Abstract
The Angelman syndrome is caused by disruption of the UBE3A gene and is clinically delineated by the combination of severe mental disability, seizures, absent speech, hypermotoric and ataxic movements, and certain remarkable behaviors. Those with the syndrome have a predisposition toward apparent happiness and paroxysms of laughter, and this finding helps distinguish Angelman syndrome from other conditions involving severe developmental handicap. Accurate diagnosis rests on a combination of clinical criteria and molecular and/or cytogenetic testing. Analysis of parent-specific DNA methylation imprints in the critical 15q11.2–q13 genomic region identifies 75–80% of all individuals with the syndrome, including those with cytogenetic deletions, imprinting center defects and paternal uniparental disomy. In the remaining group, UBE3A sequence analysis identifies an additional percentage of patients, but 5–10% will remain who appear to have the major clinical phenotypic features but do not have any identifiable genetic abnormalities. Genetic counseling for recurrence risk is complicated because multiple genetic mechanisms can disrupt the UBE3A gene, and there is also a unique inheritance pattern associated with UBE3A imprinting. Angelman syndrome is a prototypical developmental syndrome due to its remarkable behavioral phenotype and because UBE3A is so crucial to normal synaptic function and neural plasticity.
Key Words: Angelman syndrome, Imprinting, Microdeletion, 15q11.2–q13, UBE3A, Ubiquitin
History of the Syndrome
Angelman syndrome (AS) was first described by Dr. Harry Angelman in 1965 [Angelman, 1965]. It is characterized by developmental delay, absent speech, ataxic gait, seizures, and a distinctive behavioral phenotype with excitability and paroxysms of laughter [Clayton-Smith and Laan, 2003; Summers and Pittman, 2004; Williams, 2005; Dan, 2008; Van Buggenhout and Fryns, 2009; Williams et al., 2010a]. The incidence is thought to be 1/12,000 to 1/20,000 [Clayton-Smith and Pembrey, 1992; Steffenburg et al., 1996]. Cases have been reported from all over the world without racial predilection.
Clinical Features
Consensus criteria for the clinical diagnosis have been described [Williams et al., 2006] as outlined in table 1. One of the earliest distinctive behaviors of AS may be persistent social smiling beginning at 1–3 months. Chortling, giggling and constant smiling may follow. Excessive mouthing behaviors are also common in AS infants and children with active exploration of objects through manipulation and chewing. Tongue protrusion is seen in 30–50% of children associated with drooling.
Table 1.
Clinical features of AS
A. Consistent (100%)
|
B. Frequent (more than 80%)
|
C. Associated (20–80%)
|
Hyperkinetic movements of the trunk and limbs may be seen in early infancy and jitteriness or tremulousness may be present very early [Fryburg et al., 1991]. Voluntary movements may be slightly jerky or uncoordinated coarse movements that prevent walking, feeding and reaching for objects may be seen. The mildly impaired child can have almost normal walking in early childhood with only mild toe-walking or a prancing gait. More severely affected children can be extremely shaky and jerky when walking or stiff and robot-like. The legs are kept wide-based and the feet are often flat and ankles pronated and turned outward. Arms are kept uplifted with flexed elbows and downward turned hands.
Hypermotoric behaviors can be pronounced in young children and this in combination with the jerky limb movements and frequent smiling and/or laughter can give a distinctive behavioral phenotype, recently reviewed by Williams [2010]. Most have social-seeking behaviors, and their apparent happiness and laughter is often contextually appropriate [Oliver et al., 2002; Horsler and Oliver, 2006a, 2006b]. Bursts of laughter may occur in up to 70% of older individuals [Buntinx et al., 1995].
Sleep problems are well known for AS and frequent awakening at night is common [Bruni et al., 2004; Didden et al., 2004]. Dyssomnias (difficulties in initiating or maintaining sleep), irregular sleep-wake cycles, disruptive night behaviors such as periods of laughter, and sleep-related seizures have been reported [Pelc et al., 2008b]. Pelc et al. [2008b] attribute these sleep anomalies to abnormal neurodevelopmental functioning of the thalamocortical axis.
Onset of seizures of varied types (generalized tonic-clonic, absence, atonic, complex partial, and myoclonic) occurs between 1 and 3 years of age, but can occur later. Seizures are associated with specific non-epileptic EEG changes [Galvan-Manso et al., 2005]: runs of high-amplitude delta activity with intermittent spike and slow-wave discharges (at times observed as a notched delta pattern), runs of rhythmic theta activity over a wide area and runs of rhythmic sharp theta activity of 5–6/s over the posterior third of the head, forming complexes with small spikes. These are usually facilitated by or seen only with eye closure [Boyd et al., 1988; Rubin et al., 1997; Korff et al., 2005]. Nonconvulsive status epilepticus may occur [Pelc et al., 2008a]. It is believed that seizures are usually well controlled on anticonvulsants, but a recent questionnaire study by Thibert et al. [2009] suggests that the epilepsy is relatively refractory as only 15% of patients respond to the first anti-epileptic drug. Structurally the brain appears to be normal except for delayed or abnormal myelination and mild atrophy [Harting et al., 2009; Castro-Gago et al., 2010]. Peters et al. [2010] noted abnormalities in diffusor tensor imaging suggestive of dysmyelination.
Intellectual deficiency is in the severe to profound range of functioning in AS. Severe language impairment is the norm and the great majority has absent speech. Some communication via gestures and communication boards is possible [Clayton-Smith, 1993]. Accurate developmental testing is difficult because of inability to pay attention, hyperactivity and lack of speech. Psychometric testing suggests that the upper developmental potential is in the 24–30-month range [Peters et al., 2004; Trillingsgaard and Ostergaard, 2004; Didden et al., 2006].
Although there are craniofacial anomalies present in some individuals with AS, these typically do not represent significant facial dysmorphism. Individuals with AS who have prominent oral-motor behaviors associated with a protruding tongue often have deformational changes leading to some degree of mandibular prognathism, and those with microcephaly may have diminished length of the cranial base leading to midface retrusion [Frias et al., 1982]. More recently, the use of computer mapping and 3-dimensional analysis of standardized facial photographs [Hammond et al., 2005] holds promise for detecting subtle facial changes among different genetic types of AS individuals, but only a preliminary report of this has occurred (noted in Williams and Franco [2010]).
Natural History
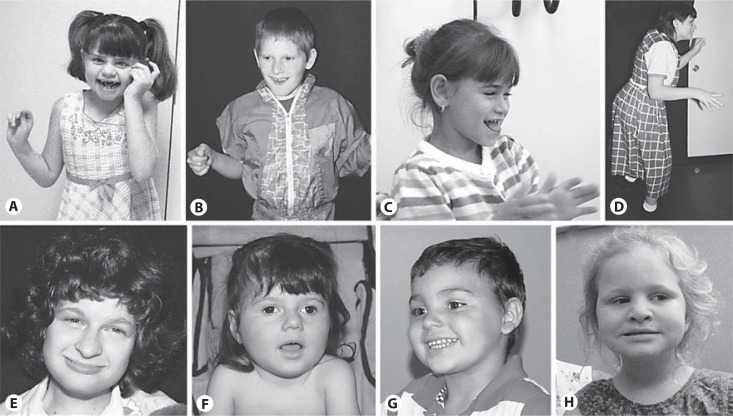
Newborns with AS appear to have had normal fetal development and have normal head circumferences at birth. It is difficult to diagnose AS in infancy since nonspecific developmental delay, hypotonia and feeding difficulties may be the only recognized features. Microcephaly becomes evident by 1 year of age in about one half and, as in Rett syndrome, acquired microcephaly is present. Most children are diagnosed by ages 3–7 years when the abnormal gait and distinctive behaviors become evident. Most walk by age 2 and a half to 6 years and have a jerky, ataxic gait associated with uplifted and pronated forearms (fig. 1) [Zori et al., 1992; Lossie et al., 2001].
Fig. 1.
Pictured are individuals who have genetic test-proven Angelman syndrome. The mechanisms identified in them are: 15q11.2–q13 deletion (A, B, D, E); paternal uniparental disomy (C); UBE3A mutation (F, G) and imprinting defect (H). Individuals A, B and C illustrate some of the gait characteristics seen in the syndrome. Protruding tongue can be a noteworthy phenotypic feature, especially in combination with laughter (as in C), but most do not have pronounced tongue protrusion. The girl H has a non-deletion, mosaic-type imprinting defect, and thus her cognitive and language skills are relatively higher than observed in the typical child with the syndrome.
