Abstract
Purpose.
Chlamydia trachomatis is the leading infectious cause of blindness. The goal of the current study was to search for biomarkers associated with C. trachomatis–induced ocular pathologies.
Methods.
We used a whole genome scale proteome array to systematically profile antigen specificities of antibody responses to C. trachomatis infection in individuals from trachoma-endemic communities with or without end-stage trachoma (trichiasis) in The Gambia.
Results.
When 61 trichiasis patients were compared with their control counterparts for overall antibody reactivity with organisms of different chlamydial species, no statistically significant difference was found. Both groups developed significantly higher titers of antibodies against C. trachomatis ocular serovars A and B than ocular serovar C, genital serovar D, or Chlamydia psittaci, whereas the titers of anti–Chlamydia pneumoniae antibodies were the highest. When antisera from 33 trichiasis and 26 control patients (with relatively high titers of antibodies to C. trachomatis ocular serovars) were reacted with 908 C. trachomatis proteins, 447 antigens were recognized by at least 1 of the 59 antisera, and 10 antigens by 50% or more antisera, the latter being designated as immunodominant antigens. More importantly, four antigens were preferentially recognized by the trichiasis group, with antigens CT414, CT667, and CT706 collectively reacting with 30% of trichiasis antisera but none from the normal group, and antigen CT695 reacting with 61% of trichiasis but only 31% of normal antisera. On the other hand, eight antigens were preferentially recognized by the control group, with antigens CT019, CT117, CT301, CT553, CT556, CT571, and CT709 together reacting with 46% of normal antisera and none from the trichiasis group, whereas antigen CT442 reacted with 35% of normal and 19% of trichiasis antisera respectively.
Conclusions.
The current study, by mapping immunodominant C. trachomatis antigens and identifying antigens associated with both ocular pathology and protection, has provided important information for further understanding chlamydial pathogenesis and the development of subunit vaccines.
Whole genome scale profiling of antigen specificities of anti–Chlamydia trachomatis antibodies in trachoma patients led to the identification of antigens associated with ocular pathology.
Introduction
The obligate intracellular bacterium Chlamydia trachomatis is the leading cause of both infectious blinding diseases1 and sexually transmitted bacterial diseases2 worldwide. Repeated or persistent ocular infection with C. trachomatis can cause inflammatory pathologies in the eye, leading to trachoma, trachomatous trichiasis, and blindness. Despite extensive efforts in defining the role of host3–7 and bacterial factors8–11 in ocular inflammatory pathologies, chlamydial pathogenic mechanisms remain unclear. The main C. trachomatis serovars responsible for trachoma are A, B, and C, whereas C. trachomatis serovars D to L3 mainly cause urogenital tract infection.
To understand the molecular mechanisms of chlamydial pathogenesis and immunity in humans, efforts have been made to identify serological markers of Chlamydia-induced pathologies in either the female upper genital tract or ocular tissues. Antibodies to chlamydial heat shock protein 60 (HSP60) have been associated with Chlamydia-induced inflammatory damage in fallopian tubes,12–14 therefore the possibility arises that chlamydial HSP60 and host immune responses to HSP60 may contribute to chlamydial pathogenicity. There is evidence that shows that chlamydial HSP60 can directly activate macrophages to secrete inflammatory cytokines15 and that chlamydial HSP60 can also be the target of16,17 or promote16,18 T-cell responses; however, because of the high degree of amino acid sequence conservation between chlamydial and human HSP60 proteins, antibodies against chlamydial HSP60 may cross-react with host HSP60.19,20 A whole genome scale protein array profiling of antigen specificities of antibody responses to C. trachomatis urogenital infection has confirmed the association of HSP60 with tubal inflammatory damage and also identified new serological markers, such as OmcB, which are preferentially recognized by serum from patients with tubal factor infertility.21 A recent study also reported high titers of antibodies to C. trachomatis HSP60, chlamydial protease/proteasome-like activity factor (CPAF), and CT795 in Nepalese trachoma patients.9 In the current study, we compared the antigen specificity profiles of antibody responses to C. trachomatis in individuals with trichiasis and healthy controls from trachoma-endemic communities. This has revealed a map of the immunodominant C. trachomatis antigens in trachoma and has identified antigens associated with both protection and pathology in trichiasis.
Materials and Methods
Ethical Permission and Study Participants
The study was conducted in accordance with the tenets of the Declaration of Helsinki. The study and its procedures were approved by the joint Gambian Government/Medical Research Council Ethics Committee (SCCL2006.10 and 18). Informed consent was obtained before the enrollment of each subject. Participants were recruited from the Western, Central, and Lower River Regions of The Gambia between May 2006 and February 2009. Trachoma was graded using the World Health Organization simplified grading system by a single experienced field supervisor. Subjects with trachomatous conjunctival scarring (TS) who also had trichiasis (TT), defined as at least one eyelash touching the globe of the eye, were identified. For each TT case, an age, sex, and location–matched control subject with normal eyes who was not a member of the same family was also recruited. Participants were age matched within 5 years (up to 45 years of age) or 10 years (participants older than 45 years). In a standardized manner, an ocular swab from the everted tarsal conjunctiva of each participant was collected as described previously.5,22 A venous blood sample was requested for the isolation of peripheral blood mononuclear cells and plasma as described previously.23 All subjects requiring a lid margin rotation procedure were offered unilamella tarsal rotation (Trabut) surgery free of charge, which was carried out by Gambian National Eye Care Programme ophthalmic nurses in regional health centers or in the homes of the patients. All other conditions requiring treatment were referred to regional centers for care.
Defibrination of Heparinized Plasma
Plasma was converted to serum using a previously described method.24 Briefly, a solution of 100 IU/mL of thrombin (Sigma, Poole, UK) was prepared by addition of 1M CaCl2. Protamine sulfate was then added to a final concentration of 5 mg/mL. Then, 10 μL of this solution was added to 1 mL of each plasma sample. After vigorous vortex mixing, the sample was incubated at room temperature (23°C) for 1 hour followed by centrifugation at 10,000g for 20 minutes at 4°C. The supernatant was carefully collected and the clot discarded. The supernatant was aliquoted and frozen at −20°C until used. Frozen samples were transported between the laboratories involved in different countries on a dry ice cold chain.
