Abstract
Background
Alcohol dehydrogenase 1C (ADH1C) is the key enzyme catalyze oxidation of alcohol to acetaldehyde, which plays vital roles in the etiology of various cancer. To date, studies investigated the association between a functional polymorphism in ADH1C, Ile350Val (rs698), and risk of cancer have shown inclusive results.
Methods
A meta-analysis based on 35 case-control studies was performed to address this issue. Odds ratios (OR) with 95% confidence intervals (CIs) were used to assess the association. The statistical heterogeneity across studies was examined with χ2-based Q-test.
Results
Overall, no significant associations between ADH1C Ile350Val polymorphism and cancer risk were observed in any genetic models (P>0.05). In the stratified analyses, there was a significantly increased cancer risk among African (Val/Val vs. Ile/Ile OR = 2.19, 95% CI = 1.29−3.73, P heterogeneity = 0.989; Ile/Val + Val/Val vs. Ile/Ile: OR = 1.79, 95%CI = 1.18−2.71, P heterogeneity = 0.761; Val/Val vs. Ile/Val + Ile/Ile: OR = 1.92, 95% CI = 1.16−3.17, P heterogeneity = 0.981) and Asian (Ile/Val vs. Ile/Ile: OR = 1.58, 95% CI = 1.32−1.90, P heterogeneity = 0.375; Val/Val vs. Ile/Ile: OR = 3.84, 95% CI = 1.74−8.49, P heterogeneity = 0.160; Ile/Val + Val/Val vs. Ile/Ile: OR = 1.65, 95% CI = 1.38−1.96, P heterogeneity = 0.330; Val/Val vs. Ile/Val + Ile/Ile: OR = 3.54, 95% CI = 1.62−7.75, P heterogeneity = 0.154) studies.
Conclusions
The results indicate that ADH1C Ile350Val polymorphism may contribute to cancer risk among Africans and Asians. Additional comprehensive system analyses are required to validate this association combined with other related polymorphisms.
Introduction
There has been convincing evidence that alcohol ingestion is carcinogenic to humans and causally related to liver, colorectal, breast and upper aerodigestive tract (UADT) cancers [1]. Although multiple mechanisms are involved in alcohol-mediated carcinogenesis, it has been shown that acetaldehyde (AA), the oxidative product of ethanol (commonly called alcohol), rather than alcohol itself is the principal carcinogenic material in alcohol metabolism [2]. AA interferes at many sites with DNA synthesis and repair and consequently has direct mutagenic and carcinogenic effects [3]. The key enzyme responsible for oxidation of ethanol to AA is alcohol dehydrogenase (ADH) [4]. Human ADH family is a well-defined system of enzymes which play important role in detoxification of alcohols and are categorized into several classes based on differences in substrate specificity, sensitivity to inhibitors, localization, electrophoretic migration and immunological properties [5]. In addition to the first-pass ethanol metabolism, ADH has shown various functions including activity towards hydroxysteroids, detoxification of endogenous and exogenous formaldehyde, retinoid transformation, etc. [6], [7], [8]. The differences of the activities of total ADH and ADH isoenzymes between cancer and healthy tissue have been demonstrated [4]. As production rate of AA is mainly modulated by ADH, it is rational that ADH activity variation may have effects on the level of AA in vivo and be one of the factors intensifying carcinogenesis.
There are seven genes that encode the seven known isozymes of human ADH. According to structural characteristics, the seven isozymes are categorized into five different classes, among which Class I isozymes account for most of the alcohol metabolism [9]. The three class I genes, ADH1A, ADH1B, and ADH1C (formerly known as ADH1, ADH2 and ADH3) are very closely related; they encode alpha (α), beta (β) and gamma (γ) subunits, respectively [10]. Functional variants (i.e., single nucleotide polymorphisms, SNPs) arousing wide concern exist in two of three genes encoding ADH enzymes (i.e., the ADH1B and ADH1C genes) [11]. The polymorphic sites for ADH1B are Arg48His in exon 3 (rs1229984) and Arg370Cys in exon 9 (rs2066702) and for ADH1C are Arg272Gln (rs1693482) and Ile350Val (rs698) [10]. The ADH1B*1 allele is a name for the reference allele encoding β1 subunit which has arginine (Arg) at positions 48 and 370. ADH1B*2 (β2) refers to a variant allele defined by histidine (His) at position 48 while ADH1B*3 encoding β3 subunit that has cysteine (Cys) at position 370 [10]. For polymorphisms in ADH1C, 272Arg and 350Ile carriers have the ADH1C*1 allele, whereas 272 Gln and 350 Val carriers have the ADH1C*2 allele [12]. It is worth noting that significant linkage disequilibrium has been detected between the ADH1B and ADH1C polymorphisms as well as the two variants in ADH1C [13], [14]. These functional variants result in the production of enzymes with different kinetic properties [10], [15] and subsequently the generation of different quantities of AA. For example, individuals with ADH1C*1 allele have an ethanol oxidizing capacity 2.5-times higher when compared to ADH1C*2 allele [12]. Thus, not only the amount of alcohol is determinant for organ injury, but also the genetic factors may modulate and determine carcinogenesis.
An increasing number of studies have investigated the association between ADH polymorphisms and cancer risk in human. Among them, studies of ADH1C Ile350Val variant accounted for more than others. Most of the ADH1C studies focused on head and neck cancer (HNC) development, and to a less extent on the cancers of breast, colorectum, etc. Although genotype frequency of Ile350Val polymorphism varies among different populations [16], evidences supporting the association between this genetic variant and risk of cancer have arisen from studies of different ethnic background [17], [18], [19]. Recently, Chang et al. conducted a meta-analysis to assess the association between ADH1B and ADH1C polymorphisms and risk of HNC [20], and they found a reduced risk for HNC associated with ADH1B*2 and ADH1C*1 alleles. However, as the studies on ADH1C polymorphism and different cancer risk have shown contradictory and inconclusive results, a pooled analysis of all studies on ADH1C and cancer risk is needed.
Here, we performed a meta-analysis on 35 eligible case-control studies to estimate the overall cancer risk and ADH1C polymorphisms. Because polymorphisms of Arg272Gln and Ile350Val were in strong linkage disequilibrium and both of them can be used to distinguish ADH1C*1 and ADH1C*2 alleles, we focused on the most commonly studied polymorphism Ile350Val.
Materials and Methods
Identification and Eligibility of Relevant Studies
PubMed and EMBASE were searched for all relevant reports (the last search update was July 18, 2011), using the search terms “ADH1C” or “ADH3”, “polymorphism” and “cancer”. The search was limited to English language papers. In addition, studies were identified by a manual search of the references of original studies. Of the articles with the overlapping data, we only selected the publication with the most extensive information. For inclusion in the meta-analysis, the identified articles had to meet the following criteria: (a) there were information on the evaluation of the ADH1C Ile350Val polymorphism and cancer risk, (b) used a case–control design, and (c) contained complete information about all genotype frequency. The exclusion criteria were as follows: (a) not for cancer research, (b) review articles, (c) reports without usable data and (d) duplicate publications.
