Abstract
Disorders related to the autosomal transcription factor MEF2C located in 5q14.3 were first described in 2009 and have since evolved to one of the more common microdeletion syndromes. Mutational screening in a larger cohort revealed heterozygous de novo mutations of MEF2C in about 1% of patients with moderate to severe intellectual disability, and the phenotype is similar in patients with intragenic deletions and multigenic microdeletions. Clinically, MEF2C-related disorders are characterized by severe intellectual disability with absent speech and limited walking abilities, hypotonia, seizures, and a variety of minor brain anomalies. The majority of patients show a similar facial gestalt with broad forehead, flat nasal bridge, hypotonic mouth, and small chin, as well as strabismus, but this phenotype is clinically not well recognized. The course of the disease is generally quite uniform, but patients with point mutations and smaller deletions seem to have a higher chance of walking skills and a lower risk of refractory seizures. Patients in whom the microdeletion also includes the RASA1 gene show features of the respective capillary and arterio-venous malformations and fistula syndrome. The phenotypic overlap with Rett syndrome is explained by a shared pathway and, accordingly, diminished MECP2 and CDKL5 expression is measureable in patients with MEF2C defects. Further research of this pathway may therefore eventually lead to a common therapeutic target.
Key Words: MEF2C, Microdeletion 5q14, Rett syndrome-like, Seizures, Severe intellectual disability, Severe mental retardation
History of the Syndrome
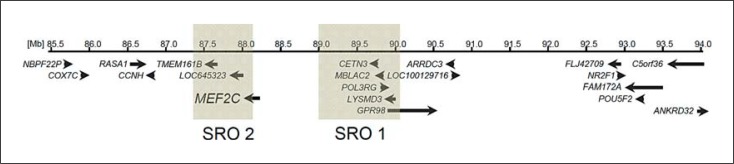
As a result of joint efforts of the German Mental Retardation Network (MRNET) and the Decipher database, Engels, Firth and Rauch initiated a study on 5q14.3q15 microdeletions and delineated a novel syndrome characterized by severe psychomotor retardation, epilepsy or febrile seizures, muscular hypotonia and variable brain and minor anomalies [Engels et al., 2009]. These deletions partially overlapped with the distally more extended deletions in 3 patients with severe mental retardation, seizures and periventricular heterotopia described by Cardoso et al. [2009]. The 1.6-Mb smallest region of overlap contained the neurodevelopmental LYSMD3 gene as a potential underlying cause of intellectual disability and the GPR98/MASS1 gene was considered potentially related to seizures [Engels et al., 2009]. However, the observation of 2 very similar patients with more proximal deletions not overlapping the 1.6-Mb smallest region of overlap led Zweier et al. [2010] to further investigate the nearby MEF2C gene as a potential monogenic cause of the 5q14.3q15 microdeletion syndrome (fig. 1). They showed that MEF2C expression levels were indeed significantly decreased in blood probes of their novel patients as well as in all patients described by Engels et al. [2009], indicating MEF2C as the underlying gene and a positional effect in the patient with a deletion breakpoint distal to MEF2C. Accordingly, subsequent mutational screening in patients with moderate to severe intellectual disability of unknown cause revealed 4 MEF2C de novo mutations in patients with similar phenotype as seen in patients with larger deletions [Zweier et al., 2010]. At the same time Le Meur et al. [2010] independently identified MEF2C as the gene underlying the 5q14.3q15 microdeletion syndrome by detection of a microdeletion limited to MEF2C only and subsequent identification of a MEF2C stop mutation in a similar patient. Both Zweier et al. [2010] and Le Meur et al. [2010] pointed out the phenotypic overlap to Rett syndrome, and Zweier et al. [2010] demonstrated molecular interaction of the respective genes. Attributable to the wide use of molecular karyotyping or array comparative genomic hybridization, by now a total of 23 patients with 5q14.3q15 microdeletions have been reported [Cardoso et al., 2009; Engels et al., 2009; Berland and Houge, 2010; Le Meur et al., 2010; Novara et al., 2010; Nowakowska et al., 2010; Zweier et al., 2010; Carr et al., 2011; Mikhail et al., 2011; Tonk et al., 2011], but no further patients with point mutations, indicating that the phenotype is not well recognized. Sixteen microdeletions involved MEF2C and additional genes, 3 multi-gene microdeletions had a proximal breakpoint close to, but distally to MEF2C [Cardoso et al., 2009; Engels et al., 2009], and 4 microdeletions involved MEF2C only [Le Meur et al., 2010; Novara et al., 2010; Nowakowska et al., 2010; Mikhail et al., 2011]. Of note, one of the balanced translocation breakpoints in a patient with severe intellectual disability, early-onset epileptic encephalopathy, and hypoplastic corpus callosum was mapped 121.5 kb upstream of MEF2C [Saitsu et al., 2011].
