Abstract
Kleefstra syndrome is characterized by the core phenotype of developmental delay/intellectual disability, (childhood) hypotonia and distinct facial features. The syndrome can be either caused by a microdeletion in chromosomal region 9q34.3 or by a mutation in the euchromatin histone methyltransferase 1 (EHMT1) gene. Since the early 1990s, 85 patients have been described, of which the majority had a 9q34.3 microdeletion (>85%). So far, no clear genotype-phenotype correlation could be observed by studying the clinical and molecular features of both 9q34.3 microdeletion patients and patients with an intragenic EHMT1 mutation. Thus, to further expand the genotypic and phenotypic knowledge about the syndrome, we here report 29 newly diagnosed patients, including 16 patients with a 9q34.3 microdeletion and 13 patients with an EHMT1 mutation, and review previous literature. The present findings are comparable to previous reports. In addition to our former findings and recommendations, we suggest cardiac screening during follow-up, because of the possible occurrence of cardiac arrhythmias. In addition, clinicians and caretakers should be aware of the regressive behavioral phenotype that might develop at adolescent/adult age and seems to have no clear neurological substrate, but is rather a so far unexplained neuropsychiatric feature.
Key Words: EHMT1, Kleefstra syndrome, 9q34.3 microdeletion, Review, 9q subtelomeric deletion syndrome
Introduction
Kleefstra syndrome (KS) (OMIM 610253), previously named 9q subtelomeric deletion syndrome (9qSTDS), is characterized by the core clinical phenotype of mostly moderate to severe developmental delay (DD)/intellectual disability (ID), (childhood) hypotonia and distinct facial features, comprising brachy(micro)cephaly, synophrys, unusual shape of the eyebrows, midface hypoplasia, full everted lower lip, cupid bowed upper lip, protruding tongue and prognathism. Additional clinical features include congenital heart and urogenital defects, epilepsy, behavioral and psychiatric disorders and overweight. The syndrome is either caused by a submicroscopic deletion in the chromosomal region 9q34.3 or an intragenic mutation of the euchromatin histone methyltransferase 1 (EHMT1) gene causing haploinsufficiency of EHMT1. So far, 85 patients, including 75 patients with a 9q34.3 deletion and 10 patients with an EHMT1 mutation, have been reported [Schimmenti et al., 1994; Ayyash et al., 1997; Dawson et al., 2002; Cormier-Daire et al., 2003; Font-Montgomery et al., 2004; Harada et al., 2004; Iwakoshi et al., 2004; Stewart et al., 2004; Neas et al., 2005; Yatsenko et al., 2005, 2009; Kleefstra et al., 2006a, b, 2009; Stewart and Kleefstra, 2007; Verhoeven et al., 2010; Willemsen et al., 2011]. EHMT1 encodes a histone H3 Lys 9 methyltransferase and is thereby involved in chromatin remodeling [Ogawa et al., 2002]. Among the genetically confirmed cases the majority of the patients have a deletion and the remaining a mutation in EHMT1 [Yatsenko et al., 2009; Kleefstra et al., 2010]. Almost all cases are sporadic, but recently we described 3 familial cases due to a subtelomeric 9q deletion present in mosaic pattern in the mothers [Willemsen et al., 2011]. Since the identification of EHMT1 as the major gene [Kleefstra et al., 2006b], an increasing number of mutations in this gene is being detected in patients with the core phenotype of KS. Genotype-phenotype studies in patients collected so far indicated that patients with an EHMT1 mutation and those with a 9q34.3 deletion smaller than 3 Mb have highly comparable clinical findings [Yatsenko et al., 2005; Kleefstra et al., 2009]. Here we report 29 additional diagnosed patients with KS that have not been published before. Sixteen cases were caused by a submicroscopic 9q34.3 deletion and 13 cases by an intragenic EHMT1 mutation. We describe the clinical and molecular characteristics of this additional cohort, provide an overview of previous patients and compare present and previous findings.
Patients and Methods
A subset of the patients included in this study was diagnosed at our diagnostic center after referral from our outpatient clinic or from clinical geneticists from national/international collaborating centers. Others were referred to our diagnostic center for follow-up (studies) after the diagnosis had been established elsewhere. The cohort included 16 patients with a 9q34.3 deletion and 13 patients with an intragenic EHMT1 mutation.
DNA was obtained from peripheral blood cells and extracted according to standardized procedures. Deletions were detected by routine subtelomeric multiplex ligation-dependent probe amplification (MLPA) (P070; MRC Holland)/FISH (probes from Vysis) or whole genome array analysis with different platforms (Agilent Technologies and Affymetrix Inc., Santa Clara, Calif., USA). In patients without a deletion in 9q34.3, subsequent gene-specific extensive MLPA analysis and direct sequencing of the coding region of EHMT1 was performed as described previously [Kleefstra et al., 2006b]. In addition, 2 novel coding exons were sequenced since the recent identification of an elongated EHMT1 transcript (NM_024757.4) [Nillesen et al., 2011].
