cancer with mutation frequencies ranging from 38–50% in some reports to as high as 75% and 96% in pancreatic adenocarcinoma and high grade serous ovarian carcinomas respectively (Hingorani et al., 2005; Cancer Genome Atlas Research Network, 2011; Petitjean et al., 2007). The majority of mutations are mis-sense mutations that occur most frequently in six “hotspot” codons within the DNA binding domain (Olivier et al., 2010). These mutant proteins are classified as either DNA contact mutants (e.g. p53R273H) when the mutation occurs in a DNA binding residue, or conformational mutants (e.g. p53R175H) when a conformational change causes a loss of WT p53 DNA binding.
Mutant p53 proteins are found at high concentrations in tumor cells relative to WT p53 mostly due to a loss of WT p53 transcription of the MDM2 gene that negatively regulates p53, as well as other tumor specific alterations such as loss of p16INK4a (Haupt et al., 1997; Midgley and Lane, 1997; Terzian et al., 2008). The concept that these mutant proteins are functional and regulate important processes relevant to tumor biology is referred to as the mutant p53 gain-of-function (GOF) phenotype (Sigal and Rotter, 2000). Properties attributed to mutant p53 GOF include enhanced tumorigenesis, invasion and metastasis (Adorno et al., 2009; Dittmer et al., 1993; Liu et al., 2000; Muller et al., 2009). Taken together, these properties make mutant p53 an attractive target for drug development.
The NCI anticancer drug screen has reported growth inhibition IC50s on 48,129 compounds tested on a panel of sixty human tumor cell lines (NCI60 screen). Given that the p53 status (WT, null, mutant) of these cell lines is known (Ikediobi et al., 2006; Shoemaker, 2006), we hypothesized this screen could be used to uncover drugs targeting p53 mutant tumors. Thus we developed a methodology to identify compounds with increased activity in a panel of tumor cell lines with p53 mutations, relative to p53 WT controls. In this study, we attempted to validate this methodology using two compounds that belong to the thiosemicarbzone family of metal ion chelators. We further investigated one of the compounds with particular toxicity to cell lines containing a p53R175 mis-sense mutation; identifying the mechanism of this toxicity and the properties of the compound that are relevant to this mechanism.
RESULTS
Identification of thiosemicarbazones with activity in cell lines expressing mutant p53
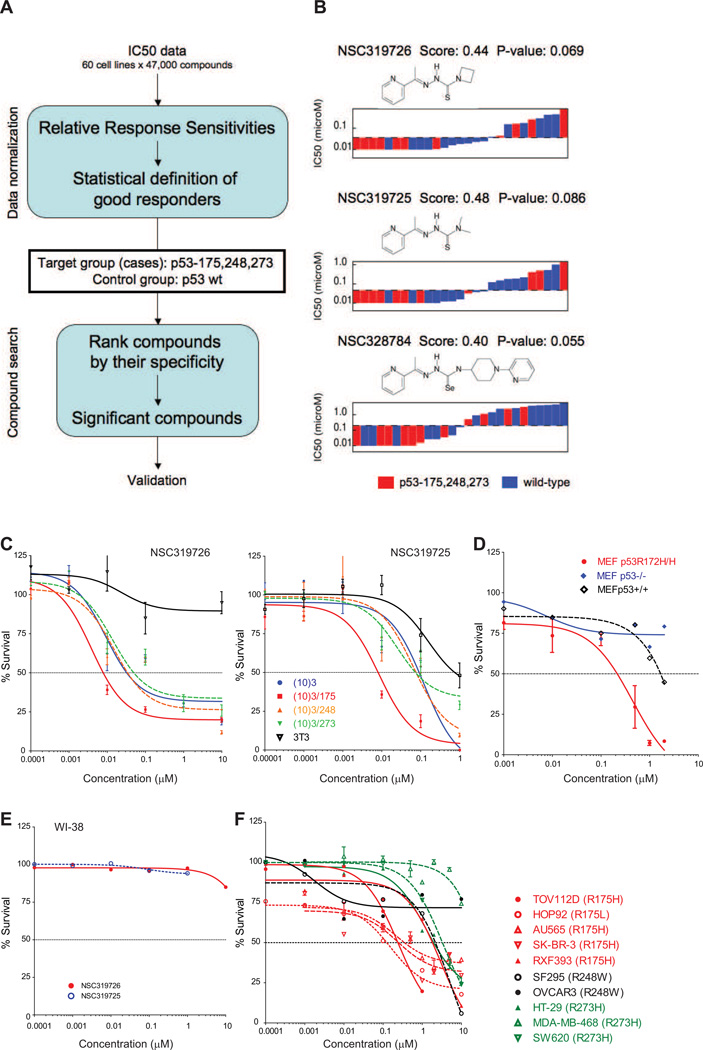
Our methodology deals with the intrinsic heterogeneities of the NCI60 screen (Figure 1A and Supplementary Methods). This takes as input IC50 data, applies data normalization to obtain a working definition of a good response; and then ranks compounds manifesting a good response using a scoring function. This score function identifies those compounds with an enrichment of good responders in the case group (mutant p53), while simultaneously having a depletion of good responders in the control group (wild-type p53). To reduce the heterogeneity in the mutant p53 group, we focused on mutations in hotspot codons 175, 248 and 273 (11 cell lines), while the control group was composed of sixteen p53 wild-type cell lines. Applying this methodology, we observed three of the highest scoring compounds belonged to the thiosemicarbazone family (NSC319725, NSC319726, and NSC328784) that preferentially inhibited p53 mutant cell lines (Figure 1B). When comparing the IC50’s of these compounds by p53 mutational status, we see the low IC50s are enriched of cells with a p53 mutation (Figure 1B, red), while the majority of p53 wild-type cells exhibit high IC50s (Figure 1B, blue). In contrast, two reported mutant p53 reactivators (PRIMA-1, MIRA-1) as well as an additional thiosemicarbazone currently in clinical trials (Triapine) scored poorly using this methodology in comparison to NSC319726 (Figure S1A–D).
Figure 1. Identification of thiosemicarbazones with activity in cell lines expressing mutant p53.
(A) Flow-diagram of the in silico screen methodology. (B) IC50 distribution and chemical structure of the thiosemicarbazones aligning lower IC50s to higher IC50s in cells with mutant p53 (red) and WT p53 (blue), analyzed with the NCI60 dataset. In the NCI60 dataset, the human tumor cell lines are treated with 5 serial dilutions of each compound for 48 hours at 37°C. (C) Validation of the sensitivity of the compounds in the cell lines expressing mutant p53. Cell growth inhibition assays using a mouse embryonic fibroblast (MEF) cell line system consisting of (10)3 (p53−/−), (10)3/175, (10)3/248, (10)3/273 and 3T3 (p53+/+). Left panel: Growth inhibition with NSC319726. Right panel: Growth inhibition with NSC319725. (D) Sensitivity to NSC319726 in a set of isogenic MEF cell lines from p53+/+, p53−/− and p53R172H/R172H mice. (E) NSC319725 and NSC319726 do not inhibit cell growth of WI38 (p53+/+) human fibroblasts. (F) The sensitivity of NSC319726 in human tumor cell lines with hotspot p53 mutations. Cell growth inhibition was analyzed by MTS assay (C) or by Guava ViaCount assay (D, E, F). In both the MTS and Guava ViaCount assays, cells were treated with five serial dilutions of the compounds (0.00001 µM to 10 µM) for 3 days. The error bars are +/− SD. See also Figure S1.