Detection of Current C. trachomatis Ocular Infection
The AMPLICOR CT/NG test (Roche Diagnostics, Indianapolis, IN) was used for detection of C. trachomatis in cases and controls as described elsewhere.3 Positive and negative samples were assigned according to the manufacturer's instructions. Sample quality, inhibition, and sufficiency for PCR were assessed by PCR amplification of human mitochondrial DNA–specific sequences as described previously.3
Cell Culture and Chlamydial Infection
HeLa cells (American Type Culture Collection, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium (DMEM; GIBCO PRL, Rockville, MD) with 10% fetal calf serum (FCS; GIBCO BRL) at 37°C with 5% CO2 as described previously.14,25,26 C. trachomatis serovars A, B, C, and D; Chlamydia psittaci 6BC; or Chlamydia pneumoniae AR39 organisms were grown, purified, and titrated as previously described.14,27–29 After titration, organisms were stored in aliquots at −80°C. For immunofluorescence assay, chlamydial organisms were used to infect HeLa cells grown on glass coverslips in 24-well plates. The subconfluent HeLa cells were treated with DMEM containing 30 μg/mL of diethylaminoethyl (DEAE)-Dextran (Sigma, St. Louis, MO) for 10 minutes at 37°C. After removal of DEAE-Dextan solution, chlamydial organisms were added to the wells for 2 hours at 37°C. The infected cells were continuously cultured in DMEM with 10% FCS and 2 μg/mL of cycloheximide (Sigma) for 30 (C. psittaci), 48 (C. trachomatis), or 72 (C. pneumoniae) hours before being processed for immunofluorescence assay as described in the following section. Infection was also carried out in tissue culture flasks for preparing whole cell lysates.
Immunofluorescence Assay
Antichlamydial antibodies in human sera were titrated using an immunofluorescence assay (IFA) as previously described.14,26,30 Briefly, HeLa cells infected with different species and serovars of chlamydia were fixed with 2% paraformaldehyde and permeabilized with 2% saponin. After blocking, human antisera was added to the chlamydia-infected cell samples. The primary Ab binding was visualized with a goat anti-human IgG conjugated with Cy3 (red; Jackson ImmunoResearch Laboratories, West Grove, PA), and DNA was labeled with Hoechst dye (blue; Sigma-Aldrich, St. Louis, MO). The highest dilution of a serum that still gave a positive reactivity was defined as the titer of the given serum sample. Serum samples were serially diluted and the appropriate dilutions were repeated multiple times based on the results obtained from prior dilutions to obtain a more accurate titer for each serum. The slides were observed under an Olympus AX70 fluorescence microscope equipped with multiple filter sets (Olympus, Melville, NY), as described previously.14,30
Chlamydial Fusion Protein-arrayed Microplate Enzyme-linked Immunosorbent Assay
Glutathione S-transferase (GST) fusion protein enzyme-linked immunosorbent assay (ELISA) for detecting human antibody recognition of chlamydial proteins was carried out as described previously.14,21,26 The C. trachomatis proteome array was established based on the 908 open reading frames (ORFs) encoded by C. trachomatis genital serovar D genome and plasmid.26 Bacterial lysates containing individual chlamydial GST fusion proteins were added to 96-well microplates precoated with glutathione (Pierce, Rockford, IL) at a 1:10 dilution in PBS with a total volume of 200 μL/well. Lysates containing GST alone, as negative, and GST-chlamydial protease/proteosomelike activity factor (CPAF), as positive controls, were also included on each plate. The plates were incubated overnight at 4°C to allow GST fusion proteins to bind to the plate-immobilized glutathione then blocked with 2.5% milk in PBS and washing with PBST (PBS with 0.05% Tween 20; Sigma-Aldrich). After blocking, human serum samples were preabsorbed with bacterial lysates containing GST alone at 4°C overnight, then incubated with glutathione beads (bioWorld, Dublin, OH) for 1 hour at room temperature. This pretreatment can significantly reduce background caused by nonspecific human antibodies. The human antibody reactivity was detected with a goat anti-human-IgG, IgA, IgM conjugated with horseradish peroxidase (HRP; Jackson ImmunoResearch Laboratories) plus the substrate 2,2′-azino-bi(2-ethylbenzothiazoline-6-sulforic acid) diammonium salt (ABTS; Sigma). The optical density (OD) was measured at 405 nm using a microplate reader (Molecular Devices Corporation, Sunnyvale, CA). To confirm the antibody binding specificity, all antisera were further absorbed with lysates made from either HeLa cells alone or Chlamydia-infected HeLa cells before reacting with the fusion protein-coated plates. The antibody binding that remained positive after HeLa-alone lysate absorption but significantly reduced by Chlamydia-HeLa lysate absorption was considered true positive. In addition, the fusion protein-based array assay is also routinely evaluated using normal human sera similarly preabsorbed as described previously for further excluding nonspecific binding.11,26,30,31 In this particular experiment, we used the proteome to assay sera from eight health individuals and no significant binding was detected between any of the 908 fusion proteins and the normal human sera (data not shown). These eight sera displayed no significant reactivity with C. trachomatis–infected cells (data not shown).
Data Analyses
Data were analyzed using SPSS v. 15.0 software (IBM, Chicago, IL) as previously described.14,21 Student's t-test was used to analyze the overall mean differences in anti-Chlamydia antibody titers between the two groups of patients or between different chlamydial organisms. Because prior analyses of antibodies against some or all Chlamydia species in trachoma patients showed no significant difference between case and control groups,10,32 a two-tailed test seems to be more appropriate for capturing all possible associations of antibody responses with a given group. Because the antibody titers had large variations within a given group, the serum titers were evaluated by ranges of less than 1:100 (Negative), 1:100 to 1:10,000 (Low), and greater than 1:10,000 (High). Chi-squared and Fisher's exact test were used to compare overall antibodies to various chlamydial species and serovars.
For ELISA results, both Student's t-test (for comparing quantitative OD value data reflecting reactivity intensity) and Fisher's exact test (for comparing the number of antisera positively reacted with a given antigen as reactivity frequency) were performed. When Student's t-test was used, the OD values after subtracting background from the same plate were used. When Fisher's exact test was used, a response was determined positive when the OD value was equal to or greater than 2 SDs above the mean calculated from the same 96-well plate as previously described.21 We accepted that there would be some false-positive associations when screening antisera by the array and did not adjust P values to account for multiple comparisons because the normal methods of correcting P values and the general applicability of adjusting for multiplicity of tests remain contentious among biostatisticians.33–35 We did not estimate the power of the study because, in the absence of other proteome array studies with sera from trachoma endemic communities, it was not possible to estimate the size of the expected differences or the number of antigens that may be recognized. Unadjusted P values after multiple testing are presented with the expected significant P value level after Bonferroni correction presented in the footnotes.35 We calculated the potential number of false-positive reactive antigen-antisera reactions at the 5% level according to the method described by Stekel.34 At a P value less than 0.05, the number of potential false-positive antigens reactive with sera is expected to be 0.05 × 19 = 1 when the 19 antigens listed Table 3 are analyzed statistically.
Table 3.
