Background: γ-Tubulin moderates the expression of E2F-regulated promoters by direct binding to DNA.
Results: RB1 and γ-tubulin proteins moderate each other's expression by binding to their respective gene promoters.
Conclusion: Reduction of γ-tubulin protein levels in tumors with nonfunctional RB1 leads to induction of apoptosis.
Significance: The RB1/γ-tubulin signal network can be considered as a new target for cancer treatment.
Keywords: Cancer Biology, Promoters, Retinoblastoma (Rb), Transcription Regulation, Tubulin
Abstract
In various tumors inactivation of growth control is achieved by interfering with the RB1 signaling pathway. Here, we describe that RB1 and γ-tubulin proteins moderate each other's expression by binding to their respective gene promoters. Simultaneous reduction of RB1 and γ-tubulin protein levels results in an E2F1-dependent up-regulation of apoptotic genes such as caspase 3. We report that in various tumors types, there is an inverse correlation between the expression levels of γ-tubulin and RB1 and that in tumor cell lines with a nonfunctioning RB1, reduction of γ-tubulin protein levels leads to induction of apoptosis. Thus, the RB1/γ-tubulin signal network can be considered as a new target for cancer treatment.
Introduction
During cell division, the commitment to divide occurs in the passage from G1 to S phase, which coincides with the hyperphosphorylation of retinoblastoma protein (RB1)2 (1). This event leads to the dissociation of E2 promoter binding factor (E2F) from RB1 and initiation of E2F transcriptional activity and S phase entry (1). Following activation of E2F during S phase, the transcriptional activity of E2F is then modulated by γ-tubulin, which forms a DNA-binding complex with E2F on E2F-regulated promoters (2). We have previously reported that reduced γ-tubulin protein levels result in an E2F-mediated up-regulation of RB1 (2). This could imply that RB1 and γ-tubulin may complement each other in the regulation of E2F activity and suggests a potential interrelationship between the two proteins, where the lack of activity/expression of one protein could be compensated by the increase in activity/expression of the other. In this way, the RB1/γ-tubulin regulatory network could assure a constant control on E2F activity (3).
Here, we demonstrate that RB1 and γ-tubulin proteins moderate each other's expression and that in various tumors there is an inverse correlation between the expression levels of γ-tubulin and RB1. In addition, reduction of γ-tubulin in tumor cell lines lacking RB1 lead to induction of caspases and initiation of apoptosis.
EXPERIMENTAL PROCEDURES
cDNA, RNA Interference, and Reagents
pSG5L-hemagglutinin (HA)-RB1 was provided by Dr. W. Sellers (4) (addgene plasmid 10720). Human γ-TUBULIN shRNA was prepared as reported previously (5). For human RB1 shRNA, the annealed oligonucleotides 5′-GATCCCGAACGATTATCCATTCAAATTCAAGAGATTTGAATGGATAATCGTTCTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGAACGATTATCCATTCAAATCTCTTGAATTTGAATGGATAATCGTTCGG-3′ were cloned into the BglII and HindIII sites of the plasmid pTER (a gift from Dr. M. van de Wetering) as described elsewhere (5).
The following antibodies and reagents were used: anti-RB1, anti-Ser(P)780 RB1, anti-E2F1, and anti-caspase 3 (Santa Cruz Biotechnology and Sigma), anti-γ-tubulin (mouse and rabbit antibodies, Sigma), anti-α-tubulin (Calbiochem), protein G PLUS-agarose (Oncogene Science), protein A/G PLUS-agarose (Santa Cruz Biotechnology), and SDS-PAGE reagents (Bio-Rad). All other reagents were obtained from Sigma.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was described elsewhere (2). ChIPs were performed using the following rabbit polyclonal antibodies: anti-RB1 C-15 (Santa Cruz Biotechnology) and anti-γ-tubulin T5192 (Sigma), on nontransfected U2OS cells. Coprecipitated chromatin was analyzed by PCR for the presence of RB1 promoter DNA between −145 and 29 bp using primers 5′-TTAGCGTCCCAGCCCGCGCACCGAC-3′ and 5′-TTCAACGTCCCCTGAGAAAAACCGG-3′, TUBG1 promoter DNA between −77 and 108 bp using primers 5′-CATGAACGAAGGCCAGTGCT-3′ and 5′-CTGTAGGGTGATGATTTCCC-3′ or the presence of TUBG2 promoter DNA between −265 and −65 bp using primers 5′-TGGACAAGCACATCCACCTG-3′ and 5′-GCATCATTTTCTCCCTCAGC-3′.