Fig. 1.
Partial scheme of the gene content of the 15q14.3q15 microdeletion region with the first smallest region of overlap (SRO1) defined by 6 cases reported by Engels et al. [2009] and Cardoso et al. [2009], and the distinct SRO2 delineated by Zweier et al. [2010] through 2 novel cases.
Clinical Features
Neurodevelopmental Signs
Apart from 1 exceptional patient, all patients with mutations or microdeletions uniformly showed severe mental retardation with absent speech, limited walking abilities and lack of gross malformations (table 1). The exceptional patient described by Tonk et al. [2011] with the largest deletion of 21 Mb only had mild intellectual disability with a global IQ of 69 and a seizure disorder. It was hypothesized that the large extent of the deletion could have a compensatory effect explaining the mild phenotype. However, hidden mosaicism may be a more likely explanation since overlapping cytogenetically visible deletions were associated with significant dysmorphism and mental disability. Seven of 9 patients with MEF2C limited mutations or deletions, 17 of 19 with larger deletions, and overall all patients older than 3 years had seizures. However, the age of onset was usually in infancy or early childhood and commonly associated with fever. The majority of patients had tonic-clonic seizures, but myoclonic or complex partial seizures as well as infantile spasms were also described. Seizures were well controlled in the majority of patients, but were refractory in 3 patients with deletions exceeding the MEF2C gene [Le Meur et al., 2010; Novara et al., 2010]. Mild to severe hypotonia was reported in 17 of 19 patients with larger deletions and in 6 of 9 patients with MEF2C defects. The ability to walk independently was reported only in the following few patients: at the age of 2 years 8 months in a boy with p.E34X mutation [Zweier et al., 2010], at the age of 3 years in a girl with p.S228X mutation [Le Meur et al., 2010], at the ages of 5 and 3 years, respectively, in 2 patients with larger deletions distally to MEF2C [Cardoso et al., 2009], and at the age of 11 years in a girl with a 1.15-Mb deletion including MEF2C and TMEM161B [Berland and Houge, 2010]. Developmental milestones of the exceptionally mildly affected patient with the 21-Mb deletion were within the normal range [Tonk et al., 2011]. Stereotypic movements including bruxism, head rocking, hand washing or clapping, hand-mouth movements, and others, as well as autistic features or poor eye contact were reported in nearly half of the patients (13 of 28 each). Episodic hyperventilation and apnea was reported in 2 patients with MEF2C limited defects only [Le Meur et al., 2010; Zweier et al., 2010].
Table 1.
Features frequently observed in patients with MEF2C defects and 5q14.3q15 microdeletions
| Features | 5q14.3q15 multigenic microdeletionsa (n = 19) | MEF2C limited defects (n = 9) | All together (n = 28) |
|---|---|---|---|
| Severe intellectual disability with absent speech | 18 (95%) | 9 (100%) | 27 (96%) |
| MRI anomalies | 17 (89%) | 7 (77%) | 24 (86%) |
| Seizures | 17 (89%) | 7 (77%) | 24 (86%) |
| Hypotonia | 17 (89%) | 6 (66%) | 23 (85%) |
| Repetitive movements | 9 (47%) | 4 (44%) | 13 (46%) |
| Autistic features or poor eye contact | 8 (42%) | 4 (44%) | 12 (43%) |
| Strabism | 6 (32%) | 3 (33%) | 9 (32%) |
| Episodic hyperventilation | 0 | 2 (22%) | 2 (7%) |
| Broad/high or bulging forehead | 13 (68%) | 6 (66%) | 19 (68%) |
Two of these deletions described by Cardoso et al. [2009] do not involve MEF2C and it was not proven if they have a positional effect on MEF2C.