Data Extraction
Two authors (Y Xue and M Wang) extracted data from all eligible publications independently and reached a consensus on all the items. For each study, the following characteristics were considered: the first author’s last name, year of publication, country of origin, ethnicity, cancer type, source of control groups (population- or hospital-based controls) and numbers of genotyped cases and controls. Different ethnic descents were categorized as African, Asian, European, or Mixed (composed of different ethnic groups). Cancers of oral cavity, oropharynx, hypopharynx, larynx, esophagus and stomach were defined as upper aerodigestive tract (UADT) cancers [21], [22]. For studies including subjects of different ethnic groups or cancer types, data were extracted separately for each ethnic group or cancer type whenever possible.
Statistical Analysis
The strength of the association between the ADH1C Ile350Val polymorphisms and cancer risk was measured by odds ratios (ORs) with their 95% confidence intervals (CIs). The statistical significance of the summary OR was determined with the Z-test. We first explored the risks of the Ile/Val and Val/Val genotypes on cancer, compared with the wild-type Ile/Ile homozygote, and then evaluated the risks of Ile/Val + Val/Val versus Ile/Ile and Val/Val versus Ile/Val + Ile/Ile on cancer, assuming dominant and recessive effects of the variant Val allele, respectively. Stratified analyses were also performed by cancer types (if one cancer type contained less than three individual studies, it was classified as other cancers group), ethnicity, source of controls and sample size (subjects >500 in both case and control groups or not).
In consideration of the possibility of heterogeneity across the studies, a statistical test for heterogeneity was performed by a χ2-based Q-test. A P-value greater than 0.10 for the Q-test indicated lack of heterogeneity among the studies, and then the fixed-effects model (the Mantel–Haenszel method) was used to calculate the summary OR estimate of each study. Otherwise, the random-effects model (DerSimonian and Laird method) was used. Sensitivity analyses were performed to assess the stability of the results, namely, a single study in the meta-analysis was deleted each time to reflect the influence of the individual data set to the pooled OR. The presence of publication bias indicates that non-significant or negative findings remain unpublished. We used Funnel plots and Egger’s linear regression test to provide diagnosis of the potential publication bias. All statistical analyses were performed with the Stata software (version 8.2; StataCorp LP, College Station, TX, USA), using two-sided P-values.
Results
Characteristics of Studies
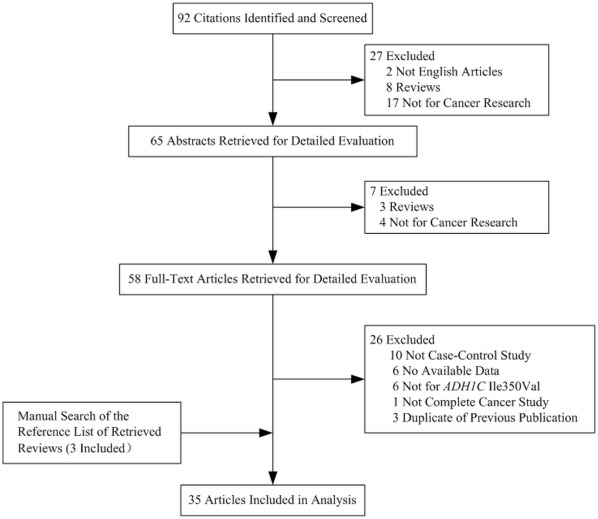
There were 35 studies retrieved on the basis of the search criteria for cancer susceptibility associated with ADH1C Ile350Val polymorphisms (Fig. 1). Totally, 19,154 cases and 26,519 controls were included in the meta-analysis. Study characteristics are summarized in Table 1. Among the 35 case–control studies, there were 5 studies of Asians, 19 studies of Europeans and 8 studies of mixed descendents. Besides, 3 studies included more than one ethnic group [14], [23], [24]. Thus, in total, 2 African groups, 5 Asian groups, 21 European groups and 10 groups of mixed descendents were recruited in our analyses. Controls were mainly matched on sex and age, of which 14 were population based [21], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], 16 were hospital based [17], [18], [19], [23], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48] and 5 studies was conducted on both population-based and hospital-based control group [14], [22], [49], [50], [51]. Furthermore, 7 studies were conducted with subjects >500 in both case and control groups [14], [26], [28], [29], [34], [38], [51]. There were 4 studies of breast cancer, 25 of UADT cancer, 3 of colorectal and 3 of other cancers. Among the 25 UADT cancer studies, Homann et al. investigated the ADH1C polymorphism and cancer risk in both UADT cancer and hepatocellular cancer groups [50]. Thus, number of studies of “other cancers” was 4. Cancers were confirmed histologically or pathologically in most studies. The distribution of genotypes in the controls of all studies was consistent with Hardy–Weinberg equilibrium (HWE) except for 10 studies, 9 of which did not mention the HWE test [24], [31], [33], [36], [37], [41], [44], [48], [49] and in one study allele distributions were not in HWE for a part of controls [43].
Figure 1. Studies identified with criteria for inclusion and exclusion.
Table 1. Characteristics of studies included in the meta-analysis.
| First author | Year | Country | Ethnicity | Cancer types | Source of controls | Sample size | |
| Case | Control | ||||||
| Coutelle | 1997 | France | European | UADT | HB | 39 | 37 |
| Harty | 1997 | Puerto Rico | Mixed | UADT | PB | 146 | 146 |
| Freudenheim | 1999 | USA | European | Breast | PB | 315 | 356 |
| Bouchardy | 2000 | France | European | UADT | HB | 244 | 167 |
| Chao | 2000 | China | Asian | UADT | HB | 88 | 434 |
| Olshan | 2001 | USA | European and African | UADT | HB | 173 | 194 |
| Sturgis | 2001 | USA | European | UADT | HB | 229 | 575 |
| Schwartz | 2001 | USA | European | UADT | PB | 333 | 541 |
| Dijk | 2001 | Netherlands | European | Bladder | HB | 115 | 131 |
| Zavras | 2002 | Greece | European | UADT | HB | 93 | 99 |
| Yokoyama | 2002 | Japan | Asian | UADT | PB | 234 | 634 |
| Freudenheim | 2003 | USA | Mixed | Lung | PB | 113 | 212 |
| Nishimoto | 2004 | Brazil | Mixed | UADT | Combined | 141 | 134 |
| Coutelle | 2004 | German | European | Breast | HB | 117 | 111 |
| Peters | 2005 | USA | European | UADT | PB | 521 | 599 |
| Wang | 2005 | USA | European | UADT | HB | 348 | 330 |
| Homann | 2006. | German | European | UADT and Hepatocellular | Combined | 293 | 729 |
| Logt | 2006 | Netherlands | European | Colorectal | PB | 320 | 385 |
| Terry | 2006 | USA | Mixed | Breast | PB | 1047 | 1101 |
| Terry | 2007 | USA | Mixed | UADT | PB | 197 | 160 |
| Zhang | 2007 | Poland | European | UADT | PB | 297 | 425 |
| Yin | 2007 | Japan | Asian | Colorectal | PB | 685 | 777 |
| Visvanathan | 2007 | USA | European | Breast | PB | 303 | 312 |
| Asakage | 2007 | Japan | Asian | UADT | HB | 96 | 642 |
| Curtin | 2007 | USA | Mixed | Colorectal | PB | 915 | 1969 |
| Solomon | 2008 | India | Mixed | UADT | HB | 126 | 100 |
| Hashibe | 2008 | Multi-Countries | European and Mixed | UADT | Combined | 3393 | 4851 |
| Li | 2008 | South Africa | African and Mixed | UADT | PB | 237 | 268 |
| Oze | 2009 | Japan | Asian | UADT | HB | 585 | 1170 |
| Garcia | 2010 | Brazil | Mixed | UADT | HB | 207 | 244 |
| Duchonova | 2010 | Czech | European | Pancreatic | HB | 235 | 264 |
| Kortunay | 2010 | Turkey | European | UADT | HB | 50 | 100 |
| Soucek | 2010 | Slav | European | UADT | HB | 121 | 121 |
| Brocic | 2011 | Serbia | European | UADT | Combined | 123 | 177 |
| Mckay | 2011 | Multi-Countries | European | UADT | Combined | 6675 | 8024 |
UADT, upper aerodigestive tract; HB, hospital based; PB, population based; Combined, studies conducted on both population-based and hospital-based control group.