Cell Culture, Transfection, Cell Cycle Analysis, and Apoptosis
NIH3T3, U2OS, and U1690 cells were cultured and transfected as described (5, 6). Y79 cells were cultured (37 °C, 5% CO2) in RPMI 1640 medium supplemented with 20% FCS, 2 mm l-glutamine, 100 units/ml penicillin, and 100 mg/ml streptomycin. The cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Cell cycle analysis was performed by flow cytometry (5). The fraction of apoptotic cells was analyzed by flow cytometry using annexin V-APC and 7AAD (BD Biosciences) staining as described (7). In addition, cell death was assayed by staining with trypan blue to analyzed morphology and cell viability. A minimum of 200 cells was counted in each sample, and the percentage of trypan blue-positive cells was calculated.
Immunoprecipitation and Western Blot Analysis
Cells extracts were obtained by incubation (4 °C, 15 min) in lysis buffer (2, 5). For immunoprecipitation, anti-RB1 or an isotype match control antibody were incubated with lysates, as described elsewhere (2). Immunoprecipitates and total lysates were boiled in sample buffer prior to resolution by SDS-PAGE. Western blotting was performed as described (8).
Real-time Quantitative PCR
Total RNA isolation (RNeasy; Qiagen), cDNA synthesis (Reverse Transcriptase kit; Applied Biosystems), and quantitative real-time PCR analysis with SYBR Green PCR master mix (Applied Biosystems) was performed according to the manufacturer's instructions. In brief, 2 μg of total RNA, extracted from transfected or not transfected U2OS, Y79, or U1690 cells, respectively, were used for cDNA synthesis with random hexamers. Obtained cDNA was diluted to 500 μl in water, and 5 μl was used as template in each 25-μl quantitative PCR. The amplification reactions were performed in a GeneAmp 5700 Sequence detector (Applied Biosystems). Each reaction was performed in triplicate, and the comparative Ct method was used for normalization with YWHAC, SDHA, and GAPDH genes (9), using the following primers: 5′-ACTTTTGGTACATTGTGGCTTCAA-3′ and 5′-CCGCCAGGACAAACCAGTAT-3′ for YWHAC, 5′-TGGGAACAAGAGGGCATCTG-3′ and 5′-CCACCACTGCATCAAATTCATG-3′ for SDHA, and 5′-TGCACCACCAACTGCTTAGC-3′ and 5′-TGGCATGGACTGTGGTCATGAG-3′ for GAPDH. For RB1 and γ-TUBULIN amplification, the following primers were used: 5′-AGATACCAGATCATGTCAGAGAG-3′ and 5′-TACAGATTCCCCACAGTTCCTTT-3′ for RB1 and 5′-AAGCTCTACAACCCAGAGAACAT-3′ and 5′-CAATGGAGTGACACAGCACAAA-3′ for γ-TUBULIN.
Gene Expression Array Analyses
Publicly available datasets were analyzed including a breast cancer dataset of 136 tumors (GEO accession number GSE12093), a bladder cancer dataset of 57 tumors (10) (dataset available at arrayexpress; accession number E-TABM-147), a colon cancer dataset consisting of 232 tumors (GEO accession number GSE17538), and a lung cancer cell line dataset containing 29 cell lines (GEO accession number GSE4127). Correlations between TUBG1 and RB1 expression were analyzed using Pearson's correlation. RB1 pathway inactivation events and pathway activity was calculated using CCND1/CDKN2A expression ratio as described (11).
Tissue Microarray Construction and Immunohistochemistry
Breast cancer and colorectal cancer samples were obtained from 144 and 118 patients, respectively, diagnosed in Malmö University Hospital, Malmö, Sweden between 1999 and 2002, as described (12–14). All cases were histopathologically reevaluated on hematoxylin and eosin-stained slides (7, 15). Paraffin-embedded human tumor specimens from 37 small cell lung cancer (SCLC) patients were histologically evaluated by a lung cancer pathologist, as described (6). Tissue microarrays were constructed as reported previously (6, 7, 15), using duplicate cores of 1 mm from nonnecrotic areas representative of invasive cancer. Ethical approval for this study was obtained from the Ethics Committee at Lund University (reference numbers 447-07, 2004/762, and 2008/702), whereby written consent was not required, and patients were offered the option to opt out.