Physical health in older AS children and in adults appears to be remarkably good. Young adults with AS continue to learn and are generally not expected to have deterioration in their mental abilities. The main adult problems are essentially a continuation of any present in childhood. Although the severity or frequency of seizures may improve with age, many still require some type of anticonvulsant medication. Prolonged disabling bouts of tremor have recently been noted in teenagers and adults, but the cause of this is unclear, and it does not appear to be a seizure manifestation nor is it representative of a Parkinson-like movement disorder. This episodic tremor problem is often overlaid on an existing long-term tremulousness. In the author's experience (C.A.W.), these tremors are present both at rest and upon intention and last 2–6 weeks before returning to baseline.
Mobility issues become a more predominant concern in teenagers and young adults, often associated with obesity. Individuals with AS who have severe ataxia may lose their ability to walk if ambulation is not encouraged. Scoliosis can develop in adolescence and is especially a problem in those who are nonambulatory [Clayton-Smith, 1993; Clayton-Smith and Laan, 2003; Dan, 2008]. Scoliosis is treated with early bracing to prevent progression, and surgical correction or stabilization may be necessary for severe cases.
Due to disruptive behaviors, those with AS may be given neuroleptic medications, and the sedating or other side effects of these agents can be a health problem.
Pubertal onset and development are generally normal in AS and procreation appears possible for both males and females. Lossie and Driscoll [1999] reported transmission of an AS deletion to a fetus by the affected mother.
Independent living is not possible for adults, and until recently, most adults probably lived in small residential facilities or larger institutional care programs. More recently, a growing number continue to live at home or near their homes in small home-like placements such a group homes. Life span does not appear to be dramatically shortened in AS but may be decreased by 10–15 years. Those who are not ambulatory and have difficulty with eating so as to require a gastrostomy tube, and/or have a severe seizure disorder are expected to have some diminution in their life span. There are reports of AS individuals living beyond 70 years; although there is, as of yet, no actuarial data about longevity in AS [Bjerre et al., 1984; Philippart and Minassian, 2005].
Molecular Biology of the UBE3A Gene
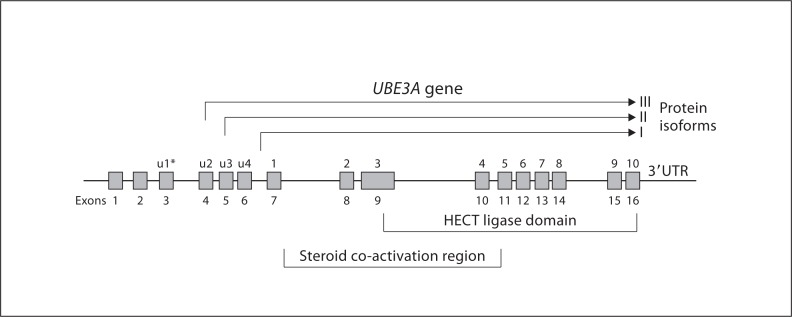
AS is caused by disruption of the function of the maternally inherited ubiquitin-protein ligase E3A (UBE3A) gene [Kishino et al., 1997; Matsuura et al., 1997; Jiang et al., 1999; Nicholls and Knepper, 2001]. The gene lies within the AS critical region 15q11.2–q13, spans approximately 120 kb of genomic DNA and contains 16 exons (see fig. 2). The coding region is 60 kb. The main 3 mRNA transcripts have 10 exons, are approximately 5 kb in size and encode 3 protein isoforms (I, II and III) [Kishino et al., 1997; Vu and Hoffman, 1997; Yamamoto et al., 1997; Kishino and Wagstaff, 1998] (fig. 2). There appears to be 8–10 other smaller transcripts of unknown function or of uncertain significance. Isoform I corresponds to the open reading frame for E6-associated protein (E6-AP). Isoform II has an additional 20 amino acids and isoform III has an additional 23 amino acids at the amino terminus. The functions of the different protein isoforms are unknown. They may interact with different substrates in different intracellular regions. The 5′ untranslated region has a somewhat complex structure with additional exons located upsteam of the initiation site. The 3′ UTR extends for about 2.0 kb [Kishino and Wagstaff, 1998].
Fig. 2.
Schematic diagram of the UBE3A gene showing exons 1–16 and 3 of the most studied protein isoforms that differ by 20 and 23 amino acids at the amino terminal aspect. Exons 9–16 constitute the HECT binding and transfer domains. The steroid coactivation region does occupy a contiguous genomic region but spans a region that contains several 5-amino acid motifs known to be receptor interacting motifs [Ramamoorthy and Nawaz, 2008]. An alternative exon numbering system for UBE3A is indicated by the asterisk, as designated by Yamamoto et al. [1997].
UBE3A produces an 865 amino acid E6-AP. This associated protein was first recognized as a protein that binds to p53 and mediates its association with human papilloma virus E6 protein. This binding leads to degradation of p53 tumor suppressor via the ubiquitin proteasome pathway and thus promotes development cervical carcinoma [Huibregtse et al., 1991; Huang et al., 1999]. E6-AP facilitates the transfer and covalent linkage of activated ubiquitin (a 76-amino acid protein) to the target protein. The polyubiquitylated substrates are then identified and degraded by the 26S proteasome pathway. E6-AP belongs to the HECT (homologous to E6-AP COOH-terminus) class of E3 enzymes that share a 40 kDa conserved COOH-terminal catalytic domain. The HECT domain of E6-AP is a bilobed structure with a broad catalytic cleft at the junction of the 2 lobes. The domain is encoded by exons 9 through 16. The E6-binding site is encoded by exon 9 and the active site cysteine residue that accepts ubiquitin from the E2 ubiquitin-conjugating enzyme is encoded within exon 16 [Yamamoto et al., 1997; Kishino and Wagstaff, 1998]. Mutations within the cleft interfere with ubiquitin-thioester bond formation. Indeed, most AS mutations due to missense or single amino acid insertion or deletion mutations in the HECT domain map to the catalytic clefts [Huang et al., 1999].
The ubiquitin-proteasome pathway is essential for cellular functioning such as signal transduction, cell-cycle progression, DNA repair, and transcriptional regulation [Ciechanover, 1998; Hershko and Ciechanover, 1998]. Several E6-AP targets have been discovered [Kühne and Banks, 1998; Kumar et al., 1999; Oda et al., 1999; Khan et al., 2006; Li et al., 2006; Reiter et al., 2006; Louria-Hayon et al., 2009; Shimoji et al., 2009], and recently 2 target proteins, ARC (activity-regulated cytoskeleton-associated protein) and Ephexin-5, have been identified that appear to be crucial components of synaptic plasticity and dendrite growth regulation [Greer et al., 2010; Margolis et al., 2010; Scheiffele and Beg, 2010]. The ARC protein is involved regulating membrane stabilization of excitatory postsynaptic receptors. The guanine exchange protein, Ephexin-5, is known to regulate activity of EphB receptor signaling that is a crucial component of dendritic growth [Margolis et al., 2010]. Eph receptors are known to be enriched at synapses and are important in regulating dendritic spine density. The EphB receptors interact with ephrin ligands and regulate dendritic development through small GTPases of the Rho family (Rho, Rac and Cdc42) by activation of guanine nucleotide exchange factors [Murai and Pasquale, 2003]. It is unknown how abnormalities in E6-AP-target proteins interaction lead to AS, but the recently identified targets strongly indicate that the UBE3A protein is crucial to development of normal synaptic development and neural plasticity. This concept is supported by AS mouse studies that demonstrate abnormal dendritic processes [Dindot et al., 2008; Lu et al., 2009] and defects in hippocampal long-term potentiation, experience-dependent maturation of excitatory cortical circuits [Jiang et al., 1998; Yashiro et al., 2009] and postsynaptic kinase pathways involving calmodulin-dependent protein kinase II [Weeber et al., 2003]. The major features of the AS mouse phenotype have been rescued by reducing the phosphorylation of calmodulin-dependent protein kinase II [van Woerden et al., 2007].
A steroid receptor coactivation domain is located upstream of the HECT region, but its role in neuronal development is uncertain. E6-AP appears to have at least 2 independent functions since the ligase region and the HECT domain are not required for function of the coactivation domain [Ramamoorthy and Nawaz, 2008].