We validated two of our screened compounds (NSC319725, NSC319726) using a mouse fibroblast cell line containing no functional TP53 gene (10(3)) from which several stable CMV-driven mutant p53 transfectants (175, 248, 273) were derived (Dittmer et al., 1993). Balb/c 3T3 fibroblasts were used as a p53 WT control as this is the same background as the 10(3) and its derived cell lines. Both compounds exhibited growth inhibition at markedly lower concentrations in cells expressing mutant p53 as compared to the WT control, particularly in the 175 allele (Figure 1C). The IC50 for the NSC319725 treated 175 mutant was ≥100 fold lower than the WT. For NSC319726, the effect was even greater as the IC50 for the 175 mutant was 8 nM while the IC50 of the WT was not reached. Similar to the Balb/c 3T3, these two compounds were remarkably nontoxic to WI38 human fibroblasts (p53 WT), as an IC50 for both compounds was not obtained (Figure 1D). Furthermore, NSC319726 did not induce WT p53 protein levels or transcriptional activity as common cytotoxic agents such as etoposide do in vitro (Figure S1E).
We further validated NSC319726 employing additional p53 mutant cell line systems. In a set of isogenic MEF cell lines from p53+/+, p53−/− and p53R172H/R172H mice, we found that NSC319726 exhibited a much higher sensitivity for the MEF-p53R172H/R172H cell line as compared to the p53+/+ and p53−/− controls (Figure 1E). We compared the sensitivities across human tumor cell lines with different p53 “hot spot” mutations (175, 248, 273). With the exception of one p53R175 cell line (RXF393), the 175 cells exhibited similar IC50’s that were approximately 10-fold and in some instances 100-fold lower than the other hotspot mutants indicating a 175 allele preference of growth inhibition by NSC319726 (Figure 1F). It is important to emphasize that the results of these assays depend to a significant degree on a number of factors including the method of assaying cell viability, the proliferation rates of the cells, the confluency at the time of drug exposure as well as differences in redox in tumor cells. To diminish intra-assay variability we used the Guava-ViaCount assay to compare the effects of NSC319726 across tumor cell lines that varied by p53 status given the heterogeneity of these cell lines. In both the MTS and Guava-ViaCount assay we found it to be important to have the cells at 50–60% confluency at the time of drug exposure. Cell growth inhibition assays carried out under different conditions can and do give different IC50’s.
Induction of p53R175-dependent apoptosis by NSC319726
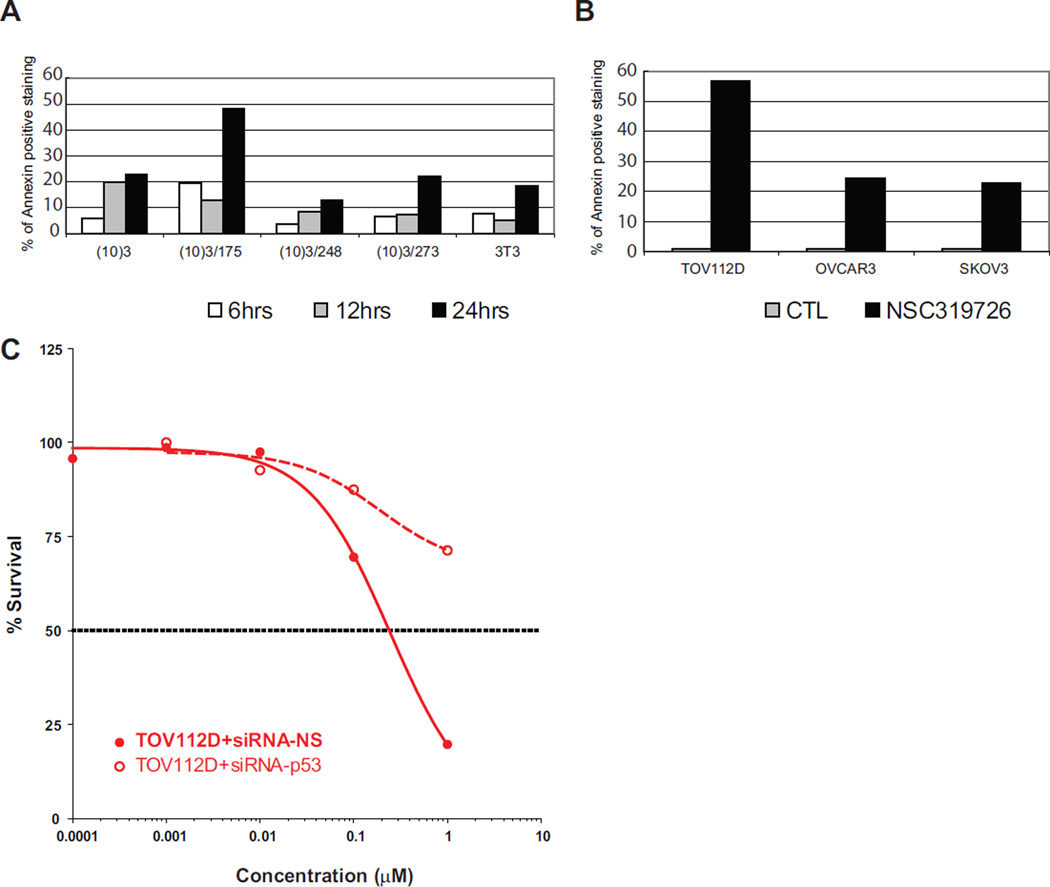
To determine if the inhibition of cell growth was mediated by apoptosis, we performed Annexin-V staining of different cell lines treated with NSC319726. We observed an increase in the number of Annexin-V stained cells with a maximum increase in the 175 mutant (Figure 2A). Similarly, treatment of three different ovarian carcinoma cell lines (TOV112D (p53R175H), OVCAR3 (p53R248W) and SKOV3 (p53−/−)) with NSC319726 resulted in an induction of apoptosis in the p53R175 mutant more than 2-fold higher than the other two mutant cell lines (Figure 2B). When we silenced expression of the p53R175 mutant protein by si-RNA, we observed a marked reduction in sensitivity to cell growth inhibition, demonstrating that the NSC319726 mechanism is at least partially dependent on the p53R175 mutant protein (Figure 2C).
Figure 2. The p53-175 mutant-dependent apoptosis induced by NSC319276.
The apoptosis is measured with Annexin-V staining using Guava Nexin reagent and Guava PCA instrument. (A) Apoptosis is measured in the (10)3-derived MEF cells. (B) Apoptosis is measured in a panel of ovarian cancer cell lines, TOV112D (p53R175H), OVCAR3 (p53R248W) and SKOV3 (p53−/−). The treatment is 1 µM NSC319726 (as in the other figures unless described differently) for 24 hours. (C) Growth inhibition by NSC319726 in TOV112D cells with siRNA knockdown of p53R175H mutant protein, measured as in Figure 1E.