Comparison of Antibodies against C. trachomatis Serovars A, B, C, and D; C. psittaci; and C. pneumoniae
|
Student'st-Test (mean ± SD) (n = 122) |
C. trachomatis |
C. psittaci | C. pneumoniae | ||||
|
Serovar A |
Serovar B |
Serovar C |
Serovar D |
6BC |
AR39 |
||
| C. trachomatis | Serovar A (23,043.4 ± 42,671.8) | 1 | 0.5468 | 0.0088 | 0.0015 | <0.0001 | 0.0567 |
| Serovar B (20,162.3 ± 30,988.4) | 1 | 0.0123 | 0.0011 | <0.0001 | 0.0039 | ||
| Serovar C (11,892.6 ± 18,755.8) | 1 | 0.3446 | 0.0006 | <0.0001 | |||
| Serovar D (9810.7 ± 15,422.8) | 1 | 0.0067 | <0.0001 | ||||
| C. psittaci | 6BC (5485.2 ± 8214.4) | 1 | <0.0001 | ||||
| C. pneumo-niae | AR39 (32,759.0 ± 36,340.3) | 1 | |||||
The antibodies against different chlamydial organisms were compared against each other using ANOVA (data not shown) and Student's t-test. The titers of anti–C. trachomatis serovar A or B antibodies in the 122 subjects are significantly higher than those of anti–C. trachomatis serovar C or D, suggesting that these 122 individuals were mainly infected with ocular serovars A and B. The titers of antibodies against C. psittaci are the lowest, whereas those of anti–C. pneumoniae antibodies were highest in the same 122 individuals, which both confirms that the titers of antibodies detected in the assay were not caused by cross-reactivity and indicates that most of the 122 individuals were also coinfected with C. pneumoniae but not C. psittaci.
Results
Patient Demographics
The study subject demographics information is listed in Table 1. Sixty-one case control pairs were screened against the different chlamydial species and serotypes used in the study (see Supplementary Material, available at: http://www.iovs.org/content/53/6/2551/suppl/DC1). Fifty-nine of these individual serum samples were then screened against the complete proteome array. Both groups had similar age, sex, and ethnicity distributions. Fifty-seven percent of the cases had TS/ TT in both eyes. Overall, 10% of the cases had corneal opacity (CO) in at least one eye and phthisis (wasting) was observed in the left eye of a single individual who also had right eye TS/TT/CO. The ethnic groups from which the samples were collected were consistent with main ethnicities present in the Gambian districts and regions in which the study was performed. All ocular swabs for these samples were positive for human mitochondrial DNA. A single sample (L120) was positive by C. trachomatis/Neisseria gonorrhoeae (CT/NG) Amplicor (F. Hoffmann-La Roche Ltd., Basel, Switzerland) for current ocular infection with C. trachomatis and this is consistent with previous findings in The Gambia and elsewhere in Africa.22,36,37 Tests for current infection with other ocular pathogens were not performed.
Table 1.
Patient Demographics
|
Individuals Screened against Whole Organisms |
Individuals Screened against 908 Fusion Proteins |
|||||
| Group | Cases | Controls | Cases | Controls | ||
| Group Size | 61 | 61 | 33 | 26 | ||
|
Age (range), y |
60 (30–85) |
55 (28–80) |
59 (30–85) |
52 (31–72) |
||
| Sex | Male | 19 | 19 | 12 | 7 | |
| Female | 42 | 42 | 21 | 19 | ||
| Ethnicity | Mandinka | 23 | 22 | 7 | 5 | |
| Jola | 22 | 23 | 15 | 9 | ||
| Manjago | 14 | 15 | 9 | 12 | ||
| Other | 2 | 1 | 1 | 0 | ||
| Pathology | Uni | TS/TT | 22 | 0 | 14 | 0 |
| +CO | 4 | 0 | 1 | 0 | ||
| Bi | TS/TT | 33 | 0 | 16 | 0 | |
| +CO | 2 | 0 | 2 | 0 | ||
| No. of CT positive (Amplicor result) | 1 | 0 | 1 | 0 | ||
Sixty-one case control pairs were first screened against the different chlamydial species and serotypes as described in the Materials and Methods section. Fifty-nine of these individual serum samples with relatively high titers of antibodies against ocular serovars were further screened against 908 C. trachomatis fusion proteins. All study groups displayed similar age, sex, and ethnicity distribution. The ethnic groups from which the samples were collected were consistent with main ethnicities present in the Gambian districts and regions in which the study was performed. A single sample (L120) was positive by CT/NG Amplicor for current ocular infection with C. trachomatis. Tests for current infection with other ocular pathogens were not performed.
Both Trichiasis and Control Subjects Had High Levels of Antibodies to Ocular C. trachomatis but Not to Genital Serovars or C. psittaciOrganisms
When antisera from 61 trichiasis subjects and 61 control counterparts were reacted with organisms from different chlamydial species, we found no statistically significant difference in the titers of Chlamydia-specific antibodies between the two groups (Table 2). Both groups developed significantly higher titers of antibodies against C. trachomatis ocular serovars A and B than serovar C (Table 3, P < 0.05), suggesting that most of the study subjects had been infected with serovars A and B but not C. Furthermore, a significantly lower level of anti–C. trachomatis genital serovar D antibody was detected in the 122 subjects (P < 0.01, compared with anti-serovar A and B antibodies). Interestingly, in all 122 subjects, the highest titers of antibodies were anti–C. pneumoniae antibodies (higher than those of antiserovars A and B [P < 0.05]). The older ages of the study subjects may contribute to the high titers of anti–C. pneumoniae antibodies as anti–C. pneumoniae antibody titers are known to increase with age. The anti–C. psittaci antibody levels were the lowest among all chlamydial antibodies assayed (P < 0.01).
Table 2.