MRI was reported to show minor anomalies in 24 patients without any consistent pattern. Observed anomalies included delayed myelinization, enlarged ventricles, hypoplastic or thickened corpus callosum, periventricular heterotopia, simplified gyral pattern, polymicrogyria, colpocephaly, and periventricular leucomalacia.
Growth Parameters and Other Features
Height, weight and head circumference are commonly within the normal range, but are abnormal in both directions in some patients. While some patients showed relative or absolute macrocephaly, others had relative or absolute microcephaly. Although most patients show a variety of minor anomalies, no easily recognizable facial phenotype evolved. However, broad and/or high, bulging forehead, upslanting palpebral fissures, flat nasal root and bridge, small, upturned nose, hypotonic small mouth, large ears with prominent lobes, and small chin, as well as strabismus are the most consistent facial features reported. Unique, special anomalies were substernal fistula, jugular pit, bilateral club feet, and postaxial polydactyly of toes, all observed in deletions involving more than the MEF2C gene. While few patients had hypermetropia or myopia, hearing loss was not reported.
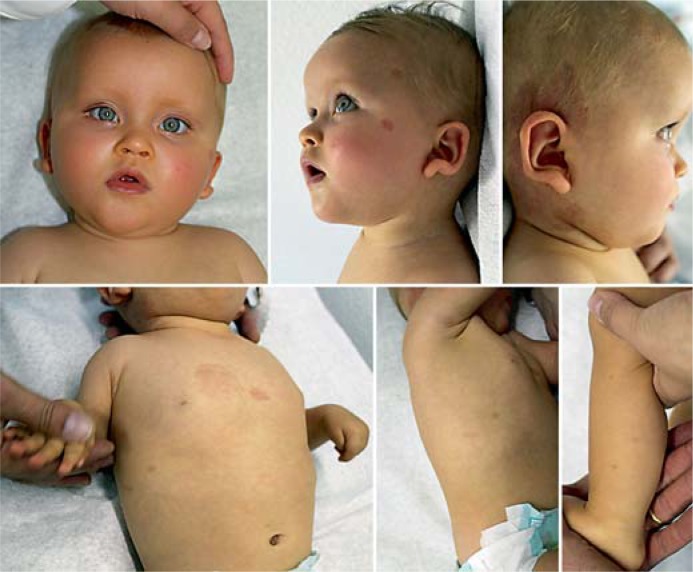
As pointed out by Carr et al. [2011] in patients with deletions including the RASA1 gene located proximally to MEF2C, capillary or arterio-venous malformations or fistulae (CMs, AVMs, AVFs) should be expected. The RASA1 associated autosomal dominant CM-AVM-syndrome is characterized by multiple pink-red, round, or oval CMs mostly localized on the face and limbs increasing in number with age (fig. 2) [Carr et al., 2011]. About 30% of affected individuals have associated AVMs and/or AVFs which are typically located in the head and neck region [Bayrak-Toydemir and Stevenson, 1993–2011; Boon et al., 2005]. These fast-flow vascular anomalies typically arise in the skin, muscle, bone, spine, and brain, and life-threatening complications may include bleeding, congestive heart failure, or neurologic symptoms which seem to occur early in life [Bayrak-Toydemir and Stevenson, 1993–2011]. Some patients have the clinical diagnosis of Parkes Weber syndrome (multiple micro-AVFs associated with a cutaneous capillary stain and excessive soft tissue and skeletal growth of an affected limb) [Bayrak-Toydemir and Stevenson, 1993–2011].
Fig. 2.
Clinical photographs of a previously unpublished patient with 15q14.3q15 microdeletion amongst others including the MEF2C and RASA1 genes, at the age of 14 months. Note the typical facial features with high, broad and bulging forehead, upslanting palpebral fissures, flat nasal root and bridge, small nose with anteverted nares, small mouth with downturned corners, mild retrognathia, large ears with up-lifted, prominent earlobes, as well as multiple round CMs of variable size.
Natural History and Disease Management
So far there is no evidence for specific internal or life-threatening complications associated with MEF2C defects. However, the oldest age of investigation in reported patients was 18 years only. Seizures seem to be benign or well controllable by standard treatment such as valproate in the majority of patients. For AVMs or AVFs, the individual risks and benefits of intervention have to be considered.