NSC319726 induction of a “WT-like” conformational change in the p53R175 mutant protein
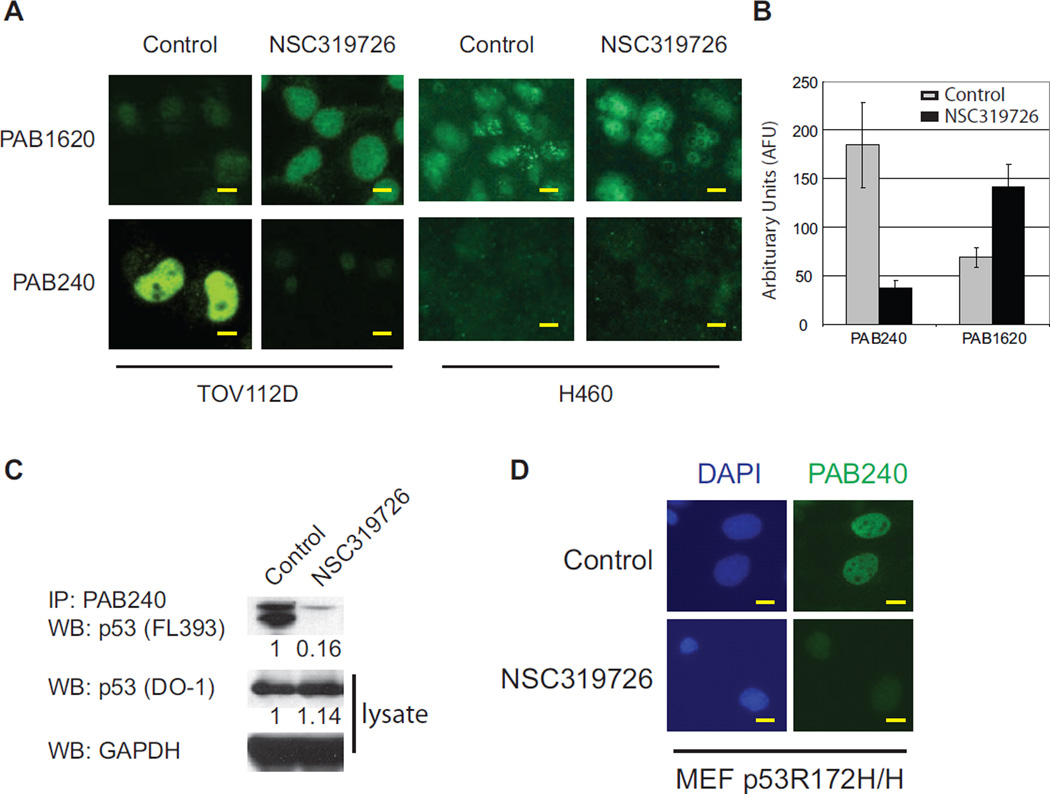
Because NSC319726 induced a p53 apoptotic function in p53R175 cells, we investigated if NSC319726 restored WT conformation to the mutant p53R175 protein. Using conformation specific antibodies by immunoflourescence, we observed that NSC319726 induced a conformation change in the p53R175 mutant to a structure that was recognized by the WT specific antibody (PAB1620) and was no longer recognized by the mutant specific antibody (PAB240) (Figure 3A). Quantification of the fluoresence intensity for PAB240 was reduced by five-fold, while that of PAB1620 was increased by two-fold (Figure 3B). We confirmed this conformation change by PAB240 immunoprecipitation of NSC319726 treated TOV112D lysates indicating a more than 85% decrease in PAB240 immunoreactivity (Figure 3C). Next we sought to confirm this conformation change in an additional cell line. NSC319726 treatment of a MEF cell line derived from p53R172H/R172H mice resulted in a loss of PAB240 immunoflouresence staining (Figure 3D). The PAB1620 antibody staining is not shown as this antibody is human specific. This demonstrates that the conformation change induced by NSC319726 occurs in both human (p53R175) and mouse (p53R172) proteins.
Figure 3. A “WT-like” conformational change in the p53-175 mutant protein induced by NSC319726.
(A) Immunoflourescence of TOV112D (p53R175H) cells using p53 conformation specific antibodies (PAB1620 for WT conformation and PAB240 for mutant conformation). The H460 cell line is used as a control to show the WT p53 conformation is not changed by NSC319726. All scale bars represent a size of 25 µm. (B) Quantification of PAB240 and PAB1620 staining indicates significant differences upon treatment with NSC319726 (p<0.0001) (t-test). The error bars are +/− SD. (C) Immunoprecipitation of mutant p53 protein in TOV112D cells using PAB240 and detected by p53 (FL393) by Western blot. (D) Immunoflourescence of MEF p53R172H/H cells using the PAB240 mutant specific antibody. In all experiments (A, C, D) cells are treated with NSC319726 (1 µM) for 6 hours before harvesting for immunostaining or immunoprecipitation.
Restoration of p53 transactivational function through the “WT-like” conformational change induced by NSC319726
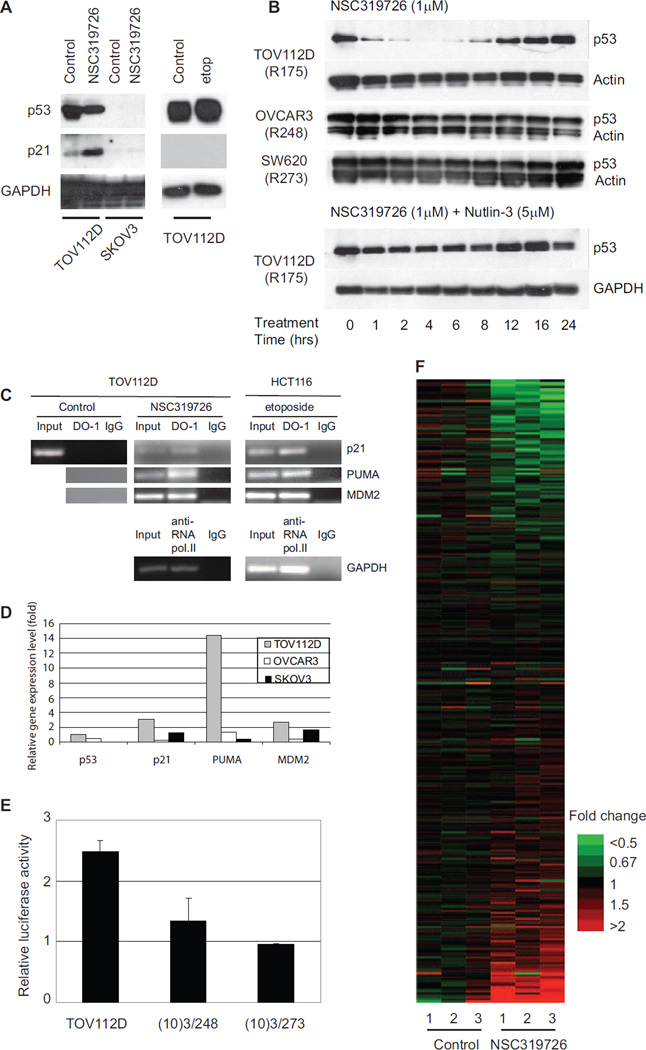
To determine if the conformation change observed with the p53R175 mutant resulted in restoration of p53 transcriptional function, we examined p21 protein levels after NSC319726 treatment in TOV112D and SKOV3 cells. We found that NSC319726 induced p21 in the TOV112D cell line but not in the SKOV3 (Figure 4A). Treatment of the TOV112D cells with the DNA damaging agent etoposide failed to induce p21 (Figure 4A). This suggests that the p21 induction in NSC319726 treated TOV112D cells was p53R175 mutant dependent.
Figure 4. Restoration of site-specific p53-175 mutant protein transactivational function by NSC319726.