Quantitative Synthesis
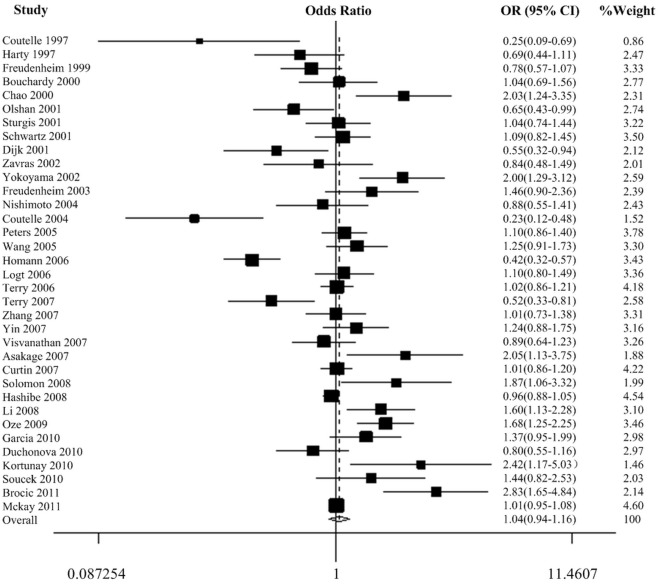
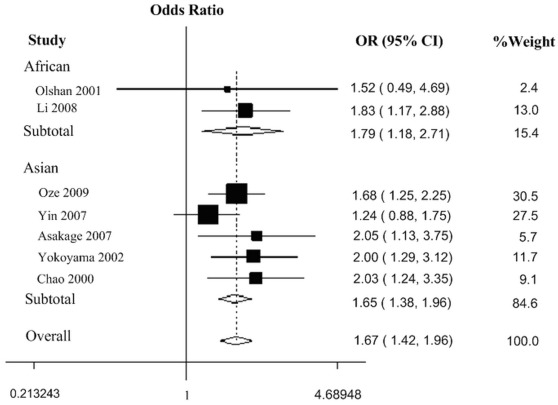
There was a wide variation of the 350 Val allele frequency among the controls across different ethnicities. The 350 Val allele frequency was the lowest in Asian populations and was the highest in European populations (0.05, 95% CI = 0.03−0.09, vs. 0.40, 95% CI = 0.36−0.44). In African and mixed populations, the allele frequency was 0.29 (95% CI = −1.15−1.58) and 0.35 (95% CI = 0.27−0.39), respectively. The difference among the four population groups was statistically significant (P<0.001). In the overall analyses, we did not observe any significant associations between the ADH1C Ile350Val polymorphism and cancer risk in all the genetic models (Table 2, Fig. 2 of dominant model). Because population admixture may be a potential cause of inconsistent results [52], we excluded studies with mixed populations to further evaluate the overall effect of ADH1C Ile350Val polymorphism and we still didn’t find any significant associations (data not shown). However, in the stratified analysis by ethnicity, significant increased risks were found for African populations (homozygote comparison: OR = 2.19, 95% CI = 1.29−3.73, P heterogeneity = 0.989; dominant model: OR = 1.79, 95% CI = 1.18−2.71, P heterogeneity = 0.761; recessive model: OR = 1.92, 95% CI = 1.16−3.17, P heterogeneity = 0.981) and Asian populations (heterozygote comparison: OR = 1.58, 95% CI = 1.32−1.90, P heterogeneity = 0.375; homozygote comparison: OR = 3.84, 95% CI = 1.74−8.49, P heterogeneity = 0.160; dominant model: OR = 1.65, 95% CI = 1.38−1.96, P heterogeneity = 0.330; recessive model: OR = 3.54, 95% CI = 1.62−7.75, P heterogeneity = 0.154) (Table 2, Fig. 3 of dominant model ). When we performed stratified analyses by cancer type, we found individuals with the Val/Val genotypes had a 0.58-fold lower breast cancer risk compared with the Ile/Ile genotype (OR = 0.58, 95% CI = 0.34−1.00, P heterogeneity = 0.001, Table 2) (P = 0.049, data not shown). We did not observe any significant associations among UADT cancer, colorectal cancer and other cancers (Table 2). However, cancer-specific analysis excluding studies with mixed populations indicated that a moderate increased risk of UADT cancer was associated with variant Val allele in dominant model (OR = 1.17, 95% CI = 1.01−1.36, P heterogeneity <0.001, data not shown). Furthermore, when we conducted stratified analyses according to source of controls and sample size, no significant associations were found in any genetic models (Table 2).
Table 2. Stratification analyses of the ADH1C Ile350Val polymorphism on cancer.