Results
Molecular Data
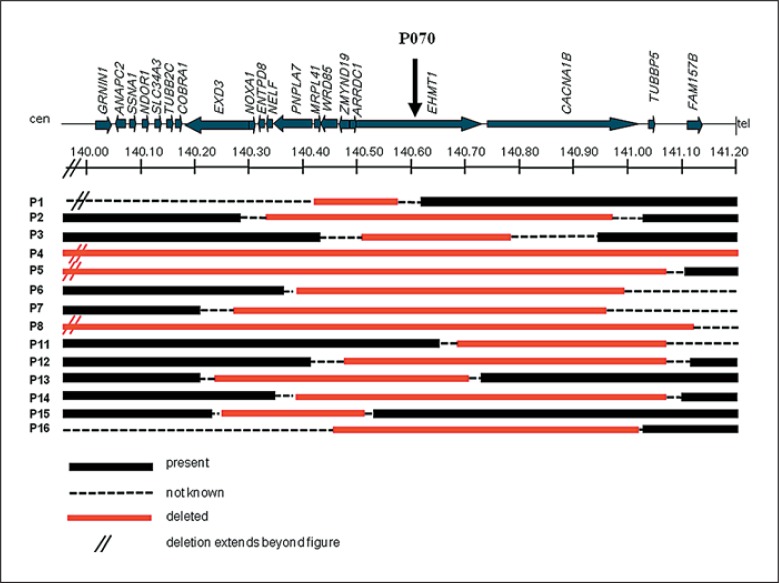
Figure 1 shows a schematic overview of the deleted regions in patients 1–8 and 11–16 diagnosed with a 9q34.3 deletion. Five deletions were initially identified by routine subtelomeric MLPA analysis (patients 1–3, 9 and 10). The deletions of patients 1–3 were fine-mapped by additional region-specific MLPA experiments with probes in EHMT1 and flanking genes, with the most proximal probe in the gene RAPGEF1, which is located approximately 6 Mb distant from EHMT1 and the most distal probe in CACNA1B. The deletion of patient 1 was shown to have a minimum size of 200 kb and a maximum size of 6 Mb and thus was poorly mapped (no probes could be tested in between RAPGEF1 exon 2 (present) and MRPL41 exon 1 (deleted)). Deletions of patients 9 and 10 are not shown, because these could not be further fine-mapped either. The deletions of patients 4–8 and 11–16 were identified and delineated by different genome-wide array platforms, including the Affymetrix 250K SNP array (patients 5 and 16), 2.7M array (patient 4) and 6.0 SNP array platforms (patients 13–15), and the Agilent 60K (patient 7), 105K (patients 11 and 12) and 180K oligoarray (patients 6 and 8) platforms. Mb positions were converted to UCSC genome browser build February 2009 (Hg 19). Present deletions vary in size from 270 kb (patient 15) to 3.85 Mb (patient 4).
Fig. 1.
Schematic overview of deleted regions in 9q34.3 in patients 1–8 and 11–16. The relative positions of involved genes and the P070 MPLA probe are indicated (UCSC genome browser build February 2009, Hg 19). Deletions of patients 1–3 were identified by MLPA. Deletions of patients 4–8 and 11–16 were detected by genome-wide array analysis. The deletion of patient 4 was identified by 2.7M array, the deletions of patients 5 and 16 by 250K SNP array, the deletions of patients 6 and 8 by 180K oligoarray, the deletion of patient 7 by 60K oligoarray and the deletions of patients 11 and 12 by 105K oligoarray. In patients 13–15, the Affymetrix 6.0 SNP array platform was used. Deletions of patients 9 and 10 are not depicted, because these were poorly delineated.
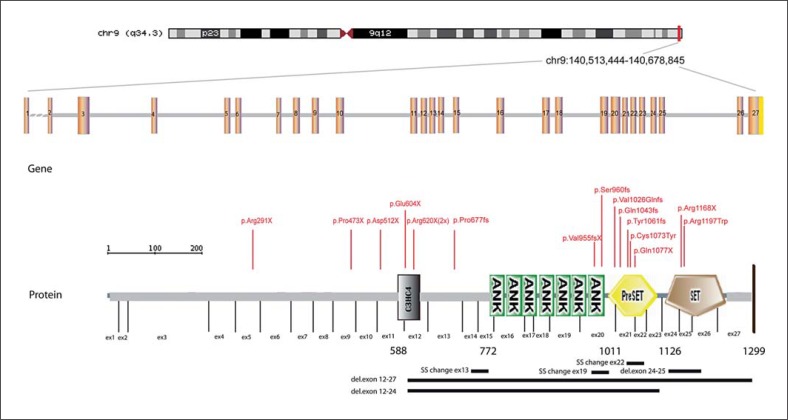
EHMT1 mutations of the present and previously reported patients are shown in figure 2. Mutations found in the present group are summarized in table 1 as well and include 6 frameshift mutations, 3 nonsense mutations (2 identical: p.Arg620X), 2 exon deletions, 1 missense mutation and 1 mutation in a donor splice site. In 5 patients de novo occurrence of the mutation was confirmed by segregation analysis in the parents (patients 21, 22, 24, 25 and 27). In the remaining cases, DNA of one or both parents was not available. Two mutations occurred in the Su(var)3–9, Enhancer-of-zetste, Trithorax (SET) domain (patients 22 and 26), 4 in the preSET domain (patients 19, 25, 27 and patient 28 for whom clinical data was not available), 2 in the ankyrin repeats (ANK) domain 7 (patients 18 and 21), 1 affected the donor splice site in exon 22 (patient 20), 2 mutations (the recurrent p.Arg620X) occurred in the C3HC4 type zinc-finger (RING finger) (C3HC4) domain (patient 17 and patient 29 without available clinical data) and 1 mutation was localized outside the characterized domains (patient 23). The intragenic deletion of patient 24 comprised all characterized domains.
Fig. 2.
Schematic diagram of the EHMT1 gene structure (including 27 exons) and the protein domain structure of the EHMT1 protein. The conserved domains include the C3HC4 type zinc-finger (RING finger) (C3HC4) domain, 7 ankyrin repeat (ANK) domains, the Su(var)3–9, Enhancer-of-zetste, trithorax (SET) domain and the PreSET domain (N-terminal to the SET domain).The relative positions of in total 22 mutations are displayed, including the 13 mutations of the present patient cohort (1 recurrent mutation (p.Arg620X) present in 2 patients) and 9 previously reported mutations. The first identified intragenic EHMT1 aberration due to a translocation X;9 is not shown. del = Deletion; SS = splice site mutation.
Table 1.