(A) TOV112D and SKOV3 cells are treated with NSC319726 or etoposide and protein extracts are analyzed by Western blot with antibodies directed against p21, p53. GAPDH is used as the internal (loading) control. (B) Stability of p53 mutant protein in TOV112D (p53R175), OVCAR3 (p53R248), and SW620 (p53R273) cells treated with NSC319726 (1 µM) with/without Nutlin-3 (5 µM) over a 24-hour time course. Protein levels are shown by Western blot using antibodies to p53 (DO-1) or either actin or GAPDH. (C) Chromatin Immunoprecipitation (ChIP) assay. Lysates from TOV112D cells treated with NSC319726 (1 µM) and HCT116 cells treated with etoposide (20 µM) for 6 hours are subject to immunoprecipitation using anti-p53 antibody (DO-1) followed by PCR of p53 recognition elements (p53REs) of p21, PUMA and MDM2. HCT116 treated with etoposide is used as a positive control for functional p53 DNA-binding property. ChIP with anti-RNA polymerase II antibody and PCR for GAPDH is also used as a positive control. (D) qRT-PCR of p21, MDM2 and PUMA in TOV112D, OVCAR3 and SKOV3 cells with treatment of NSC319726 (1 µM) for 24 hours. The RNA is extracted from the cells using Qiagen Rneasy kit and the gene expression level is measured by quantitative RT-PCR using TaqMan gene expression assays. (E) TOV112D cells are transfected with a plasmid bearing the 20-bp p53 response element (p53RE) in p21 promoter region, and then treated with NSC319726 (1 µM) for 24 hours, followed by luciferase reporter assay. The error bars are +/− SD. (F) Microarray analysis of NSC319726 (1 µM, 24 hours) treated and control TOV112D cells comparing gene expression levels of p53 targets. The list of the genes is presented in Table S1. The heatmap is shown with the color scale indicating fold change. The data from the microarrays have been deposited in the GEO database with accession number GSE35972.
We note NSC319726 caused a reduction in the levels of the mutant protein (Figure 4A). We examined this further by measuring the mutant protein levels at different time points over a 24 hour period. Treatment of TOV112D cells with NSC319726 decreased the stabilization of the mutant protein with the lowest levels seen at six hours with a return to pre-treatment levels by 24 hours. This p53 mutant protein destabilization was not seen when we treated two cell lines containing the p53 DNA contact mutants R248 (OVCAR3), and R273 (SW620) with NSC319726. This indicates an allele specificity to the NSC319726 mediated effect on p53 mutant protein levels. We hypothesized that the decrease in p53R175 protein stability was due to restoration of Mdm2 negative regulation of the “WT-like” p53R175 mutant. To test this, we performed this experiment in the presence of Nutlin-3 (small molecule antagonist of Mdm2) and found that Nutlin-3 abrogated the decrease in stability of the p53-175 mutant induced by NSC319726 (Figure 4B). This restoration of Mdm2 negative regulation is the result of transactivation of the MDM2 promoter by the “WT” like p53R175 mutant (Figure 4C and 4D).
We confirmed the ability of NSC319726 to restore DNA binding properties to the p53R175 mutant protein by chromatin immunoprecipitation (ChIP). Using the p53 antibody, DO-1, ChIP analysis of NSC319726 treated TOV112D cells revealed the restoration of site-specific DNA binding of p53R175 mutant to the promoters of p21, PUMA and MDM2 (Figure 4C). HCT116 cells treated with etoposide as well as RNA polymerase II binding of the GAPDH promoter were used as controls (Figure 4C). We compared the mRNA levels of several p53 targets (p21, PUMA, MDM2) in the TOV112D (p53R175H), OVCAR3 (p53R248W) and SKOV3 (p53−/−) cells upon treatment and found that NSC319726 increased the levels of all three p53 target genes in TOV112D cells, particularly the apoptotic gene PUMA (Figure 4D). To provide further evidence that NSC319726 restored site-specific p53 transactivational function we transfected the TOV112D cells with a luciferase reporter plasmid containing 20 base pairs of the p53 response element in the p21 promoter. NSC319726 treatment induced a 2.5 fold increase in luciferase activity that was not seen in MEF cells expressing the 248 and 273 alleles, indicating that the restoration of transcriptional function is allele specific (Figure 4E). We next utilized gene expression microarrays to examine the transcriptional activity of a larger cohort of p53 targets in TOV112D cells. NSC319726 treatment produced a p53 target expression signature remarkably different from the untreated controls, confirming that the conformation change of the p53R175 mutant results in a transcriptionally active protein (Figure 4F). Table S1 reports the average expression log2-ratios between cells treated with NSC319726 and untreated controls, focusing on probes that map to p53 target genes. The list of p53 targets were selected from a literature review, providing a comprehensive list of experimentally verified targets of p53 (Riley et al., 2008).
In vivo allele-specific p53R175H mutant reactivation by NSC319726
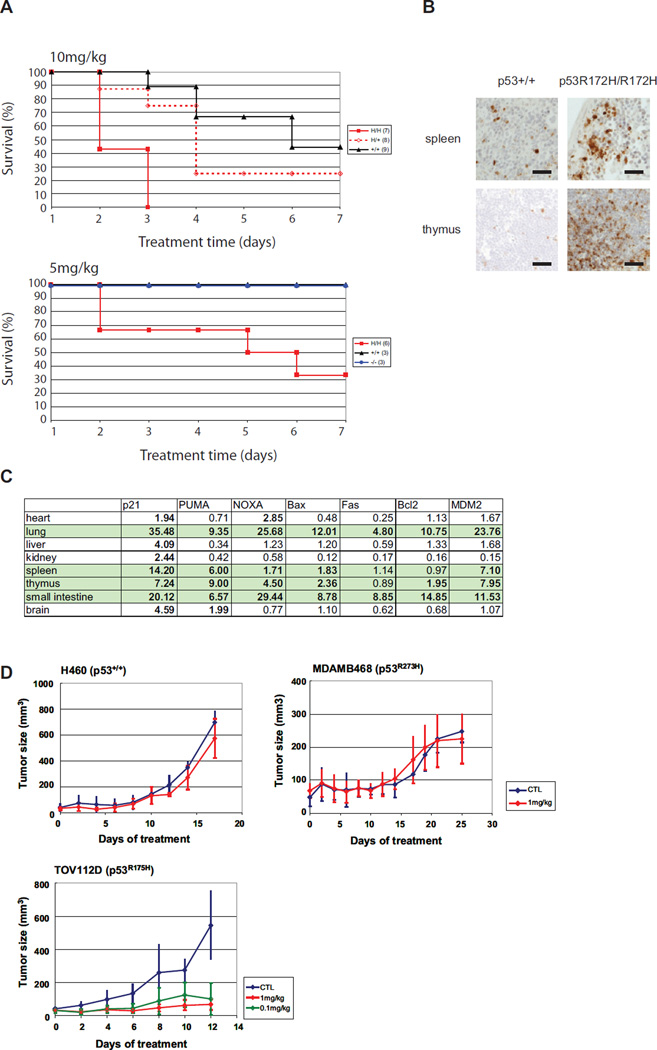
To investigate the p53 mutant reactivation in vivo we performed toxicity assays in p53+/+, p53−/− and p53R172H knock-in mice (Lang et al., 2004). We hypothesized that p53R172H mice would experience greater toxicity for a given dose of NSC319726 as compared to p53+/+ mice. By day 3 of treatment (10 mg/kg/day), all seven of the p53R172H/R172H mice had died while only 1/9 p53+/+ mice had died. By day 4, the survival of the p53+/+ mice was 70% while the survival of p53+/R172H was 30%, suggesting a clear dosage effect that is dependent on the TP53R175 genotype (Figure 5A). Next we lowered the dose to 5mg/kg and found that by day seven, the p53+/+ and p53−/− mice exhibited a 100% survival compared to only 30% of the p53R172H/R172H mice (Figure 5A).