Titers of Human Antibodies against C. trachomatis, C. psittaci, and C. pneumoniae
| Antigens |
C. trachomatis |
C. psittaci |
C. pneumoniae |
||||||||||
|
Ocular Biovar |
Genital Biovar |
||||||||||||
|
Serovar A |
Serovar B |
Serovar C |
Serovar D |
||||||||||
|
Groups |
Trichiasis (n = 61) |
Control (n = 61) |
Trichiasis (n = 61) |
Control (n = 61) |
Trichiasis (n = 61) |
Control (n = 61) |
Trichiasis (n = 61) |
Control (n = 61) |
Trichiasis (n = 61) |
Control (n = 61) |
Trichiasis (n = 61) |
Control (n = 61) |
|
| Mean | 20,293.4 | 25,793.4 | 19,904.9 | 20,419.7 | 12,809.8 | 10,975.4 | 9806.6 | 9814.7 | 6018.0 | 4952.5 | 28,670.5 | 36,847.5 | |
| SD | 31,157.0 | 51,826.5 | 29,187.4 | 32,932.2 | 20,903.9 | 16,454.1 | 15,823.2 | 15,143.1 | 8818.5 | 7598.2 | 32,955.2 | 39,283.8 | |
| Student's t-test | P = 0.479 | P = 0.927 | P = 0.591 | P = 0.998 | P = 0.476 | P = 0.215 | |||||||
|
Categorization of serum samples into negative-, low-, and high-titer groups | |||||||||||||
| Negative titers (<100) | 1 (2%) | 4 (6%) | 2 (3%) | 3 (5%) | 2 (3%) | 1 (2%) | 2 (3%) | 4 (6%) | 10 (16%) | 9 (15%) | 0 (0%) | 0 (0%) | |
| Low titers (1:100-1:10,000) | 34 (56%) | 36 (59%) | 34 (56%) | 37 (61%) | 43 (70%) | 48 (79%) | 48 (79%) | 44 (72%) | 43 (70%) | 44 (72%) | 26 (43%) | 26 (43%) | |
| High titers (>10,000) | 26 (43%) | 21 (34%) | 25 (41%) | 21 (34%) | 16 (26%) | 12 (20%) | 11 (18%) | 13 (21%) | 8 (13%) | 8 (13%) | 35 (57%) | 35 (57%) | |
| χ2 test | P = 0.303 | P = 0.741 | P = 0.554 | P = 0.604 | P = 0.968 | P = 1 | |||||||
Serum samples from Individuals with (61) or without (61) trichiasis were twofold serially diluted starting with 1:10 and reacted with HeLa cells infected with C. trachomatis ocular serovars A, B, or C or genital serovar D; C. psittaci; or C. pneumoniae organisms. The highest dilution that still gave a positive reactivity was defined as the serum titer. Each serum sample was titrated in triplicate and the average was used as the geometric titer of a given serum sample. Student's t-test was used to analyze the differences between the two groups of patients. There was no statistically significant difference in titers of antibodies against any of the chlamydial organisms between the trichiasis and control groups. When the serum samples were divided into 3 categories (Negative, Low, and High) based on antibody titers, Fisher's Exact test showed no significance difference in any categories between the two groups.
Mapping Chlamydial Immunodominant Antigens in Individuals from a Trachoma-Endemic Area
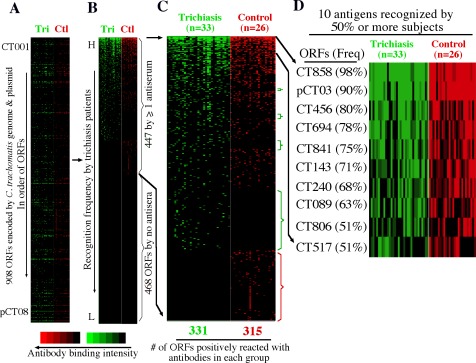
Antisera from 59 individuals (33 trichiasis and 26 control sera) with the highest titers of antibodies to C. trachomatis ocular serovars were further reacted with 908 C. trachomatis proteins. As shown, a total of 447 C. trachomatis-encoded proteins were recognized by at least 1 of the 59 antisera (Figs. 1A, 1B, 1C) and 10 of these were positive in 50% or more serum samples (Fig. 1D). These 10 antigens were designated as immunodominant antigens. Interestingly, 5 of the 10 immunodominant antigens are chlamydial proteins known to be secreted into the host cell cytosol (CT858, pCT03, CT456, CT694, and CT089). CT858, also called CPAF, is a serine protease that is secreted into host cell cytosol via a sec-dependent secretion pathway for manipulating host signaling pathways.25,38–40 pCT03, also called Pgp3, is a plasmid-encoded 28-kDa glycoprotein that is secreted into host cell cytosol with unknown function and by unknown mechanisms.27 CT456, known as translocated actin-recruiting phosphoprotein (TARP), is an effector molecule that is secreted into host cell cytosol via a type III secretion pathway to facilitate internalization of chlamydial elementary bodies (EBs) by inducing actin polymerization.41,42 Interestingly, TARP appears to be highly variable between different serovars of the same species.11,43 CT694 is also a type III secretion effector that modulates host cytoskeletal protein organization during EB invasion,44 whereas CT089 or CopN is a type III secretion pathway regulator.45 Among the remaining five immunodominant antigens, CT143 is a hypothetical protein (HP), whereas CT841 (FtsH) and CT806 (Ptr) are proteolytic enzymes. CT240 is a recombinase (RecR), whereas CT517 is the 50S ribosomal protein L24. Although many of these immunodominant antigens overlap with those recognized by patients who are urogenitally infected with C. trachomatis, the sera from those with ocular exposure appear to selectively target type III secretion effectors for antibody production, suggesting that type III secretion activity may be more prominent or a more important requirement in ocular infection.
Figure 1.
Reactivity of 33 trichiasis and 26 normal control antisera with 908 ORFs of C. trachomatis. Each of the human antibodies (displayed along the x-axis on top of each panel) after 1/1000 dilution was reacted with each of the 908 ORF GST fusion proteins (listed along the y-axis) immobilized onto the 96-well microplates. Each colored bar represents a positive reactivity between a given ORF and an antiserum from trichiasis (Tri, green) or normal control (Ctl, red) group. The results are expressed as OD readings obtained at the wavelength of 405 nm. Any reaction with an OD ≥ mean + 2 SDs calculated from the same plate is defined positive. The positive OD values are expressed as binding intensity in increasing brightness of fluorescent color. The negative OD values are black. The 908 proteins are listed first in order of the ORFs from CT001 to pCT08 (A). The 908 proteins were then reordered based on binding frequency by the trichiasis antisera from high (top) to low (bottom; see [B]). As indicated on the right of the panel, a total of 447 ORFs were recognized by one or more antisera. (C) By visual inspection of the expanded view of the positive antigens, the fusion proteins preferentially recognized by either trichiasis (green) or control (red) antisera were marked with brackets on the right of the panel. (D) A total of 10 antigens were recognized by 50% or more of the 59 antisera.
