Abstract
The components in plant signal transduction pathways are intertwined and affect each other to coordinate plant growth, development, and defenses to stresses. The role of ubiquitination in connecting these pathways, particularly plant innate immunity and flowering, is largely unknown. Here, we report the dual roles for the Arabidopsis (Arabidopsis thaliana) Plant U-box protein13 (PUB13) in defense and flowering time control. In vitro ubiquitination assays indicated that PUB13 is an active E3 ubiquitin ligase and that the intact U-box domain is required for the E3 ligase activity. Disruption of the PUB13 gene by T-DNA insertion results in spontaneous cell death, the accumulation of hydrogen peroxide and salicylic acid (SA), and elevated resistance to biotrophic pathogens but increased susceptibility to necrotrophic pathogens. The cell death, hydrogen peroxide accumulation, and resistance to necrotrophic pathogens in pub13 are enhanced when plants are pretreated with high humidity. Importantly, pub13 also shows early flowering under middle- and long-day conditions, in which the expression of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 and FLOWERING LOCUS T is induced while FLOWERING LOCUS C expression is suppressed. Finally, we found that two components involved in the SA-mediated signaling pathway, SID2 and PAD4, are required for the defense and flowering-time phenotypes caused by the loss of function of PUB13. Taken together, our data demonstrate that PUB13 acts as an important node connecting SA-dependent defense signaling and flowering time regulation in Arabidopsis.
The ubiquitin/26S proteasome-mediated pathway is involved in selective degradation of proteins in cells of eukaryotic organisms. In plants, ubiquitination has been implicated in a variety of processes, including cell cycle, circadian rhythm control, hormone signaling, senescence, disease resistance, and photomorphogenesis/flowering (Jang et al., 2005; Vega-Sánchez et al., 2008; Henriques et al., 2009; Farmer et al., 2010). The critical role of ubiquitination in disease resistance has been demonstrated in many different plant species in the last several years. Identification and characterization of rice (Oryza sativa) SPL11, a U-box protein with E3 ligase activity, provided the first direct evidence that ubiquitination controls resistance and programmed cell death (PCD) in plants (Zeng et al., 2004). The spl11 mutation is characterized by spontaneous cell death in leaves and enhanced disease resistance to bacterial and fungal pathogens in rice (Yin et al., 2000; Zeng et al., 2004). Thus, SPL11 serves as a negative regulator of PCD and defense in rice. In tomato (Solanum lycopersicum), a set of E3 ligase genes such as CMPG1 and ACRE276, which are rapidly induced in Avr9-treated Cf-9 tobacco (Nicotiana tabacum) cell cultures, were identified as positive regulators of the hypersensitive response (González-Lamothe et al., 2006; Yang et al., 2006; van den Burg et al., 2008). In addition, knocking out of Plant U-box protein17 (PUB17), an Arabidopsis (Arabidopsis thaliana) ortholog of ACRE276, causes compromised RPM1- and RPS4-mediated resistance against Pseudomonas syringae pv tomato containing the avirulence genes AvrB and AvrRPS4, respectively (Yang et al., 2006). Similarly, silencing of the F-box gene ACIF1 compromises the hypersensitive response triggered by various elicitors and by the activation of different R genes in tobacco plants (van den Burg et al., 2008).
Flowering is a well-defined plant development process that involves transition from vegetative maturity to the reproductive stage. Multiple external and internal signals, including photoperiod, temperature, hormone, and age-related signals, have been shown to regulate plant flowering. These signals ultimately converge at the floral pathway integrators, a group of genes that are turned on or off to determine the flowering time. Among these flowering pathway integrators, FT and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) have been well characterized (Kardailsky et al., 1999; Borner et al., 2000). SOC1 is regulated by two antagonistic flowering regulators, FLOWERING LOCUS C (FLC) and CO, which act as a floral repressor and a floral activator, respectively. CO protein is unstable in the morning and in the dark under long-day (LD) conditions, but the treatment with proteasome inhibitors stabilizes CO, suggesting that CO is targeted for degradation via the 26S proteasome (Valverde et al., 2004). Furthermore, an E3 ligase responsible for the degradation of CO in the dark was identified as the photomorphogenesis-related RING finger protein COP1 (Liu et al., 2008). The light receptor PhyB was shown to be responsible for CO protein instability in the morning, while the blue light receptor CRY2 contributes to the stabilization of CO in the evening and in the dark (Valverde et al., 2004; Liu et al., 2008). CRY2 is ubiquitinated in response to blue light and that ubiquitinated CRY2 is degraded by the 26S proteasome in the nucleus, where it acts as a blue light receptor to promote flowering under LD conditions (Yu et al., 2007). Taken together, these studies suggest that ubiquitination plays a pivotal role in flowering time regulation in Arabidopsis.
The rice spl11 mutant displays late flowering under LD conditions (Vega-Sánchez et al., 2008). Genetic and molecular analyses showed that SPL11 regulates flowering via interaction with SPL11-interacting protein1 (SPIN1), a member of the STAR family. SPIN1 inhibits flowering by suppressing Hd3a (an ortholog of FT) via Hd1 (an ortholog of CO)-dependent mechanisms under short-day (SD) conditions and by targeting Hd1-independent factors in LD conditions, suggesting that rice SPL11 regulates flowering time probably through ubiquitination of SPIN1, a component associated with rice flowering signaling. However, how SPL11 regulates both defense responses and flowering time in rice is unclear.
Salicylic acid (SA) plays a critical role in plant disease resistance. In addition, SA also regulates plant development (Martínez et al., 2004; Wada et al., 2010). Recently, WIN3 was found to regulate both plant innate immunity and flowering time in Arabidopsis (Wang et al., 2011). WIN3 is a type II SA regulator belonging to the firefly luciferase family that consists of 19 members (Staswick et al., 2005). It plays multiple roles, including conferring broad-spectrum disease resistance to biotrophic and necrotrophic pathogens, modulating cell death in the SA signaling mutant acd6-1, and contributing to flg22-induced pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI; Wang et al., 2011). Additionally, WIN3 negatively regulates flowering time under LD conditions via the regulation of FLC and FT (Wang et al., 2011). These data indicate that signaling in the control of plant defense and plant flowering time is interconnected at WIN3.
In this study, we show that the U-box-type E3 ubiquitin ligase PUB13, the ortholog of rice SPL11 in Arabidopsis, regulates cell death, broad-spectrum disease resistance to various pathogens, and flowering time. Our data indicate that PUB13 negatively regulates SA-mediated defense and flowering time in a SID2- and PAD4-dependent manner, which implies that PUB13 plays dual roles in the regulation of both plant defense and development via SA-mediated signaling.