Genetics
MEF2C Gene and Protein Function
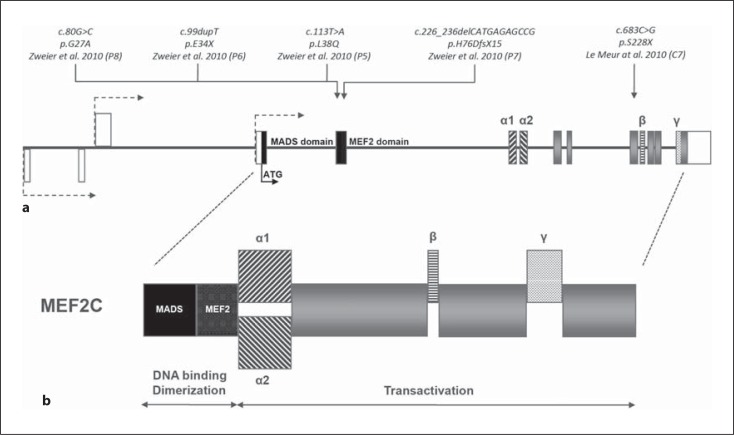
The MEF2C gene, first identified by Leifer et al. [1993], is located within the microdeletion syndrome region on chromosome 5q14.3. Three transcriptional start sites with variable 5′-untranslated regions are annotated and MEF2C contains up to 11 coding exons spanning approximately 100 kb of genomic DNA (fig. 3). Up to now, 6 transcript variants in humans are annotated (Ref Seq: NM_002397, NM_001131005, NM_001193347, NM_001193348, NM_001193349, NM_001193350). The longest of these variants encodes for a 483-amino-acid protein, the shortest one for 393 amino acids.
Fig. 3.
Genomic and protein structure of MEF2C and the respective location of all point mutations published so far. a Schematic representation of the MEF2C gene. The 3 annotated transcription start sites are shown as dashed arrows, the non-coding exons for the untranslated region are depicted as white boxes, and the start codon as a black arrow. The universal coding exons are shown as grey boxes, black-marked regions are coding for the MADS domain, dark grey regions for the MEF2 domain, and sequences coding for alternatively spliced elements α1, α2, β and γ are represented by differently dashed boxes. b Schematic representation of the protein structure of MEF2C and its alternatively spliced transcripts. MADS and MEF2 domains are required for DNA binding and dimerization, while the transactivation domains are required for transcriptional activation via protein-protein interactions. Alternative splicing occurs in the region immediately adjacent to the MEF2 domain (α1, α2; unclear function), in the transactivation region (β; inclusion is brain specific) [Leifer et al., 1993; Janson et al., 2001; Zweier et al., 2010] and within the last coding exons (γ; suggested transcriptional repression domain) [Janson et al., 2001].
Like all members of the MADS family (MCM1-agamous-deficiens-serum response factor) MEF2C, which belongs to the myocyte enhancer factor 2 (MEF2) subfamily, is characterized by a highly conserved N-terminal MADS box. Together with the directly adjacent subfamily-specific MEF2 domain motif it mediates dimerization, DNA binding, and cofactor interaction [Potthoff and Olson, 2007]. It is also known that MEF2 family members possess strong nuclear localization signals, for MEF2C it is reported in the N-terminal region [Janson et al., 2001]. The C-terminal region of MEF2 proteins contains the transcriptional activation domains, differs between family members, and is subject to complex patterns of alternative splicing (fig. 3). Whereas vertebrates have 4 MEF2 genes, MEF2A–D, yeast, Drosophila, and Caenorhabditis elegans possess only a single Mef2 gene. MEF2 factors bind to the A/T rich consensus DNA sequence YTA(A/T)4TAR as homo- and heterodimers. Their transcriptional activity relies on the recruitment of and cooperation with many other transcription factors, as well as on translational and posttranslational modifications [Potthoff and Olson, 2007]. For instance, phosphorylation of a highly conserved site enhances the DNA binding activity of MEF2C [Molkentin et al., 1996]. MEF2 proteins are known to act as central regulators of diverse developmental programs [Potthoff and Olson, 2007]. The 4 vertebrate MEF2 genes display overlapping, but distinct temporal and spatial expression patterns during embryonic development and in adult tissues with highest expression in striated muscles and brain [Edmondson et al., 1994].