Intragenic EHMT1 mutations identified in patients 17–29
| Patient | Mutation NM_024757.4 | Alteration NP_079033.4 |
|---|---|---|
| 17 | c.1858C>T | p.Arg620X |
| 18 | c.2863_2864del | p.Val955fsX221 |
| 19 | c.3229C>T | p.Gln1077X |
| 20 | c.3180+1G>T | (donor splice site) |
| 21 | c.2878_2881 | p.Ser960GlyfsX7 |
| 22 | c.3375–?_3541+?del | (exon 24+25 deletion) |
| 23 | c.2029insG | p.Pro677fs |
| 24 | exon 12–27 deletion | |
| 25 | c.3181–80_3233del | p.Tyr1061fs |
| 26 | c.3589C>T | p.Arg1197Trp |
| 27 | c.3072_3073del | p.Val1026fs |
| 28 | c.3126_3127del | p.Gln1043fs |
| 29 | c.1858C>T | p.Arg620X |
Clinical Data
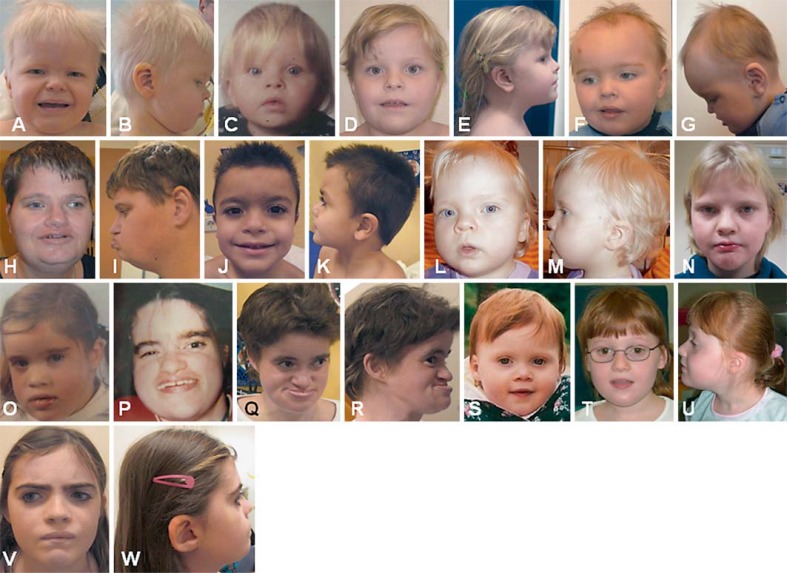
The clinical features of the patients in this study are summarized in table 2 (9q34.3 deletions) and table 3 (EHMT1 mutations). Information on the clinical features of the patient with the p.Gln1043fs mutation (patient 28) and one of the patients with the p.Arg620X mutation (patient 29) was not available and therefore not included in table 3 and table 4. Clinical features were systematically obtained by a standardized clinical data sheet. The age of patients with a microdeletion ranged from 1 to 25 years (average age 9.5 years/median age 5.5 years, including 3 adults and 13 children) and from 1 to 41 years among patients with an EHMT1 mutation (average age 11.8 years/median age 7.5 years, including 2 adults and 9 children). All patients had the core KS phenotype, including DD/ID, childhood hypotonia and distinct facial features. Clinical photographs of patients with a submicroscopic 9q34.3 deletion are shown in figure 3. Patients with EHMT1 mutations are shown in figure 4. Patient 20 with an EHMT1 mutation is described in more detail elsewhere in this issue (see chapter ‘Adult Phenotypes in Angelman- and Rett-Like Syndromes’ by Willemsen et al., this vol., DOI:101159/000335661).
Table 2.
Clinical features of present and previous patients with a 9q34.3 deletion
| Clinical features | Present patients (corresponding photographs in fig. 3) |
Total | Previous | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | n = 16 (%) | n = 75 (%) | |
| (A, B) | (C–E) | (no) | (F, G) | (H, I) | (J, K) | (L) | (M–O) | (no) | (P, Q) | (R–U) | (V) | (no) | (no) | (no) | (no) | |||
| Deletion size, Mb | 0.2–6 | ˜0.6 | 0.27–0.41 | 3.85 | 1.2 | 0.6 | 0.7 | 3.1 | STD | STD | 0.4 | 0.6 | 0.5 | 0.7 | 0.27 | 056–1.32 | 0.2–6 | 0.04–CV |
| Age, years | 5 | 6 | 2 9/12 | 5 | 11 | 2 | 1 | 10 | 1 4/12 | 25 | 5 | 5 | 14 | 15 | 25 | 18 | 1–25 | 0–59 |
| Gender | M | M | F | F | F | F | F | M | M | F | M | F | M | M | F | M | 8 M, 8 F | M = F |
| Growth parameters | ||||||||||||||||||
| High birth weight | − | + | − | − | NR | − | − | − | − | − | − | − | − | + | − | − | 2/15 (13) | 4/51 (8) |
| Microcephaly | + | − | + | NR | + | NR | + | − | + | + | + | + | − | − | − | − | 8/14 (57) | 24/50 (48) |
| Short stature | + | − | − | NR | − | − | − | − | − | + | − | − | − | − | − | − | 2/15 (13) | 16/41 (39) |
| Overweight (BMI>25) | − | − | − | NR | − | − | − | + | − | − | − | − | − | + | + | − | 3/15 (20) | 15/49 (31) |
| DD/ID | sev. | mild | sev. | sev. | sev. | mild | yes | sev. | mild | sev. | mod. | yes | mild | sev. | mod. | sev. | 16/16 (100) | 75/75 (100) |
| Heart defect | − | − | − | + | + | − | − | − | + | − | − | − | − | + | + | − | 5/16 (31) | 22/50 (44) |
| Genital anomaly | − | + | − | − | − | − | − | − | + | − | + | − | + | − | − | + | 5/8 M (63) | 12/45 (27) |
| Renal anomaly (including VUR) | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 0/16 (0) | 7/43 (16) |
| Recurrent infections | − | − | − | + | − | − | + | + | − | − | − | − | − | − | − | + | 4/16 (25) | 5/19 (26) |
| Hearing deficit | − | − | − | − | − | − | − | − | + | + | + | − | + | − | − | − | 5/16 (31) | 7/36(19) |
| Gastro-esophageal reflux | − | + | − | − | + | − | + | + | − | − | − | − | − | + | − | + | 6/16 (38) | 4/36 (11) |
| Epilepsy | − | − | − | − | − | − | − | − | − | − | + | + | + | + | + | + | 7/16 (44) | 14/43 (33) |
| Behavioral/psychiatric | − | + | + | + | + | − | − | + | − | + | + | + | − | − | + | + | 10/16 (63) | 22/43 (51) |
| Anomalies on brain imaging | NR | NR | − | + | + | NR | NR | − | NR | NR | − | + | + | + | − | − | 5/10 (50) | 20/33 (61) |
| Tracheomalacia | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 0/16 (0) | 6/37 (16) |
| Umbilical/inguinal hernia | − | − | − | − | − | − | − | + | + | − | − | − | − | − | − | + | 3/16 (19) | 6/38 (16) |
| Anal atresia | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 0/16 (0) | 2/51 (4) |
| Musculoskeletal anomaly | − | − | − | + | + | − | − | − | − | − | + | − | − | − | − | + | 4/16 (25) | 9/35 (26) |
| Respiratory complications* | − | − | − | + | − | − | + | − | − | + | − | − | − | − | − | − | 3/16 (19) | 5/42 (12) |
| Other | − | − | − | − | T | − | − | E | − | − | − | − | − | − | − | E | − | − |
CV = Cytogenetically visible; DD/ID = developmental delay/intellectual disability: mild = mild ID; mod. = moderate ID; sev. = severe ID; yes = ID present, level not known; E = eye anomalies: coloboma in patient 8, severe hypermetropia in patient 16; NR = not reported; STD = subtelomeric deletion; T = tooth anomalies; VUR = vesicoureteral reflux; + = yes; – = no; +/– = uncertain.