Figure 5. In vivo evidence of NSC319726 mediated p53-175 mutant reactivation.
(A) Toxicity assays of NSC319726 in p53R172H/R172H, p53+/+ and p53−/− mice. Mice in the top panel are administered 10mg/kg by daily intraperitoneal (IP) injection for up to 7 days. Mice in the bottom panel are administered 5 mg/kg daily (IP) for up to 7 days. (B) Immunohistochemical staining with cleaved caspase-3 antibody of spleen and thymic tissues in p53+/+ and p53R172H/R172H mice. All scale bars represent a size of 100 µm. (C) qRT-PCR of mRNA levels of several p53-regulated genes in p53+/+ and p53R172H/R172H mouse tissues. The inductions of over 1.7× are highlighted in bold. (D) Efficacy assays of NSC319726. Xenograft tumors are generated using H460, MDAMB468 and TOV112D cells and allowed to grow to 60 mm3 prior to daily intravenous administration of NSC319726 at 1 mg/kg or 0.1 mg/kg (TOV112D only). Tumor measurements are mean ± SD. The following number of animals were used, H460 (Control-5, Treatment-5), MDAMB468 (Control-9, Treatment-8), TOV112D (Control-7, Treatment 1mg/kg-7, 0.1 mg/kg-7). CTL is Control. The error bars are +/− SD.
We examined the tissues of p53+/+ and p53R172H/R172H mice after treatment for evidence of apoptosis as well as gene expression of a panel of p53 targets. We detected abundantly more apoptotic cells in the sections of the spleen and thymus of p53R172H/R172H mice as compared with the p53+/+ controls (Figure 5B). We also detected elevated mRNA levels of a number of p53 targets in a tissue specific fashion in the p53R172H/R172H mice as compared with the p53+/+ controls, most notably in the lung, spleen, thymus and small intestine (Figure 5C).
We tested the ability of NSC319726 to inhibit the growth of xenograft mouse tumors derived from human tumor cell lines carrying different p53 alleles. At a dose of 1mg/kg, tumor growth of the H460 (p53+/+) and MDAMB468 (p53R273W) xenografts was not inhibited relative to the vehicle control whereas tumor growth was significantly inhibited in the TOV112D (p53R175H) xenografts (Figure 5D). When we lowered the dose ten-fold to 0.1 mg/kg in the TOV112D mice, we observed only a small difference in tumor growth inhibition demonstrating both a dosage effect of the drug and a larger therapeutic window. Taken together, these findings provide in vivo evidence for allele specific p53 mutant reactivation.
Zinc ion chelation and redox changes are important for the NSC319726 mediated p53R175 reactivation mechanism
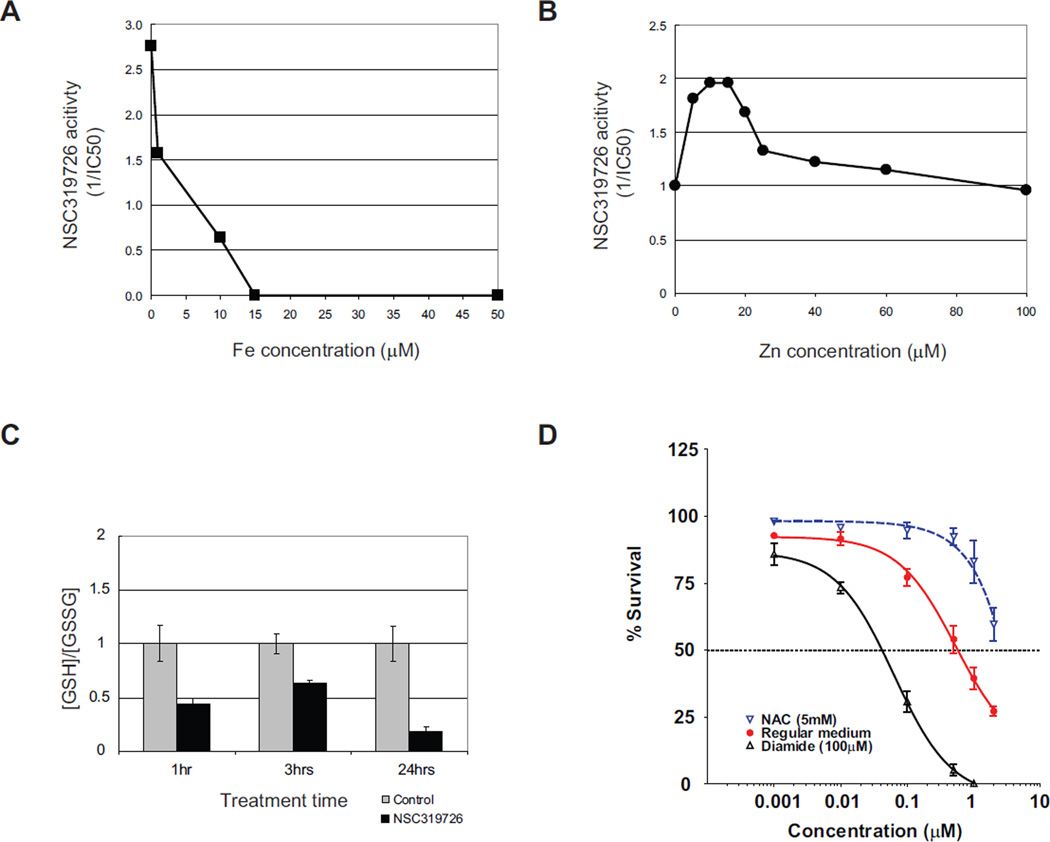
Thiosemicarbazones are metal ion chelators with strong affinity for iron, copper, and zinc (Yu et al., 2009). They have been investigated as anticancer agents and have been shown to inhibit DNA synthesis by inhibiting the iron dependent enzyme ribonucleotide reductase (RR), but at much higher concentrations than are employed to inhibit the growth of p53R175 mutant cells. To determine if the metal ion chelating property of NSC319726 is important to its p53R175 mutant activity we added NSC319726 to TOV112D cells in the presence of various concentrations of FeSO4. We found that FeSO4 at concentrations above 15 µM completely abrogated the activity of NSC319726. At concentrations below 15 µM, the activity of NSC319726 was inhibited in a dose-dependent manner (Figure 6A).
Figure 6. Zinc ion chelation and redox changes are important for the NSC319726 mediated p53-175 mutant reactivation.
(A) TOV112D cells are treated with NSC319726 with or without the presence of various concentrations of FeSO4, followed by measurement of growth inhibition. The NSC319726 activity is shown as 1/IC50 for cell growth inhibition. (B) TOV112D cells are treated with NSC319726 with or without addition of various concentrations of ZnCl2, followed by measurement of growth inhibition. The NSC319726 activity is shown as 1/IC50 for cell growth inhibition. (C) Ratio of reductant GSH and oxidative GSSG in the p53R175H cells upon NSC319726 (1 µM) treatment is measured at several time points (p=0.0057 at 1hr, p=0.0027 at 3hrs, and p=0.001 at 24hrs, t test). The error bars are +/− SD. (D) TOV112D cells are treated with NSC319726 using six serial dilutions (0.001 µM to 2 µM) with either N-acetyl cysteine (NAC, 5 mM) or diamide (100µM) for three days. The growth inhibition was analyzed by MTS assay as in Figure 1C. The error bars are +/− SD. See also Figure S2.