RESULTS
PUB13, Encoding a U-Box/ARM Protein, Is the Closest Arabidopsis Ortholog of Rice SPL11
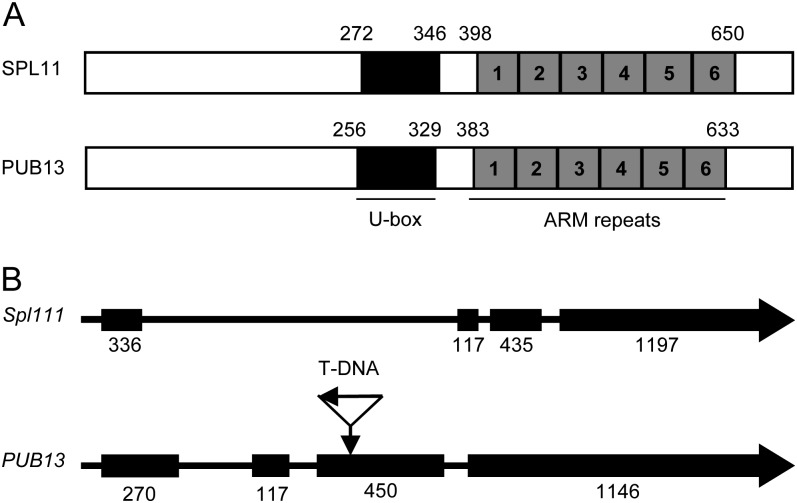
To identify the putative orthologous gene of Spl11 in Arabidopsis, we performed BLAST searches against Arabidopsis genome sequences using the amino acid sequence of SPL11 as a query. The data mining identified Arabidopsis PUB13 (At3g46510) as the closet ortholog of SPL11 (Azevedo et al., 2001; Zeng et al., 2008). The protein sequence of PUB13 is highly similar to SPL11, sharing 73% identity of amino acids. The PUB13 protein contains a conserved U-box domain spanning amino acid residues 256 to 329, which is highly similar to that in SPL11 (Fig. 1A; Supplemental Fig. S1A). The highly conserved amino acid residues Val-273, Cys-297, and Pro-298 in known U-box proteins are present in both PUB13 and SPL11. Like SPL11, PUB13 contains six tandem repeats of Armadillo (ARM) motifs in its central region and C terminus, and the PUB13 gene possesses a similar gene structure to Spl11 (Fig. 1B). Because of the existence of the ARM repeat domain, PUB13 is classified into PUB family class II (Azevedo et al., 2001). Two other members of the same class, PUB14 and PUB17, have been implicated in cell death and disease resistance, respectively (Yang et al., 2006; Yee and Goring, 2009).
Figure 1.
The protein and gene structures between PUB13 and SPL11. A, Schematic representation of PUB13 and SPL11 proteins. The black box indicates the U-box domain, and the individual ARM repeats are indicated by numbered gray boxes. Numbers above the schematic representation indicate the positions of amino acid residues. B, Gene structures of PUB13 and Spl11. Exons are represented as black boxes. The number below each exon indicates the length of the exon in bp. The position of the T-DNA insertion in the pub13 mutant is marked by an arrow.
PUB13 Possesses E3 Ligase Activity in Vitro, and the U-Box Domain Is Required for E3 Ligase Activity
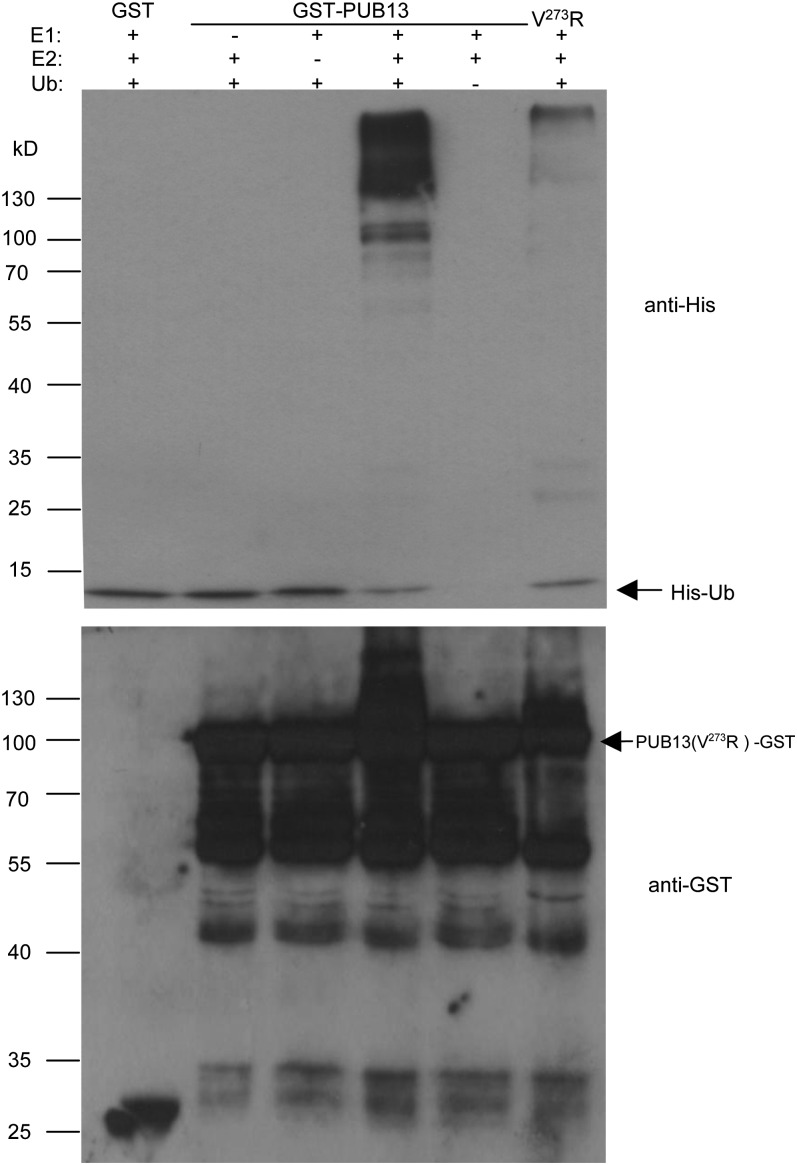
Possession of E3 ubiquitin ligase activity is an important feature of U-box-containing proteins (Hatakeyama et al., 2001; Mudgil et al., 2004). To analyze E3 ubiquitin ligase activity of PUB13 in vitro, we fused the full-length cDNA of the PUB13 gene to glutathione S-transferase (GST) and expressed it in Escherichia coli. In the E3 ligase activity assay, the purified GST-PUB13 was incubated in the presence or absence of wheat E1 (GI: 136632), human E2 UBCH5b, and/or His-tagged ubiquitin. Ubiquitination activity was detected by immunoblot with nickel-horseradish peroxidase. In the presence of E1, E2, and ubiquitin, GST-PUB13 showed a strong ligase activity by forming high-molecular-mass polyubiquitin (Fig. 2, lane 4, top panel).
Figure 2.
Analysis of E3 ubiquitin ligase activity for PUB13. An in vitro ubiquitination assay was performed with GST-PUB13/V273R fusion proteins in the presence or absence of wheat E1, human E2 (UBCH5b), and His-tagged ubiquitin. The numbers on the left denote the molecular masses of marker proteins in kD. The ubiquitination signal was detected using nickel-horseradish peroxidase (top panel). The expression of GST-fused proteins in the assay was detected with an anti-GST immunoblot (bottom panel).
To find out whether an intact U-box domain is necessary for the E3 ligase activity of PUB13, we made the mutant construct GST-PUB13 (V273R) by substituting Val-273 to Arg in the U-box domain. This Val is highly conserved in different U-box proteins and was demonstrated to be important for the biological and biochemical functions of U-box proteins (Ohi and Gould, 2002; Zeng et al., 2004). Under the same analysis conditions, GST-PUB13 (V273R) showed much reduced ligase activity compared with wild-type PUB13 (Fig. 2Fig. 2, lane 6, top panel). The anti-GST immunoblot was used to confirm the presence of the similar amount of GST-fused proteins in the assay. As shown in Figure 2 (bottom panel), GST-PUB13 fusions were presented in reactions as expected, and self-ubiquitination of GST-PUB13 was also detected in the anti-GST immunoblot in the presence of E1, E2, and ubiquitin (Fig. 2, lane 4, bottom panel). Consistent with the nickel-horseradish peroxidase immunoblot, anti-GST also showed that the self-ubiquitination of PUB13 (V273R) was significantly suppressed (Fig. 2, lane 6, bottom panel). These results indicate that PUB13 possesses E3 ligase activity and that the intact U-box domain is required for its activity.