Including cardiorespiratory failure, apnea.
Table 3.
Clinical features of present and previous patients with an intragenic EHMT1 mutation
| Clinical features | Present patients (corresponding photographs in fig. 4) | Total | Previous | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 17(A, B) | 18(C–E) | 19(F, G) | 20(H, I) | 21(J, K) | 22(no) | 23(no) | 24(L–N) | 25(O–R) | 26(S–U) | 27(V, W) | n = 11 (%) | n = 75 (%) | |
| Mutation (NM_024757.4) | p.Arg620X | p.Val955fs | p.Gln 1077X | SSexon 22 | p.Ser929GlyfsX | del. exon 24–25 | p.Pro 677fs | del. exon12–27 | p.Tyr 1061fs | p.Arg 1197Trp | p.Val 1026fs | ||
| Age, years | 2 2/12 | 6 2/12 | 2 9/12 | 41 | 5 5/12 | 7 5/12 | 2 5/12 | 9 2/12 | 32 | 10 | 11 5/12 | 2 9/12–41 | 2 6/12–16 |
| Gender | M | F | M | F | M | F | F | F | F | F | F | 3 M, 8 F | 4 M, 6 F |
| Growth parameters | |||||||||||||
| High birth weight | – | – | – | – | – | NR | – | – | + | – | – | 1/10(10) | 3/9 (33) |
| Microcephaly | + | – | – | – | – | – | + | – | – | – | – | 2/11 (18) | 2/10 (20) |
| Short stature | – | – | – | – | – | NR | – | – | + | – | – | 1/10(10) | 2/8 (25) |
| Overweight (BMI >25) | – | – | + | + | – | + | – | – | NR | + | – | 4/10 (40) | 4/9 (44) |
| DD/ID | yes | mod. | yes | mod. | yes | mild | mild | mild | mod. | yes | mod. | 11/11 (100) | 10/10 (100) |
| Heart defect | + | – | + | + | – | – | + | + | – | – | – | 5/11 (45) | 4/10 (40) |
| Genital anomaly | + | – | – | – | – | – | – | – | – | – | – | 1/3 M (33) | 2/4 M (50) |
| Renal anomaly (including VUR) | – | – | + | – | – | – | – | + | – | – | – | 2/11 (18) | 1/10 (10) |
| Recurrent infections | – | + | + | + | + | – | + | – | + | – | + | 7/11 (64) | NR |
| Hearing deficit | – | – | – | – | + | – | – | – | + | + | + | 4/11 (36) | 1/10 (10) |
| Gastro–esophageal reflux | – | – | + | – | + | – | – | – | – | – | + | 3/11 (27) | 0/10 (0) |
| Epilepsy | + | – | – | – | + | – | – | – | – | – | – | 2/11 (18) | 3/10 (30) |
| Behavioral/psychiatric problems | – | + | – | + | + | – | + | + | + | + | + | 8/11 (73) | 7/9 (78) |
| Anomalies on brain imaging | – | – | + | + | + | + | NR | + | + | + | – | 7/10 (70) | 5/9 (56) |
| Tracheomalacia | – | – | – | – | – | – | – | – | – | – | – | 0/11 (0) | 1/10 (10) |
| Umbilical/inguinal hernia | – | – | – | – | – | – | + | – | – | – | – | 1/11 (9) | 1/10 (10) |
| Anal atresia | – | – | – | – | – | – | – | + | – | – | – | 1/11 (9) | 0/10 (0) |
| Musculoskeletal anomaly | – | – | – | – | – | – | + | + | + | – | – | 3/11 (27) | 1/10 (10) |
| Respiratory complications | – | – | – | – | – | – | + | – | – | – | – | 1/11 (9) | 0/10 (0) |
| Other | – | – | T | – | – | – | S | E | H | – | – |
DD/ID = Developmental delay/intellectual disability: mild = mild ID; mod. = moderate ID; yes = ID present, level not known; E = eye anomaly: congenital cataract; H = hormonal: central pubertas praecox; NR = not reported; S = hyperelastic skin; SS = splice site; T = tooth anomalies; VUR = vesicoureteral reflux; + = yes; – = no.
Table 4.