Identification of C. trachomatis Antigens Differentially Recognized by Trichiasis or Control Groups
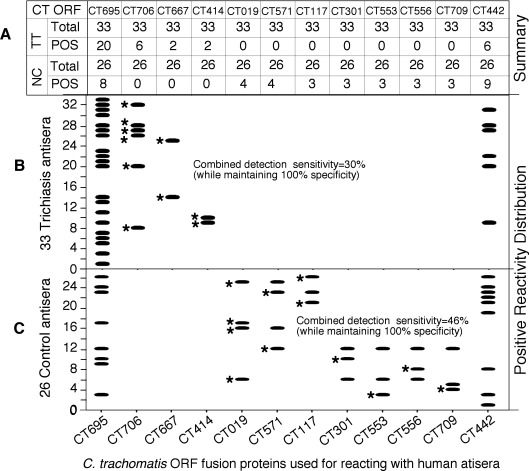
Although most of the 447 antigens were recognized by the 59 antisera from both groups, filtering the results using statistical analyses allowed us to identify 19 antigens that were associated with differential reactivity and preferentially recognized by individual groups (Table 4). A positive reaction between an antigen and an antiserum was defined as a reaction with an OD value equal to or above the mean plus 2 SDs derived from the same microplate and used for calculating reactivity frequency for a given antigen. Both the reactivity intensity (expressed as absolute OD values) and reactivity frequency of an antigen were statistically compared between triachiasis and normal control groups. We found that sera from trichiasis patients preferentially recognized four antigens. Antigens CT414, CT667, and CT706 reacted with 30% of antisera from trichiasis but none from the normal group. Antigen CT695 reacted with 61% of trichiasis but only 31% of normal group antisera (Fig. 2). These four antigens were designated as pathology-associated antigens. CT414 is a polymorphic outer membrane protein designated as PmpC. CT667 is predicted to be a type III secretion effector with unknown function. CT706 is an ATP-dependent ClpP endopeptidase subunit. CT695 is a hypothetical protein encoded by an ORF downstream of CT694, a known type III secretion effector.
Table 4.
Antigens Preferentially Recognized by Trichiasis or Control Subjects
|
ORFs |
Trichiasis (n = 33) | Control (n = 26) |
PValue |
Antigen Affiliation |
|
|
X ± SD (Freq) |
X ± SD (Freq) |
t-Test |
Fisher's |
||
| CT695 | 0.281 ± 0.300 (61%) | 0.189 ± 0.157 (31%) | 0.163 | 0.016 | Pathology-associated |
| CT706 | 0.096 ± 0.165 (18%) | 0.027 ± 0.043 (0%) | 0.044 | 0.024 | |
| CT667 | 0.066 ± 0.084 (6%) | 0.021 ± 0.034 (0%) | 0.012 | 0.201 | |
| CT414 | 0.055 ± 0.088 (6%) | 0.016 ± 0.025 (0%) | 0.034 | 0.201 | |
| CT019 | 0.059 ± 0.058 (0%) | 0.075 ± 0.075 (15%) | 0.337 | 0.019 | Protection-associated |
| CT571 | 0.044 ± 0.048 (0%) | 0.075 ± 0.121 (15%) | 0.185 | 0.019 | |
| CT117 | 0.032 ± 0.049 (0%) | 0.075 ± 0.211 (12%) | 0.256 | 0.045 | |
| CT301 | 0.024 ± 0.036 (0%) | 0.065 ± 0.124 (12%) | 0.077 | 0.045 | |
| CT553 | 0.029 ± 0.030 (0%) | 0.139 ± 0.398 (12%) | 0.123 | 0.045 | |
| CT556 | 0.025 ± 0.041 (0%) | 0.068 ± 0.116 (12%) | 0.054 | 0.045 | |
| CT709 | 0.061 ± 0.053 (0%) | 0.101 ± 0.202 (12%) | 0.281 | 0.045 | |
| CT442 | 0.114 ± 0.167 (18%) | 0.317 ± 0.503 (35%) | 0.034 | 0.087 | |
| CT656 | 0.003 ± 0.007 (0%) | 0.019 ± 0.036 (0%) | 0.012 | NA | The reactivity is too low to be biologically significant. |
| CT404 | 0.028 ± 0.031 (0%) | 0.012 ± 0.016 (0%) | 0.026 | NA | |
| CT279 | 0.008 ± 0.018 (0%) | 0.023 ± 0.030 (0%) | 0.029 | NA | |
| CT368 | 0.022 ± 0.025 (0%) | 0.039 ± 0.039 (0%) | 0.031 | NA | |
| CT651 | 0.043 ± 0.053 (0%) | 0.019 ± 0.023 (0%) | 0.033 | NA | |
| CT817 | 0.010 ± 0.021 (0%) | 0.025 ± 0.031 (0%) | 0.037 | NA | |
| CT864 | 0.009 ± 0.020 (0%) | 0.023 ± 0.030 (0%) | 0.049 | NA | |
When both the reactivity intensity (comparison of raw OD values using Student's t-test) and frequency (Fisher's exact test) listed in Figure 2 were statistically compared between the trichiasis and control groups, a total of 19 antigens (as listed in the first column) showed significant differences in either reactivity intensity or frequency or both between the two groups. However, the reactivity of seven antigens is too low to be biologically significant (bottom portion). Among the remaining 12 antigens, 4 were preferentially recognized by sera from the trichiasis group (top portion) and were designated as pathology-associated antigens, whereas 8 were recognized by the normal control group (middle portion), thus designated as protection-associated antigens.
Figure 2.
Distribution of the reactivity patterns of 12 C. trachomatis antigens with 59 patient antisera. Number of trichiasis (TT) and control (NC) antisera that positively (POS) reacted with each of the 12 antigens (listed at both top and bottom of the figure) is summarized in (A). The reactivity distribution patterns of the 12 antigens with 33 trichiasis and 26 control antisera are revealed in (B) and (C), respectively. Each horizontal bar indicates a positive reactivity. The pathology-associated antigens CT706, CT667, and CT414 reacted with 10 unique antisera from the trichiasis group (each marked with a star, representing 30% of the total trichiasis antisera) but failed to react with any normal control antisera (100% specificity), whereas the protection-associated antigens CT019, CT571, CT117, CT301, CT553, CT556, and CT709 reacted with 12 of the normal antisera (each unique antiserum was marked with a star, representing 46% of the total control antisera) without reacting with any of the trichiasis antisera. However, the pathology-associated antigen CT695 and the protection-associated antigen CT442 reacted with both TT and NC antisera. Nevertheless, CT695 was statistically significantly recognized by TT whereas CT442 by NC respectively as revealed in Table 3.
We also found that eight other antigens were preferentially recognized by the normal control subject group with antigens CT019, CT117, CT301, CT553, CT556, CT571, and CT709 reacting with 46% of antisera from normal but none from trichiasis groups, and CT442 reacting with 35% of the normal and 19% of the trichiasis antisera respectively. Most of these proteins (CT019 [isoleucyl-tRNA synthetase or ileS], CT301 [probable serine/threonine kinase or pknD], CT553 [FMU/SUN-related methyltransferase or fmu], CT709 [rod shape determining protein MREB/HSP70 sugar kinase or mreB]) are metabolic or modifying enzymes. CT117 and CT556 are hypothetical proteins. CT571 is related to the general secretion pathway (GspE), whereas CT442 is a cysteine-rich 15-kDa outer membrane protein.