Regarding MEF2C in particular, high expression levels were detected in skeletal muscle, cardiac muscle and brain in both humans and mice [Edmondson et al., 1994; Zweier et al., 2010]. It is reported to be highly expressed in the embryonic cerebral cortex, hippocampus, amygdala, midbrain, olfactory bulb, and cerebellum, as well as in the adult frontal cortex, dentate gyrus, hippocampus, thalamus, and cerebellum in the development of mouse CNS [Lyons et al., 1995]. Alternatively spliced MEF2C transcripts differ significantly in both expression pattern and transactivation functions, some of them shown to be brain-specific [Leifer et al., 1993; Janson et al., 2001; Zweier et al., 2010].
Previous studies revealed an important role of Mef2c in several differentiation and developmental processes like myogenesis, the development of the anterior heart field, neural crest and craniofacial development, chondrocyte hypertrophy and vascularization, endothelial cell proliferation and survival, lymphoid development, neurogenesis, and synaptic formation [Potthoff and Olson, 2007; Li et al., 2008a, b; Stehling-Sun et al., 2009]. Mef2c homozygous knockout in mice results in embryonic lethality due to cardiovascular defects even before brain development [Lin et al., 1997], resembling the indicated crucial role of MEF2C in developmental processes. However, in vivo analysis of the neuronal function of MEF2C using a conditional homozygous deletion of murine Mef2c in radial glial cells during late embryogenesis and expression of a superactive form of Mef2c in neurons indicated an essential role in hippocampus-dependent learning and memory by suppressing the number of excitatory synapses and thus regulating basal and evoked synaptic transmission [Barbosa et al., 2008]. Interestingly, Li et al. [2008a] reported that mice with conditional Mef2c knockout in neural progenitors have abnormal aggregation and compaction of neurons migrating into the lower layers of the neocortex during development. This manifested in smaller brain size with smaller, less mature neurons in adulthood, with resultant aberrant electrophysiology and severe behavioral anomalies resembling those seen in mouse models of Rett syndrome-like altered anxiety and paw-clasping [Li et al., 2008a]. Supported by a study showing that activated MEF2C drives the formation of neurons from murine stem cells [Li et al., 2008b], this work indicates the pivotal role of MEF2C in early neuronal differentiation and offers a phenotypic link to Rett syndrome.
The phenotype of murine models of Mef2c inactivation and the biological function in neuronal pathways supports the causal role of defects in MEF2C for the severe mental retardation phenotype observed in human patients with MEF2C haploinsufficiency. Of note, in the human phenotype the impairment seems to be restricted to its central nervous functions [Le Meur et al., 2010; Zweier et al., 2010]. Trying to explain the observed phenotypic overlap of patients with MEF2C mutations and atypical Rett syndrome and Pitt-Hopkins syndrome due to the involvement of a common pathway, Zweier et al. [2010] found diminished MECP2 and CDKL5 expression in vivo in blood of MEF2C-deficient patients. Supporting evidence was given by transcriptional reporter assays indicating that MEF2C truncating and missense mutations diminish synergistic transactivation of E-box promoters including that of MECP2 and CDKL5. These results are in line with other molecular findings indicating involvement of MECP2 and CDKL5 in a common pathway [Mari et al., 2005]. Of note, MECP2 binding to the murine Mef2c promoter as a repressor was shown by [Chahrour et al., 2008]. In contrast, consistently altered expression levels of TCF4, mutations which cause Pitt-Hopkins syndrome [Zweier et al., 2007], were not found in MEF2C-deficient patients. Therefore, a molecular link between MEF2C and TCF4 at this level was not obvious [Zweier et al., 2010].
MEF2 proteins like MEF2C are reported to cooperate with many different cofactors and promote gene expression of many different genes, some of which are responsible for intellectual disability themselves. There is evidence that fragile X mental retardation protein (FMRP) is required to enable MEF2 proteins to eliminate excitatory synapses in hippocampal neurons of mice [Pfeiffer et al., 2010].
Expression profiling in hippocampal neurons of rats indicated that MEF2 proteins also regulate several transcripts like Dia1, Pcdh10, and Ube3a in which defects are known to cause neurodevelopmental features such as intellectual disability, epilepsy and autism [Flavell et al., 2008; Morrow et al., 2008].