| Variables | na | Sample size (case/control) | Ile/Val vs. Ile/Ile | Val/Val vs. Ile/Ile | Ile/Val + Val/Val vs. Ile/Ile (dominant) | Val/Val vs. Ile/Val + Ile/Ile (recessive) | ||||
| OR (95% CI) | P b | OR (95% CI) | P b | OR (95% CI) | P b | OR (95% CI) | P b | |||
| Total | 35 | 19154/26519 | 1.02 (0.92−1.14) | <0.001 | 1.01 (0.87−1.16) | <0.001 | 1.04 (0.94−1.16) | <0.001 | 1.03 (0.93−1.15) | <0.001 |
| Cancer Type | ||||||||||
| Breast | 4 | 1782/1880 | 0.78 (0.54−1.12) | 0.003 | 0.58 (0.34−1.00) | 0.001 | 0.73 (0.50−1.07) | 0.001 | 0.72 (0.51−1.01) | 0.048 |
| UADT | 25 | 14903/20901 | 1.09 (0.95−1.24) | <0.001 | 1.10 (0.93−1.31) | <0.001 | 1.12 (0.98−1.28) | <0.001 | 1.09 (0.96−1.24) | 0.004 |
| Colorectal | 3 | 1920/3131 | 1.05 (0.92−1.21)c | 0.537 | 1.07 (0.87−1.31)c | 0.504 | 1.06 (0.93−1.21)c | 0.566 | 1.06 (0.88−1.27)c | 0.445 |
| Other | 4 | 549/1336 | 0.72 (0.44−1.17) | 0.008 | 0.76 (0.42−1.39) | 0.018 | 0.73 (0.44−1.20) | 0.003 | 0.93 (0.71−1.24)c | 0.199 |
| Race* | ||||||||||
| African | 2 | 204/198 | 1.47 (0.89−2.44)c | 0.867 | 2.19 (1.29−3.73) c | 0.989 | 1.79 (1.18−2.71) c | 0.761 | 1.92 (1.16−3.17) c | 0.981 |
| Asian | 5 | 1688/3657 | 1.58 (1.32−1.90) c | 0.375 | 3.84 (1.74−8.49) c | 0.160 | 1.65 (1.38−1.96) c | 0.330 | 3.54 (1.62−7.75) c | 0.154 |
| European | 21 | 12964/17476 | 0.93 (0.81−1.06) | <0.001 | 0.98 (0.82−1.17) | <0.001 | 0.94 (0.82−1.08) | <0.001 | 1.03 (0.91−1.17) | 0.002 |
| Mixed | 10 | 4294/5186 | 1.04 (0.90−1.20) | 0.052 | 1.05 (0.83−1.34) | 0.013 | 1.05 (0.89−1.23) | 0.006 | 1.02 (0.90−1.150) c | 0.110 |
| Source of controls | ||||||||||
| PB | 14 | 5663/7885 | 1.05 (0.93−1.18) | 0.017 | 0.98 (0.83−1.17) | 0.019 | 1.05 (0.92−1.19) | 0.001 | 0.97 (0.88−1.08)c | 0.116 |
| HB | 16 | 2866/4719 | 1.01 (0.79−1.29) | <0.001 | 1.16 (0.79−1.69) | <0.001 | 1.06 (0.82−1.37) | <0.001 | 1.24 (0.93−1.66) | 0.002 |
| Combined | 5 | 10625/13915 | 0.95 (0.74−1.21) | <0.001 | 0.89 (0.65−1.21) | <0.001 | 0.93 (0.73−1.20) | <0.001 | 0.96 (0.82−1.12) | 0.049 |
| Sample Sized | ||||||||||
| <500 | 28 | 5333/8028 | 1.00 (0.83−1.19) | <0.001 | 1.00 (0.79−1.27) | <0.001 | 1.02 (0.85−1.22) | <0.001 | 1.05 (0.89−1.23) | 0.001 |
| >500 | 7 | 13821/18491 | 1.04 (0.96−1.13) | 0.094 | 1.01 (0.89−1.15) | 0.069 | 1.06 (0.97−1.16) | 0.026 | 1.01 (0.89−1.14) | 0.049 |
: Number of studies.
P b: The value of heterogeneity test.
: Fix-effects model was used when P value for heterogeneity test >0.10; otherwise, random-effects model was used.
: Stratified according to subjects >500 in both case and control groups or not.
Combined, studies conducted on both population-based and hospital-based control group.
The sum of sample size of each race group was less than total sample size because in Olshan’s study, the sum of sample size of each race group was less than its total sample size.
Figure 2. Forest plot of cancer risk associated with the ADH1C Ile350Val polymorphism (dominant model).
The squares and horizontal lines correspond to the study-specific OR and 95% CI. The area of the squares reflects the weight (inverse of the variance). The diamond represents the summary OR and 95% CI.
Figure 3. Forest plot of cancer risk associated with the ADH1C Ile350Val polymorphism in African and Asian populations (dominant model).
The squares and horizontal lines correspond to the study-specific OR and 95% CI. The area of the squares reflects the weight (inverse of the variance). The diamond represents the summary OR and 95% CI.
Test for Heterogeneity
There was significant heterogeneity for heterozygote comparison (Ile/Val versus Ile/Ile: P heterogeneity <0.001), homozygote comparison (Val/Val versus Ile/Ile: P heterogeneity <0.001), dominant model comparison (Val/Val + Ile/Val versus Ile/Ile: P heterogeneity <0.001) and recessive model comparison (Val/Val versus Ile/Val + Ile/Ile: P heterogeneity <0.001). Then, we assessed the source of heterogeneity for heterozygote comparison (Ile/Val versus Ile/Ile) by ethnicity, cancer type, source of controls and sample size. As a result, ethnicity (χ2 = 28.01, df = 3, P<0.001) and cancer type (χ2 = 8.39, df = 3, P = 0.039) but not source of controls (χ2 = 3.54, df = 2, P = 0.171) or sample size (χ2 = 0.52, df = 1, P = 0.470) were found to contribute to substantial heterogeneity.
Sensitivity Analyses
Sensitivity analyses indicated that two independent studies by Hashibe et al. in 2008 and Homann et al. in 2006 were the main origin of heterogeneity [14], [50]. In addition, no other single study influenced the pooled OR qualitatively, as indicated by sensitivity analyses, suggesting that the results of this meta-analysis are stable.
Furthermore, when we performed cancer-specific and population-specific sensitivity analyses we found studies conducted by Terry et al. in 2006 [34], Hashibe et al. in 2008 [14], Homann et al. in 2006 [50] and Terry et al. in 2007 [21] were the main origin of heterogeneity in subgroup of breast cancer, UADT cancer, European population and mixed population, respectively. Moreover, no single study influenced the pooled OR in each subgroup, which indicated that results of stratified analyses were also stable.
Publication Bias
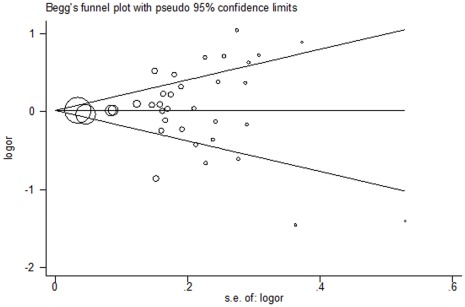
Begg’s funnel plot and Egger’s test were performed to evaluate the publication bias of literatures. As shown in Fig. 4, the shape of the funnel plots seemed symmetrical in the dominant model comparison. Then, the Egger’s test was adopted to provide statistical evidence of funnel plot symmetry. The results still did not show any evidence of publication bias (t = 0.42, P = 0.674 for Val/Val + Ile/Val versus Ile/Ile).
Figure 4. Begg’s funnel plot for publication bias test (dominant model).
Each point represents a separate study for the indicated association. Log[or], natural logarithm of OR. Horizontal line, mean effect size.
Discussion
Genetic variation in carcinogen metabolism pathway may exert influence on the risk of exposure-related cancer [9]. Based on the vital role of ADH1C in ethanol oxidation to AA, numerous studies have investigated the association of the functional ADH1C polymorphism with types of cancers. Several studies have observed significant association between ADH1C*1 allele and cancer risk [21], [24], [38], [41], [43]–[45], [48]–[50]. It has been widely known that ADH1C*1 allele encode isozymes with higher catalytic activity than the one encoded by ADH1C*2 allele and result in more production of AA, which is a major part in ethanol-related carcinogenesis [3] and alcoholism [53].