Discussion
In the current study, we have demonstrated that the overall antibody responses to chlamydial organisms are indistinguishable between individuals with or without trichiasis in trachoma-endemic communities. Most individuals developed high titers of antibodies against both C. trachomatis ocular serovars A and B and C. pneumoniae organisms regardless of the trichiasis status. It is likely that the antibodies detected using C. trachomatis ocular serovar A and B organisms are induced by ocular infection with C. trachomatis serovars A and B in the study subjects and are not as a result of cross-reactivity between C. pneumoniae–induced antibodies and C. trachomatis ocular organisms. This is because these same individuals displayed significantly lower levels of antibodies to C. trachomatis ocular serovar C, genital serovar D, and C. psittaci organisms, although the C. pneumoniae genome is believed to share similar levels of homology with all C. trachomatis serovars46,47 and an even higher level of homology with C. psittaci.48,49 The observation that the antiocular serovar A and B antibody titers were significantly higher than that of antiserovar C antibodies suggests that C. trachomatis ocular serovars A and B were most prevalent in the communities from which our study subjects were recruited. This is in keeping with the results of previous studies in The Gambia, in which serovars A and B were found to be predominant.50,51
To understand the molecular mechanisms of C. trachomatis–induced ocular pathologies and to identify biomarkers for trichiasis, we used a whole genome scale proteome array established based on the 908 ORFs encoded by C. trachomatis genital serovar D genome and plasmid26 to profile the antigen specificities of the anti–C. trachomatis ocular serovar antibodies in the study subjects. The C. trachomatis genital serovar and ocular serovar genomes are highly conserved. For example, there is 99.6% identity between serovars D and A.52 Major variations between the genital and ocular strain genomes are in the regions containing genes coding for tryptophan synthase A/B,47 cytotoxins,53 TARP,52 and polymorphic membrane proteins or Pmps.52 In most cases, reported nucleic acid polymorphisms result in silent mutations or variations that lead to ocular serovar proteins with shorter sequences or loss of function. No ocular serovar–unique proteins have been identified. Thus, the genital serovar D proteins can be used to detect antibodies that recognize the vast majority of antigens from the ocular serovars. We recognize that the serovar D proteins-based proteome array has some limitations and would not detect antibodies that react only with ocular strain–specific epitopes; however, this deficiency should not significantly alter the major findings in the current study.
Among the 908 C. trachomatis proteins measured, 447 antigens were recognized by at least one of the human antisera and 10 antigens were recognized by 50% or more antisera, the latter thus designated as immunodominant antigens. Interestingly, 5 of the 10 immunodominant antigens are proteins known to be secreted into host cell cytoplasm,25,27,41,45 suggesting that chlamydial secretion proteins have enhanced immunogenicity. These immunodominant antigens have also been shown to be preferentially recognized by individuals infected with C. trachomatis urogenital strains,26 suggesting that these proteins may be essential for C. trachomatis infection in humans, even though some of these proteins are not highly expressed in chlamydial EBs.25,26
Importantly, the proteome array has also allowed us to identify four antigens that were preferentially recognized by sera from the trichiasis group, thus these were designated as pathology-associated antigens. The four pathology-associated antigens are CT414, CT667, CT706, and CT695. Expression of these antigens by Chlamydia and immune responses to these antigens during chlamydial infection may be biomarkers associated with the trichiasis phenotype or could be mechanistically contributing to ocular pathologies. In women with tubal factor infertility, chlamydial anti-HSP60 and OmcB antibodies have been identified as biomarkers.14,21 Moreover, immune responses to HSP60 have been implicated in the pathogenesis of tubal damage. Anti-chlamydial HSP60 antibodies may cross-react with host HSP6019,20 or anti-HSP60 T-cell responses may be directed against host tissues.16,17 Surprisingly, neither the chaperon HSP60 nor the outer membrane protein OmcB was identified by trichiasis antisera reactivity in our current study subjects. Instead, another chaperone with proteolytic activity, CT706 (an ATP-dependent ClpP endopeptidase subunit) and another outer membrane protein CT414 (PmpC) plus two putative type III secretion effectors (CT667 and CT695) were significantly associated with trichiasis. At present, it remains unknown whether these four proteins and immune responses induced by these antigens can mechanistically contribute to ocular pathology during C. trachomatis infection.
Equally important, the same proteome array has also allowed us to identify eight antigens that were preferentially recognized by sera from the control group, thus designated as protection-associated antigens. The eight protection antigens are CT019, CT117, CT301, CT442, CT553, CT556, CT571, and CT709. Most of these proteins are highly conserved metabolic or modifying enzymes (CT019, CT301, CT553, CT709) or related to the general secretion pathway (CT571). The remaining three antigens are hypothetical proteins unique to Chlamydia. CT117, consisting of 104 amino acids (aa) with a bilobal hydrophobic region and a pI of 8.21, is a putative inclusion membrane protein designated as IncF,54,55 and IncF is highly immunogenic in women urogenitally infected with C. trachomatis.30,31 CT556, consisting of 159 aa with a putative transmembrane domain at its C-terminus and an acidic pI of 4.95, shares a low level of homology with a thiol:disulfide interchange protein DsbC of Pseudomonas aeruginosa.56 CT442 has been characterized as a cysteine-rich 15-kDa outer membrane protein, designated as CrpA.57 CT442 is predicted to localize in the inclusion membrane54; moreover, it contains a murine CD8+ T-cell epitope that can elicit partial protective immunity to subsequent challenge with C. trachomatis.58 These three antigens (CT117, 442, and 556) appear to be suitable candidates for vaccine development, as they possess properties ideal for constructing an antitrachoma subunit vaccine: each are Chlamydia-specific, likely localized in chlamydial organism or inclusion membrane, immunogenic in humans, and able to induce protective immunity. More than half a century ago, vaccination with formalin-fixed whole chlamydial organisms not only failed to induce long-lasting protective immunity against trachoma but also exacerbated ocular pathologies when some immunized children were exposed to natural infection.59–62 These early observations might suggest that the formalin-fixed organisms not only had their protective antigen conformation altered by formalin fixation but also carried pathogenic antigens. Thus, a subunit vaccine by selecting protective antigens in their native conformation and excluding the pathogenic determinants may represent a promising approach for development of a vaccine to prevent trachomatous disease. Further evaluation of these Chlamydia-specific protection-associated antigens for their ability to induce protective immunity in an appropriate model system should provide important information for developing a subunit vaccine against C. trachomatis ocular infection and C. trachomatis–induced ocular diseases.
Supplementary Material
Acknowledgments
The authors thank the manager and ophthalmic nurses of the Gambian National Eye Care Programme. They also thank the field staff from the Medical Research Council Laboratories for their hard work and dedication.