For immunohistochemical analysis of Ser(P)780 RB1 pRB1/γ-tubulin the cohorts were stained in a Autostainer Plus (DAKO, Glostrup, Denmark) with anti-γ-tubulin, anti-RB1, and anti-pRB1 antibodies diluted 1:2000, 1:2000, and 1:50, respectively, and for cell lines anti-γ-tubulin, anti-α-tubulin, anti-RB1, and anti-pRB1 antibodies were diluted 1:4000, 1:2000, 1:2000, and 1:250, respectively. The specificity of the anti-Ser(P)780 pRB1 was assay by treating U2OS cells with λ-phosphatase (New England Biolabs), as described (8), prior to staining of U2OS cells with anti-Ser(P)780 RB1.
Statistical Analyses
All data are expressed as mean ± S.D., and Student's paired t test was used to analyze the differences. Cell cycle profiles were assessed using FlowJo (Tree Star, Inc.). For gene expression array analyses, Pearson's product moment correlation coefficient was used to examine correlations between expressions of genes of interest. Corresponding p values (based on Fisher's Z transformation) were provided with a p value < 0.05 being considered significant. For tissue microarray analysis, Spearman's Rho test was used for comparison of pRB1, phospho-pRB1, and γ-tubulin protein expression. Corresponding p values (based on Edgeworth series approximation) were provided with a p value < 0.05 being considered significant.
RESULTS
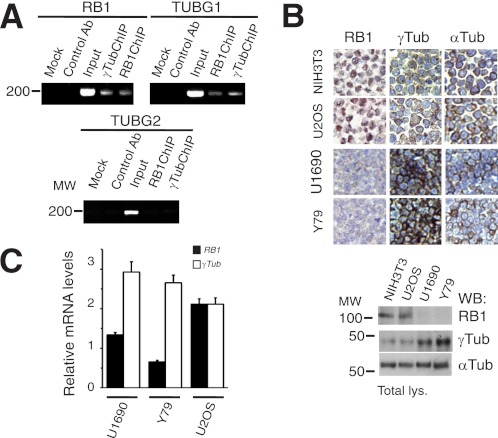
To test a potential interrelationship between RB1 and γ-tubulin, we searched for E2F DNA binding sites in the promoters of RB1 and γ-tubulin isoforms 1 and 2 (RB1, TUBG1, and TUBG2 gene promoters). The E2F DNA binding sites tttggcgc and tttcccgc were found 34 bp and 49 bp upstream of the transcription start site of the TUBG1 (GenBank nucleotide NM_001070) and RB1 gene (GenBank nucleotide NM_000321), respectively; however, no E2F DNA binding site was found in TUBG2 gene promoter (GenBank nucleotide NM_016437). To study the ability of γ-tubulin and RB1 to bind to each other's promoters, ChIPs using γ-tubulin or RB1-targeting antibodies were performed (2). In U2OS cells, both endogenous γ-tubulin and endogenous RB1 proteins were present on their own promoters and on RB1 and TUBG1 promoters, respectively; however, neither RB1 nor γ-tubulin was immunoprecipitated on the TUBG2 promoter (Fig. 1A). These results indicate that alterations in the protein levels of RB1 or γ-tubulin may alter the transcription of TUBG1 and RB1.
FIGURE 1.
γ-Tubulin and RB1 regulate the transcriptional activity of each other's promoters. A, U2OS cells analyzed by ChIP using γ-tubulin or RB1 antibodies. The PCR primers amplified the RB1, TUBG1, or TUBG2 promoter region from −145 to 29 bp, −77 to 108 bp and −265 to −65 bp, respectively (n = 3). B, upper, immunocytochemistry of NIH3T3, U2OS, Y79, and U1690 cells using antibodies against RB1, γ-tubulin, and α-tubulin (Tub). Lower, total lysate from 1 × 106 of the indicated cell lines prepared and examined by Western blotting (WB) using the indicated antibodies (n = 3). C, reverse transcription-PCR in U2OS, Y79, and U1690 cells, measuring the relative mRNA expression of RB1 (filled bars) and γ-TUBULIN (open bars). The data are expressed as mean ± S.D. (error bars) of experiments performed in triplicate.