However, in view of the linkage disequilibrium between ADH1B and ADH1C and the fact that the kinetic differences among ADH1B isozymes are much more striking than those among the ADH1C isozymes [54], some studies ascribed the association between ADH1C polymorphism and cancer risk to the reflects of effect of ADH1B polymorphism, especially in East Asian [18], [32]. But a matter of particular note was the significant difference of allele frequency between ADH1B and ADH1C polymorphism, that is, the minor allele frequency (MAF) for ADH1B was 0–0.025 in European (ADH1B*2 allele)and 0.223–0.261 in Asian (ADH1B*1 allele) populations while the MAF for ADH1C was 0.473–0.483 in European (ADH1C*2 allele) and 0.023–0.081 in Asian (ADH1C*2 allele) populations (data from www. ncbi. nlm. nih. gov/projects/SNP/snp_ref. cgi), which made the general attribution of ADH1C effect to its linkage with ADH1B not reasonable. In other words, as the two more-active alleles, ADH1B*2 and ADH1C*1 were linked and the frequency of ADH1B*2 was too low in European, the explanation of ADH1C effect was not well founded, especially in European. Furthermore, there were also some studies suggesting that the influence of ADH1C polymorphism on cancer risk was independent of that of ADH1B in both European and Asian populations [14], [20], [55], [56]. Thus, the detail mechanism underlying ADH1C polymorphism and cancer risk remains controversial and the hypothesis that the variant of ADH1C exert an independent influence on cancer risk by changing ethanol oxidizing capacity [10] was better founded.
Although many studies have investigated the association between the ADH1C polymorphism and cancer risk, the results were inconsistent. In order to resolve this conflict, we conducted a meta-analysis of 35 case-control studies. Because data could be confounded by the differences between subgroups, we subsequently conducted stratified analysis by cancer type, ethnicity, source of controls and sample size. Moreover, as Deng et al. suggested in 2001 that population admixture may potentially elevate type I error rate of association studies and lead to inconsistent results [52], we also conducted overall and cancer-specific analyses excluding studies with mixed populations to confirm the effect of this polymorphism and the impact of mixed populations.
Generally speaking, we did not find any association between Ile350Val polymorphism and overall cancer risk. This result indicated that individuals with the ADH1C genotype leading to more exposure to acetaldehyde from alcohol were not at statistically different risk of cancers. When we further performed analyses excluding mixed populations, there were still no associations between this polymorphism and overall cancer risk. To a certain extent, analyses excluding mixed populations confirmed the negative result of initial overall analyses.
In the analysis stratified by cancer type, we still did not find any significant associations among studies of breast cancer, UADT cancer, colorectal cancer and other cancers in any genetic model. However, a similar meta-analysis reported recently had shown that ADH1C*1 allele was associated with a significantly decreased risk of pharynx cancer in dominant model [20]. Probably the discrepancy arose because they collect data of either Arg272Gln or Ile350Val polymorphism which were in perfect linkage disequilibrium but may have minor differences of genotype distribution [14], [25], [27] and relatively small number of studies (22 studies) they included. Interestingly, we found the effect of variant 350 Val allele on breast and other cancer risk was contrary to that on UADT and colorectal cancer, although all the effects were not significant. As heterogeneity among different cancers may interfere the authenticity of result in “other cancers”, the inverse result of breast cancer studies called more attention. A possible explanation is that carcinogenesis involved in different cancers is extremely diverse. Thus, specific role of ADH1C in carcinogenic mechanisms of breast cancer [27], [57] as well as interaction between special risk factors of breast cancer [58] and ADH1C gene may contribute to the inconsistent results.
Furthermore, when we performed cancer-specific analyses excluding studies with mixed populations, major results were nearly the same except that a moderate increased UADT cancer risk was founded in individuals carrying Val allele (i.e. dominant model). However, as the lower limit of the 95% CI was 1.01 in that comparison and the removing of some studies with relatively large sample size (although they were with mixed populations) may also decrease the reliability of result, we didn’t think this result was sufficient to support the risk effect of Val allele in UADT cancer. Therefore, studies with more samples randomly selected from one homogeneous population are needed to further determine the association between this variant and specific cancer risk.
Subsequently, we found an increased risk of cancer in variant homozygote (Val/Val) carriers among Africans and in variant allele (350 Val) carriers among Asians. Studies have indicated that 350 Val allele increases the risk for alcoholism [59], which may lead to accumulated exposure to the highly toxic and carcinogenic material, AA [4]. Thus, it is plausible that the presence of 350 Val allele puts one at a greater cancer risk through susceptibility to alcoholism. Although a few studies of Europeans suggested this variation might be significantly associated with risk of cancer [21], [41], [43], [44], [45], [49], [50], the overall difference was not significant. We presume that the difference among ethnic groups might be a reflection of different genetic backgrounds and environmental context. As a number of studies attributed the effect of ADH1C variant in East Asian to its linkage disequilibrium with ADH1B, it would be better for us to adjust the association found in Asian for ADH1B polymorphism. However, among the five studies conducted in Asian populations, only one [18] provided detailed data of ADH1C genotype adjusted for ADH1B genotype. Thus, the independent effect of ADH1C polymorphism in Asians could not be directly estimated in the present analysis, which to some extent was a flaw. In addition, other factors such as relatively small sample size (204 VS. 198 of African studies and 1688 VS. 3657 of Asian studies), selection bias and different matching criteria may also be a possible explanation to this result.
Although hospital-based studies may have inherent selection biases, we did not find any positive result in the stratified analysis by population-based and hospital-based controls, indicating that the different source of controls did not influence the association. In addition, because studies with small sample size may have insufficient statistical power or may have generated a fluctuated risk estimate, we performed stratified analyses according to subjects more than 500 in both case and control groups or not and no significant association was detected. These results suggested that there was no substantial impact of study sample size on this meta-analysis.
Because identification of the source of heterogeneity was very important in a meta-analysis, we subsequently detected source of heterogeneity by stratifying studies according to ethnicity, cancer type, source of control and sample size. Results showed the sources of heterogeneity were from ethnicity and cancer type, suggesting that certain effects of genetic variant were population and cancer specific.
Our meta-analysis had some advantages. First, substantial number of cases and controls were pooled from different studies, which significantly increased statistical power of the analysis. Second, studies included in our present meta-analysis strictly met our selection criteria. Third, we did not detect any publication bias indicating that the whole pooled result may be unbiased.