Cell Death and Hydrogen Peroxide Accumulation Are Elevated in pub13
We identified a T-DNA insertion mutant, pub13, in the Arabidopsis Biological Resource Center mutant collection, in which the T-DNA insertion was located at the third exon of PUB13 (Fig. 1B). Gene expression analysis showed that the transcription of PUB13 is completely abolished in pub13 (Supplemental Fig. S1B). Under LD growth conditions (23°C, 70% relative humidity [RH], 16 h of daylight/8 h of dark), we found that the lower leaves of the pub13 mutant displayed chlorosis and lesion-mimic phenotypes spontaneously, and the chlorosis was formed from leaf edge to main vein (Fig. 3A). To check whether this lesion-mimic phenotype is associated with cell death, the leaves of pub13 were subjected to trypan blue staining. The lower leaves of pub13 with chlorosis were stained blue, whereas the leaves at the same position in wild-type ecotype Columbia (Col-0) were not (Fig. 3B, top panel). In the middle-aged green leaves of pub13 (i.e. the seventh or eighth leaf of 4-week-old plants), cell death was also detected, although no macroscopic death was visible (Supplemental Fig. S2B, top panel).
Figure 3.
Cell death and H2O2 accumulation in pub13, complemented pub13, and PUB13 RNAi transgenic plants. A, Lesion-mimic phenotype of pub13. Plants were 4 weeks old and grown under LD conditions. B, Cell death and H2O2 accumulation in pub13, complemented pub13, and PUB13 RNAi transgenic plants. Cell death and H2O2 accumulation were examined in the lower leaves of pub13 with chlorosis and leaves at the same positions of other lines. Plants were 4 weeks old and grown under LD conditions. The leaves in the top panel were stained with trypan blue for cell death. H2O2 accumulation was detected by DAB staining in the bottom panel. EV, Empty vector used for the RNAi construct.
To confirm whether the cell death is caused by the knockout of PUB13, we transformed the intact PUB13 coding sequence with 35S promoter into pub13 and a PUB13 RNA interference (RNAi) construct into Col-0. Trypan blue staining assays showed that the non-cell-death phenotype was restored in the PUB13-complemented pub13 transgenic plants (Fig. 3B, top panel) while the lower leaves of PUB13 RNAi lines showed clear cell death like that in pub13 (Fig. 3B, top panel). These results demonstrate that PUB13 negatively controls cell death in Arabidopsis.
Since reactive oxygen species are key players in the regulation of PCD, we measured the accumulation of hydrogen peroxide (H2O2) in pub13, complemented pub13, and RNAi lines. The lower leaves of these lines grown under LD conditions were stained by 3,3′-diaminobenzidine tetrahydrochloride (DAB). In agreement with the cell death phenotypes, the H2O2 content in lower leaves of pub13 and PUB13 RNAi lines was markedly increased, but this increase was restored to the wild-type level in complemented pub13 plants (Fig. 3B, bottom panel). Interestingly, no difference was observed in the H2O2 level for the middle-aged leaves of pub13 and Col-0 (Supplemental Fig. S2B, bottom panel).
The Increase of Cell Death and H2O2 Accumulation in pub13 Depends on the SA Signal
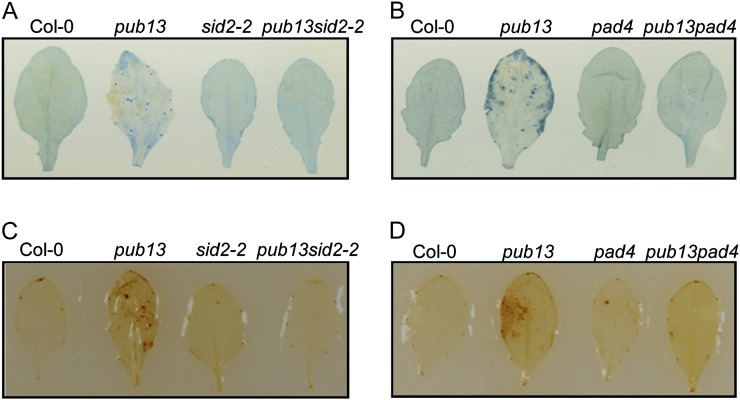
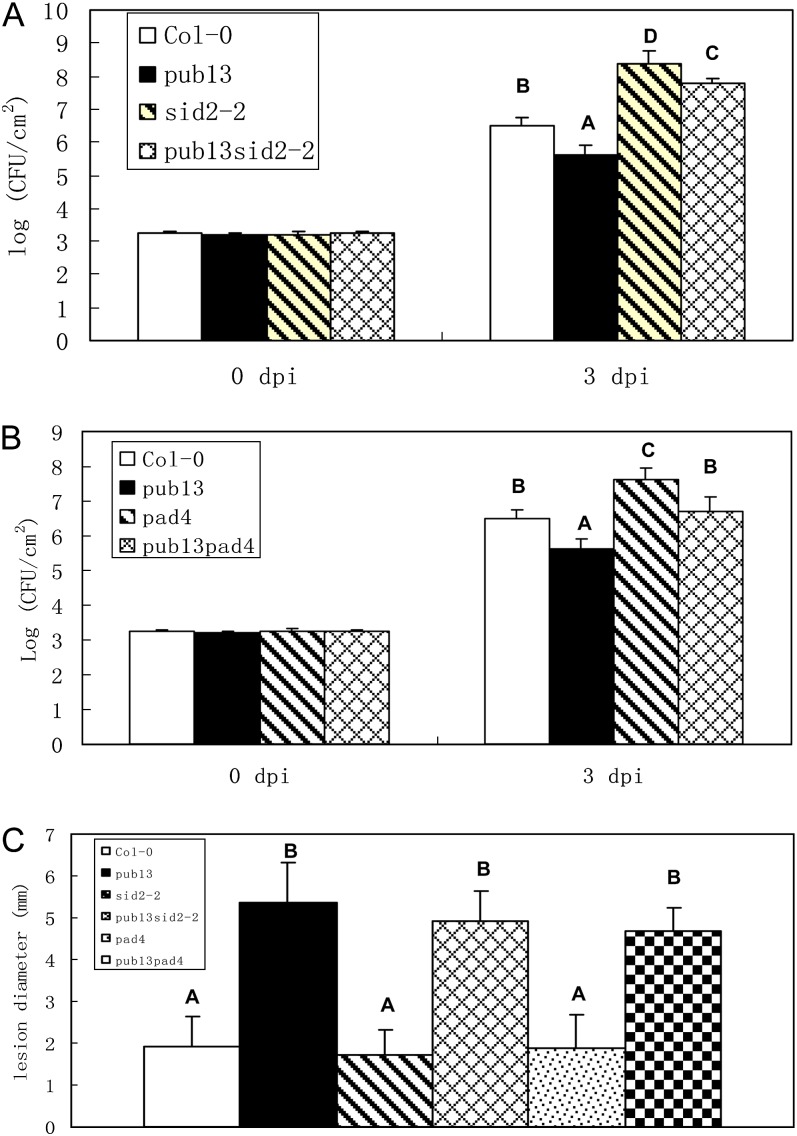
The PCD is closely associated with SA via an unclear mechanism (Ludwig and Tenhaken, 2000; Kawai-Yamada et al., 2004). We introduced the SA induction-deficient mutant sid2-2 into pub13 by genetic cross and analyzed the role of SA for cell death increase in pub13. Leaves of 4-week-old plants grown under LD and high-humidity conditions (see details below) were collected for trypan blue staining. As shown in Figure 4A, cell death was suppressed significantly in the pub13sid2-2 double mutant compared with the clear cell death phenotype in pub13, and no visible cell death was observed in sid2-2 and Col-0. Another SA-deficient mutant, pad4, was also introduced into pub13 to confirm the pub13sid2-2 result. Similarly, the cell death in the pub13pad4 double mutant was considerably suppressed under LD and high-humidity conditions (Fig. 4B).