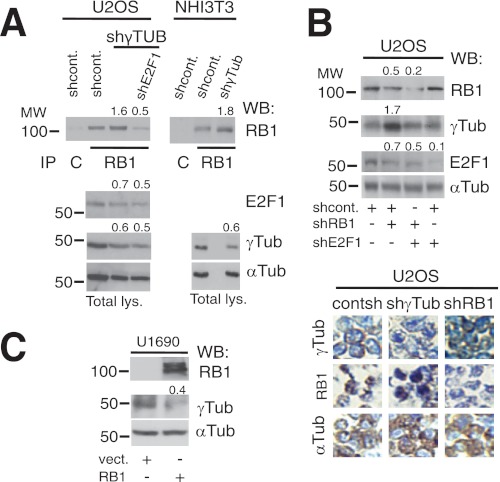
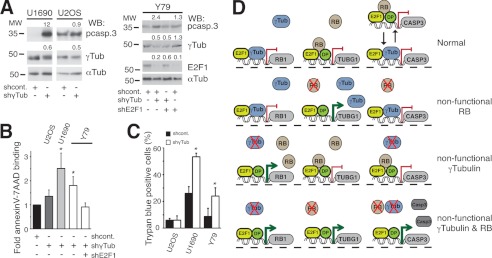
Inactivation of the RB1 gene by loss of heterozygosity, various deletions, or hyperphosphorylation often impairs its expression or function in tumors, such as retinoblastoma, SCLC, and breast cancer (16). Taking into consideration the potential compensatory actions of RB1 and γ-tubulin, malignancies with low RB1 activity should present higher γ-tubulin protein levels. We therefore examined the expression levels of RB1 and γ-tubulin in cell lines derived from tumors lacking RB1 expression and function (17), i.e. retinoblastoma Y79 (18) and SCLC U1690 (19) cell lines. Indeed, we found that U1690 and Y79 cell lines lacking RB1 expression (18) displayed substantially elevated protein and mRNA levels of γ-tubulin compared with cell lines expressing RB1 (Fig. 1, B and C). However, α-tubulin protein levels were unchanged (Fig. 1B). In addition, as reported previously (2), in γ-TUBULIN-shRNA- and γ-TUBULIN-shRNA-transfected U2OS and NIH3T3 cells there was an increased protein (Fig. 2A) and mRNA levels of RB1 (2). Moreover, in the RB1-expressing cell line U2OS, reduction of RB1 expression resulted in increased γ-tubulin protein levels in an E2F1-dependent manner (Fig. 2B). Finally, expression of recombinant RB1 (4) reduced the γ-tubulin protein levels in the RB1-negative cell line, U1690 (Fig. 2C). Together these data imply the existence of a γ-tubulin and RB1 concentration-dependent regulatory mechanism that controls the transcriptional activity of the RB1 and TUBG1 promoters.
FIGURE 2.
γ-Tubulin and RB1 proteins moderate each other's expression. A, using extracts from U2OS and NIH3T3 cells transfected with control-shRNA (shcont.), E2F1-shRNA (shE2F1), or γ-TUBULIN-shRNA (shγTUB) vectors, RB1 was immunoprecipitated (IP) with a monoclonal anti-RB1 antibody and developed by Western blotting (WB) with a polyclonal anti-RB1 antibody (top). Total lysate (Total lys.) from 1 × 106 of the indicated cell lines was run in parallel and examined by Western blotting using antibodies against E2F1, γ-tubulin, and α-tubulin (Tub) (n = 3). B and C, U2OS or U1690 cells were transfected with the control-shRNA, RB1-shRNA (shRB1), E2F1-shRNA, pSG5 (vect.), or HA-RB1 (RB1) vectors, as indicated; after 48 h (B) or 24 h (C), total lysate from 1 × 106 was prepared and examined by Western blotting using antibodies against RB1, E2F1, γ-tubulin, and α-tubulin (n = 3). B, lower, immunocytochemistry of U2OS cells transfected with γ-TUBULIN-shRNA or RB1-shRNA vectors using the indicated antibodies is shown. A–C, numbers on Western blots indicate variations on protein expression relative to control. To adjust for differences in protein loading, the protein concentration of RB1, E2F1, and γ-tubulin was determined by their ratio with α-tubulin for each treatment. The protein ratio in control treatment was set to 1.