Frequency of features in the present and in previously reported patients with KS
| Clinical features of the total number of KS cases | Mutation | |
|---|---|---|
| 9q34 deletions n (%) | EHMT1 mutationsn (%) | |
| Age range, years | 0–59 | 2.5–41 |
| High birth weight (>P90) | 6/66 (9) | 4/19 (21) |
| Microcephaly | 32/64 (50) | 4/21 (19) |
| Short stature | 18/56 (32) | 3/18 (17) |
| Overweight (BMI >25) | 18/64 (28) | 8/19 (42) |
| DD/ID | 91/91 (100) | 21/21 (100) |
| Heart defect | 27/66 (41) | 9/21 (43) |
| Genital anomaly (in males) | 17/53 (32) | 3/7 (43) |
| Renal anomaly | 7/59 (12) | 3/21 (14) |
| Recurrent infections | 9/35 (26) | 7/11 (64) |
| Hearing deficit | 12/52 (23) | 5/21 (24) |
| Gastro–esophageal reflux | 10/52 (19) | 3/21 (14) |
| Epilepsy | 21/59 (36) | 5/21 (24) |
| Behavioral/psychiatric problems | 32/59 (54) | 15/20 (75) |
| Anomalies on brain imaging | 25/43 (58) | 12/19 (63) |
| Tracheomalacia | 6/53 (11) | 1/21 (5) |
| Umbilical/inguinal hernia | 9/54 (17) | 2/21 (10) |
| Anal atresia | 2/67 (3) | 1/21 (5) |
| Musculoskeletal anomaly | 13/51 (25) | 4/21 (19) |
| Respiratory complications | 8/58 (14) | 1/21 (5) |
Study group included 91 patients with a 9q34.3 deletion (75 previous and 16 present) and 21 patients with an EHMT1 mutation (10 previous and 11 present). Two patients with an EHMT1 mutation are not included, because clinical data were not available.
Fig. 3.
A–V Facial profiles of patients with a 9q34.3 deletion. Patient 1 at age 2 years (A) and 5 years (B), patient 2 at age 9 months (C), 2 years (D) and 6 years (E), patient 4 at age 5 years (F, G), patient 5 at age 11 years (H, I), patient 6 at age 1 year (J) and 2 years (K), patient 7 at age 9 months (L), patient 8 at different childhood ages (M–O), patient 10 at young childhood age (P, Q), patient 11 at age 3 years (R, S) and 5 years (T, U) and patient 12 at age 5 years (V). The highly recognizable facial features comprise hypertelorism, midface hypoplasia, prognathism, prominent eyebrows, cupid bow or tented upper lip and everted lower lip.
Fig. 4.
A–W Facial appearance of patients with an EHMT1 mutation. Patient 17 at age 2 years (A, B), patient 18 at age 1 year (C) and 6 years (D, E), patient 19 at age 3 years (F, G), patient 20 at age 41 years (H, I), patient 21 at age 5 years (J, K), patient 24 at age 2 years (L, M) and 10 years (N), patient 25 at childhood age (O), during teenage (P), and age 32 years (Q, R), patient 26 at age 2 years (S) and age 10 years (T, U), patient 27 at age 11 years (V, W). Facial characteristics are similar to Kleefstra syndrome patients with a 9q34.3 deletion.
Among the patients with a deletion, the degree of ID ranged from mild to severe, but the majority of the patients had severe ID. Five patients were not yet able to walk (patients 1, 3, 4, 7 and 9 at 5 years, 33 months, 5 years, 12 months and 16 months of age, respectively). Most patients, except for patients 4, 5, 8, 10 and 16, developed some speech, though mostly very primitive (single words). One patient had a remarkably mild phenotype and was even able to read and write (patient 13). In the cohort of patients with an intragenic EHMT1 mutation, cognitive performance varied from mild ID with speaking in full sentences and ability to read and write (patient 24) to moderate ID with regression at adult age (patients 20 and 25). Speech ability ranged from a few single words to long sentences with a vocabulary of more than 100 words (patients 23 and 24). All were able to walk.
Microcephaly was present in 8 out of 14 patients with a microdeletion and in 2 out of 11 patients with an intragenic EHMT1 mutation. Overweight (body mass index (BMI) >25) was reported among 3 of 15 patients with a deletion and was more frequent among patients with a mutation (4 of 10). Reported heart anomalies were mainly structural defects, including atrial septal defects, ventricular septal defects, valve anomalies comprising pulmonary valve stenosis and bicuspid aortic valve, persistent open foramen ovale and ductus arteriosus anomalies. One patient (patient 20) was diagnosed with atrial flutter and 1 patient had an aberrant muscle bundle in the left ventricle (patient 14). Genital defects were only observed in males and included cryptorchidism, hypospadias and micropenis (in 5 of 8 males with a microdeletion and in 1 out of 3 males with a mutation, respectively). Musculoskeletal anomalies were reported in 4 of 16 patients with a microdeletion and in 3 of 11 patients with a mutation, respectively, and comprised joint hypermobility, scoliosis and club feet. Renal anomalies were only twice reported in this cohort. Both patients had an EHMT1 mutation. One was diagnosed with vesicoureteral reflux (patient 19) and the other was shown to have a hydronephrosis (patient 24). Epilepsy was present in 7 of 16 patients with a microdeletion and in 2 of 11 patients with a mutation. Behavioral problems were often reported (in 10 of 16 microdeletion patients and in 8 of 11 patients with an intragenic EHMT1 mutation) and comprised autistic features, attention deficit problems, self-mutilation, aggressive and emotional outbursts/crises and severe sleep disturbance. In 2 of the 5 patients who had reached adult age at the time of the examination, a behavioral change at adult age was reported. They showed alternating periods with apathy and catatonia-like behaviors with a general decline in functioning (patient 20 and 25). These neuropsychiatric conditions were recently described in detail [Verhoeven et al., 2011].
In 10 of the 16 patients with a microdeletion and in 10 of the 11 patients with an intragenic EHMT1 aberration, cerebral imaging by MRI had been performed. Variable anomalies were reported in 5 and 7 patients, respectively, and included dilated ventricles, white matter anomalies, corpus callosum hypoplasia and cerebellar hypoplasia.
Tables 2 and 3 also include a summary of the clinical features of patients reported in previous reports. Table 4 summarizes the frequency of clinical features in the total (present and previously reported patients) cohort of patients with KS, including 91 patients with a 9q34.3 deletion and 21 patients with an EHMT1 mutation.