Except for the lacking of evaluation of independent effect of ADH1C adjusted for ADH1B in Asian, we also had a limitation of the present study. It has been identified that after generated from oxidization of alcohol by ADH enzymes, AA was further oxidized to acetate by aldehyde dehydrogenase (ALDH) enzymes and ALDH2 contributed most to the process [60]. Thus, besides ADH, activity of ALDH2 can also exert impact on accumulation of AA. A functional polymorphism in ALDH2 has been identified (rs671) to be associated with cancer risk [61], [62], which lead to different activity of ALDH2 enzyme and is prevalent in Asians [60]. Although polymorphisms of ALDH2 were not in linkage disequilibrium with ADH, it might influence the effect of polymorphisms of ADH1C through its impact on AA elimination. Therefore, ALDH2 polymorphism was a potential confounder of the present study, especially of Asian studies. Independent and combined effect of ADH1B, ADH1C and ALDH2 variants should be evaluated in further meta-analyses.
In conclusion, our results suggested that the ADH1C Ile350Val polymorphism is not a candidate for susceptibility to overall cancers. However, an increased cancer risk was observed in populations among African and Asian, but not in European and mixed race, which may be a reflection of ethnic differences. Additional larger studies assessing gene-gene and gene-environment interactions should be performed to further clarify the association of ADH genetic variants and cancer risk.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was partly supported by National Natural Science Foundation of China (30972444 and 81102089), the Key Program of Natural Science Foundation of Jiangsu Province (BK2010080), National Natural Science Foundation of Jiangsu Province (BK2011773 and BK2011775), the Key Program for Basic Research of Jiangsu Provincial Department of Education (11KJB330002), the Qin Lan Project of Jiangsu Provincial Department of Education, and the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.IARC. Alcohol drinking. Epidemiological studies of cancer in humans. IARC Monogr Eval Carcinog Risks Hum. 1988;44:153–250. [PMC free article] [PubMed] [Google Scholar]
- 2.Seitz HK, Matsuzaki S, Yokoyama A, Homann N, Vakevainen S, et al. Alcohol and cancer. Alcohol Clin Exp Res. 2001;25:137S–143S. doi: 10.1097/00000374-200105051-00024. [DOI] [PubMed] [Google Scholar]
- 3.Seitz HK, Meier P. The role of acetaldehyde in upper digestive tract cancer in alcoholics. Transl Res. 2007;149:293–297. doi: 10.1016/j.trsl.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Jelski W, Szmitkowski M. Alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) in the cancer diseases. Clin Chim Acta. 2008;395:1–5. doi: 10.1016/j.cca.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Holmes RS. Alcohol dehydrogenases: a family of isozymes with differential functions. Alcohol. 1994;Alcohol(Suppl 2):127–130. [PubMed] [Google Scholar]
- 6.Staab CA, Hellgren M, Hoog JO. Medium- and short-chain dehydrogenase/reductase gene and protein families : Dual functions of alcohol dehydrogenase 3: implications with focus on formaldehyde dehydrogenase and S-nitrosoglutathione reductase activities. Cell Mol Life Sci. 2008;65:3950–3960. doi: 10.1007/s00018-008-8592-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoog JO, Hedberg JJ, Stromberg P, Svensson S. Mammalian alcohol dehydrogenase - functional and structural implications. J Biomed Sci. 2001;8:71–76. doi: 10.1007/BF02255973. [DOI] [PubMed] [Google Scholar]
- 8.Chou CF, Lai CL, Chang YC, Duester G, Yin SJ. Kinetic mechanism of human class IV alcohol dehydrogenase functioning as retinol dehydrogenase. J Biol Chem. 2002;277:25209–25216. doi: 10.1074/jbc.M201947200. [DOI] [PubMed] [Google Scholar]
- 9.Seitz HK, Stickel F. Genes Nutr; 2009. Acetaldehyde as an underestimated risk factor for cancer development: role of genetics in ethanol metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- 11.Seitz HK, Becker P. Alcohol metabolism and cancer risk. Alcohol Res Health 30: 38–41, 2007;44–37 [PMC free article] [PubMed] [Google Scholar]
- 12.Hoog JO, Heden LO, Larsson K, Jornvall H, von Bahr-Lindstrom H. The gamma 1 and gamma 2 subunits of human liver alcohol dehydrogenase. cDNA structures, two amino acid replacements, and compatibility with changes in the enzymatic properties. Eur J Biochem. 1986;159:215–218. doi: 10.1111/j.1432-1033.1986.tb09855.x. [DOI] [PubMed] [Google Scholar]
- 13.Yokoyama A, Tsutsumi E, Imazeki H, Suwa Y, Nakamura C, et al. Polymorphisms of alcohol dehydrogenase-1B and aldehyde dehydrogenase-2 and the blood and salivary ethanol and acetaldehyde concentrations of Japanese alcoholic men. Alcohol Clin Exp Res. 2010;34:1246–1256. doi: 10.1111/j.1530-0277.2010.01202.x. [DOI] [PubMed] [Google Scholar]
- 14.Hashibe M, McKay JD, Curado MP, Oliveira JC, Koifman S, et al. Multiple ADH genes are associated with upper aerodigestive cancers. Nat Genet. 2008;40:707–709. doi: 10.1038/ng.151. [DOI] [PubMed] [Google Scholar]
- 15.Tiemersma EW, Wark PA, Ocke MC, Bunschoten A, Otten MH, et al. Alcohol consumption, alcohol dehydrogenase 3 polymorphism, and colorectal adenomas. Cancer Epidemiol Biomarkers Prev. 2003;12:419–425. [PubMed] [Google Scholar]
- 16.Brennan P, Lewis S, Hashibe M, Bell DA, Boffetta P, et al. Pooled analysis of alcohol dehydrogenase genotypes and head and neck cancer: a HuGE review. Am J Epidemiol. 2004;159:1–16. doi: 10.1093/aje/kwh003. [DOI] [PubMed] [Google Scholar]
- 17.Soucek P, Susova S, Mohelnikova-Duchonova B, Gromadzinska J, Moraviec-Sztandera A, et al. Polymorphisms in metabolizing enzymes and the risk of head and neck squamous cell carcinoma in the Slavic population of the central Europe. Neoplasma. 2010;57:415–421. doi: 10.4149/neo_2010_05_415. [DOI] [PubMed] [Google Scholar]
- 18.Asakage T, Yokoyama A, Haneda T, Yamazaki M, Muto M, et al. Genetic polymorphisms of alcohol and aldehyde dehydrogenases, and drinking, smoking and diet in Japanese men with oral and pharyngeal squamous cell carcinoma. Carcinogenesis. 2007;28:865–874. doi: 10.1093/carcin/bgl206. [DOI] [PubMed] [Google Scholar]