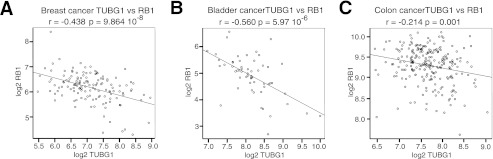
To determine whether the same interplay between RB1 and γ-TUBULIN was present in different tumor types, we made use of multiple publicly available datasets. Consistent with our functional results in cell lines, a significant negative correlation between γ-TUBULIN and RB1 expression was observed in all datasets examined including 136 breast (GEO accession number GSE12093; Fig. 3A), 57 bladder (10) (dataset available at arrayexpress; accession number: E-TABM-147; Fig. 3B), and 232 colon cancers (GEO accession number GSE17538; Fig. 3C).
FIGURE 3.
γ-TUBULIN expression negatively correlates with RB1 expression in multiple tumor types. A–C, scatter plots of γ-TUBULIN expression versus RB1 expression in breast tumors (n = 136, r = −0.438, p value = 9.864e-08; A) in bladder tumors (n = 57, r = −0.560, p value = 5.97e-06; B), and in colon tumors (n = 395, r = −0.214, p value = 0.001; C). In the displayed plots, r represents the Pearson product moment correlation coefficient between γ-tubulin and RB1 expression with respective p values. p values displayed represent the significance of the Pearson correlation with a significant value cutoff of <0.05.
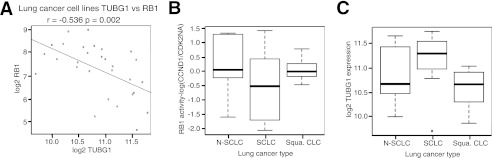
In addition, a dataset consisting of multiple lung cancer cell lines (10 non-small cell adenocarcinoma, 10 SCLC, and 9 squamous cell cancer; GEO accession number GSE4127) was examined, and again a significant negative correlation was found between γ-TUBULIN and RB1 expression (Fig. 4A). Based on the high frequency of RB1 mutations in SCLCs (11, 16), we hypothesized that γ-TUBULIN levels should be highest in this subgroup of lung cancers. To examine first whether inactivating mutations in the RB1 pathway were overrepresented in SCLCs compared with the other subgroups, we employed the CCND1/CDKN2A (11) ratio for determining the RB1 activity and the presence of inactivating mutations in the RB1 pathway in multiple cell and tumor types (11). As expected, RB1 activity was found to be lower in SCLC cell lines compared with both non-small cell and squamous cell lung cancers (Fig. 4B). Accompanying the low RB1 activity observed in SCLCs, this subgroup displayed the highest expression of γ-TUBULIN (Fig. 4C).
FIGURE 4.
γ-TUBULIN expression negatively correlates with RB1 expression in multiple lung cancer cell lines. A and B, scatter plot of γ-TUBULIN expression (A) versus RB1 expression in various lung cancer cell lines (n = 29, r = −0.536, p value = 0.002; B). B and C, box plot displaying log(CCND1/CDK2NA) ratio (B) or γ-TUBULIN expression (C) in the three represented lung cancer subtypes. In the displayed plots, r represents the Pearson product moment correlation coefficient between γ-tubulin and RB1 expression with respective p values. p values displayed represent the significance of the Pearson correlation with a significant value cutoff of <0.05.