Figure 4.
Cell death and H2O2 accumulation in the pub13sid2-2 and pub13pad4 mutants. Four-week-old plants grown under LD conditions were treated with high humidity (95% RH) for 48 h, and then cell death and H2O2 accumulation were detected in the middle-aged leaves (i.e. the seventh or eighth leaves in 4-week-old plants). A, Cell death in Col-0, pub13, sid2-2, and pub13sid2-2. B, Cell death in Col-0, pub13, pad4, and pub13pad4. C, H2O2 accumulation in Col-0, pub13, sid2-2, and pub13sid2-2. D, H2O2 accumulation in Col-0, pub13, pad4, and pub13pad4.
The role of sid2-2 and pad4 for the H2O2 accumulation in pub13 was also determined. After high-humidity treatment, the H2O2 level in pub13 increased significantly compared with that of Col-0 but the H2O2 content in sid2-2 was at a similar level to Col-0 (Fig. 4C). Notably, the H2O2 accumulation in the pub13sid2-2 double mutant was reduced to a background level. Similarly, introduction of pad4 into pub13 reduced the high H2O2 accumulation in pub13 to the normal level under LD and high-humidity conditions (Fig. 4D). The elimination of cell death and H2O2 accumulation in pub13sid2-2 and pub13pad4 suggests that SA is required for the spontaneous cell death and elevated H2O2 accumulation in pub13.
The pub13 Mutant Confers Enhanced Resistance against Biotrophic Pathogens
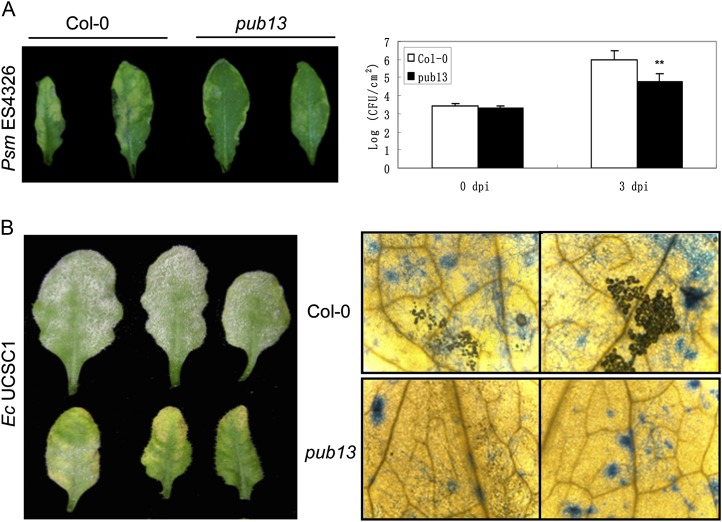
The PCD phenotype and H2O2 accumulation in pub13 led us to investigate the function of PUB13 in plant defense. We first inoculated the Col-0 and pub13 plants grown under LD conditions with the virulent strain ES4326 of the biotrophic pathogen Pseudomonas syringae pv maculicola (Psm). Three days after inoculation, the water-soaked lesions of pub13 leaves were smaller than those in Col-0 leaves (Fig. 5A, left panel). To monitor the growth of bacteria, Psm ES4326 was infiltrated into pub13 and Col-0 plants at a low titer. The bacterial growth analysis showed that the amount of bacteria in Col-0 was more than that in pub13 (Fig. 5A, right panel), suggesting that the pub13 mutation confers elevated resistance to the biotrophic bacterial pathogen Psm ES4326.
Figure 5.
Disease resistance of pub13 against biotrophic pathogens. A, Disease symptoms and bacterial growth assays of Psm ES4326 in Col-0 and pub13 under LD conditions. Disease symptoms (left panel) were photographed on day 3 after spraying with 1 × 108 CFU mL−1 ES4326. Bacterial growth (right panel) was assessed in the leaves injected with 1 × 105 CFU mL−1 ES4326. Student’s t test was carried out to determine the significance of the difference between Col-0 and pub13 plants. ** Significant difference at P < 0.01. B, Phenotypes of Col-0 and pub13 grown under LD conditions against E. cichoracearum (Ec) UCSC1. Plants were inoculated with conidia (approximately 100 conidia mm−2) of UCSC1. Inoculated plants were photographed at 7 dpi (left panel) or photographed using a microscope after staining with trypan blue at 10 dpi (right panel). These experiments were repeated at least twice with similar results.
To test whether pub13 confers elevated resistance to biotrophic fungal pathogens as well, we inoculated both pub13 and Col-0 with Erysiphe cichoracearum UCSC1, which causes powdery mildew diseases in many plants. Abundant conidiophores and conidiophore peduncles spread all over the Col-0 leaves but much less on the pub13 leaves, and visible cell death appeared in the infected pub13 leaves at 7 d post inoculation (dpi; Fig. 5B, left panel). To further confirm the resistance of pub13 to UCSC1, we stained the inoculated Col-0 and pub13 leaves with trypan blue at 10 dpi. As shown in Figure 5B (right panel), a large number of hyphae and conidiophores existed in Col-0 while only a few hyphae were observed in pub13. These results indicate that pub13 confers enhanced resistance to the biotrophic fungus strain UCSC1.
In addition to examining the resistance of pub13 against biotrophic pathogens, we also tested whether pub13 confers resistance to the necrotrophic fungal pathogens Botrytis cinerea BO5-10 and Alternaria brassicicola AB. In contrast to biotrophic pathogens, no significant difference in disease symptom and pathogen growth was observed when the mutant and wild-type plants were inoculated with these two fungal pathogens under LD and normal-humidity conditions (70% RH; Supplemental Fig. S3, C and D).
Cell Death, H2O2 Content, and Disease Resistance of pub13 Are Increased under High Humidity
It is known that some lesion-mimic mutants are sensitive to environmental stress conditions such as high humidity (Mosher et al., 2010). To test the effect of humidity on the occurrence of the phenotypes on pub13 plants, 4-week-old pub13 plants grown under regular conditions (approximately 70% RH) were subjected to high-humidity (95% RH) treatment. After 48 h of high-humidity treatment, the lesion-mimic phenotype of pub13 was more severe than that in wild-type plants (Supplemental Fig. S2A). Consistent with the lesion-mimic phenotype, cell death in middle-aged leaves of pub13 after high-humidity treatment was much stronger compared with that in untreated plants (Supplemental Fig. S2B, top panel). In addition, the middle-aged leaves from humidity-treated pub13 but not from treated Col-0 exhibited abundant H2O2 accumulation, while there was no obvious H2O2 accumulation in the middle-aged leaves of untreated pub13 (Supplemental Fig. S2B, bottom panel). Humidity, therefore, can enhance cell death and H2O2 accumulation in pub13 plants.