UBE3A displays predominant maternal expression in human fetal brain and adult frontal cortex [Rougeulle et al., 1997; Vu and Hoffman, 1997; Herzing et al., 2001]. Detailed studies of imprinting regions in the human brain are limited; however, in the mouse model maternal allele-specific expression is detected in the hippocampus, cerebellum, olfactory bulb, and visual cortex [Albrecht et al., 1997; Jiang et al., 1998; Yashiro et al., 2009]. It is possible that there is widespread, if not global, UBE3A allele-specific expression in the mouse and in the human brain neurons. Primary cell cultures from fetal mouse brain also reveal that UBE3A imprinting is limited to neurons and that glial cells show biallelic expression [Yamasaki et al., 2003]. UBE3A has a large 5′ CpG island, but its DNA methylation does not differ between the maternal and paternal alleles [Lossie et al., 2001], both are unmethylated. Because no differentially methylated promoter region is present in UBE3A, it has been proposed that the imprinted expression of UBE3A may be regulated indirectly through a paternally expressed antisense transcript [Rougeulle et al., 1998]. Runte et al. [2001] have shown that a long SNURF-SNRPN sense/UBE3A antisense RNA transcript exists in the AS/PWS region, starting from the SNURF-SNRPN IC and extending more than 460 kb to at least the 5′ end of UBE3A. It has been proposed that paternally active UBE3A antisense transcripts block paternal UBE3A transcription (fig. 3).
Fig. 3.
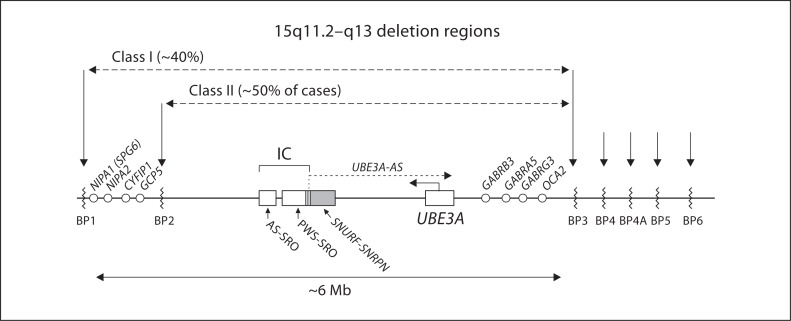
Schematic drawing of chromosome region 15q11.2–q13 indicating the breakpoint regions BP1–BP6. Low-copy repeat elements are located within these breakpoint regions (see text for details). Approximately 90% of chromosome deletions resulting in Angelman syndrome initiate at BP1 or BP2 and terminate in region BP3 (class I and class II). Approximately 10% of deletions are larger, typically spanning from BP1 to BP5, rarely beyond BP5. Genes that are not imprinted and thus biparentally expressed are noted by the open circles. The 2 critical imprinting center (IC) elements, the AS-SRO and the PWS-SRO, are drawn as open boxes. The shaded box for the SNURF-SNRPN gene is shown with some overlap with the PWS-SRO. The SNURF-SNRPN sense/UBE3A antisense transcript is labeled UBE3A-AS. Distances are not to scale particularly between SNRPN and UBE3A; not pictured are the paternally expressed snoRNAs that are transcribed as part of the long antisense transcript between these 2 genes.
Molecular Classes of Angelman Syndrome
In normal neurons, UBE3A is transcriptionally inactivated (imprinted) on the paternally derived allele of chromosome 15 and is active only on the maternally derived allele. All other somatic cells have biallelic transcription. The syndrome can occur by 4 different mutational mechanisms affecting the maternally derived chromosome 15: intragenic mutation, deletion of the gene (e.g. via chromosome microdeletion), paternal uniparental disomy (UPD) with absence of maternal chromosome 15, and a defect in the imprinting center (IC) that controls UBE3A transcription. These mechanisms are reviewed below.
Chromosome Microdeletions
Three chromosome 15q11.2–q13 breakpoints (proximal BP1, BP2 and the more distal BP3) are involved in about 90% of AS causing de novo deletion events, and these deletions span approximately 5–7 Mb [Knoll et al., 1990; Amos-Landgraf et al., 1999; Christian et al., 1999]. Class I deletions account for 40% of deletion cases and extend from BP1–BP3. Class II deletions extend from BP2–BP3 and account for 50% of cases. Fewer than 10% of individuals with AS may have deletions extending from the BP1/BP2 region to regions more distal at BP4, BP4A, BP5, or BP6 (fig. 3) [Sahoo et al., 2007]. The BP1, BP2 and BP3 regions are characterized by low-copy repeats, typically in direct orientation, that contain repeats of several pseudogenes and other expressed sequences. One of the most noteworthy elements in these low-copy repeats is derived from the HERC2 gene (HEct domains and RCc1 domain protein 2) [Pujana et al., 2002]. The BP sites distal to BP3 contain low-copy repeats without HERC2 duplications, but they share other chromosome 15-derived repeat elements. Microdeletions have been described that flank the typical deletion region and include areas between BP1 and BP2 [Doornbos et al., 2009], BP3 and BP4 [Rosenfeld et al., 2011] and the more distal microdeletion syndrome involving region 15q13.3 [Masurel-Paulet et al., 2010]. Individuals with these deletions do not exhibit features of the AS. Interstitial duplications of 15q11.2–q13 on the maternally derived chromosome cause a disorder involving autism and/or intellectual deficiency, but it is clinically distinct from the phenotype of AS [Boyar et al., 2001]. A proportion of mothers who have a child with an AS deletion have molecular inversions in the 15q11.2–q13 region [Gimelli et al., 2003]. The significance of this is uncertain and needs further study. A kindred in which 2 first cousins had deletions (one deletion causing Prader-Willi syndrome (PWS) and the other causing AS, inherited from a brother and sister, respectively) has been reported to be associated with an inherited inverted intrachromosomal insertion of 15q11.2–q13 [Collinson et al., 2004]. It is thus possible that in otherwise normal individuals, such preexisting genomic abnormalities may predispose to deletion of 15q11.2–q13 in the germ line resulting in offspring with AS.
Paternal Uniparental Disomy 15
Paternal UPD of chromosome 15 causes 3–7% of AS; these individuals have a milder presentation with a lower incidence of seizures. Robinson et al. [2000] reported that a somatic segregation error is the most likely mechanism leading to the paternal UPD. Paternal UPD cases of meiotic origin do occur, but this mechanism is less common than is seen in the maternal UPD cases associated with PWS. Individuals with UPD should have chromosomal analysis to ensure that they do not have a paternally inherited Robertsonian translocation. In individuals with paternal UPD and no Robertsonian translocation, the risk to sibs of having AS is less than 1%. The recurrence risk is not zero, as recurrent meiotic nondisjunction of maternal chromosome 15 has been reported [Harpey et al., 1998]. If an individual has paternal UPD with a normal karyotype, a maternal chromosomal analysis could be performed to rule out a Robertsonian translocation or a marker chromosome that may generate a maternal gamete nullisomic for chromosome 15. This situation could theoretically lead to a postzygotic correction to paternal disomy.
Imprinting Defects
About 2–4% of AS individuals have a defect in the resetting/maintaining of imprints during gametogenesis or after fertilization. Genetic (small deletions) and epigenetic (abnormal DNA methylation pattern but no deletion) defects in the AS IC within 15q11.2–q13 change the DNA methylation and expression patterns along 15q11.2–q13. Even though these individuals have biparental inheritance of chromosome 15, the maternal 15q11.2–q13 region has a paternal epigenotype and is therefore transcriptionally incompetent for the maternal-only expressed UBE3A gene in this region [Glenn et al., 1993; Buiting et al., 2001, 2003].
The IC has a bipartite structure and regulates in cis imprint resetting and maintenance within the 15q11.2–q13 imprinted domain [Sutcliffe et al., 1994; Buiting et al., 1995, 1999]. About 8–15 % of those with an imprinting defect will have deletions that disrupt the IC, and mapping of these deletions (as well as mapping of the IC deletions that are associated with PWS) has delineated 2 smallest regions of deletion overlap (SRO) that define 2 critical elements in the IC region, the AS-SRO and the PWS-SRO [Buiting et al., 1995]. The PWS-SRO is 4.3 kb in size and overlaps with the SNURF-SNRPN exon1/promoter region [Ohta et al., 1999] (fig. 3). IC deletions found in patients with AS affect the more centromeric SNURF-SNRPN promoter/exon 1 region. The smallest region of overlap in patients with AS and an IC deletion (AS-SRO) is 880 bp in size and maps 35 kb proximal to SNURF-SNRPN exon 1 [Buiting et al., 1999; Horsthemke and Buiting, 2008]. Two out of the 13 IC deletions described so far have occurred de novo on the maternal chromosome, but in most of the cases they have been inherited from the mother [Horsthemke and Buiting, 2008]. The deletions are without any phenotypic effect when transmitted through the male germ line, but lead to an incorrect paternal imprint when transmitted through the female germ line. It appears that the AS-SRO has an important role in the establishment of the maternal imprint in the female germ line, possibly by interacting with the PWS-SRO and that a deletion of this element prevents maternal imprinting of the deletion chromosome.