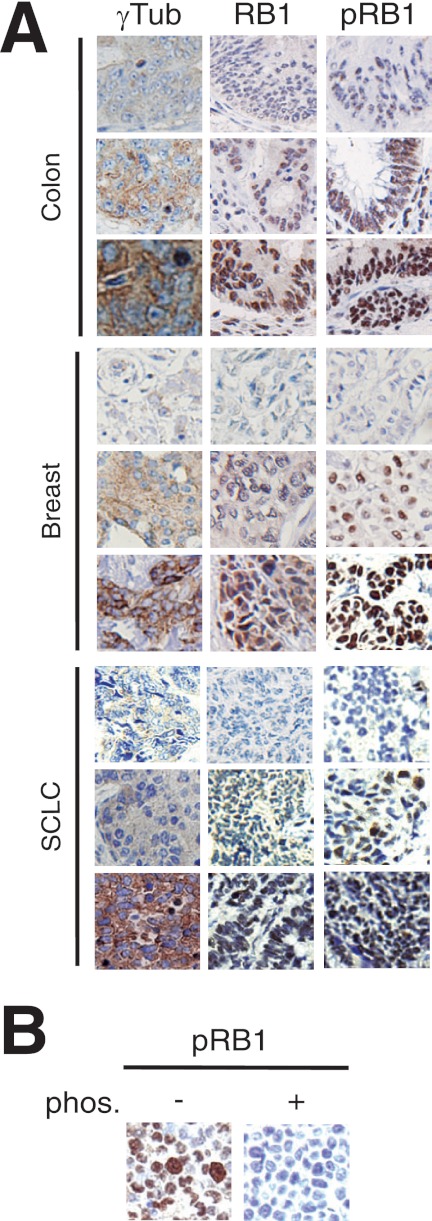
Although significant negative correlations were observed in multiple systems, the degree of this correlation varied depending on tumor type. This could be attributed to mRNA expression not representing RB1 regulation at the protein level or defects in its phosphorylation. We therefore studied the protein expression of γ-tubulin, RB1, and phosphorylated (p)RB1 on Ser780 (Fig. 5) in three cohorts of breast, colorectal, and small cell lung tumors (Table 1). We used an anti-RB1 antibody that recognized the C terminus of the protein to exclude all tumors expressing RB1 lacking this region (Fig. 5A). In addition, we used anti-pRB1 antibody as phosphorylation of RB1 on Ser780 was shown to be sufficient for disruption of the RB1-E2F1 complex (20) (Fig. 5). In all three cohorts, γ-tubulin trended toward a negative correlation with RB1 protein levels, however, only reaching significance in the breast cancer cohort (Table 1). In contrast, when examining γ-tubulin correlation with pRB1, a significant positive correlation was detected in all three cohorts. Considering that phosphorylation of RB1 results in inhibition of RB1 activity, the positive correlation of γ-tubulin with pRB1 indicates a negative association between RB1 activity and γ-tubulin protein expression. This negative correlation is in accordance with gene expression data analyzed in these cancer types (Figs. 3 and 4).
FIGURE 5.
Protein expression of γ-tubulin, RB1, and phosphorylated RB1 in various tumors. A, immunohistochemical staining of γ-tubulin (γTub), RB1, and Ser(P)780 RB1 (pRB1) in breast, colon, and SCLC tumors. Stainings are denoted as low, intermediate, and strong. B, immunohistochemical staining of phosphorylated Ser780 RB1 in U2OS cells treated with or without λ-phosphatase (phos.).
TABLE 1.
Associations between γ-tubulin (γ-tub), RB,1 and phosphorylated Ser780 RB1 (pRB1) expression in breast, colorectal, and small cell lung carcinoma tumors
R, Spearman's correlations coefficient. p < 0.005 in bold. n, number of tumor samples.
| Variable | γ-tub | RB1 | pRB1 | Variable | γ-tub | RB1 | pRB1 | Variable | γ-tub | RB1 | pRB1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| γ-tub | γ-tub | γ-tub | |||||||||
| R | −0.241 | 0.345 | R | −0.100 | 0.388 | R | −0.095 | 0.339 | |||
| p | 0.015 | 0.000 | p | 0.335 | 0.000 | p | 0.598 | 0.050 | |||
| n | 129 | 131 | n | 95 | 98 | n | 33 | 34 | |||
| RB1 | RB1 | RB1 | |||||||||
| R | −0.214 | 0.168 | R | −0.100 | 0.313 | R | −0.095 | 0.261 | |||
| p | 0.015 | 0.057 | p | 0.335 | 0.002 | p | 0.598 | 0.143 | |||
| n | 129 | 129 | n | 95 | 95 | n | 33 | 33 |
Based on our results, we postulated that if the RB/γ-tubulin regulatory network could ensure a constant control on E2F1 activity, then inhibition of both RB/γ-tubulin might lead to an uninhibited E2F1 activation and an induction of cell death (21). To finally test this hypothesis, we reduced the γ-tubulin protein levels in tumor cells carrying a defective RB1 signal pathway (Fig. 6). Although real-time quantitative PCR measurements demonstrated that the expression of γ-TUBULIN mRNA in U2OS, U1690, and Y79 cells transfected with γ-TUBULIN-shRNA was 54 ± 5%, 36 ± 8%, and 58 ± 15% of the expression in control cells (n = 3), respectively, this partial reduction of γ-tubulin levels triggered an E2F1-dependent increase of the expression of caspase 3 in U1690 and Y79 RB1-negative cells, but not in U2OS RB1-positive cells (Fig. 6A). Annexin V staining and the trypan blue exclusion test also confirmed the increase in number of death cells upon reduced γ-tubulin levels in U1690 and Y79 cells (Fig. 6, B and C).