To test whether the enhanced cell death and H2O2 accumulation in pub13 is correlated with its elevated resistance to pathogen infections, we monitored the level of resistance of pub13 and Col-0 plants pretreated with high humidity for 24 h. Treated and untreated plants were inoculated with the biotrophic pathogens P. syringae pv tomato DC3000 and Hyaloperonospora parasitica Noco2. Under regular humidity, the disease symptoms of pub13 inoculated with P. syringae pv tomato DC3000 (1 × 108 colony-forming units [CFU] mL−1) were milder than those of Col-0 at 5 dpi (Supplemental Fig. S3A, top panel). Analysis of bacterial growth also showed that pub13 plants were more resistant against DC3000 than Col-0 (Supplemental Fig. S3A, bottom panel). Interestingly, a much larger difference of resistance against DC3000 was observed between the high-humidity-pretreated pub13 and Col-0 (Supplemental Fig. S3A). High-humidity-pretreated leaves of Col-0 displayed an atrophic and chlorosis phenotype, whereas only some local chlorosis existed on pub13 leaves. Similar to the inoculation with DC3000, pub13 was slightly more resistant than Col-0 to Noco2 without humidity treatment (Supplemental Fig. S3B). After pretreatment of high humidity, however, the resistance of pub13 was much increased, as revealed by less chlorosis symptoms and smaller lesions on the leaves (Supplemental Fig. S3B).
The resistance of pub13 against the necrotrophic pathogens B. cinerea and A. brassicicola after high-humidity treatment was also examined. The detached leaves of pub13 and Col-0 did not show any difference in lesion size under regular humidity after inoculation with B. cinerea BO5-1 (Supplemental Fig. S3C). Nevertheless, after high-humidity treatment, leaves of pub13 plants displayed severe rotting symptoms and large lesions, while no visible necrotic lesion occurred in leaves of Col-0 plants (Supplemental Fig. S3C). Similar to challenge by B. cinerea, there was no visible symptom difference from A. brassicicola AB between pub13 and Col-0 under normal humidity (Supplemental Fig. S3D). Conversely, pub13 was much more susceptible to A. brassicicola AB than Col-0 when the plants were pretreated with high humidity (Supplemental Fig. S3D), corroborating the observation that high humidity promotes the susceptibility of pub13 against necrotrophic pathogens.
PAD4 and SID2 Are Required for the PUB13-Mediated Resistance to Psm ES4326 But Not to B. cinerea BO5-10
As mentioned above, the cell death and H2O2 accumulation in pub13 are dependent on the SA signaling components SID2 and PAD4. To test the function of SID2 and PAD4 genes in the PUB13-mediated defense pathway, we inoculated the pub13sid2-2 and pub13pad4 plants with the biotrophic pathogen ES4326. Under regular-humidity and LD conditions, pub13 displayed elevated resistance but sid2-2 showed reduced resistance against ES4326 (Fig. 6A). As expected, the resistance of the pub13sid2-2 double mutant against ES4326 was markedly reduced to a level even more susceptible than Col-0. Similarly, the pub13pad4 plants were slightly more susceptible than Col-0 (Fig. 6B). In contrast to the biotrophic pathogens, the resistance level of pub13sid2-2 and pub13pad4 plants against the necrotrophic pathogen BO5-10 was comparable to that of pub13 plants after high-humidity treatment (Fig. 6C). Taken together, these results demonstrated that SID2 and PAD4 are required for PUB13-mediated resistance to the biotrophic pathogen ES4326 but are dispensable for resistance to the necrotrophic pathogen BO5-10.
Figure 6.
Disease reaction of the pub13sid2-2 and pub13pad4 double mutants against Psm ES4326 and B. cinerea BO5-10. A, Resistance of pub13sid2-2 against ES4326. B, Resistance of pub13pad4 to ES4326. Four-week-old plants in A and B were grown under LD conditions and injected with 1 × 105 CFU mL−1 ES4326. C, Resistance of pub13sid2-2 and pub13pad4 double mutants to BO5-10. Detached leaves from high-humidity-pretreated plants grown under LD conditions were inoculated with 5 μL of conidia suspension (1 × 104 conidia mL−1) of BO5-10, and lesion diameter was measured at 3 dpi. All the experiments were repeated at least twice with similar results. Student’s t test was carried out to determine the significance of the difference. Uppercase letters indicate significant difference at P < 0.01. [See online article for color version of this figure.]
The pub13 Mutant Contains Elevated Levels of SA
The intimate involvement of PUB13 in host defense against biotrophic and necrotrophic pathogens prompted us to determine the expression levels of known defense-related genes in pub13. The transcriptional levels of PR1, a marker gene in the SA signaling pathway, and PDF1.2, a marker gene of the jasmonic acid (JA) and ethylene (ET) signaling pathways, were examined in Col-0 and pub13 plants grown under LD conditions using real-time PCR. As shown in Supplemental Figure S4A, the transcriptional level of PR1 was significantly induced in pub13 compared with Col-0, whereas PDF1.2 was markedly suppressed in pub13. We then determined the SA levels in Col-0 and pub13 plants by HPLC. Total SA was extracted from 0.2 g (fresh weight) of leaves of 4-week-old Col-0 and pub13 plants grown under LD conditions. As expected, the SA content in pub13 was 63% higher than in Col-0 (Supplemental Fig. S4B). Although the SA level was undetectable in both SA signal deficiency mutants sid2-2 and pad4, the increased SA level in pub13 was greatly reduced in the pub13sid2-2 and pub13pad4 double mutant plants, only 10% and 32% of that of Col-0, respectively (Supplemental Fig. S4B), suggesting that PUB13 negatively regulates SA accumulation via both SID2 and PAD4.
PUB13 Is a Negative Regulator of Flowering Time
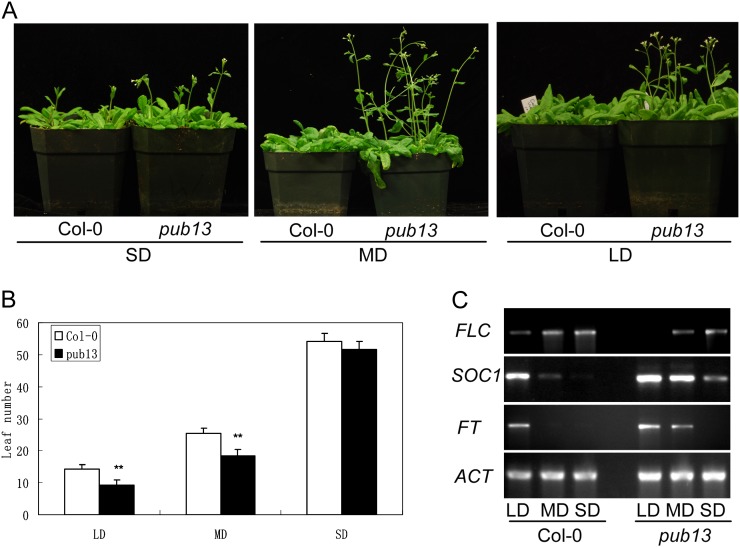
During the reproductive development stage of the pub13 mutant, we found that the mutant displayed altered flowering time. We grew the pub13 plants under different photoperiod conditions: SD (8 h of daylight/16 h of dark), MD (for middle day; 12 h of daylight/12 h of dark), and LD (16 h of daylight/8 h of dark). Under SD conditions, there was no significant difference between Col-0 and pub13 in flowering time, except that a few pub13 plants exhibited slightly earlier flowering than Col-0 (Fig. 7A, left panel). However, under LD conditions, pub13 flowered about 4 d earlier than Col-0 (Fig. 7A, right panel). The early-flowering phenotype of pub13 under MD conditions was even more significant compared with Col-0 (Fig. 7A, middle panel). The leaf number of pub13 before flowering was four and six less than Col-0 under LD and MD conditions, respectively, while the leaf number of pub13 was slightly less than Col-0 under SD conditions (Fig. 7B). Under LD conditions, the PUB13 RNAi plants also exhibited early flowering; however, the early-flowering phenotype was abolished in the complemented pub13 plants (Supplemental Fig. S5). These results indicate that PUB13 acts as a suppressor of flowering time.