IC deletions are found only in a small fraction of AS patients with imprinting defects. In the vast majority of patients (>90%), the imprinting defect represents a primary epimutation [Buiting et al., 2003; Horsthemke and Buiting, 2008] without any changes in the DNA sequence. In AS patients with a primary epimutation, the maternal chromosome carrying a wrong paternal epigenotype can be inherited from either the maternal grandmother or grandfather, suggesting that in these patients the imprinting defect results from an error in the imprint establishment in the female germ line or in imprint maintenance in the early embryo, leading to somatic mosaicism [Horsthemke, 2010]. The postzygotic loss of the maternal imprint is a significant cause of AS with an imprinting defect as more than 40% of AS patients with an imprinting defect are found to have somatic mosaicism [Buiting, unpubl. data]. These patients were found to have a small amount of methylated alleles, as they show a weak maternal band in methylation analysis for the SNURF-SNRPN locus using various techniques.
UBE3A Mutations
The majority of UBE3A mutations found in AS are protein truncating mutations [Kishino et al., 1997; Matsuura et al., 1997; Malzac et al., 1998; Lossie et al., 2001]. More than 60 mutations have been reported and 60–70% of these involve small deletions and duplications leading to frameshift mutations [Abaied et al., 2009; Camprubi et al., 2009; Stenson et al., 2009]. Another approximate 25% involve missense and nonsense mutations with the remainder representing splicing defects, gross deletions and complex rearrangements [Stenson et al., 2009]. All mutations noted thus far are predicted to disrupt the HECT ligase domain. Exons 9 and 16, which code for part of the HECT domain, account for a high percentage of all mutations, but these coding regions are disproportionately large, so this high percentage probably does not represent true hot spots for mutation. It is possible that individuals with milder effect mutations (e.g. certain missense and inframe deletions or duplications) may show some, but not all, of the clinical features associated with AS. Novel missense mutations, especially de novo ones, can be problematic in establishing pathogenicity, but determination of the parental origin of the mutation can be helpful since the mutation would need to arise from the maternal chromosome [Horsthemke et al., 2011] Complete or partial overlapping deletions of UBE3A and intragenic deletions have also been identified. These can be missed by sequencing but can be detected using various methods, such as quantitative PCR, real time PCR and MLPA. In some instances, array-CGH has detected multi-exonic or whole gene deletions [Govoni et al., 1985; Lawson-Yuen et al., 2006; Sato et al., 2007]. Detection of such deletions may vary by laboratory and methodology [Lawson-Yuen et al., 2006; Sato et al., 2007; Ramsden et al., 2010].
Diagnostic Testing for AS
Laboratory diagnostic testing for AS can be complicated. One approach to evaluation starts with a DNA methylation analysis of the AS/PWS IC region. If the DNA methylation test is abnormal, one of 3 AS mechanisms is present: the large common deletion, uniparental disomy or defects in the IC. If the methylation test is abnormal, additional studies are needed to define the specific genetic mechanism. In such situations, the next step typically is to perform a microsatellite, FISH or microarray chromosome study to determine if the common 15q11.2–q13 deletion is present (other methods such as MS-MLPA can test for the deletion concurrently with the methylation assay) [Ramsden et al., 2010]. If a deletion is excluded, the next step is to rule out paternal UPD by additional microsatellite or microarray testing. Individuals with an abnormal AS DNA methylation study without a deletion and with biparental inheritance of chromosomes 15 are then presumed to have an imprinting defect. The imprinting defect can be further studied to determine if there is a deletion of the IC. Molecular testing for IC region deletions is available clinically from a small number of laboratories. Finally, if the methylation test is normal, mutation analysis of the UBE3A gene is the next step and may detect a significant percentage of individuals.
Genotype to Phenotype Correlation
Identifying significant genotype to phenotype correlations has been an ongoing issue in AS since the initial discovery of the deletion in 1987. Soon after this discovery, those with the deletion were compared to those without the microdeletion. As additional AS-causing mechanisms were identified, the clinical correlations became more complicated because of overlapping symptoms among all mechanisms [Saitoh et al., 1994; Smith et al., 1997; Fridman et al., 2000; Lossie et al., 2001; Nazlican et al., 2004; Saitoh et al., 2005]. It soon became evident that the core features of the syndrome can be attributed solely to disruption of the UBE3A gene, regardless of the mechanism, but some differences did exist among the genetic types. Those with the large chromosome deletion appeared to have more severe symptoms and this was presumably due to haploinsufficiency of genes adjacent to UBE3A such as the downstream GABA genes (GABRB3, GABRA5 and GABRG3) or those located upstream in the BP1 to BP2 breakpoint region (NIPA1, NIPA2, CYFIP1, and GCP5). Individuals with the large deletions (class I [BP1–BP3] or class II [BP2–BP3]) are more likely to have microcephaly, seizures and more severe language impairment compared to those with UPD, UBE3A mutations or imprinting defects. Also, most large deletions cause haploinsufficiency of OCA2 leading to relatively hypopigmented irides, skin and hair. OCA2 plays a role in tyrosine metabolism and is important for the development of pigment in the skin, hair and irides. Tan et al. [2011] presented clinical data from 92 children with a molecular diagnosis of AS established between 5 and 60 months of age. They noted that individuals with the larger deletions have diminished weight compared to the general population and to those with UPD/imprinting defects. This could be in part related to diminished muscle mass in those with the large deletion.
From the perspective of having a relatively higher verbal speech ability and cognitive understanding, it appears that individuals with the epigenetic type of imprintingdefect associated with some degree of somatic mosaicism are higher functioning. These individuals may speak up to 50–60 words and use simple sentences [Nazlican et al., 2004]. However, this degree of expressive language in AS is rare. AS patients with IDs or UPD have relatively higher developmental and language ability, but there is much overlap between all of the genetic categories. Individuals with larger class I deletions may have more language impairment or autistic traits [Sahoo et al., 2007] than those with smaller class II deletions.
Individuals with UPD appear to have better physical growth and fewer movement abnormalities, ataxia and seizures and are less likely to have microcephaly than the other classes [Lossie et al., 2001; Saitoh et al., 2005], but the reason for this is unclear.
Differential Diagnosis
AS presents in infancy with nonspecific features, such as psychomotor delay and seizures. This can lead to the descriptive labels of cerebral palsy or static encephalopathy. Hypotonia and seizures may raise the possibility of an inborn error of metabolism or a defect in oxidative phosphorylation, but metabolic and mitochondrial testing is normal. Infants with AS may have feeding difficulties, hypotonia, and developmental delay, features which overlap with PWS. Parent-specific DNA methylation analysis at the SNRPN locus can distinguish between these 2 syndromes.
The Phelan-McDermid syndrome (22q13.3 deletion) can mimic some of the features of AS [Precht et al., 1998], as it presents with absent or minimal speech, moderate-to-severe developmental delay and autistic features. The 2q23.1 microdeletion results in severe speech delay, seizures, microcephaly, and behavioral abnormalities that may overlap with AS [van Bon et al., 2010; Williams et al., 2010b]. Newer microdeletion conditions discovered by microarray may be associated with some of the features of AS [Brunetti-Pierri et al., 2008; Sharkey et al., 2009]. Microduplication involving MECP2 (typically encompassing an approximate 500-kb region at Xq28) causes severe developmental impairment, absent speech, seizures, and ataxic gait with spastic paraparesis in males. Adult males are typically nonambulatory and are prone to infectious illnesses, but presentation in childhood may be relatively nonspecific and include features of mental retardation with autism, absent speech and unstable gait [Van Esch et al., 2005; Friez et al., 2006; Lugtenberg et al., 2009].
Affected female infants with seizures, acquired microcephaly and severe speech impairment can resemble girls with Rett syndrome. Girls with Rett syndrome do not have a distinctive happy-demeanor and girls with AS do not have a neuroregressive course and do not lose purposeful use of their hands. Older girls with undiagnosed Rett syndrome may have features that resemble AS [Watson et al., 2001].
Mowat-Wilson syndrome can present with happy affect, prominent mandible, diminished speech, microcephaly, and constipation [Zweier et al., 2005]. Mowat-Wilson syndrome results from a de novo dominant mutation or deletions in ZEB2. Individuals with Christianson syndrome have seizures, severe developmental delay, ataxia, microcephaly, and a happy disposition [Christianson et al., 1999; Gilfillan et al., 2008; Schroer et al., 2010]. Christianson syndrome is X-linked and is caused by mutations in the SLC9A6 gene. Pitt-Hopkins syndrome is caused by mutations/deletions of the TCF4 gene. This syndrome presents with mental retardation, wide mouth and distinctive facial features, intermittent hyperventilation followed by apnea, microcephaly, seizures, ataxic gait, and happy personality [Peippo et al., 2006; Zweier et al., 2007].