Inheritance and Genotype-Phenotype Correlation
All reported MEF2C mutations and microdeletions occurred de novo and have been heterozygous. Although no recurrence within siblings was observed so far, an approximately 1% recurrence risk is likely due to the possibility of parental germ line mosaicism. Similarity of the phenotype in microdeletions and point mutations indicates haploinsufficiency as the underlying genetic mechanism. Moreover, Zweier et al. [2010] demonstrated significantly diminished MEF2C expression levels in patients with microdeletions and truncating mutations. In contrast, MEF2C expression levels in patients with missense mutations were unaltered or significantly increased, but downstream effects on the expression levels of MECP2 and CDKL5 was equally in patients with missense or truncating mutations and deletions. Both reported missense mutations are located in the MADS domain and like truncating mutations abolish the transcriptional activity of MEF2C in vitro [Zweier et al., 2010].
Despite the contiguous gene deletion phenotype associated with the RASA1 gene deletion, no striking genotype-phenotype correlation evolved so far. However, patients with point mutations and smaller deletions seem to have a higher chance of walking skills and a lower risk of refractory seizures.
The only large scale mutational screening study identified MEF2C mutations in 1.1% of patients with moderate to severe intellectual disability of unknown cause and in about 2% of patients fitting into the spectrum of Rett syndrome-like disorders [Zweier et al., 2010]. Thus, MEF2C-related disorders represent one of the more common causes of intellectual disability.
Research towards Disease-Specific Therapeutic Approaches
So far no disease-specific therapeutic targets are known. However, due to the at least partially shared pathway with typical and atypical Rett syndromes, further elucidation of this pathway may eventually lead to a common therapeutic approach in this group of disorders.
References
- Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, et al. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci USA. 2008;105:9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayrak-Toydemir P, Stevenson D. RASA1-related disorders. In: Pagon RA, Bird TD, Dolan CR, Stephens K, editors. GeneReviews. Seattle: University of Washington; 1993–2011. [Google Scholar]
- Berland S, Houge G. Late-onset gain of skills and peculiar jugular pit in an 11-year-old girl with 5q14.3 microdeletion including MEF2C. Clin Dysmorphol. 2010;19:222–224. doi: 10.1097/MCD.0b013e32833dc589. [DOI] [PubMed] [Google Scholar]
- Boon LM, Mulliken JB, Vikkula M. RASA1: variable phenotype with capillary and arteriovenous malformations. Curr Opin Genet Dev. 2005;15:265–269. doi: 10.1016/j.gde.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Cardoso C, Boys A, Parrini E, Mignon-Ravix C, McMahon JM, et al. Periventricular heterotopia, mental retardation, and epilepsy associated with 5q14.3–q15 deletion. Neurology. 2009;72:784–792. doi: 10.1212/01.wnl.0000336339.08878.2d. [DOI] [PubMed] [Google Scholar]
- Carr CW, Zimmerman HH, Martin CL, Vikkula M, Byrd AC, Abdul-Rahman OA. 5q14.3 neurocutaneous syndrome: a novel continguous gene syndrome caused by simultaneous deletion of RASA1 and MEF2C. Am J Med Genet A. 2011;155A:1640–1645. doi: 10.1002/ajmg.a.34059. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Lyons GE, Martin JF, Olson EN. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994;120:1251–1263. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- Engels H, Wohlleber E, Zink A, Hoyer J, Ludwig KU, et al. A novel microdeletion syndrome involving 5q14.3–q15: clinical and molecular cytogenetic characterization of three patients. Eur J Hum Genet. 2009;17:1592–1599. doi: 10.1038/ejhg.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson CG, Chen Y, Li Y, Leifer D. Functional regulatory regions of human transcription factor MEF2C. Brain Res Mol Brain Res. 2001;97:70–82. doi: 10.1016/s0169-328x(01)00187-5. [DOI] [PubMed] [Google Scholar]