We suspected iron was not the relevant metal ion as iron is not associated with the p53 protein and the treatment of the TOV112D cells with another iron chelator, desferrioxamine (DFO), had no apoptotic effect (Figure S2A). However, zinc is required for proper folding of WT p53, and the 175 mutant is classified as a non-zinc binding mutant because it fails to coordinate zinc (Butler and Loh, 2003; Joerger and Fersht, 2007). When we added NSC319726 to TOV112D cells in the presence of different concentrations of ZnCl2, we found that there was an optimal zinc concentration range (5–15 µM) in which the activity of NSC319726 increased 2-fold (Figure 6B). We did not test higher concentrations over 100 µM of ZnCl2 in combination with NSC319726 because we found that concentrations above 100 µM of ZnCl2 (by itself) were toxic to cells. This toxicity was independent of p53 status (Figure S2B–C).
Another property of thiosemicarbazones is their effect on the redox state of the cell. Thiosemicarbazone:Fe complexes cause oxidative stress by the creation of hydroxyl radicals through Fenton chemistry (Kalinowski and Richardson, 2007; Richardson et al., 2009; Richardson et al., 2006). This is relevant to the mechanism of NSC319726 as redox changes have been reported to influence WT p53 folding (Hainaut and Milner, 1993a). In support of this, we observed a statistically significant decrease in the levels of the cellular reductant glutathione upon NSC319726 treatment of TOV112D cells at 1, 3 and 24 hours (Figure 6C). To determine the importance of these redox changes to the activity of NSC319726 we treated TOV112D cells in the presence of the reducing agent N-acteyl-cysteine (NAC) and the oxidizing agent diamide. We found that 5mM NAC inhibited the apoptotic activity of NSC319726 while diamide (100 µM) enhanced it (Figure 6D). These data suggest that ROS changes are important for the apoptotic mechanism of NSC319726 on p53R175 mutant cells.
DISCUSSION
The reactivation of p53 in mouse tumor models has been shown to be a highly effective therapeutic strategy (Martins et al., 2006; Ventura et al., 2007; Xue et al., 2007). Several small molecules have been claimed to reactivate mutant p53, including CP-31398, WR-1065, PRIMA- 1 and MIRA-1 (Bykov et al., 2002; Bykov et al., 2005; Foster et al., 1999; North et al., 2002). With the exception of one compound, WR1065, all have been identified using traditional chemical screens (Bykov et al., 2002; Bykov et al., 2005). Traditional chemical screens favor the use of matched case/control cell lines derived from the same parental cell line, engineered such that the case cell line carries the molecular alteration under consideration. This can be a fundamental caveat, since cancers are known to be heterogeneous in nature. Here we demonstrate our methodology to screen for compounds manifesting increased sensitivity in a panel of cell lines carrying p53 mutations independently of their diverse genetic backgrounds and cell type specificity, which is a more realistic model of what is observed in the clinic. Applying this methodology to the NCI60 screen we identified three compounds from the thiosemicarbazone family. Follow up experiments with two of these compounds (NSC319725 and NSC319726) corroborated the predicted p53 mutant specific growth inhibitory properties. It is possible that this methodology could be used to identify compounds with increased sensitivity in tumor cell lines carrying mutations in other major oncogene/tumor suppressor pathways.
It is important to note that cell viability assays in Figure 1C and the apoptosis assays in Figure 2A indicate that there is an apoptotic mechanism that is independent of p53 mutational status (null, mis-sense mutant). What initiates this apoptosis is unclear but may be related to either an increase in ROS levels or ribonucleotide reductase (RR) inhibition, two reported mechanisms of action for thiosemicarbazones. Non-tumor cell lines with a WT p53 gene (Balb/c 3T3, WI38) showed relatively little to no growth inhibition by NSC319726 at these same doses, which would argue against the inhibition of RR as the explanation. If increased ROS levels are the reason, we speculate that this apoptosis may be due to the inability to compensate for these oxidative changes in a cell lacking a functional p53 transcription factor.
In distinction, the mechanism of apoptosis in a p53R175 mutant cell is dependent on the mutant p53R175 mutant protein. NSC319726 treatment induces a WT-like conformational change in the p53R175 mutant protein that restores sequence-specific p53 transcription. This is best observed in the in vivo experiments employing both knock-in p53 mutant mice and xenografts.
We have demonstrated that the metal ion chelating properties of NSC319726 are required for the mutant p53 mediated apoptotic activity. Interestingly, metal ion chelation has been shown to induce p53 conformational changes (Hainaut and Milner, 1993b; Yu et al., 2009). The fact that the 175 mutant fails to bind zinc and that zinc chloride at low concentrations (5–15 µM) enhances the activity, leads us to hypothesize that NSC319726 may serve as a source of zinc to allow the 175 mutant to refold. Such a metallochaperone function was demonstrated for another zinc chelator, Nitriloacetate, which facilitated refolding of the p53 WT DNA binding domain (that was previously unfolded by removing the zinc) (Butler and Loh, 2007). Further biophysical studies are needed to confirm this.
Structural studies of the p53 DNA binding domain indicate that the zinc ion is coordinated by four amino acids (C176, H179, C238 and C242) (Cho et al., 1994; Joerger et al., 2005; Wong et al., 1999). Mutations in any of these residues result in the inability to coordinate zinc. In contrast, the R175H mutant is not directly involved in zinc binding. It is generally believed that a histidine residue at this location induces structural distortions in the protein that prevent it from binding zinc (Joerger and Fersht, 2007). If the metallochaperone hypothesis is correct then it is plausible that NSC319726 may reactivate other zinc binding mutants.
The question of what activates mutant p53 to become a better transcription factor and induce an apoptotic mechanism after a WT conformational change is an important one. Most likely this is due to the elevated oxidative state in the mutant p53 cell. This oxidative state is the result of the combination of NSC319726 treatment and elevated ROS levels in p53 mutant cells (due to loss of p53 mediated regulation of the redox state) (Sablina et al., 2005). In support of this is the observation that an oxidizing agent, diamide, enhances the apoptotic activity of NSC319726, while NAC inhibits it.
While inhibition of RR activity is a known mechanism of action of thiosemicarbazones, we feel this is unlikely involved in the p53R175 reactivation mechanism because 1) the drug is completely nontoxic to human fibroblasts at the IC90 for p53R175 mutant cells and 2) the dose that inhibited p53R175 mutant xenograft tumor growth was completely nontoxic to mice. The doses we used in our mouse toxicity experiments were considerably higher however, and RR inhibition may explain some of the toxicity observed in WT mice.
NSC319726 is an attractive lead compound for drug development for three reasons: 1) in vivo p53R175 mutant reactivation can be observed at doses that are nontoxic to WT animals, 2) the compound exhibits a wide therapeutic window when given intravenously and 3) the target (p53 mutant protein) is found at high levels in cells. The pool of potential patients for such a drug would be fairly large given the fact that the 175 mutant is the third most commonly found p53 mis-sense mutant, making up an estimated 5.5% of all mis-sense mutants (Olivier et al., 2010). Using the IARC TP53 database (http://www-p53.iarc.fr/), we estimate the annual incidence in the United States of cancer patients carrying the TP53R175 allele to be more than 32,000. Our findings support the growing trend in Developmental Therapeutics in which the efficacy of future cancer drugs will depend upon the knowledge of the patient’s tumor genotype.