Adenylosuccinate lyase deficiency results in accumulation of succinylpurines leading to psychomotor retardation, autistic features, hypotonia, and seizures [Spiegel et al., 2006]. Motor apraxia, severe speech deficits, excessive laughter, a very happy disposition, hyperactivity, a short attention span, mouthing of objects, tantrums, and stereotyped movements have been reported in female sibs by Gitiaux et al. [2009]. Diagnostic testing involves detection of succinylaminoimidazole carboxamide riboside (SAICA riboside) and succinyladenosine (S-Ado) in cerebrospinal fluid, urine and, to a lesser extent, in plasma. The rare metabolic disorder of severe methylene-tetrahydrofolate-reductase deficiency (MTHFR) associated with low methionine and elevated homocysteine blood levels was reported in an boy who presented with happy demeanor, ataxic gait, absent speech, and flattened occiput [Arn et al., 1998].
Genetic Counseling in AS
Genetic counseling to address recurrence risk for families who have one child with AS can be a complicated issue and often requires expert consultation. Fortunately, the great majority of genetic mechanisms originates by spontaneous mutations and has low recurrence risk, as is the case usually for the chromosome deletions and for UPD. However, even in these groups rare genetic mechanisms can lead to an increased recurrence risk within a family. IC deletions and UBE3A mutations can be inherited and carry as high as a 50% recurrence risk. Because of the unique aspects of imprinting inheritance, it is possible for such mutations to be transmitted asymptomatically in kindred but then become manifest depending on the parental origin of the transmission. Details of this counseling are beyond the scope of this review but have been addressed elsewhere [Buiting et al., 1998, 2000, 2001; Stalker and Williams, 1998; Stalker et al., 1998; Buiting, 2010; Ramsden et al., 2010; Williams et al., 2010a].
Conclusion
It has been almost 15 years since the UBE3A gene and its protein, E6-AP, were linked to the causation of AS. Since then, incremental progress has identified interacting proteins and substrate targets for E6-AP. Much of this discovery has led to a focus on synaptic impairment as a fundamental problem underlying the intellectual deficiency and other manifestations of the syndrome. New discoveries are leading to new ideas about how to ameliorate and potentially cure some of the symptoms of AS, but as of yet, no meaningful successes have occurred. Possibilities for future therapies include UBE3A gene insertion into neurons, attempts to induce neuronal expression of the normal paternally silenced UBE3A gene by use of artificial transcription factors or use of epigenetic modulating drugs. We may also soon see attempts to manipulate E6-AP protein targets and interacting proteins via pharmaceutical approaches. Further research into UBE3A targets is also likely to identify additional neuronal mechanisms that could lead to viable druggable targets or to other therapeutic strategies.
References
- Abaied L, Trabelsi M, Chaabouni M, Kharrat M, Kraoua L, et al. A novel UBE3A truncating mutation in large Tunisian Angelman syndrome pedigree. Am J Med Genet A. 2009;152A:141–146. doi: 10.1002/ajmg.a.33179. [DOI] [PubMed] [Google Scholar]
- Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, et al. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet. 1997;17:75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- Amos-Landgraf JM, Ji Y, Gottlieb W, Depinet T, Wandstrat AE, et al. Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet. 1999;65:370–386. doi: 10.1086/302510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelman H. ‘Puppet' children. A report of three cases. Dev Med Child Neurol. 1965;7:681–688. [Google Scholar]
- Arn PH, Williams CA, Zori RT, Driscoll DJ, Rosenblatt DS. Methylenetetrahydrofolate reductase deficiency in a patient with phenotypic findings of Angelman syndrome. Am J Med Genet. 1998;77:198–200. [PubMed] [Google Scholar]
- Bjerre I, Fagher B, Ryding E, Rosen I. The Angelman or ‘happy puppet' syndrome. Clinical and electroencephalographic features and cerebral blood flow. Acta Paediatr Scand. 1984;73:398–402. doi: 10.1111/j.1651-2227.1994.tb17755.x. [DOI] [PubMed] [Google Scholar]
- Boyar FZ, Whitney MM, Lossie AC, Gray BA, Keller KL, et al. A family with a grand-maternally derived interstitial duplication of proximal 15q. Clin Genet. 2001;60:421–430. doi: 10.1034/j.1399-0004.2001.600604.x. [DOI] [PubMed] [Google Scholar]
- Boyd SG, Harden A, Patton MA. The EEG in early diagnosis of the Angelman (happy puppet) syndrome. Eur J Pediatr. 1988;147:508–513. doi: 10.1007/BF00441976. [DOI] [PubMed] [Google Scholar]
- Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruni O, Ferri R, D'Agostino G, Miano S, Roccella M, Elia M. Sleep disturbances in Angelman syndrome: a questionnaire study. Brain Dev. 2004;26:233–240. doi: 10.1016/S0387-7604(03)00160-8. [DOI] [PubMed] [Google Scholar]
- Buiting K. Prader-Willi syndrome and Angelman syndrome. Am J Med Genet C Semin Med Genet. 2010;154C:365–376. doi: 10.1002/ajmg.c.30273. [DOI] [PubMed] [Google Scholar]
- Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, et al. Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nat Genet. 1995;9:395–400. doi: 10.1038/ng0495-395. [DOI] [PubMed] [Google Scholar]
- Buiting K, Dittrich B, Gross S, Lich C, Farber C, et al. Sporadic imprinting defects in Prader-Willi syndrome and Angelman syndrome: implications for imprint-switch models, genetic counseling, and prenatal diagnosis. Am J Hum Genet. 1998;63:170–180. doi: 10.1086/301935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buiting K, Lich C, Cottrell S, Barnicoat A, Horsthemke B. A 5-kb imprinting center deletion in a family with Angelman syndrome reduces the shortest region of deletion overlap to 880 bp. Hum Genet. 1999;105:665–666. doi: 10.1007/s004399900196. [DOI] [PubMed] [Google Scholar]
- Buiting K, Farber C, Kroisel P, Wagner K, Brueton L, et al. Imprinting centre deletions in two PWS families: implications for diagnostic testing and genetic counseling. Clin Genet. 2000;58:284–290. doi: 10.1034/j.1399-0004.2000.580406.x. [DOI] [PubMed] [Google Scholar]
- Buiting K, Barnicoat A, Lich C, Pembrey M, Malcolm S, Horsthemke B. Disruption of the bipartite imprinting center in a family with Angelman syndrome. Am J Hum Genet. 2001;68:1290–1294. doi: 10.1086/320120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buiting K, Gross S, Lich C, Gillessen-Kaesbach G, el-Maarri O, Horsthemke B. Epimutations in Prader-Willi and Angelman syndromes: a molecular study of 136 patients with an imprinting defect. Am J Hum Genet. 2003;72:571–577. doi: 10.1086/367926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buntinx IM, Hennekam RC, Brouwer OF, Stroink H, Beuten J, et al. Clinical profile of Angelman syndrome at different ages. Am J Med Genet. 1995;56:176–183. doi: 10.1002/ajmg.1320560213. [DOI] [PubMed] [Google Scholar]
- Camprubi C, Guitart M, Gabau E, Coll MD, Villatoro S, et al. Novel UBE3A mutations causing Angelman syndrome: different parental origin for single nucleotide changes and multiple nucleotide deletions or insertions. Am J Med Genet A. 2009;149A:343–348. doi: 10.1002/ajmg.a.32659. [DOI] [PubMed] [Google Scholar]
- Castro-Gago M, Gomez-Lado C, Eiris-Punal J, Rodriguez-Mugico VM. Abnormal myelination in Angelman syndrome. Eur J Paediatr Neurol. 2010;14:292. doi: 10.1016/j.ejpn.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH. Large genomic duplicons map to sites of instability in the Prader-Willi/ Angelman syndrome chromosome region (15q11–q13) Hum Mol Genet. 1999;8:1025–1037. doi: 10.1093/hmg/8.6.1025. [DOI] [PubMed] [Google Scholar]
- Christianson AL, Stevenson RE, van der Meyden CH, Pelser J, Theron FW, et al. X linked severe mental retardation, craniofacial dysmorphology, epilepsy, ophthalmoplegia, and cerebellar atrophy in a large South African kindred is localised to Xq24–q27. J Med Genet. 1999;36:759–766. doi: 10.1136/jmg.36.10.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 1998;17:7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton-Smith J. Clinical research on Angelman syndrome in the United Kingdom: observations on 82 affected individuals. Am J Med Genet. 1993;46:12–15. doi: 10.1002/ajmg.1320460105. [DOI] [PubMed] [Google Scholar]
- Clayton-Smith J, Laan L. Angelman syndrome: a review of the clinical and genetic aspects. J Med Genet. 2003;40:87–95. doi: 10.1136/jmg.40.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton-Smith J, Pembrey ME. Angelman syndrome. J Med Genet. 1992;29:412–415. doi: 10.1136/jmg.29.6.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson MN, Roberts SE, Crolla JA, Dennis NR. A familial balanced inverted insertion ins(15)(q15q13q11.2) producing Prader-Willi syndrome, Angelman syndrome and duplication of 15q11.2–q13 in a single family: importance of differentiation from a paracentric inversion. Am J Med Genet A. 2004;126A:27–32. doi: 10.1002/ajmg.a.26565. [DOI] [PubMed] [Google Scholar]
- Dan B (ed): Angelman Syndrome. (Mac Keith Press, Brussels 2008).