- 19.Solomon PR, Selvam GS, Shanmugam G. Polymorphism in ADH and MTHFR genes in oral squamous cell carcinoma of Indians. Oral Dis. 2008;14:633–639. doi: 10.1111/j.1601-0825.2007.01437.x. [DOI] [PubMed] [Google Scholar]
- 20.Chang JS, Straif K, Guha N. Mutagenesis; 2011. The role of alcohol dehydrogenase genes in head and neck cancers: a systematic review and meta-analysis of ADH1B and ADH1C. doi: 10.1093/mutage/ger073. [DOI] [PubMed] [Google Scholar]
- 21.Terry MB, Gammon MD, Zhang FF, Vaughan TL, Chow WH, et al. Alcohol dehydrogenase 3 and risk of esophageal and gastric adenocarcinomas. Cancer Causes Control. 2007;18:1039–1046. doi: 10.1007/s10552-007-9046-0. [DOI] [PubMed] [Google Scholar]
- 22.Nishimoto IN, Pinheiro NA, Rogatto SR, Carvalho AL, de Moura RP, et al. Alcohol dehydrogenase 3 genotype as a risk factor for upper aerodigestive tract cancers. Arch Otolaryngol Head Neck Surg. 2004;130:78–82. doi: 10.1001/archotol.130.1.78. [DOI] [PubMed] [Google Scholar]
- 23.Olshan AF, Weissler MC, Watson MA, Bell DA. Risk of head and neck cancer and the alcohol dehydrogenase 3 genotype. Carcinogenesis. 2001;22:57–61. doi: 10.1093/carcin/22.1.57. [DOI] [PubMed] [Google Scholar]
- 24.Li DP, Dandara C, Walther G, Parker MI. Genetic polymorphisms of alcohol metabolising enzymes: their role in susceptibility to oesophageal cancer. Clin Chem Lab Med. 2008;46:323–328. doi: 10.1515/CCLM.2008.073. [DOI] [PubMed] [Google Scholar]
- 25.Zhang FF, Hou L, Terry MB, Lissowska J, Morabia A, et al. Genetic polymorphisms in alcohol metabolism, alcohol intake and the risk of stomach cancer in Warsaw, Poland. Int J Cancer. 2007;121:2060–2064. doi: 10.1002/ijc.22973. [DOI] [PubMed] [Google Scholar]
- 26.Yin G, Kono S, Toyomura K, Moore MA, Nagano J, et al. Alcohol dehydrogenase and aldehyde dehydrogenase polymorphisms and colorectal cancer: the Fukuoka Colorectal Cancer Study. Cancer Sci. 2007;98:1248–1253. doi: 10.1111/j.1349-7006.2007.00519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visvanathan K, Crum RM, Strickland PT, You X, Ruczinski I, et al. Alcohol dehydrogenase genetic polymorphisms, low-to-moderate alcohol consumption, and risk of breast cancer. Alcohol Clin Exp Res. 2007;31:467–476. doi: 10.1111/j.1530-0277.2006.00334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters ES, McClean MD, Liu M, Eisen EA, Mueller N, et al. The ADH1C polymorphism modifies the risk of squamous cell carcinoma of the head and neck associated with alcohol and tobacco use. Cancer Epidemiol Biomarkers Prev. 2005;14:476–482. doi: 10.1158/1055-9965.EPI-04-0431. [DOI] [PubMed] [Google Scholar]
- 29.Curtin K, Slattery ML, Ulrich CM, Bigler J, Levin TR, et al. Genetic polymorphisms in one-carbon metabolism: associations with CpG island methylator phenotype (CIMP) in colon cancer and the modifying effects of diet. Carcinogenesis. 2007;28:1672–1679. doi: 10.1093/carcin/bgm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Logt EM, Bergevoet SM, Roelofs HM, Te Morsche RH, Dijk Y, et al. Role of epoxide hydrolase, NAD(P)H:quinone oxidoreductase, cytochrome P450 2E1 or alcohol dehydrogenase genotypes in susceptibility to colorectal cancer. Mutat Res. 2006;593:39–49. doi: 10.1016/j.mrfmmm.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 31.Freudenheim JL, Ram M, Nie J, Muti P, Trevisan M, et al. Lung cancer in humans is not associated with lifetime total alcohol consumption or with genetic variation in alcohol dehydrogenase 3 (ADH3). J Nutr. 2003;133:3619–3624. doi: 10.1093/jn/133.11.3619. [DOI] [PubMed] [Google Scholar]
- 32.Yokoyama A, Kato H, Yokoyama T, Tsujinaka T, Muto M, et al. Genetic polymorphisms of alcohol and aldehyde dehydrogenases and glutathione S-transferase M1 and drinking, smoking, and diet in Japanese men with esophageal squamous cell carcinoma. Carcinogenesis. 2002;23:1851–1859. doi: 10.1093/carcin/23.11.1851. [DOI] [PubMed] [Google Scholar]
- 33.Freudenheim JL, Ambrosone CB, Moysich KB, Vena JE, Graham S, et al. Alcohol dehydrogenase 3 genotype modification of the association of alcohol consumption with breast cancer risk. Cancer Causes Control. 1999;10:369–377. doi: 10.1023/a:1008950717205. [DOI] [PubMed] [Google Scholar]
- 34.Terry MB, Gammon MD, Zhang FF, Knight JA, Wang Q, et al. ADH3 genotype, alcohol intake and breast cancer risk. Carcinogenesis. 2006;27:840–847. doi: 10.1093/carcin/bgi285. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz SM, Doody DR, Fitzgibbons ED, Ricks S, Porter PL, et al. Oral squamous cell cancer risk in relation to alcohol consumption and alcohol dehydrogenase-3 genotypes. Cancer Epidemiol Biomarkers Prev. 2001;10:1137–1144. [PubMed] [Google Scholar]
- 36.Harty LC, Caporaso NE, Hayes RB, Winn DM, Bravo-Otero E, et al. Alcohol dehydrogenase 3 genotype and risk of oral cavity and pharyngeal cancers. J Natl Cancer Inst. 1997;89:1698–1705. doi: 10.1093/jnci/89.22.1698. [DOI] [PubMed] [Google Scholar]
- 37.Garcia SM, Curioni OA, de Carvalho MB, Gattas GJ. Polymorphisms in alcohol metabolizing genes and the risk of head and neck cancer in a Brazilian population. Alcohol Alcohol. 2010;45:6–12. doi: 10.1093/alcalc/agp078. [DOI] [PubMed] [Google Scholar]
- 38.Oze I, Matsuo K, Suzuki T, Kawase T, Watanabe M, et al. Impact of multiple alcohol dehydrogenase gene polymorphisms on risk of upper aerodigestive tract cancers in a Japanese population. Cancer Epidemiol Biomarkers Prev. 2009;18:3097–3102. doi: 10.1158/1055-9965.EPI-09-0499. [DOI] [PubMed] [Google Scholar]
- 39.Mohelnikova-Duchonova B, Vrana D, Holcatova I, Ryska M, Smerhovsky Z, et al. CYP2A13, ADH1B, and ADH1C gene polymorphisms and pancreatic cancer risk. Pancreas. 2010;39:144–148. doi: 10.1097/MPA.0b013e3181bab6c2. [DOI] [PubMed] [Google Scholar]