Figure 7.
Flowering phenotypes and RT-PCR analysis of flowering marker genes in pub13. A, Flowering phenotypes of Col-0 and pub13 grown under SD, MD, and LD conditions. B, Leaf number of Col-0 and pub13 under SD, MD, and LD conditions. The leaf number was counted when the first flower bud appeared. Statistical analysis was carried out as for Figure 5. C, Expression of flowering marker genes in Col-0 and pub13 under SD, MD, and LD conditions. Gene expression of FLC, SOC1, and FT in Col-0 and pub13 was determined by RT-PCR. Actin (ACT) was used as a control for loading.
To understand how PUB13 regulates the floral transition, we analyzed the transcript levels of the floral repressor FLC and the floral activators FT and SOC1 in pub13 under SD, MD, and LD conditions. As shown in Figure 7C, the transcript level of FLC in Col-0 was higher than in pub13 under LD and MD conditions, and there was no visible difference under SD conditions. On the contrary, the transcriptional level of SOC1 in Col-0 was lower than in pub13 under LD, MD, and SD conditions. Similarly, the transcript level of FT in Col-0 was also lower than in pub13 under LD and especially under MD conditions. Taken together, these data suggested that PUB13 regulates flowering time probably through the SOC1-mediated flowering pathway.
PUB13 Regulates Flowering Time Mainly through a SA-Dependent Pathway
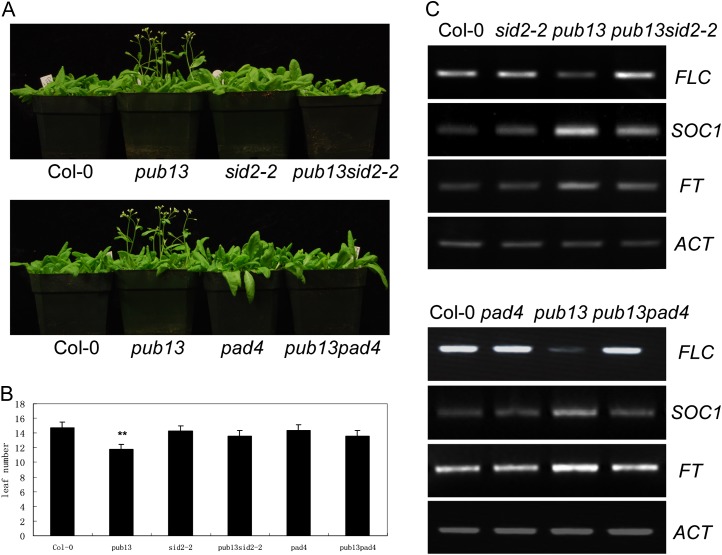
SA is not only a critical regulator of plant defense but also is involved in the regulation of plant flowering time (Martínez et al., 2004). Thus, we investigated the relationship between PUB13-mediated flowering time and the SA pathway. To this purpose, the flowering time of pub13sid2-2 and pub13pad4 was examined under LD conditions. As shown in Figure 8A, the early flowering observed in pub13 was suppressed in both pub13sid2-2 and pub13pad4, while the flowering time of sid2-2 and pad4 single mutants was the same as in Col-0. The leaf number of pub13sid2-2 and pub13pad4 before the appearance of the first floral bud was almost the same as in Col-0, which was four to five leaves more than pub13 (Fig. 8B). Analysis of the expression level of flowering marker genes in these plants under LD conditions revealed that the expression of FLC was suppressed in pub13 but was restored in pub13sid2-2 and pub13pad4, and the expression level of FLC in sid2-2 and pad4 was comparable to that in Col-0 (Fig. 8C). On the contrary, the transcript levels of SOC1 and FT were increased in pub13 but were similar in pad4, sid2-2, and Col-0. However, the expression levels of SOC1 and FT were reduced in pub13sid2-2 and pub13pad4 compared with those in pub13 (Fig. 8C). These results suggest that PUB13 regulates flowering time in a SA signaling-dependent manner.
Figure 8.
Flowering phenotypes and expression of flowering marker genes in the pub13sid2-2 and pub13pad4 double mutants. A, Flowering phenotypes of pub13, pub13sid2-2, and pub13pad4 grown under LD conditions. B, Leaf number of Col-0, pub13, sid2-2, pub13sid2-2, pad4, and pub13pad4 grown under LD conditions. Leaf number for each genotype was counted once the first flower bud appeared. Statistical analysis was carried out as for Figure 5. C, Flowering marker gene transcription levels. Gene expression of FLC, SOC1, and FT in 4-week-old plants grown under LD conditions was detected with RT-PCR. Actin (ACT) was used as a loading control.
DISCUSSION
The ubiquitin proteasome system (UPS)-mediated protein modification and degradation has been recognized as a critical mechanism in the regulation of numerous cellular processes in plants. The importance of the UPS in plant innate immunity and flowering has been well documented in plants (Henriques et al., 2009; Trujillo and Shirasu, 2010). Many UPS-related components have been implicated in either of the two biological processes. Nevertheless, the interconnection between the signaling pathways underlying these two processes via the UPS has not yet been reported. In this study, we extensively analyzed the functions of the Arabidopsis PUB13 gene in both innate immunity and flowering. We found that PUB13 encodes a U-box/ARM repeat protein endowed with E3 ligase activity. Genetic and physiological analysis revealed that PUB13 negatively regulates cell death, H2O2 accumulation, and defense against biotrophs but positively regulates the resistance to necrotrophic pathogens. We discovered that PUB13 is a negative regulator of flowering time under MD and LD conditions and that the PUB13-mediated regulation of flowering time is probably through the SOC1-mediated signaling pathway. Our results revealed dual roles for PUB13 and provided novel evidence that innate immunity and development are interconnected via the UPS in Arabidopsis.
SA is a critical signaling molecule in the pathways of local and systemic resistance in plants. In pub13, the elevated SA level is associated with enhanced defense responses. After suppressing SA in pub13 through the introduction of sid2-2 or pad4, the enhanced defense responses of pub13 are largely repressed. Therefore, PUB13 regulates plant defense responses through a SA-dependent pathway. Stresses usually can promote plant early flowering and SA accumulation (Wada and Takeno, 2010; Wada et al., 2010). Previous research showed that SA is a positive regulator of flowering not only in stressed plants but also in nonstressed plants (Martínez et al., 2004). We found that the SA level, cell death, and H2O2 accumulation, which are usually altered as responses to stress, are elevated in pub13 in a nonstressed environment, suggesting that these responses in pub13 trigger early flowering, perhaps by mimicking stress signaling.
Genetic and biochemical analyses indicated that early flowering is suppressed when the SA level in pub13 is reduced by knocking out SID2 or PAD4. Thus, the genes involved in SA-mediated defense signaling, PUB13, SID2, and PAD4, are also involved in floral transition control, which suggests that plant innate immunity and development are intimately linked at SA-mediated signaling. However, it is still unclear how PUB13 regulates flowering through the SID2/PAD4-mediated SA signaling pathway. A previous study suggested that SA regulates flowering time probably through the photoperiod and vernalization flowering pathways that are independent on FLC, CO, and FCA (Martínez et al., 2004). Consistent with this notion, we found in this study that the transcript levels of the flowering marker genes FLC, SOC1, and FT remain unchanged in sid2-2 and pad4 single mutants. Therefore, further study is needed to find out whether PUB13-SID2/PAD4 also function in other flowering pathways and how the corresponding flowering pathway interconnects with plant innate immunity through PUB13-SID2/PAD4.