- Didden R, Korzilius H, Duker P, Curfs L. Communicative functioning in individuals with Angelman syndrome: a comparative study. Disabil Rehabil. 2004;26:1263–1267. doi: 10.1080/09638280412331280271. [DOI] [PubMed] [Google Scholar]
- Didden R, Korzilius H, Kamphuis A, Sturmey P, Lancioni G, Curfs LM. Preferences in individuals with Angelman syndrome assessed by a modified Choice Assessment Scale. J Intellect Disabil Res. 2006;50:54–60. doi: 10.1111/j.1365-2788.2005.00731.x. [DOI] [PubMed] [Google Scholar]
- Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum Mol Genet. 2008;17:111–118. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]
- Doornbos M, Sikkema-Raddatz B, Ruijvenkamp CA, Dijkhuizen T, Bijlsma EK, et al. Nine patients with a microdeletion 15q11.2 between breakpoints 1 and 2 of the Prader-Willi critical region, possibly associated with behavioural disturbances. Eur J Med Genet. 2009;52:108–115. doi: 10.1016/j.ejmg.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Frias JL, King GJ, Williams CA. Cephalometric assessment of selected malformation syndromes. Birth Defects Orig Artic Ser. 1982;18:139–150. [PubMed] [Google Scholar]
- Fridman C, Varela MC, Kok F, Diament A, Koiffmann CP. Paternal UPD15: further genetic and clinical studies in four Angelman syndrome patients. Am J Med Genet. 2000;92:322–327. doi: 10.1002/1096-8628(20000619)92:5<322::aid-ajmg6>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Friez MJ, Jones JR, Clarkson K, Lubs H, Abuelo D, et al. Recurrent infections, hypotonia, and mental retardation caused by duplication of MECP2 and adjacent region in Xq28. Pediatrics. 2006;118(6):e1687–e1695. doi: 10.1542/peds.2006-0395. [DOI] [PubMed] [Google Scholar]
- Fryburg JS, Breg WR, Lindgren V. Diagnosis of Angelman syndrome in infants. Am J Med Genet. 1991;38:58–64. doi: 10.1002/ajmg.1320380114. [DOI] [PubMed] [Google Scholar]
- Galvan-Manso M, Campistol J, Conill J, Sanmarti FX. Analysis of the characteristics of epilepsy in 37 patients with the molecular diagnosis of Angelman syndrome. Epileptic Disord. 2005;7:19–25. [PubMed] [Google Scholar]
- Gilfillan GD, Selmer KK, Roxrud I, Smith R, Kyllerman M, et al. SLC9A6 mutations cause X-linked mental retardation, microcephaly, epilepsy, and ataxia, a phenotype mimicking Angelman syndrome. Am J Hum Genet. 2008;82:1003–1010. doi: 10.1016/j.ajhg.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelli G, Pujana MA, Patricelli MG, Russo S, Giardino D, et al. Genomic inversions of human chromosome 15q11–q13 in mothers of Angelman syndrome patients with class II (BP2/3) deletions. Hum Mol Genet. 2003;12:849–858. doi: 10.1093/hmg/ddg101. [DOI] [PubMed] [Google Scholar]
- Gitiaux C, Ceballos-Picot I, Marie S, Valayannopoulos V, Rio M, et al. Misleading behavioural phenotype with adenylosuccinate lyase deficiency. Eur J Hum Genet. 2009;17:133–136. doi: 10.1038/ejhg.2008.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn CC, Nicholls RD, Robinson WP, Saitoh S, Niikawa N, et al. Modification of 15q11–q13 DNA methylation imprints in unique Angelman and Prader-Willi patients. Hum Mol Genet. 1993;2:1377–1382. doi: 10.1093/hmg/2.9.1377. [DOI] [PubMed] [Google Scholar]
- Govoni S, Padovani A, Magnoni MS, Battaini F, Rius RA, et al. Effect of chronic ethanol consumption on human and animal receptor plasticity during aging. Alcohol Drug Res. 1985;6:441–448. [PubMed] [Google Scholar]
- Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond P, Hutton TJ, Allanson JE, Buxton B, Campbell LE, et al. Discriminating power of localized three-dimensional facial morphology. Am J Hum Genet. 2005;77:999–1010. doi: 10.1086/498396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpey JP, Heron D, Prudent M, Lesourd S, Henry I, et al. Recurrent meiotic nondisjunction of maternal chromosome 15 in a sibship. Am J Med Genet. 1998;76:103–104. [PubMed] [Google Scholar]
- Harting I, Seitz A, Rating D, Sartor K, Zschocke J, et al. Abnormal myelination in Angelman syndrome. Eur J Paediatr Neurol. 2009;13:271–276. doi: 10.1016/j.ejpn.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Herzing LB, Kim SJ, Cook EH, Jr., Ledbetter DH. The human aminophospholipid-transporting ATPase gene ATP10C maps adjacent to UBE3A and exhibits similar imprinted expression. Am J Hum Genet. 2001;68:1501–1505. doi: 10.1086/320616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsler K, Oliver C. The behavioural phenotype of Angelman syndrome. J Intellect Disabil Res. 50;2006a:33–53. doi: 10.1111/j.1365-2788.2005.00730.x. [DOI] [PubMed] [Google Scholar]
- Horsler K, Oliver C. Environmental influences on the behavioral phenotype of Angelman syndrome. Am J Ment Retard. 2006b;111:311–321. doi: 10.1352/0895-8017(2006)111[311:EIOTBP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Horsthemke B. Mechanisms of imprint dysregulation. Am J Med Genet C Semin Med Genet. 2010;154C:321–328. doi: 10.1002/ajmg.c.30269. [DOI] [PubMed] [Google Scholar]
- Horsthemke B, Buiting K. Genomic imprinting and imprinting defects in humans. Adv Genet. 2008;61:225–246. doi: 10.1016/S0065-2660(07)00008-9. [DOI] [PubMed] [Google Scholar]
- Horsthemke B, Wawrzik M, Gross S, Lich C, Sauer B, et al. Parental origin and functional relevance of a de novo UBE3A variant. Eur J Med Genet. 2011;54:19–24. doi: 10.1016/j.ejmg.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, et al. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Lev-Lehman E, Bressler J, Tsai TF, Beaudet AL. Genetics of Angelman syndrome. Am J Hum Genet. 1999;65:1–6. doi: 10.1086/302473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, et al. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- Khan OY, Fu G, Ismail A, Srinivasan S, Cao X, et al. Multifunction steroid receptor coactivator, E6-associated protein, is involved in development of the prostate gland. Mol Endocrinol. 2006;20:544–559. doi: 10.1210/me.2005-0110. [DOI] [PubMed] [Google Scholar]
- Kishino T, Wagstaff J. Genomic organization of the UBE3A/E6-AP gene and related pseudogenes. Genomics. 1998;47:101–107. doi: 10.1006/geno.1997.5093. [DOI] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Knoll JH, Nicholls RD, Magenis RE, Glatt K, Graham JM, Jr, et al. Angelman syndrome: three molecular classes identified with chromosome 15q11q13-specific DNA markers. Am J Hum Genet. 1990;47:149–155. [PMC free article] [PubMed] [Google Scholar]
- Korff CM, Kelley KR, Nordli DR., Jr Notched delta, phenotype, and Angelman syndrome. J Clin Neurophysiol. 2005;22:238–243. doi: 10.1097/01.wnp.0000167930.90824.0f. [DOI] [PubMed] [Google Scholar]
- Kühne C, Banks L. E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box. J Biol Chem. 1998;273:34302–34309. doi: 10.1074/jbc.273.51.34302. [DOI] [PubMed] [Google Scholar]
- Kumar S, Talis AL, Howley PM. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J Biol Chem. 1999;274:18785–18792. doi: 10.1074/jbc.274.26.18785. [DOI] [PubMed] [Google Scholar]
- Lawson-Yuen A, Wu BL, Lip V, Sahoo T, Kimonis V. Atypical cases of Angelman syndrome. Am J Med Genet A. 2006;140:2361–2364. doi: 10.1002/ajmg.a.31481. [DOI] [PubMed] [Google Scholar]
- Li L, Li Z, Howley PM, Sacks DB. E6AP and calmodulin reciprocally regulate estrogen receptor stability. J Biol Chem. 2006;281:1978–1985. doi: 10.1074/jbc.M508545200. [DOI] [PubMed] [Google Scholar]