Discussion
In this study 29 novel identified patients with KS are described (clinical data were not available for patients 28 and 29) and compared to 85 previously reported patients. The majority of the deletions described here (11/16) were identified by genome-wide array analysis. A large number of previous deletions were reported before genome-wide array platforms became common use in routine diagnostic settings, and were either detected by routine FISH or MLPA [Dawson et al., 2002; Cormier-Daire et al., 2003; Font-Montgomery et al., 2004; Iwakoshi et al., 2004; Neas et al., 2005]. Some studies reported fine-mapping of deletions with additional specific 9q probes [Harada et al., 2004; Stewart et al., 2004]. Only 2 previously reported deletions were cytogenetically visible, indicating that large cytogenetically visible deletions are a relatively rare cause of KS [Schimmenti et al., 1994; Ayyash et al., 1997]. More recent reports also included 9q34.3 deletions identified and delineated by genome-wide array analysis [Kleefstra et al., 2009; Yatsenko et al., 2009]. Genome-wide array analysis generally enables a more precise delineation of the breakpoints, though many array platforms have poor coverage of the 9q subtelomeric region. The use of different methods for molecular characterization in previous and present studies makes exact genotype-phenotype comparison more difficult. In agreement with the more recent reports, the sizes and breakpoints of the present deletions are heterogeneous [Kleefstra et al., 2009; Yatsenko et al., 2009]. Previously, it was indicated that patients with microdeletions smaller than 3 Mb in size show highly similar clinical findings. In the present study we observed a tendency to a more severe ID in patients with deletions of more than 1 Mb in size. Three to seven patients had a deletion of more than 1 Mb (patients 4, 5 and 8; in patients 1, 9, 10 and 16 deletion size was not well defined, but possibly more than 1 Mb). In 6 of these 7 patients a severe level of ID was documented. In 9 patients a deletion smaller than 1 Mb was identified. Three of them had mild ID and in 2 patients severe ID was reported. However, the reported level of ID has to be considered with caution, because we noticed more often that at very young age the severity of the delay is sometimes underestimated and tends to become more obvious at school age when a higher level of performance is expected. In this cohort 5 out of 16 patients with a 9q34.3 microdeletion were diagnosed with a structural heart defect. Two of them had a deletion of more than 1 Mb in size. The deletion of patient 9 with a heart defect was delineated poorly and is possibly more than 1 Mb in size. The deletion of patient 15 was relatively small (270 kb). She had a perimembraneous ventricle septal defect for which an operation was not necessary. These numbers are too low to conclude that the more severe structural cardiac defects are more common in deletions sized >1 Mb. Furthermore, structural heart defects are equally present in patients with intragenic EHMT1 mutations. Of note, in patient 4 the deletion encompasses both COL5A1, associated with Ehlers-Danlos type I [OMIM 130000], and NOTCH1, shown to be associated with aortic valve anomalies [OMIM 109730; Garg et al., 2005]. Therefore, the heterozygous deletion of both COL5A1 and NOTCH1 likely has contributed to the cardiac valve phenotype in this patient presenting with aortic and pulmonary valve defects in addition to cardiac septal defects. Deletion of COL5A1 likely contributed to the generalized joint hypermobility that was observed in patient 4. However, in patient 8, who also has a deletion comprising these genes, neither a heart defect nor signs of connective tissue disease were observed, so penetrance seems incomplete.
Of note, 13 of the 16 identified deletions comprised the gene CACNA1B as well. We did not observe differences in phenotype between patients with a deletion comprising CACNA1B (patients 2–12, 14 and 16) and patients with a deletion excluding CACNA1B (patients 1, 13 and 15). CACNA1B is a voltage-dependent calcium channel and despite the demonstrated importance of the a1B subunit for neurotransmission in most neuronal cells during embryogenesis and after birth, the null-mutant mice showed no gross abnormalities and normal motor coordination. These data suggest that the a1B gene is not essential for synaptogenesis and neuronal differentiation during embryonic and postnatal development [Kim et al., 2001]. This might underscore why a heterozygous loss of this gene does not contribute significantly to the observed phenotype in 9q34.3 deletions. Moreover, Yatsenko et al. [2009] described a familial case with a heterozygous loss of CACNA1B but not EHMT1. Affected family members had a normal development and did not have the KS phenotype.
Previous reports suggested a higher prevalence of respiratory complications in patients with deletions ≥3 Mb [Kleefstra et al., 2010]. In the present study respiratory complications were present in patient 4 with a large deletion of 3.85 Mb, in patient 10 with a poorly defined deletion, but possibly larger than 1 Mb in size, and in patient 7 with a 700-kb deletion. However, the respiratory problems in these patients were of different origin. Patient 4 suffered from central apnea and patient 7 and 10 had respiratory problems secondary to a viral bronchiolitis and aspiration, respectively. Central apnea has been reported in 1 previously published patient who had a cytogenetically visible deletion [Neas et al., 2005]. From a personal communication we know an additional patient with central apnea who has a 9q subtelomeric deletion of more than 3 Mb as well. So, it might be that central apneas are associated with deletions over 3 Mb.
Since the identification of EHMT1 in 2005 as the gene responsible for the phenotype [Kleefstra et al., 2005, 2006b], mutations in EHMT1 are increasingly being detected. So far, 23 intragenic EHMT1 defects have been reported (including this report). Except for one (c.1858C>T), all present mutations were unique and not reported before. Presence of distinct clinical features is comparable between the present and previously reported cohorts. Mutations are scattered throughout the gene, but are more frequent on the 3′ end of the gene and occur mostly in the PreSET domain (fig. 2). All mutations in this cohort, except for 1 missense mutation and 1 splice donor site mutation, are truncating mutations suggesting loss-of-function. We did not observe a correlation between the severity of the phenotype, indicated by the degree of ID and presence of other major medical problems, and the respective domain localizations and type of the mutations.