- Leifer D, Krainc D, Yu YT, McDermott J, Breitbart RE, et al. MEF2C, a MADS/MEF2-family transcription factor expressed in a laminar distribution in cerebral cortex. Proc Natl Acad Sci USA. 1993;90:1546–1550. doi: 10.1073/pnas.90.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Meur N, Holder-Espinasse M, Jaillard S, Goldenberg A, Joriot S, et al. MEF2C haploinsufficiency caused by either microdeletion of the 5q14.3 region or mutation is responsible for severe mental retardation with stereotypic movements, epilepsy and/or cerebral malformations. J Med Genet. 2010;47:22–29. doi: 10.1136/jmg.2009.069732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci USA. 2008a;105:9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, McKercher SR, Cui J, Nie Z, Soussou W, et al. Myocyte enhancer factor 2C as a neurogenic and antiapoptotic transcription factor in murine embryonic stem cells. J Neurosci. 2008b;28:6557–6568. doi: 10.1523/JNEUROSCI.0134-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons GE, Micales BK, Schwarz J, Martin JF, Olson EN. Expression of Mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J Neurosci. 1995;15:5727–5738. doi: 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari F, Azimonti S, Bertani I, Bolognese F, Colombo E, et al. CDKL5 belongs to the same molecular pathway of MeCP2 and it is responsible for the early-onset seizure variant of Rett syndrome. Hum Mol Genet. 2005;14:1935–1946. doi: 10.1093/hmg/ddi198. [DOI] [PubMed] [Google Scholar]
- Mikhail FM, Lose EJ, Robin NH, Descartes MD, Rutledge KD, et al. Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am J Med Genet A. 2011;155A:2386–2396. doi: 10.1002/ajmg.a.34177. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Li L, Olson EN. Phosphorylation of the MADS-box transcription factor MEF2C enhances its DNA binding activity. J Biol Chem. 1996;271:17199–17204. doi: 10.1074/jbc.271.29.17199. [DOI] [PubMed] [Google Scholar]
- Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novara F, Beri S, Giorda R, Ortibus E, Nageshappa S, et al. Refining the phenotype associated with MEF2C haploinsufficiency. Clin Genet. 2010;78:471–477. doi: 10.1111/j.1399-0004.2010.01413.x. [DOI] [PubMed] [Google Scholar]
- Nowakowska BA, Obersztyn E, Szymanska K, Bekiesinska-Figatowska M, et al. Severe mental retardation, seizures, and hypotonia due to deletions of MEF2C. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1042–1051. doi: 10.1002/ajmg.b.31071. [DOI] [PubMed] [Google Scholar]
- Pfeiffer BE, Zang T, Wilkerson JR, Taniguchi M, Maksimova MA, et al. Fragile X mental retardation protein is required for synapse elimination by the activity-dependent transcription factor MEF2. Neuron. 2010;66:191–197. doi: 10.1016/j.neuron.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Saitsu H, Igarashi N, Kato M, Okada I, Kosho T, et al. De novo 5q14.3 translocation 121.5-kb upstream of MEF2C in a patient with severe intellectual disability and early-onset epileptic encephalopathy. Am J Med Genet A. 2011;155A:2879–2884. doi: 10.1002/ajmg.a.34289. [DOI] [PubMed] [Google Scholar]
- Stehling-Sun S, Dade J, Nutt SL, DeKoter RP, Camargo FD. Regulation of lymphoid versus myeloid fate ‘choice' by the transcription factor Mef2c. Nat Immunol. 2009;10:289–296. doi: 10.1038/ni.1694. [DOI] [PubMed] [Google Scholar]
- Tonk V, Kyhm JH, Gibson CE, Wilson GN. Interstitial deletion 5q14.3q21.3 with MEF2C haploinsufficiency and mild phenotype: when more is less. Am J Med Genet A. 2011;155A:1437–1441. doi: 10.1002/ajmg.a.34012. [DOI] [PubMed] [Google Scholar]
- Zweier C, Peippo MM, Hoyer J, Sousa S, Bottani A, et al. Haploinsufficiency of TCF4 causes syndromal mental retardation with intermittent hyperventilation (Pitt-Hopkins syndrome) Am J Hum Genet. 2007;80:994–1001. doi: 10.1086/515583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweier M, Gregor A, Zweier C, Engels H, Sticht H, et al. Mutations in MEF2C from the 5q14.3q15 microdeletion syndrome region are a frequent cause of severe mental retardation and diminish MECP2 and CDKL5 expression. Hum Mutat. 2010;31:722–733. doi: 10.1002/humu.21253. [DOI] [PubMed] [Google Scholar]