- Lossie AC, Whitney MM, Amidon D, Dong HJ, Chen P, et al. Distinct phenotypes distinguish the molecular classes of Angelman syndrome. J Med Genet. 2001;38:834–845. doi: 10.1136/jmg.38.12.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossie AM, Driscoll DJ. Transmission of Angelman syndrome by an affected mother. Genet Med. 1999;1:262–266. doi: 10.1097/00125817-199909000-00004. [DOI] [PubMed] [Google Scholar]
- Louria-Hayon I, Alsheich-Bartok O, Levav-Cohen Y, Silberman I, Berger M, et al. E6AP promotes the degradation of the PML tumor suppressor. Cell Death Differ. 2009;16:1156–1166. doi: 10.1038/cdd.2009.31. [DOI] [PubMed] [Google Scholar]
- Lu Y, Wang F, Li Y, Ferris J, Lee JA, Gao FB. The Drosophila homologue of the Angelman syndrome ubiquitin ligase regulates the formation of terminal dendritic branches. Hum Mol Genet. 2009;18:454–462. doi: 10.1093/hmg/ddn373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg D, Kleefstra T, Oudakker AR, Nillesen WM, Yntema HG, et al. Structural variation in Xq28: MECP2 duplications in 1% of patients with unexplained XLMR and in 2% of male patients with severe encephalopathy. Eur J Hum Genet. 2009;17:444–453. doi: 10.1038/ejhg.2008.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malzac P, Webber H, Moncla A, Graham JM, Kukolich M, et al. Mutation analysis of UBE3A in Angelman syndrome patients. Am J Hum Genet. 1998;62:1353–1360. doi: 10.1086/301877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis SS, Salogiannis J, Lipton DM, Mandel-Brehm C, Wills ZP, et al. EphB-mediated degradation of the RhoA GEF Ephexin5 relieves a developmental brake on excitatory synapse formation. Cell. 2010;143:442–455. doi: 10.1016/j.cell.2010.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masurel-Paulet A, Andrieux J, Callier P, Cuisset JM, Le Caignec C, et al. Delineation of 15q13.3 microdeletions. Clin Genet. 2010;78:149–161. doi: 10.1111/j.1399-0004.2010.01374.x. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, et al. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- Murai KK, Pasquale EB. ‘Eph'ective signaling: forward, reverse and crosstalk. J Cell Sci. 2003;116:2823–2832. doi: 10.1242/jcs.00625. [DOI] [PubMed] [Google Scholar]
- Nazlican H, Zeschnigk M, Claussen U, Michel S, Boehringer S, et al. Somatic mosaicism in patients with Angelman syndrome and an imprinting defect. Hum Mol Genet. 2004;13:2547–2555. doi: 10.1093/hmg/ddh296. [DOI] [PubMed] [Google Scholar]
- Nicholls RD, Knepper JL. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet. 2001;2:153–175. doi: 10.1146/annurev.genom.2.1.153. [DOI] [PubMed] [Google Scholar]
- Oda H, Kumar S, Howley PM. Regulation of the Src family tyrosine kinase Blk through E6AP-mediated ubiquitination. Proc Natl Acad Sci USA. 1999;96:9557–9562. doi: 10.1073/pnas.96.17.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Buiting K, Kokkonen H, McCandless S, Heeger S, et al. Molecular mechanism of Angelman syndrome in two large families involves an imprinting mutation. Am J Hum Genet. 1999;64:385–396. doi: 10.1086/302232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver C, Demetriades L, Hall S. Effects of environmental events on smiling and laughing behavior in Angelman syndrome. Am J Ment Retard. 2002;107:194–200. doi: 10.1352/0895-8017(2002)107<0194:EOEEOS>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Peippo MM, Simola KO, Valanne LK, Larsen AT, Kahkonen M, et al. Pitt-Hopkins syndrome in two patients and further definition of the phenotype. Clin Dysmorphol. 2006;15:47–54. doi: 10.1097/01.mcd.0000184973.14775.32. [DOI] [PubMed] [Google Scholar]
- Pelc K, Boyd SG, Cheron G, Dan B. Epilepsy in Angelman syndrome. Seizure. 2008a;17:211–217. doi: 10.1016/j.seizure.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Pelc K, Cheron G, Boyd SG, Dan B. Are there distinctive sleep problems in Angelman syndrome? Sleep Med. 2008;9:434–441. doi: 10.1016/j.sleep.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Peters SU, Goddard-Finegold J, Beaudet AL, Madduri N, Turcich M, Bacino CA. Cognitive and adaptive behavior profiles of children with Angelman syndrome. Am J Med Genet A. 2004;128A:110–113. doi: 10.1002/ajmg.a.30065. [DOI] [PubMed] [Google Scholar]
- Peters SU, Bird LM, Kimonis V, Glaze DG, Shinawi LM, et al. Double-blind therapeutic trial in Angelman syndrome using betaine and folic acid. Am J Med Genet A. 2010;152A:1994–2001. doi: 10.1002/ajmg.a.33509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippart M, Minassian BA. Angelman syndrome from infancy to old age (abstract) Am J Hum Genet. 2005;79(Suppl):605. [Google Scholar]
- Precht KS, Lese CM, Spiro RP, Huttenlocher PR, Johnston KM, et al. Two 22q telomere deletions serendipitously detected by FISH. J Med Genet. 1998;35:939–942. doi: 10.1136/jmg.35.11.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujana MA, Nadal M, Guitart M, Armengol L, Gratacos M, Estivill X. Human chromosome 15q11–q14 regions of rearrangements contain clusters of LCR15 duplicons. Eur J Hum Genet. 2002;10:26–35. doi: 10.1038/sj.ejhg.5200760. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy S, Nawaz Z. E6-associated protein (E6-AP) is a dual function coactivator of steroid hormone receptors. Nucl Recept Signal. 2008;6:e006. doi: 10.1621/nrs.06006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden SC, Clayton-Smith J, Birch R, Buiting K. Practice guidelines for the molecular analysis of Prader-Willi and Angelman syndromes. BMC Med Genet. 2010;11:70. doi: 10.1186/1471-2350-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Seagroves TN, Bowers M, Bier E. Expression of the Rho-GEF Pbl/ECT2 is regulated by the UBE3A E3 ubiquitin ligase. Hum Mol Genet. 2006;15:2825–2835. doi: 10.1093/hmg/ddl225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WP, Christian SL, Kuchinka BD, Penaherrera MS, Das S, et al. Somatic segregation errors predominantly contribute to the gain or loss of a paternal chromosome leading to uniparental disomy for chromosome 15. Clin Genet. 2000;57:349–358. doi: 10.1034/j.1399-0004.2000.570505.x. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JA, Stephens LE, Coppinger J, Ballif BC, Hoo JJ, et al. Deletions flanked by breakpoints 3 and 4 on 15q13 may contribute to abnormal phenotypes. Eur J Hum Genet. 2011 doi: 10.1038/ejhg.2010.237. E-pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet. 1997;17:14–15. doi: 10.1038/ng0997-14. [DOI] [PubMed] [Google Scholar]
- Rougeulle C, Cardoso C, Fontes M, Colleaux L, Lalande M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- Rubin DI, Patterson MC, Westmoreland BF, Klass DW. Angelman's syndrome: clinical and electroencephalographic findings. Electroencephalogr Clin Neurophysiol. 1997;102:299–302. doi: 10.1016/s0013-4694(96)96105-2. [DOI] [PubMed] [Google Scholar]
- Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- Sahoo T, Bacino CA, German JR, Shaw CA, Bird LM, et al. Identification of novel deletions of 15q11q13 in Angelman syndrome by array-CGH: molecular characterization and genotype-phenotype correlations. Eur J Hum Genet. 2007;15:943–949. doi: 10.1038/sj.ejhg.5201859. [DOI] [PubMed] [Google Scholar]
- Saitoh S, Harada N, Jinno Y, Hashimoto K, Imaizumi K, et al. Molecular and clinical study of 61 Angelman syndrome patients. Am J Med Genet. 1994;52:158–163. doi: 10.1002/ajmg.1320520207. [DOI] [PubMed] [Google Scholar]
- Saitoh S, Wada T, Okajima M, Takano K, Sudo A, Niikawa N. Uniparental disomy and imprinting defects in Japanese patients with Angelman syndrome. Brain Dev. 2005;27:389–391. doi: 10.1016/j.braindev.2003.12.013. [DOI] [PubMed] [Google Scholar]