EXPERIMENTAL PROCEDURES
Cell lines and culture conditions
The mouse embryonic fibroblasts (MEFs) (10)3 and its derived cell lines with various human p53 mutations (R175, R248 and R273) were previously derived (Dittmer et al., 1993). 3T3 is derived from Balb/c and has p53+/+ alleles. MEF p53R172H/R172H, MEF p53+/+ and MEF p53−/− cells are derived from C57BL/6 mice. All MEF cells are cultured in DMEM with 10% FBS. TOV112D and WI38 are cultured in DMEM with 10% FBS. SKOV3 is cultured in McCoy’s 5A with 5% FBS. H460 and MCF7 are cultured in RPMI with 10% FBS. HCT116 is cultured in McCoy’s 5A with 10% FBS. OVCAR3 is cultured in RPMI1640 with 20% FBS.
MTS assay and Viability assay
MTS assay is done according to the manufacture’s instructions (Promega). In brief, 5000 cells of TOV112D cells (5000 cells/well, in 100 µl culture) are cultured in 96-well plate to reach the 50–60% confluence on the second day when treated with serial dilutions of the compounds. The growth is measured by MTS reagent and Victor Plate reader instrument (PerkinElmer) after incubation for 3 days. Viability assays are done according to the manufacture instruction of Guava ViaCount (Millipore). In brief, the cells (5×104 cells/well, in 1 ml culture) are cultured in a 12-well plate to reach the 50–60% confluence on the second day when treated with serial dilutions of the compound. The growth is measured by Guava ViaCount reagent and Guava PCA instrument after incubation for 3 days.
Apoptosis assay (Annexin staining)
The Annexin staining is done according to the manufacture’s instructions (Millipore). In brief, the cells are cultured in 12-well plate, followed by treatment with the compound for different time periods. The cells are stained with the Nexin staining reagent and Annexin positive cells are detected with the Guava PCA instrument.
Transfection of p53 siRNA
The siRNA transfection is done with Lipofectamine 2000 (Invitrogen), following the manufacture’s instuctions. The p53 siRNA is from SMARTpool (Thermo Scientific/Dharmacon).
Immunofluorescent staining
The cells are grown on the coverslip, followed by various treatments. The coverslips are fixed with 4% paraformaldehyde for 10 minutes and then permealized with 0.5% Triton-X100 for 5 minutes. The conformation of the mutant and WT p53 proteins can be recognized specifically by the antibodies PAB1620 (1:50, recognizing WT conformation) and PAB240 (1:200, recognizing mutant conformation) stained overnight, respectively. The secondary antibody, goat anti-mouse IgG is incubated for 40 minutes. PAB1620 and PAB240 are from EMD Chemicals. Fluorescent staining intensity was quantified using Adobe Photoshop.
Immunoprecipitation
The cell lysates (500 µg) with various treatments are subjected to immunoprecipitation with ExctaCruz matrix (Santa Cruz Biotechnology) using the antibody PAB240 (4 µg). The pulldown is detected by Western blot with p53 (FL393) (Santa Cruz Biotechnology) that recognizes all formats of the p53 protein. The density of the image relative to the Control is determined using Adobe Photoshop.
Immunohistochemistry staining
The mouse tissues are harvested and are subjected to immunohistochemistry (IHC) staining with cleaved caspase-3 (CC3, 1:100) (Cell Signaling).
Western blot
The lysates (or immunoprecipitated products) are run on SDS-PAGE and transferred onto PVDF membranes. The detection of the protein level is done with the manufacture instruction (ECL, GE healthcare). The p21 antibody is from EMD Chemicals. The GAPDH antibody and the p53 (DO-1) are from Santa Cruz Biotechnology. The density is determined using Adobe Photoshop and is expressed as the ratio to the loading control (GAPDH) relative to the Control.
Luciferase reporter assay
The p53 recognition element (p53RE) in the p21 promoter region, constructed in pGL3 vector, is from Dr. Prives laboratory (Columbia University). It is transfected into the cells, followed by the treatment of NSC319726. The cell lysate is made and the luciferase reporter assay is done according to the manufacture’s instructions (Promega).
Chromatin Immunoprecipitation (ChIP) and PCR
ChIP experiments are done using EpiTect Chip One-Day Kit according to the manufacture’s instructions (Qiagen). The recovered ChIP DNA is subject to PCR using primers flanking the p53 response elements (p53REs) in the p21, PUMA and MDM2 genes. They are: p21-F: 5’-GTGGCTCTGATTGGCTTTCTG, p21-R: 5’-CTGAAAACAGGCAGCCCAAGG; PUMA-F: 5’-TCCTTGCCTTGGGCTAGGCC, PUMA-R: 5’-CGCGGACAAGTCAGGACTTG; MDM2-F: 5’-GGTTGACTCAGCTTTTCCTCTTG, MDM2-R: 5’-GGAAAATGCATGGTTTAAATAGCC. The control primers are used for PCR of GAPDH provided by the kit.
RNA extraction and quantitative RT-PCR
RNA is extracted from the cells or mouse tissues using Qiagen RNeasy kit and the gene expression level is measured by quantitative RT-PCR using TaqMan gene expression assays (Applied BioSciences). The gene expression level is normalized with β-actin and the average is presented with standard deviation from duplicates or triplicates of repeated experiments.
Microarray assay
GeneChip Human Genome U133 Plus 2.0 array is from Affymetrix. The RNA is extracted from the cells with or without treatment with NSC319726 and subject to Microarray assay. The list of p53 targets were selected from a literature review, providing a comprehensive list of experimentally verified targets of p53 (Riley et al., 2008).
Measurement of level of Glutathione
The levels of reduced glutathione (GSH) and oxidized glutathione (GSSG) are measured using the GSH-Glo Glutathione Assay, according to the manufacture’s instruction (Promega).
Mouse experiments
Mice are housed and treated according to guidelines and all the mouse experiments are done with the approval of Institutional Animal Care and Use Committee (IACUC) of UMDNJ-Robert Wood Johnson Medical School. The mice with p53R172H/+ are a gift of Dr. Lozano (MD Anderson Cancer Center, Texas). The nude mice NCR nu/nu are purchased from Taconic. For the toxicity assays, 6–10 week old mice are of the following genotypes, p53R172H/R172H (n=8), p53R172H/+ (n=8), p53+/+ (n=9) and p53−/− (n=3). Mice are treated with either NSC319726 at 10mg/kg or 5mg/kg or vehicle control (DMSO) by intraperitoneal injection daily for up to 7 days, followed by harvesting the mouse tissues. Xenografts tumor assays are derived from the human tumor cell lines, TOV112D, H460 and MDAMB468 (6×106 cells/mouse). Tumor dimensions are measured every other day and their volumes are calculated by length (L) and width (W) by using the formula: volume = L×W2×π/6. Tumors are allowed to grow to 60 mm3 prior to daily intravenous administration of NSC319726 at 1 mg/kg or 0.1mg/kg (TOV112D only). The following number of animals were used, H460 (Control-5, Treatment-5), MDAMB468 (Control-9, Treatment-8), TOV112D (Control-7, Treatment 1mg/kg-7, 0.1 mg/kg-7). The experiment was repeated two to three times with similar results.