Comparison of observed clinical features between the total (including present and previous patients) 9q34.3 deletion group and the total intragenic EHMT1 mutation group revealed only a few remarkable differences between both groups (table 4). This is in agreement with previous observations that EHMT1 is responsible for the core phenotype of KS and most of other associated features. EHMT1 is an epigenetic regulator that affects gene transcription by histone modification. It is one of a rapidly growing list of ID genes that are implicated in chromatin remodeling [Stewart and Kleefstra, 2007; Kramer et al., 2011; van Bokhoven, 2011]. It is likely that other associated features besides ID can be explained by the effect of EHMT1 mutations on the expression of a variety of target genes.
In the present mutation group the mean age is higher than in the deletion group. There are no indications that patients with a deletion die at a younger adult age than patients with a mutation. There is only 1 death (cause unknown) at adult age reported. This concerns a 21-year-old male patient with a deletion [Kleefstra et al., 2009]. The increased average age in the present mutation group compared to the present deletion group might be due to an older age of diagnosis in the mutation group, because deletions can be detected by standard genome-wide array analysis and thus are more easily recognized than mutations for which analysis has to be specifically requested on the base of the clinical phenotype.
Microcephaly and short stature were more frequent in the 9q34.3 deletion group (50 vs. 19% and 32 vs. 17%, respectively). Respiratory complications and tracheomalacia were also more frequently observed in the 9q34.3 deletion group (14 vs. 5% and 11 vs. 5%, respectively). Features that were observed more often in the EHMT1 mutation group included high birth weight (9% in the deletion group vs. 21% in the mutation group) and overweight (28 and 42%, respectively). Recurrent infections and behavioral problems were also more often reported in patients with an EHMT1 mutation (in 26 vs. 64% and in 54 vs. 75%, respectively). These observations are highly consistent with our report in 2009 [Kleefstra et al., 2009]. The explanation for the observed differences in frequency of certain features is unclear, but might be partly explained by inconsistent report of some features in previous studies among 9q34.3 microdeletion patients and relatively low numbers of patients with an EHMT1 mutation which influences the figures. The distinct pattern of behavior problems observed in several teenage and adult patients with KS has recently gained more attention [Kleefstra et al., 2009; Verhoeven et al., 2010, 2011] and thus behavior problems are likely more consistently reported in recent reports.
The present report confirms to a large extent the phenotypic spectrum described in earlier reports. Most frequently observed clinical features in KS apart from the core phenotype included behavioral problems (∼50–75%), various brain anomalies (∼60%), heart defects (∼40–45%), male genital defects (∼30–40%), microcephaly (∼20–50%) and overweight (∼30–40%). Interestingly, cardiac defects that we observed in this cohort included structural anomalies but also other defects. In patient 14 an aberrant muscle bundle in the left ventricle was observed. Patient 20 and one of the previously reported patients with an intragenic EHMT1 mutation were both diagnosed with cardiac arrhythmias, including atrial flutter (pers. commun.). Epilepsy (∼25–35%), musculoskeletal anomalies (∼20–25%), recurrent infections (>25%), hearing problems (20–25%) and constipation (pers. observation) are quite common as well. Features that are observed more rarely are renal anomalies and other congenital anomalies, gastro-esophageal reflux and respiratory complications (table 4). As concluded previously, only minor indications for phenotype-genotype correlations could be observed within the group of deletion patients (between larger (>1 Mb) and smaller deletions), as well as between the 9q34 deletion and EHMT1 mutation group. Based on the phenotypic data presented here, we recommend cardiac screening during follow-up of KS patients, because of the possible occurrence of cardiac arrhythmias, even if no structural heart defect is present. In addition, clinicians and caretakers should be aware of the regressive behavioral phenotype that might develop at adolescent/adult age and seems to have no clear neurological substrate, but is rather a so far unexplained neuropsychiatric feature.
Identification of increasing numbers of patients with (smaller) deletions and EHMT1 mutations will probably further increase our knowledge of the phenotypic and genotypic spectrum of KS. Furthermore, application of novel sequence technologies might lead to the identification of other genes involved in phenotypes compatible with KS. Interestingly, our group recently published that the learning and memory defects in EHMT mutant Drosophila melanogaster could be rescued when EHMT expression was restored [Kramer et al., 2011]. These studies indicate that learning and memory are dynamic processes that might be rescued at least to some extent even when caused by genetic defects such as mutated EHMT. This opens the window in understanding the mechanisms of ID and strongly emphasizes the need for further investigations on possible intervention strategies that might become available in future for some aspects of cognitive processes.
Acknowledgements
We thank the participating patients and their families. This work was supported by grants from the Dutch Consortium ‘Stronger on your own feet’ (to T.K. and M.H.W.), and GENCODYS, an EU FP7 large-scale integrating project grant (grant agreement No. 241995) (to T.K., H.v.B. and A.T.V.-v.S.) and ZonMW (clinical fellowship grant 90700365 to T.K.).