Footnotes
Supported by grants from the National Institutes of Health (GZ) and from the Wellcome Trust (079246/Z/06/Z). Additional support in The Gambia was provided by the Medical Research Council Laboratories, UK, The Gambia.
Disclosure: C. Lu, None; M.J. Holland, None; S. Gong, None; B. Peng, None; R.L. Bailey, None; D.W. Mabey, None; Y. Wu, None; G. Zhong, None
References
- 1. Hu VH, Harding-Esch EM, Burton MJ, Bailey RL, Kadimpeul J, Mabey DC. Epidemiology and control of trachoma: systematic review. Trop Med Int Health. 2010;15:673–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Starnbach MN, Roan NR. Conquering sexually transmitted diseases. Nat Rev Immunol. 2008;8:313–317 [DOI] [PubMed] [Google Scholar]
- 3. Holland MJ, Jeffries D, Pattison M, et al. Pathway-focused arrays reveal increased matrix metalloproteinase-7 (matrilysin) transcription in trachomatous trichiasis. Invest Ophthalmol Vis Sci. 2010;51:3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abu el-Asrar AM, Geboes K, Missotten L Immunology of trachomatous conjunctivitis. Bull Soc Belge Ophtalmol. 2001;280:73–96 [PubMed] [Google Scholar]
- 5. Holland MJ, Bailey RL, Hayes LJ, Whittle HC, Mabey DC. Conjunctival scarring in trachoma is associated with depressed cell-mediated immune responses to chlamydial antigens. J Infect Dis. 1993;168:1528–1531 [DOI] [PubMed] [Google Scholar]
- 6. Watkins NG, Hadlow WJ, Moos AB, Caldwell HD. Ocular delayed hypersensitivity: a pathogenetic mechanism of chlamydial-conjunctivitis in guinea pigs. Proc Natl Acad Sci U S A. 1986;83:7480–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu VH, Weiss HA, Ramadhani AM, et al. Innate immune responses and modified extra-cellular matrix regulation characterize bacterial infection and cellular/connective tissue changes in scarring trachoma. Infect Immun. 2012;80:121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hessel T, Dhital SP, Plank R, Dean D. Immune response to chlamydial 60-kilodalton heat shock protein in tears from Nepali trachoma patients. Infect Immun. 2001;69:4996–5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Skwor T, Kandel RP, Basravi S, Khan A, Sharma B, Dean D. Characterization of humoral immune responses to chlamydial HSP60, CPAF, and CT795 in inflammatory and severe trachoma. Invest Ophthalmol Vis Sci. 2010;51:5128–5136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peeling RW, Bailey RL, Conway DJ, et al. Antibody response to the 60-kDa chlamydial heat-shock protein is associated with scarring trachoma. J Infect Dis. 1998;177:256–259 [DOI] [PubMed] [Google Scholar]
- 11. Wang J, Chen L, Chen F, et al. A chlamydial type III-secreted effector protein (Tarp) is predominantly recognized by antibodies from humans infected with Chlamydia trachomatis and induces protective immunity against upper genital tract pathologies in mice. Vaccine. 2009;27:2967–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morrison RP. Chlamydial hsp60 and the immunopathogenesis of chlamydial disease. Semin Immunol. 1991;3:25–33 [PubMed] [Google Scholar]
- 13. Toye B, Laferriere C, Claman P, Jessamine P, Peeling R. Association between antibody to the chlamydial heat-shock protein and tubal infertility. J Infect Dis. 1993;168:1236–1240 [DOI] [PubMed] [Google Scholar]
- 14. Rodgers AK, Wang J, Zhang Y, et al. Association of tubal factor infertility with elevated antibodies to Chlamydia trachomatis caseinolytic protease P. Am J Obstet Gynecol. 2010;203:494.e7–494.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bulut Y, Faure E, Thomas L, et al. Chlamydial heat shock protein 60 activates macrophages and endothelial cells through Toll-like receptor 4 and MD2 in a MyD88-dependent pathway. J Immunol. 2002;168:1435–1440 [DOI] [PubMed] [Google Scholar]
- 16. Ausiello CM, Palazzo R, Spensieri F, et al. 60-kDa heat shock protein of Chlamydia pneumoniae is a target of T-cell immune response. J Biol Regul Homeost Agents. 2005;19:136–140 [PubMed] [Google Scholar]
- 17. Kinnunen A, Paavonen J, Surcel HM. Heat shock protein 60 specific T-cell response in chlamydial infections. Scand J Immunol. 2001;54:76–81 [DOI] [PubMed] [Google Scholar]
- 18. Ausiello CM, Fedele G, Palazzo R, Spensieri F, Ciervo A, Cassone A. 60-kDa heat shock protein of Chlamydia pneumoniae promotes a T helper type 1 immune response through IL-12/IL-23 production in monocyte-derived dendritic cells. Microbes Infect. 2006;8:714–720 [DOI] [PubMed] [Google Scholar]
- 19. Cappello F, Conway de Macario E, Di Felice V, Zummo G, Macario AJ. Chlamydia trachomatis infection and anti-Hsp60 immunity: the two sides of the coin. PLoS Pathog. 2009;5:e1000552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Domeika M, Domeika K, Paavonen J, Mardh PA, Witkin SS. Humoral immune response to conserved epitopes of Chlamydia trachomatis and human 60-kDa heat-shock protein in women with pelvic inflammatory disease. J Infect Dis. 1998;177:714–719 [DOI] [PubMed] [Google Scholar]
- 21. Rodgers AK, Budrys NM, Gong S, et al. Genome-wide identification of Chlamydia trachomatis antigens associated with tubal factor infertility. Fertil Steril. 2011;96:715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burton MJ, Rajak SN, Bauer J, et al. Conjunctival transcriptome in scarring trachoma. Infect Immun. 2011;79:499–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gall A, Horowitz A, Joof H, et al. Systemic effector and regulatory immune responses to chlamydial antigens in trachomatous trichiasis. Front Microbiol. 2011;2:10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hudson L, Hay FC. Practical Immunology. 3rd ed. Oxford: Blackwell Scientific Publications; 1989. [Google Scholar]
- 25. Zhong G, Fan P, Ji H, Dong F, Huang Y. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J Exp Med. 2001;193:935–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang J, Zhang Y, Lu C, Lei L, Yu P, Zhong GA. Genome-wide profiling of the humoral immune response to Chlamydia trachomatis infection reveals vaccine candidate antigens expressed in humans. J Immunol. 2010;185:1670–1680 [DOI] [PubMed] [Google Scholar]
- 27. Li Z, Chen D, Zhong Y, Wang S, Zhong G. The chlamydial plasmid-encoded protein pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect Immun. 2008;76:3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luo J, Liu G, Zhong Y, et al. Characterization of hypothetical proteins Cpn0146, 0147, 0284 & 0285 that are predicted to be in the Chlamydia pneumoniae inclusion membrane. BMC Microbiol. 2007;7:38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Greene W, Zhong G. Inhibition of host cell cytokinesis by Chlamydia trachomatis infection. J Infect. 2003;47:45–51 [DOI] [PubMed] [Google Scholar]