Supplementary Material
Highlights.
In-Silico screen of the NCI60 database for compounds that target mutant p53
Identification of two Thiosemicarbazones with activity in p53 mutant cells
NSC319726 restores “WT” like structure and function to the p53R175 mutant
Zinc chelation and ROS are important for NSC319726’s p53R175 mutant reactivation
Significance.
The next generation of anticancer drugs will be defined by compounds that selectively kill cancer cells while leaving normal cells undisturbed. We developed an in-silico screening methodology that identified such a compound that selectively kills cancer cells with a p53R175 mutation. The mechanism of action restores WT structure and function to the p53R175 protein using its zinc-chelating and redox properties. This unique mechanism may allow medicinal chemistry to design other compounds that renature other p53 mutants that fail to coordinate zinc. TP53 is the most commonly mutated gene in human cancer, and the p53R175 mutant is the third most frequently found mis-sense mutant. The potential pool of patients in the United States for this drug would approximate 32,000 annually.
ACKNOWLEDGEMENTS
We would like to thank Dr. Guillermina Lozano (MD Anderson Cancer Center) for the p53R172H mice, Dr. Carol Prives (Columbia University) for the p21RE-luciferase reporter plasmid, and NCI (Developmental Therapeutics Program (DTP)) for the compounds NSC319725 and NSC319726. This work was supported by grants from the Cancer Institute of New Jersey (to D.R.C.), The Breast Cancer Research Foundation (BCRF to A.J.L.) and the NIH (P01CA87497 to A.J.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The data from the microarrays have been deposited in the GEO database with accession number GSE35972.
REFERENCES
- Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, Solari A, Bobisse S, Rondina MB, Guzzardo V, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- Butler JS, Loh SN. Structure, function, and aggregation of the zinc-free form of the p53 DNA binding domain. Biochemistry. 2003;42:2396–2403. doi: 10.1021/bi026635n. [DOI] [PubMed] [Google Scholar]
- Butler JS, Loh SN. Zn(2+)-dependent misfolding of the p53 DNA binding domain. Biochemistry. 2007;46:2630–2639. doi: 10.1021/bi062106y. [DOI] [PubMed] [Google Scholar]
- Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, Bergman J, Wiman KG, Selivanova G. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8:282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- Bykov VJ, Issaeva N, Zache N, Shilov A, Hultcrantz M, Bergman J, Selivanova G, Wiman KG. Reactivation of mutant p53 and induction of apoptosis in human tumor cells by maleimide analogs. J Biol Chem. 2005;280:30384–30391. doi: 10.1074/jbc.M501664200. [DOI] [PubMed] [Google Scholar]
- Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- Dittmer D, Pati S, Zambetti G, Chu S, Teresky AK, Moore M, Finlay C, Levine AJ. Gain of function mutations in p53. Nat Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- Foster BA, Coffey HA, Morin MJ, Rastinejad F. Pharmacological rescue of mutant p53 conformation and function. Science. 1999;286:2507–2510. doi: 10.1126/science.286.5449.2507. [DOI] [PubMed] [Google Scholar]
- Hainaut P, Milner J. Redox modulation of p53 conformation and sequence-specific DNA binding in vitro. Cancer Res. 1993a;53:4469–4473. [PubMed] [Google Scholar]
- Hainaut P, Milner J. A structural role for metal ions in the "wild-type" conformation of the tumor suppressor protein p53. Cancer Res. 1993b;53:1739–1742. [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Ikediobi ON, Davies H, Bignell G, Edkins S, Stevens C, O'Meara S, Santarius T, Avis T, Barthorpe S, Brackenbury L, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5:2606–2612. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger AC, Ang HC, Veprintsev DB, Blair CM, Fersht AR. Structures of p53 cancer mutants and mechanism of rescue by second-site suppressor mutations. J Biol Chem. 2005;280:16030–16037. doi: 10.1074/jbc.M500179200. [DOI] [PubMed] [Google Scholar]
- Joerger AC, Fersht AR. Structure-function-rescue: the diverse nature of common p53 cancer mutants. Oncogene. 2007;26:2226–2242. doi: 10.1038/sj.onc.1210291. [DOI] [PubMed] [Google Scholar]
- Kalinowski DS, Richardson DR. Future of toxicology--iron chelators and differing modes of action and toxicity: the changing face of iron chelation therapy. Chem Res Toxicol. 2007;20:715–720. doi: 10.1021/tx700039c. [DOI] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Liu G, McDonnell TJ, Montes de Oca Luna R, Kapoor M, Mims B, El-Naggar AK, Lozano G. High metastatic potential in mice inheriting a targeted p53 missense mutation. Proc Natl Acad Sci U S A. 2000;97:4174–4179. doi: 10.1073/pnas.97.8.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Midgley CA, Lane DP. p53 protein stability in tumour cells is not determined by mutation but is dependent on Mdm2 binding. Oncogene. 1997;15:1179–1189. doi: 10.1038/sj.onc.1201459. [DOI] [PubMed] [Google Scholar]
- Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, Lukashchuk N, Gillespie DA, Ludwig RL, Gosselin P, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North S, Pluquet O, Maurici D, El-Ghissassi F, Hainaut P. Restoration of wild-type conformation and activity of a temperature-sensitive mutant of p53 (p53(V272M)) by the cytoprotective aminothiol WR1065 in the esophageal cancer cell line TE-1. Mol Carcinog. 2002;33:181–188. doi: 10.1002/mc.10038. [DOI] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harbor perspectives in biology. 2010;2 doi: 10.1101/cshperspect.a001008. a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- Richardson DR, Kalinowski DS, Richardson V, Sharpe PC, Lovejoy DB, Islam M, Bernhardt PV. 2-Acetylpyridine thiosemicarbazones are potent iron chelators and antiproliferative agents: redox activity, iron complexation and characterization of their antitumor activity. J Med Chem. 2009;52:1459–1470. doi: 10.1021/jm801585u. [DOI] [PubMed] [Google Scholar]
- Richardson DR, Sharpe PC, Lovejoy DB, Senaratne D, Kalinowski DS, Islam M, Bernhardt PV. Dipyridyl thiosemicarbazone chelators with potent and selective antitumor activity form iron complexes with redox activity. J Med Chem. 2006;49:6510–6521. doi: 10.1021/jm0606342. [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res. 2000;60:6788–6793. [PubMed] [Google Scholar]
- Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, Van Pelt CS, Lozano G. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008;22:1337–1344. doi: 10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez A. Optimal drug combinations and minimal hitting sets. BMC Syst Biol. 2009;3:81. doi: 10.1186/1752-0509-3-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Wong KB, DeDecker BS, Freund SM, Proctor MR, Bycroft M, Fersht AR. Hot-spot mutants of p53 core domain evince characteristic local structural changes. Proc Natl Acad Sci U S A. 1999;96:8438–8442. doi: 10.1073/pnas.96.15.8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Kalinowski DS, Kovacevic Z, Siafakas AR, Jansson PJ, Stefani C, Lovejoy DB, Sharpe PC, Bernhardt PV, Richardson DR. Thiosemicarbazones from the old to new: iron chelators that are more than just ribonucleotide reductase inhibitors. J Med Chem. 2009;52:5271–5294. doi: 10.1021/jm900552r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.