- Sato K, Iwakoshi M, Shimokawa O, Sakai H, Ohta T, et al. Angelman syndrome caused by an identical familial 1,487-kb deletion. Am J Med Genet A. 2007;143:98–101. doi: 10.1002/ajmg.a.31550. [DOI] [PubMed] [Google Scholar]
- Scheiffele P, Beg AA. Neuroscience: Angelman syndrome connections. Nature. 2010;468:907–908. doi: 10.1038/468907a. [DOI] [PubMed] [Google Scholar]
- Schroer RJ, Holden KR, Tarpey PS, Matheus MG, Griesemer DA, et al. Natural history of Christianson syndrome. Am J Med Genet A. 2010;152A:2775–2783. doi: 10.1002/ajmg.a.33093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey FH, Morrison N, Murray R, Iremonger J, Stephen J, et al. 17q21.31 microdeletion syndrome: further expanding the clinical phenotype. Cytogenet Genome Res. 2009;127:61–66. doi: 10.1159/000279260. [DOI] [PubMed] [Google Scholar]
- Shimoji T, Murakami K, Sugiyama Y, Matsuda M, Inubushi S, et al. Identification of annexin A1 as a novel substrate for E6AP-mediated ubiquitylation. J Cell Biochem. 2009;106:1123–1135. doi: 10.1002/jcb.22096. [DOI] [PubMed] [Google Scholar]
- Smith A, Marks R, Haan E, Dixon J, Trent RJ. Clinical features in four patients with Angelman syndrome resulting from paternal uniparental disomy. J Med Genet. 1997;34:426–429. doi: 10.1136/jmg.34.5.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel EK, Colman RF, Patterson D. Adenylosuccinate lyase deficiency. Mol Genet Metab. 2006;89:19–31. doi: 10.1016/j.ymgme.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Stalker HJ, Williams CA. Genetic counseling in Angelman syndrome: the challenges of multiple causes. Am J Med Genet. 1998;77:54–59. [PubMed] [Google Scholar]
- Stalker HJ, Williams CA, Wagstaff J. Genetic counseling in Angelman syndrome: gonadal mosaicism. Am J Med Genet. 1998;78:482. [PubMed] [Google Scholar]
- Steffenburg S, Gillberg CL, Steffenburg U, Kyllerman M. Autism in Angelman syndrome: a population-based study. Pediatr Neurol. 1996;14:131–136. doi: 10.1016/0887-8994(96)00011-2. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, et al. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers JA, Pittman D. Angelman syndrome. In: Griffiths D, King R, editors. Demystifying Syndromes: Clinical and Educational Implications of Common Syndromes Associated with Persons with Intellectual Disabilities. New York: NADD Press; 2004. pp. 161–188. [Google Scholar]
- Sutcliffe JS, Nakao M, Christian S, Orstavik KH, Tommerup N, et al. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- Tan WH, Bacino CA, Skinner SA, Anselm I, Barbieri-Welge R, et al. Angelman syndrome: mutations influence features in early childhood. Am J Med Genet A. 2011;155A:81–90. doi: 10.1002/ajmg.a.33775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibert RL, Conant KD, Braun EK, Bruno P, Said RR, et al. Epilepsy in Angelman syndrome: a questionnaire-based assessment of the natural history and current treatment options. Epilepsia. 2009;50:2369–2376. doi: 10.1111/j.1528-1167.2009.02108.x. [DOI] [PubMed] [Google Scholar]
- Trillingsgaard A, Ostergaard JR. Autism in Angelman syndrome: an exploration of comorbidity. Autism. 2004;8:163–174. doi: 10.1177/1362361304042720. [DOI] [PubMed] [Google Scholar]
- van Bon BW, Koolen DA, Brueton L, McMullan D, Lichtenbelt KD, et al. The 2q23.1 microdeletion syndrome: clinical and behavioural phenotype. Eur J Hum Genet. 2010;18:163–170. doi: 10.1038/ejhg.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buggenhout G, Fryns JP. Angelman syndrome (AS, MIM 105830) Eur J Hum Genet. 2009;17:1367–1373. doi: 10.1038/ejhg.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Esch H, Bauters M, Ignatius J, Jansen M, Raynaud M, et al. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am J Hum Genet. 2005;77:442–453. doi: 10.1086/444549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Woerden GM, Harris KD, Hojjati MR, Gustin RM, Qiu S, et al. Rescue of neurological deficits in a mouse model for Angelman syndrome by reduction of alphaCaMKII inhibitory phosphorylation. Nat Neurosci. 2007;10:280–282. doi: 10.1038/nn1845. [DOI] [PubMed] [Google Scholar]
- Vu TH, Hoffman AR. Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nat Genet. 1997;17:12–13. doi: 10.1038/ng0997-12. [DOI] [PubMed] [Google Scholar]
- Watson P, Black G, Ramsden S, Barrow M, Super M, et al. Angelman syndrome phenotype associated with mutations in MECP2, a gene encoding a methyl CpG binding protein. J Med Genet. 2001;38:224–228. doi: 10.1136/jmg.38.4.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeber EJ, Jiang YH, Elgersma Y, Varga AW, Carrasquillo Y, et al. Derangements of hippocampal calcium/calmodulin-dependent protein kinase II in a mouse model for Angelman mental retardation syndrome. J Neurosci. 2003;23:2634–2644. doi: 10.1523/JNEUROSCI.23-07-02634.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, Franco L. Angelman syndrome at the synapse: meeting report of the Angelman Syndrome Foundation's 2009 scientific symposium. J Child Neurol. 2010;25:254–261. doi: 10.1177/0883073809353450. [DOI] [PubMed] [Google Scholar]
- Williams CA. Neurological aspects of the Angelman syndrome. Brain Dev. 2005;27:88–94. doi: 10.1016/j.braindev.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Williams CA. The behavioral phenotype of the Angelman syndrome. Am J Med Genet C Semin Med Genet. 2010;154C:432–437. doi: 10.1002/ajmg.c.30278. [DOI] [PubMed] [Google Scholar]
- Williams CA, Beaudet AL, Clayton-Smith J, Knoll JH, Kyllerman M, et al. Angelman syndrome 2005: updated consensus for diagnostic criteria. Am J Med Genet A. 2006;140:413–418. doi: 10.1002/ajmg.a.31074. [DOI] [PubMed] [Google Scholar]
- Williams CA, Driscoll DJ, Dagli AI. Clinical and genetic aspects of Angelman syndrome. Genet Med. 2010a;12:385–395. doi: 10.1097/GIM.0b013e3181def138. [DOI] [PubMed] [Google Scholar]
- Williams SR, Mullegama SV, Rosenfeld JA, Dagli AI, Hatchwell E, et al. Haploinsufficiency of MBD5 associated with a syndrome involving microcephaly, intellectual disabilities, severe speech impairment, and seizures. Eur J Hum Genet. 2010b;18:436–441. doi: 10.1038/ejhg.2009.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Huibregtse JM, Howley PM. The human E6-AP gene (UBE3A) encodes three potential protein isoforms generated by differential splicing. Genomics. 1997;41:263–266. doi: 10.1006/geno.1997.4617. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Joh K, Ohta T, Masuzaki H, Ishimaru T, et al. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum Mol Genet. 2003;12:837–847. doi: 10.1093/hmg/ddg106. [DOI] [PubMed] [Google Scholar]
- Yashiro K, Riday TT, Condon KH, Roberts AC, Bernardo DR, et al. Ube3a is required for experience-dependent maturation of the neocortex. Nat Neurosci. 2009;12:777–783. doi: 10.1038/nn.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zori RT, Hendrickson J, Woolven S, Whidden EM, Gray B, Williams CA. Angelman syndrome: clinical profile. J Child Neurol. 1992;7:270–280. doi: 10.1177/088307389200700307. [DOI] [PubMed] [Google Scholar]
- Zweier C, Thiel CT, Dufke A, Crow YJ, Meinecke P, et al. Clinical and mutational spectrum of Mowat-Wilson syndrome. Eur J Med Genet. 2005;48:97–111. doi: 10.1016/j.ejmg.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Zweier C, Peippo MM, Hoyer J, Sousa S, Bottani A, et al. Haploinsufficiency of TCF4 causes syndromal mental retardation with intermittent hyperventilation (Pitt-Hopkins syndrome) Am J Hum Genet. 2007;80:994–1001. doi: 10.1086/515583. [DOI] [PMC free article] [PubMed] [Google Scholar]