FIGURE 6.
Cells with reduced expression of γ-tubulin and RB1 undergo apoptosis. A, Y79, U1690, or U2OS cells were transfected with control-shRNA (shcont.), γ-TUBULIN-shRNA (shγTub), or E2F1-shRNA (shE2F1) vectors; after 48 h, total lysate from 1 × 106 were prepared and examined by Western blotting (WB) using antibodies against pro-caspase 3 (pcasp.3), E2F1, γ-tubulin, and α-tubulin (Tub) (n = 3). Numbers above Western blots indicate variations on protein expression relative to control. To adjust for differences in protein loading, the protein concentration of RB1, E2F1, and γ-tubulin was determined by their ratio with α-tubulin for each treatment. The protein ratio in control treatment was set to 1. B and C, aliquots of different cell lines (as indicated), treated as in A for 72 h, were stained with annexin V-FITC and 7AAD and analyzed by flow cytometry (B) or stained with trypan blue and analyzed by morphology (C). The number of annexin V- and 7AAD-positive cells transfected with control construct was set as 1, and relative changes were calculated (mean ± S.D. (error bars), n = 3–6). D, γ-tubulin (γTub) and RB1 (RB) protein levels regulate each other's expression by direct binding to respective promoters. From left to right along each line RB1, TUBG1, and caspase 3 (CASP3) genes are represented with their respective promoter regions. Red lines and green arrows represent repressed and activated genes, respectively. Red cross denotes the absence of protein expression of the represented protein. In a cell, the presence of γ-tubulin and RB1 represses transcription from the RB1 and TUBG1 genes, respectively. The absence of RB1 increases the transcriptional activity of the TUBG1 gene, and accordingly, the absence of γ-tubulin increases the transcriptional activity of the RB1 gene. Finally, concomitant reduced levels of γ-tubulin and RB1 will lead to increase expression of CASP3. In promoter regions of repressed genes, RB1 is in complex with the transcription factor heterodimer E2F1-Dp. Alternatively, the inhibiting protein complex could be E2F–γ-tubulin. Black arrows point toward the two possible protein complexes that may repress expression of the CASP3.
In summary, the mechanism that allows γ-tubulin and RB1 to moderate each other's expression involves direct binding to E2F binding sites on TUBG1 and RB1 promoter regions (Fig. 6D). In the absence of γ-tubulin and RB1, the E2F1 activity elevates E2F1-mediated expression of caspase 3 resulting in induction of cellular apoptosis (Fig. 6D).
DISCUSSION
We have shown previously that in G1 to S phase transition, nuclear γ-tubulin governs E2F transcriptional activities by directly binding to E2Fs on E2F-regulated promoters. In doing so, γ-tubulin is involved in coordinating cell cycle progression (2). Specifically, such interaction represses the activities of E2Fs and thereby assures a transient expression of genes necessary for G1 to S phase entry (2). Upon reduction of γ-tubulin protein levels, there was an observed increase in RB1 protein levels resulting in G1 phase arrest. It can also be noted that both γ-tubulin and RB1 interact with E2F on E2F-regulated promoters (1, 2). In this way γ-tubulin and RB1 moderate the expression of genes involved in a wide range of cellular processes, including cell cycle progression, DNA replication, DNA repair, differentiation, and apoptosis (22). However, the mechanism of action of RB1 and γ-tubulin differs between the two molecules. RB1 inhibits the transcriptional activity of E2Fs by forming an inhibitory complex with E2F and Database of Rice transcription factor Polypeptide (Dp) on E2F-regulated promoters (22), whereas γ-tubulin binds to the Dp binding domain on E2F once E2F is released from RB1, and in this way γ-tubulin directly interacts with DNA (2). During cell cycle, we found that both deactivation of RB1 by hyperphosphorylation and the translocation of γ-tubulin to the nucleus occur simultaneously when E2F activity triggers S phase entry (2). This finding implies that RB1 as well as γ-tubulin play different functions during cell cycle. In addition, it may also suggest that upon RB1 hyperphosphorylation, γ-tubulin takes over the modulation of E2Fs to assure cell cycle progression (2).