Numerous studies have shown that effective plant resistance to biotrophs is largely dependent on PCD and the activation of defense responses regulated by the SA pathway. On the contrary, necrotrophs benefit from the cell death of host plants, and the host defense responses are mainly modulated by the JA/ET pathways. We found that the pub13 mutant contains elevated levels of SA and confers enhanced resistance against four biotrophs (Psm ES4326, E. cichoracearum UCSC1, P. syringae pv tomato DC3000, and H. parasitica Noco2). When the SA levels are reduced in pub13 through introducing sid2-2 or pad4, elevated disease resistance to the biotrophs is abolished, indicating that SA signaling is indispensable for PUB13-regulated resistance to biotrophs. Conceivably, due to antagonism between SA and JA/ET, the JA/ET signal and PDF1.2 expression are suppressed in pub13, which displays susceptibility to two necrotrophs (B. cinerea BO5-10 and A. brassicicola AB) under high-humidity conditions.
A role of PUB proteins in PTI has been reported in recent years. For example, a homologous triplet of PUB proteins, PUB22, PUB23, and PUB24, in Arabidopsis negatively regulates PTI in response to multiple PAMPs (Trujillo et al., 2008). A more recent study also showed that PUB13 is involved in preventing prolonged/excessive activation of FLS2-mediated PTI (Lu et al., 2011). PUB13 and its close homolog PUB12 were found to polyubiquitinate their substrate FLS2, a pattern-recognition receptor of bacterial flagellin. PUB13 and PUB12 promote FLS2 degradation after the cells are stimulated by flg22, a 22-amino acid peptide derived from the conserved region of bacterial flagellin (Lu et al., 2011). The receptor-like kinase BAK1, which forms a receptor complex with FLS2 immediately upon flagellin stimulation, can phosphorylate both PUB13 and PUB12, and this phosphorylation is required for the flagellin-induced FLS2-PUB13/PUB12 association (Chinchilla et al., 2007; Lu et al., 2011). Furthermore, flagellin-induced transient reactive oxygen species burst is increased in the pub12 and pub13 single mutants compared with the wild type, although FLS2 is not constitutively accumulated in pub12/pub13 (Lu et al., 2011). To further confirm the function of PUB13 in PAMP signaling, we examined pretreated flg22-induced resistance in pub13 against subsequent infection of the virulent strain DC3000. The flg22-induced resistance in pub13 is not significantly different compared with the wild type (data not shown), probably due to functional redundancy between PUB13 and PUB12. Since PUB13 ubiquitinates FLS2 and plays a negative role in FLS2 signaling, we also investigated whether FLS2 functions in flowering. We planted fls2 mutant and FLS2 transgenic lines in the Col-0 background under MD and LD conditions, respectively, and found that the flowering phenotypes of fls2 and FLS2 transgenic plants are the same as in the wild type (data not shown), suggesting that FLS2 does not relate to flowering.
Sequence and gene structure analyses revealed that PUB13 is the putative ortholog of the rice E3 ligase gene SPL11. SPL11 was found to negatively regulate cell death and defense but to positively regulate flowering time under LD conditions, probably through ubiquitination of SPIN1 in rice (Zeng et al., 2004; Vega-Sánchez et al., 2008). In this study, we report that PUB13 regulates cell death, defense, as well as flowering time through a SA-dependent pathway. Although PUB13 plays a similar role to SPL11 in defense, it acts as a negative regulator of flowering time in Arabidopsis under LD and MD conditions. This difference is due to the fact that Arabidopsis is a LD plant and rice is a SD plant. Interestingly, our genetic complementation showed that the early-flowering and cell death phenotypes of pub13 were restored when the rice Spl11 gene was expressed in the pub13 plants under the control of the 35S promoter (Supplemental Fig. S6), indicating that the functions of PUB13/SPL11 are highly conserved in dicot and monocot plants. Further characterization of PUB13/SPL11 and other associated components, therefore, should provide exciting insights into the interconnection and coordination of innate immunity and development in plants.
MATERIALS AND METHODS
Plants, Growth Conditions, and High-Humidity Treatment
Arabidopsis (Arabidopsis thaliana) wild-type (Col-0), pub13 (Salk_093164), sid2-2 (Wildermuth et al., 2001), pad4 (Glazebrook et al., 1996), and fls2 (Xiang et al., 2008) were used in this study. pub13 was obtained from the Arabidopsis Biological Resource Center and confirmed with PCR primers: PUB13-1F (5′-ATGGAGGAAGAGAAAGCTTC-3′), LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′), and PUB13-1074R (5′-CATAAGATCTTCAATCTTGTTCGC-3′). pub13sid2-2 and pub13pad4 were made by genetic crosses. To generate PUB13-complemented pub13 (35S:PUB13/pub13), the full-length cDNA of PUB13 was amplified from Col-0 cDNA and cloned into pEarleyGate 101. Then, the Agrobacterium tumefaciens-mediated flower-dipping method of transformation was performed according to a standard protocol to get 35S:PUB13/pub13 transgenic plants. To generate PUB13 RNAi plants, an artificial microRNA targeting PUB13 was cloned into the binary vector pKANNIBAL as described previously (Schwab et al., 2006), and the new construct was transformed into Col-0. FLS2 transgenic plants were made as described (Xiang et al., 2008). Plants were grown under 22°C, 70% RH, and light intensity of 250 μmol photons m−2 s−1 with 8 h of daylight/16 h of dark as SD, 12 h of daylight/12 h of dark as MD, or 16 h of daylight/8 h of dark as LD. For high-humidity treatment, plants were transferred to a dew chamber and a humidifier was used to keep the humidity about 95% RH.
Trypan Blue and DAB Staining
Trypan blue staining was performed for cell death assay as described previously (Bowling et al., 1994). To determine the accumulation of H2O2, we stained the selected leaves with DAB as described previously (Wohlgemuth et al., 2002). Briefly, leaves were stained with 0.1% (w/v) DAB for 8 h in the dark, destained with 95% ethanol, and preserved in 50% ethanol. The leaves for trypan blue or DAB staining were pretreated with high humidity for 48 h.
Pathogen Inoculation
Four-week-old plants grown under LD conditions were inoculated with different pathogens. To detect the humidity effect on disease resistance, the plants were treated with high humidity (95% RH) for 24 h, and then the treated plants or detached leaves were used for inoculation with different pathogens. Pseudomonas syringae pv maculicola ES4326 and Pseudomonas syringae pv tomato DC3000 were sprayed on the plants with 1 × 108 CFU mL−1 for disease symptoms or injected with 5 × 105 CFU mL−1 for bacterial growth assay. The flg22 protection analysis was performed as described previously (Zhang et al., 2010). Erysiphe cichoracearum UCSC1 was inoculated as described (Vorwerk et al., 2007), and the macroscopic symptoms were observed at 7 dpi and the microscopic symptoms were checked at 10 dpi after trypan blue staining. For Botrytis cinerea inoculation, the fungus was cultured on a potato dextrose agar plate for 2 weeks at 24°C with a 12-h photoperiod. The fungal culture was washed with water and filtered with a nylon mesh. Then, the conidia were resuspended in potato dextrose broth and the concentration was adjusted to 1 × 104 conidia mL−1. The detached rosette leaves were placed in petri dishes containing 0.8% agar, and 5 μL of conidia suspension was dropped on the leaf surface. The petri dishes with the inoculated leaves were incubated at 22°C with a 12-h photoperiod. The diameter of the lesion was measured at 3 dpi. Inoculation of detached leaves with Alternaria brassicicola (1 × 105 spores mL−1) was performed as described previously (van Wees et al., 2003). Photographs were taken and lesion diameter was measured at 3 dpi. For the inoculation of Hyaloperonospora parasitica Noco2, plants were sprayed with 1 × 105 spores mL−1, then the inoculated plants were kept under 90% RH humidity and 17°C conditions. Symptoms were observed and lesion diameter was measured at 7 dpi.