- 40.Zavras AI, Wu T, Laskaris G, Wang YF, Cartsos V, et al. Interaction between a single nucleotide polymorphism in the alcohol dehydrogenase 3 gene, alcohol consumption and oral cancer risk. Int J Cancer. 2002;97:526–530. doi: 10.1002/ijc.1642. [DOI] [PubMed] [Google Scholar]
- 41.Coutelle C, Ward PJ, Fleury B, Quattrocchi P, Chambrin H, et al. Laryngeal and oropharyngeal cancer, and alcohol dehydrogenase 3 and glutathione S-transferase M1 polymorphisms. Hum Genet. 1997;99:319–325. doi: 10.1007/s004390050365. [DOI] [PubMed] [Google Scholar]
- 42.Bouchardy C, Hirvonen A, Coutelle C, Ward PJ, Dayer P, et al. Role of alcohol dehydrogenase 3 and cytochrome P-4502E1 genotypes in susceptibility to cancers of the upper aerodigestive tract. Int J Cancer. 2000;87:734–740. [PubMed] [Google Scholar]
- 43.van Dijk B, van Houwelingen KP, Witjes JA, Schalken JA, Kiemeney LA. Alcohol dehydrogenase type 3 (ADH3) and the risk of bladder cancer. Eur Urol. 2001;40:509–514. doi: 10.1159/000049827. [DOI] [PubMed] [Google Scholar]
- 44.Coutelle C, Hohn B, Benesova M, Oneta CM, Quattrochi P, et al. Risk factors in alcohol associated breast cancer: alcohol dehydrogenase polymorphism and estrogens. Int J Oncol. 2004;25:1127–1132. [PubMed] [Google Scholar]
- 45.Kortunay S, Koseler A, Orhan Kara C, Topuz B, Omer Atalay E. Frequencies of ADH1C alleles and genotypes in a Turkish head and neck cancer population. Methods Find Exp Clin Pharmacol. 2010;32:187–191. doi: 10.1358/mf.2010.32.3.1440739. [DOI] [PubMed] [Google Scholar]
- 46.Wang D, Ritchie JM, Smith EM, Zhang Z, Turek LP, et al. Alcohol dehydrogenase 3 and risk of squamous cell carcinomas of the head and neck. Cancer Epidemiol Biomarkers Prev. 2005;14:626–632. doi: 10.1158/1055-9965.EPI-04-0343. [DOI] [PubMed] [Google Scholar]
- 47.Sturgis EM, Dahlstrom KR, Guan Y, Eicher SA, Strom SS, et al. Alcohol dehydrogenase 3 genotype is not associated with risk of squamous cell carcinoma of the oral cavity and pharynx. Cancer Epidemiol Biomarkers Prev. 2001;10:273–275. [PubMed] [Google Scholar]
- 48.Chao YC, Wang LS, Hsieh TY, Chu CW, Chang FY, et al. Chinese alcoholic patients with esophageal cancer are genetically different from alcoholics with acute pancreatitis and liver cirrhosis. Am J Gastroenterol. 2000;95:2958–2964. doi: 10.1111/j.1572-0241.2000.02328.x. [DOI] [PubMed] [Google Scholar]
- 49.Brocic M, Supic G, Zeljic K, Jovic N, Kozomara R, et al. Genetic polymorphisms of ADH1C and CYP2E1 and risk of oral squamous cell carcinoma. Otolaryngol Head Neck Surg. 2011;145:586–593. doi: 10.1177/0194599811408778. [DOI] [PubMed] [Google Scholar]
- 50.Homann N, Stickel F, Konig IR, Jacobs A, Junghanns K, et al. Alcohol dehydrogenase 1C*1 allele is a genetic marker for alcohol-associated cancer in heavy drinkers. Int J Cancer. 2006;118:1998–2002. doi: 10.1002/ijc.21583. [DOI] [PubMed] [Google Scholar]
- 51.McKay JD, Truong T, Gaborieau V, Chabrier A, Chuang SC, et al. A genome-wide association study of upper aerodigestive tract cancers conducted within the INHANCE consortium. PLoS Genet. 2011;7:e1001333. doi: 10.1371/journal.pgen.1001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng HW, Chen WM, Recker RR. Population admixture: detection by Hardy-Weinberg test and its quantitative effects on linkage-disequilibrium methods for localizing genes underlying complex traits. Genetics. 2001;157:885–897. doi: 10.1093/genetics/157.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen YC, Peng GS, Wang MF, Tsao TP, Yin SJ. Polymorphism of ethanol-metabolism genes and alcoholism: correlation of allelic variations with the pharmacokinetic and pharmacodynamic consequences. Chem Biol Interact. 2009;178:2–7. doi: 10.1016/j.cbi.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 54.Eriksson CJ, Fukunaga T, Sarkola T, Chen WJ, Chen CC, et al. Functional relevance of human adh polymorphism. Alcohol Clin Exp Res. 2001;25:157S–163S. doi: 10.1097/00000374-200105051-00027. [DOI] [PubMed] [Google Scholar]
- 55.Chen WJ, Loh EW, Hsu YP, Chen CC, Yu JM, et al. Alcohol-metabolising genes and alcoholism among Taiwanese Han men: independent effect of ADH2, ADH3 and ALDH2. Br J Psychiatry. 1996;168:762–767. doi: 10.1192/bjp.168.6.762. [DOI] [PubMed] [Google Scholar]
- 56.Risch A, Ramroth H, Raedts V, Rajaee-Behbahani N, Schmezer P, et al. Laryngeal cancer risk in Caucasians is associated with alcohol and tobacco consumption but not modified by genetic polymorphisms in class I alcohol dehydrogenases ADH1B and ADH1C, and glutathione-S-transferases GSTM1 and GSTT1. Pharmacogenetics. 2003;13:225–230. doi: 10.1097/00008571-200304000-00007. [DOI] [PubMed] [Google Scholar]
- 57.Jelski W, Chrostek L, Markiewicz W, Szmitkowski M. Activity of alcohol dehydrogenase (ADH) isoenzymes and aldehyde dehydrogenase (ALDH) in the sera of patients with breast cancer. J Clin Lab Anal. 2006;20:105–108. doi: 10.1002/jcla.20109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iwasaki M, Tsugane S. Risk factors for breast cancer: epidemiological evidence from Japanese studies. Cancer Sci. 2011;102:1607–1614. doi: 10.1111/j.1349-7006.2011.01996.x. [DOI] [PubMed] [Google Scholar]
- 59.Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, et al. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1991;48:677–681. [PMC free article] [PubMed] [Google Scholar]
- 60.Druesne-Pecollo N, Tehard B, Mallet Y, Gerber M, Norat T, et al. Alcohol and genetic polymorphisms: effect on risk of alcohol-related cancer. Lancet Oncol. 2009;10:173–180. doi: 10.1016/S1470-2045(09)70019-1. [DOI] [PubMed] [Google Scholar]
- 61.Yang CX, Matsuo K, Ito H, Hirose K, Wakai K, et al. Esophageal cancer risk by ALDH2 and ADH2 polymorphisms and alcohol consumption: exploration of gene-environment and gene-gene interactions. Asian Pac J Cancer Prev. 2005;6:256–262. [PubMed] [Google Scholar]
- 62.Chen YJ, Chen C, Wu DC, Lee CH, Wu CI, et al. Interactive effects of lifetime alcohol consumption and alcohol and aldehyde dehydrogenase polymorphisms on esophageal cancer risks. Int J Cancer. 2006;119:2827–2831. doi: 10.1002/ijc.22199. [DOI] [PubMed] [Google Scholar]