- 30. Sharma J, Zhong Y, Dong F, Piper JM, Wang G, Zhong G. Profiling of human antibody responses to Chlamydia trachomatis urogenital tract infection using microplates arrayed with 156 chlamydial fusion proteins. Infect Immun. 2006;74:1490–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Z, Chen C, Chen D, Wu Y, Zhong Y, Zhong G. Characterization of fifty putative inclusion membrane proteins encoded in the Chlamydia trachomatis genome. Infect Immun. 2008;76:2746–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dean D, Kandel RP, Adhikari HK, Hessel T. Multiple Chlamydiaceae species in trachoma: implications for disease pathogenesis and control. PLoS Med. 2008;5:e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stekel DJ, Sarti D, Trevino V, et al. Analysis of host response to bacterial infection using error model based gene expression microarray experiments. Nucleic Acids Res. 2005;33:e53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stekel DJ. Microarray Bioinformatics. 1st ed. New York, NY: Cambridge University Press; 2003. [Google Scholar]
- 35. Feise RJ. Do multiple outcome measures require p-value adjustment? BMC Med Res Methodol. 2002;2:8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hu VH, Massae P, Weiss HA, et al. Bacterial infection in scarring trachoma. Invest Ophthalmol Vis Sci. 2011;52:2181–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burton MJ, Bailey RL, Jeffries D, et al. Conjunctival expression of matrix metalloproteinase and proinflammatory cytokine genes after trichiasis surgery. Invest Ophthalmol Vis Sci. 2010;51:3583–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhong G, Liu L, Fan T, Fan P, Ji H. Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in Chlamydia-infected cells. J Exp Med. 2000;191:1525–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhong G, Fan T, Liu L. Chlamydia inhibits interferon gamma-inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor 1. J Exp Med. 1999;189:1931–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pirbhai M, Dong F, Zhong Y, Pan KZ, Zhong G. The secreted protease factor CPAF is responsible for degrading pro-apoptotic BH3-only proteins in Chlamydia trachomatis-infected cells. J Biol Chem. 2006;281:31495–31501 [DOI] [PubMed] [Google Scholar]
- 41. Clifton DR, Fields KA, Grieshaber SS, et al. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A. 2004;101:10166–10171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Engel J. Tarp and arp: how Chlamydia induces its own entry. Proc Natl Acad Sci U S A. 2004;101:9947–9948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lutter EI, Bonner C, Holland MJ, et al. Phylogenetic analysis of Chlamydia trachomatis Tarp and correlation with clinical phenotype. Infect Immun. 2010;78:3678–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hower S, Wolf K, Fields KA. Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol Microbiol. 2009;72:1423–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fields KA, Hackstadt T. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol Microbiol. 2000;38:1048–1060 [DOI] [PubMed] [Google Scholar]
- 46. Kalman S, Mitchell W, Marathe R, et al. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat Genet. 1999;21:385–389 [DOI] [PubMed] [Google Scholar]
- 47. Caldwell HD, Wood H, Crane D, et al. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J Clin Invest. 2003;111:1757–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gaydos CA, Palmer L, Quinn TC, Falkow S, Eiden JJ. Phylogenetic relationship of Chlamydia pneumoniae to Chlamydia psittaci and Chlamydia trachomatis as determined by analysis of 16S ribosomal DNA sequences. Int J Syst Bacteriol. 1993;43:610–612 [DOI] [PubMed] [Google Scholar]
- 49. Grinblat-Huse V, Drabek EF, Creasy HH, et al. Genome sequences of the zoonotic pathogens Chlamydia psittaci 6BC and Cal10. J Bacteriol. 2011;193:4039–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Andreasen AA, Burton MJ, Holland MJ, et al. Chlamydia trachomatis ompA variants in trachoma: what do they tell us? PLoS Negl Trop Dis. 2008;2:e306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hayes LJ, Pecharatana S, Bailey RL, et al. Extent and kinetics of genetic change in the omp1 gene of Chlamydia trachomatis in two villages with endemic trachoma. J Infect Dis. 1995;172:268–272 [DOI] [PubMed] [Google Scholar]
- 52. Carlson JH, Porcella SF, McClarty G, Caldwell HD. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect Immun. 2005;73:6407–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Carlson JH, Hughes S, Hogan D, et al. Polymorphisms in the Chlamydia trachomatis cytotoxin locus associated with ocular and genital isolates. Infect Immun. 2004;72:7063–7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bannantine JP, Griffiths RS, Viratyosin W, Brown WJ, Rockey DD. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell Microbiol. 2000;2:35–47 [DOI] [PubMed] [Google Scholar]
- 55. Bannantine JP, Rockey DD, Hackstadt T. Tandem genes of Chlamydia psittaci that encode proteins localized to the inclusion membrane. Mol Microbiol. 1998;28:1017–1026 [DOI] [PubMed] [Google Scholar]
- 56. Roy PH, Tetu SG, Larouche A, et al. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS One. 2010;5:e8842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang YX, Watkins NG, Stewart S, Caldwell HD. The low-molecular-mass, cysteine-rich outer membrane protein of Chlamydia trachomatis possesses both biovar- and species-specific epitopes. Infect Immun. 1987;55:2570–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Starnbach MN, Loomis WP, Ovendale P, et al. An inclusion membrane protein from Chlamydia trachomatis enters the MHC class I pathway and stimulates a CD8+ T cell response. J Immunol. 2003;171:4742–4749 [DOI] [PubMed] [Google Scholar]
- 59. Clements C, Dhir SP, Grayston JT, Wang SP. Long term follow-up study of a trachoma vaccine trial in villages of Northern India. Am J Ophthalmol. 1979;87:350–353 [DOI] [PubMed] [Google Scholar]
- 60. Grayston JT, Woolridge RL, Wang S. Trachoma vaccine studies on Taiwan. Ann N Y Acad Sci. 1962;98:352–367 [DOI] [PubMed] [Google Scholar]
- 61. Sowa S, Sowa J, Collier LH, Blyth WA. Trachoma vaccine field trials in The Gambia. J Hyg (Lond). 1969;67:699–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Woolridge RL, Grayston JT, Chang IH, Yang CY, Cheng KH. Long-term follow-up of the initial (1959–1960) trachoma vaccine field trial on Taiwan. Am J Ophthalmol. 1967; 63:Suppl:1650–1655 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.