References
- Ayyash H, Mueller R, Maltby E, Horsfield P, Telford N, Tyler R. A report of a child with a deletion (9)(q34.3): a recognisable phenotype? J Med Genet. 1997;34:610–612. doi: 10.1136/jmg.34.7.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier-Daire V, Molinari F, Rio M, Raoul O, de Blois MC, et al. Cryptic terminal deletion of chromosome 9q34: a novel cause of syndromic obesity in childhood? J Med Genet. 2003;40:300–303. doi: 10.1136/jmg.40.4.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson AJ, Putnam S, Schultz J, Riordan D, Prasad C, et al. Cryptic chromosome rearrangements detected by subtelomere assay in patients with mental retardation and dysmorphic features. Clin Genet. 2002;62:488–494. doi: 10.1111/j.1399-0004.2002.tb02255.x. [DOI] [PubMed] [Google Scholar]
- Font-Montgomery E, Weaver DD, Walsh L, Christensen C, Thurston VC. Clinical and cytogenetic manifestations of subtelomeric aberrations: report of six cases. Birth Defects Res A Clin Mol Teratol. 2004;70:408–415. doi: 10.1002/bdra.20036. [DOI] [PubMed] [Google Scholar]
- Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- Harada N, Visser R, Dawson A, Fukamachi M, Iwakoshi M, et al. A 1-Mb critical region in six patients with 9q34.3 terminal deletion syndrome. J Hum Genet. 2004;49:440–444. doi: 10.1007/s10038-004-0166-z. [DOI] [PubMed] [Google Scholar]
- Iwakoshi M, Okamoto N, Harada N, Nakamura T, Yamamori S, et al. 9q34.3 deletion syndrome in three unrelated children. Am J Med Genet A. 2004;126A:278–383. doi: 10.1002/ajmg.a.20602. [DOI] [PubMed] [Google Scholar]
- Kim C, Jun K, Lee T, Kim SS, McEnery MW, et al. Altered nociceptive response in mice deficient in the alpha-1B subunit of the voltage-dependent calcium channel. Mol Cell Neurosci. 2001;18:235–245. doi: 10.1006/mcne.2001.1013. [DOI] [PubMed] [Google Scholar]
- Kleefstra T, Smidt M, Banning MJ, Oudakker AR, Van Esch H, et al. Disruption of the gene Euchromatin Histone Methyl Transferase1 (Eu-HMTase1) is associated with the 9q34 subtelomeric deletion syndrome. J Med Genet. 2005;42:299–306. doi: 10.1136/jmg.2004.028464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, Koolen DA, Nillesen WM, de Leeuw N, Hamel BC, et al. Interstitial 2.2 Mb deletion at 9q34 in a patient with mental retardation but without classical features of the 9q subtelomeric deletion syndrome. Am J Med Genet A. 2006a;140:618–623. doi: 10.1002/ajmg.a.31123. [DOI] [PubMed] [Google Scholar]
- Kleefstra T, Brunner HG, Amiel J, Oudakker AR, Nillesen WM, et al. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet. 2006b;79:370–377. doi: 10.1086/505693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, van Zelst-Stams WA, Nillesen WM, Cormier-Daire V, Houge G, et al. Further clinical and molecular delineation of the 9q subtelomeric deletion syndrome supports a major contribution of EHMT1 haploinsufficiency to the core phenotype. J Med Genet. 2009;46:598–606. doi: 10.1136/jmg.2008.062950. [DOI] [PubMed] [Google Scholar]
- Kleefstra T, Nillesen WM, Yntema HG: Kleefstra Syndrome; in Pagon RA, Bird TD, Dolan CR, Stephens K (eds): GeneReviews [Internet] (University of Washington, Seattle 1993–2011). http://www.ncbi.nlm.nih.gov/books/NBK47079/ (2010).
- Kramer JM, Kochinke K, Oortveld MA, Marks H, Kramer D, et al. Epigenetic regulation of learning and memory by Drosophila EHMT/G9a. PLoS Biol. 2011;9:e1000569. doi: 10.1371/journal.pbio.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neas KR, Smith JM, Chia N, Huseyin S, St Heaps L, et al. Three patients with terminal deletions within the subtelomeric region of chromosome 9q. Am J Med Genet A. 2005;132:425–430. doi: 10.1002/ajmg.a.30496. [DOI] [PubMed] [Google Scholar]
- Nillesen WM, Yntema HG, Moscarda M, Verbeek NE, Wilson LC, et al. Characterization of a novel transcript of the EHMT1 gene reveals important diagnostic implications for Kleefstra syndrome. Hum Mutat. 2011;32:583–859. doi: 10.1002/humu.21523. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- Schimmenti LA, Berry SA, Tuchman M, Hirsch B. Infant with multiple congenital anomalies and deletion (9)(q34.3) Am J Med Genet. 1994;51:140–142. doi: 10.1002/ajmg.1320510211. [DOI] [PubMed] [Google Scholar]
- Stewart D, Kleefstra T. The chromosome 9q subtelomere deletion syndrome. Am J Med Genet C Semin Med Genet. 2007;145:383–392. doi: 10.1002/ajmg.c.30148. [DOI] [PubMed] [Google Scholar]
- Stewart DR, Huang A, Faravelli F, Anderlid BM, Medne L, et al. Subtelomeric deletions of chromosome 9q: a novel microdeletion syndrome. Am J Med Genet A. 2004;128:340–351. doi: 10.1002/ajmg.a.30136. [DOI] [PubMed] [Google Scholar]
- van Bokhoven H. Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet. 2011;45:81–104. doi: 10.1146/annurev-genet-110410-132512. [DOI] [PubMed] [Google Scholar]
- Verhoeven WM, Kleefstra T, Egger JI. Behavioral phenotype in the 9q subtelomeric deletion syndrome: a report about two adult patients. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:536–541. doi: 10.1002/ajmg.b.31015. [DOI] [PubMed] [Google Scholar]
- Verhoeven WM, Egger JI, Vermeulen K, van de Warrenburg BP, Kleefstra T. Kleefstra syndrome in three adult patients: further delineation of the behavioral and neurological phenotype shows aspects of a neurodegenerative course. Am J Med Genet A. 2011;155:2409–2415. doi: 10.1002/ajmg.a.34186. [DOI] [PubMed] [Google Scholar]
- Willemsen M, Beunders G, Callaghan M, de Leeuw N, Nillesen W, et al. Familial Kleefstra syndrome due to maternal somatic mosaicism for interstitial 9q34.3 microdeletions. Clin Genet. 2011;80:31–38. doi: 10.1111/j.1399-0004.2010.01607.x. [DOI] [PubMed] [Google Scholar]
- Yatsenko SA, Cheung SW, Scott DA, Nowaczyk MJ, Tarnopolsky M, et al. Deletion 9q34.3 syndrome: genotype-phenotype correlations and an extended deletion in a patient with features of Opitz C trigonocephaly. J Med Genet. 2005;42:328–335. doi: 10.1136/jmg.2004.028258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko SA, Brundage EK, Roney EK, Cheung SW, Chinault AC, Lupski JR. Molecular mechanisms for subtelomeric rearrangements associated with the 9q34.3 microdeletion syndrome. Hum Mol Genet. 2009;18:1924–1936. doi: 10.1093/hmg/ddp114. [DOI] [PMC free article] [PubMed] [Google Scholar]