E3 Ubiquitin Ligase Activity Assay
The full-length coding sequence of PUB13 (1,983 bp) was cloned into the pGEX-6P-1 vector, which contains a GST tag. PUB13 carrying the mutation of Val-273 to Arg (V273R) was generated using the QuikChange Site-Directed Mutagenesis kit (Stratagene; no. 200518) with the following primers: M1F (5′-TCGCTGGAAATGATGAGAGATCCACGTATTGTTTCATCAG-3′) and M1R (5′-CTGATGAAACAATACGTGGATCTCTCATCATTTCCAGCGA-3′). The GST-PUB13/V273R fusion protein was expressed in Escherichia coli BL21, and 1.0 μg of GST-PUB13/V273R was used for each reaction. Arabidopsis ubiquitin (an ubiquitin monomer of UBQ14 [At4g02890]; approximately 2.0 μg) fused with a His tag, wheat (Triticum aestivum) E1 (GI: 136632; approximately 40.0 ng), and human E2 (UBCH5b; approximately 40.0 ng) were used in the in vitro E3 ligase activity assays as described (Xie et al., 2002). The reaction samples were separated by 10% SDS-PAGE and detected by western blot with the nickel-horseradish peroxidase or anti-GST antibody.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number At3g46510.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid comparison of the U-box domain in PUB13 and SPL11, and identification of the T-DNA mutant pub13.
Supplemental Figure S2. Cell death and H2O2 accumulation in pub13 after high-humidity treatment.
Supplemental Figure S3. Disease resistance of pub13 under high humidity.
Supplemental Figure S4. Defense-related gene expression and SA level in pub13.
Supplemental Figure S5. Flowering phenotypes of PUB13 RNAi and complemented pub13 plants.
Supplemental Figure S6. Flowering phenotypes of Spl11-complemented pub13 transgenic plants.
Supplemental Table S1. Primers used for real-time PCR and RT-PCR.
Supplementary Material
Acknowledgments
We are grateful to Drs. Xinnian Dong and Mohan Rajinikanth at Duke University for providing sid2-2 and pad4 seeds.
References
- Azevedo C, Santos-Rosa MJ, Shirasu K. (2001) The U-box protein family in plants. Trends Plant Sci 6: 354–358 [DOI] [PubMed] [Google Scholar]
- Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S. (2000) A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J 24: 591–599 [DOI] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X. (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6: 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Farmer LM, Book AJ, Lee KH, Lin YL, Fu H, Vierstra RD. (2010) The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis. Plant Cell 22: 124–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM. (1996) Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143: 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Lamothe R, Tsitsigiannis DI, Ludwig AA, Panicot M, Shirasu K, Jones JD. (2006) The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18: 1067–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. (2001) U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem 276: 33111–33120 [DOI] [PubMed] [Google Scholar]
- Henriques R, Jang IC, Chua NH. (2009) Regulated proteolysis in light-related signaling pathways. Curr Opin Plant Biol 12: 49–56 [DOI] [PubMed] [Google Scholar]
- Jang IC, Yang JY, Seo HS, Chua NH. (2005) HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kawai-Yamada M, Ohori Y, Uchimiya H. (2004) Dissection of Arabidopsis Bax inhibitor-1 suppressing Bax-, hydrogen peroxide-, and salicylic acid-induced cell death. Plant Cell 16: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ. (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L. (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Tenhaken R. (2000) Defence gene expression in soybean is linked to the status of the cell death program. Plant Mol Biol 44: 209–218 [DOI] [PubMed] [Google Scholar]
- Martínez C, Pons E, Prats G, León J. (2004) Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J 37: 209–217 [DOI] [PubMed] [Google Scholar]
- Mosher S, Moeder W, Nishimura N, Jikumaru Y, Joo SH, Urquhart W, Klessig DF, Kim SK, Nambara E, Yoshioka K. (2010) The lesion-mimic mutant cpr22 shows alterations in abscisic acid signaling and abscisic acid insensitivity in a salicylic acid-dependent manner. Plant Physiol 152: 1901–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgil Y, Shiu SH, Stone SL, Salt JN, Goring DR. (2004) A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol 134: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Gould KL. (2002) Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA 8: 798–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M, Ichimura K, Casais C, Shirasu K. (2008) Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr Biol 18: 1396–1401 [DOI] [PubMed] [Google Scholar]
- Trujillo M, Shirasu K. (2010) Ubiquitination in plant immunity. Curr Opin Plant Biol 13: 402–408 [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- van den Burg HA, Tsitsigiannis DI, Rowland O, Lo J, Rallapalli G, Maclean D, Takken FL, Jones JD. (2008) The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell 20: 697–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees SC, Chang HS, Zhu T, Glazebrook J. (2003) Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol 132: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Sánchez ME, Zeng L, Chen S, Leung H, Wang GL. (2008) SPIN1, a K homology domain protein negatively regulated and ubiquitinated by the E3 ubiquitin ligase SPL11, is involved in flowering time control in rice. Plant Cell 20: 1456–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorwerk S, Schiff C, Santamaria M, Koh S, Nishimura M, Vogel J, Somerville C, Somerville S. (2007) EDR2 negatively regulates salicylic acid-based defenses and cell death during powdery mildew infections of Arabidopsis thaliana. BMC Plant Biol 7: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada KC, Takeno K. (2010) Stress-induced flowering. Plant Signal Behav 5: 944–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada KC, Yamada M, Shiraya T, Takeno K. (2010) Salicylic acid and the flowering gene FLOWERING LOCUS T homolog are involved in poor-nutrition stress-induced flowering of Pharbitis nil. J Plant Physiol 167: 447–452 [DOI] [PubMed] [Google Scholar]
- Wang GF, Seabolt S, Hamdoun S, Ng G, Park J, Lu H. (2011) Multiple roles of WIN3 in regulating disease resistance, cell death, and flowering time in Arabidopsis. Plant Physiol 156: 1508–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Wohlgemuth H, Mittelstrass K, Kschieschan S, Bender J, Weigel HJ, Overmyer K, Kangasjarvi J, Sandermann H, Langebartels C. (2002) Activation of an oxidative burst is a general feature of sensitive plants exposed to the air pollutant ozone. Plant Cell Environ 25: 717–726 [Google Scholar]
- Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, et al. (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol 18: 74–80 [DOI] [PubMed] [Google Scholar]
- Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH. (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170 [DOI] [PubMed] [Google Scholar]
- Yang CW, González-Lamothe R, Ewan RA, Rowland O, Yoshioka H, Shenton M, Ye H, O’Donnell E, Jones JD, Sadanandom A. (2006) The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18: 1084–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee D, Goring DR. (2009) The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot 60: 1109–1121 [DOI] [PubMed] [Google Scholar]
- Yin Z, Chen J, Zeng L, Goh M, Leung H, Khush GS, Wang GL. (2000) Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol Plant Microbe Interact 13: 869–876 [DOI] [PubMed] [Google Scholar]
- Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, Yang H, Lee J, Liu X, Lopez J, Lin C. (2007) Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19: 3146–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LR, Park CH, Venu RC, Gough J, Wang GL. (2008) Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol Plant 1: 800–815 [DOI] [PubMed] [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL. (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.