Although a complementary function of γ-tubulin and RB1 has been described, if expression of one gene modulates the expression of the other has not yet been examined. In the present study, we noted that γ-tubulin and RB1 regulated each other's expression by direct binding to respective promoters. Moreover, the lack of activity of one of them is replaced by the increase activity of the other, suggesting that this network is part of a cell cycle checkpoint that assures a constant control of E2F activity. The existence of a compensatory relationship between RB1 and γ-tubulin gives a mechanistic explanation of how the lost of RB1 function in various tumors leads to an increased protein expression of γ-tubulin. The RB1 pathway is frequently silenced in multiple malignancies, such as retinoblastoma, prostate, colon, small cell lung and breast cancer, osteosarcoma (16), neuroblastoma (23), and lymphoblastic leukemia (24). Thus, it is tempting to speculate that upon inactivation of both RB1 and γ-tubulin checkpoint proteins, there will be an increased expression of E2F regulated genes such as caspase 3 that could result in induction of apoptosis (3). To ascertain whether γ-tubulin depletion could play a role in activation of apoptotic pathways, we applied shRNA techniques to reduce γ-tubulin protein levels in cell lines lacking RB1 expression. Despite only a partial reduction in γ-tubulin protein levels in the transfected cells, these effects resulted in tumor cell death. Consequently, our data reveal a link between γ-tubulin and RB1 expression that opens up a new strategy for targeted therapy in cancers harboring silencing alterations to the RB1 pathway. In addition, we present evidence that inhibition of a ubiquitous and nonaltered protein can be used alone to treat a wide variety of tumor types (1, 16, 17, 23, 24). Due to the high frequency of alterations to the RB1 pathway in multiple malignancies (1, 16, 17, 23, 24), the scope for the application of using γ-tubulin as a molecular target is extremely broad.
Acknowledgments
We thank W. Sellers and M. van de Wetering for reagents.
This work was supported by the Swedish Research Council, the Swedish Cancer Society, the Royal Physiographic Society in Lund, Per-Eric and Ulla Schyberg, Cradfoord, Gunnar Nilsson, H. and G. Jeanssons and O. and E. Ericsson Foundations; Gyllenstiernska Krapperupsstiftelsen; and the Skane University Hospital in Malmö Cancer Research Fund.
- RB1
- retinoblastoma
- Dp
- database of Rice transcription factor polypeptide
- E2F
- E2 promoter-binding factor
- SCLC
- small cell lung cancer
- APC
- allophycocyanin
- 7AAD
- 7-Aminoactinomycin D.
REFERENCES
- 1. Hanahan D., Weinberg R. A. (2000) The hallmarks of cancer. Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 2. Höög G., Zarrizi R., von Stedingk K., Jonsson K., Alvarado-Kristensson M. (2011) Nuclear localization of γ-tubulin affects E2F transcriptional activity and S-phase progression. FASEB J. 25, 3815–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nahle Z., Polakoff J., Davuluri R. V., McCurrach M. E., Jacobson M. D., Narita M., Zhang M. Q., Lazebnik Y., Bar-Sagi D., Lowe S. W. (2002) Direct coupling of the cell cycle and cell death machinery by E2F. Nat. Cell Biol. 4, 859–864 [DOI] [PubMed] [Google Scholar]
- 4. Sellers W. R., Novitch B. G., Miyake S., Heith A., Otterson G. A., Kaye F. J., Lassar A. B., Kaelin W. G., Jr. (1998) Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 12, 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alvarado-Kristensson M., Rodríguez M. J., Silió V., Valpuesta J. M., Carrera A. C. (2009) SADB phosphorylation of γ-tubulin regulates centrosome duplication. Nat. Cell Biol. 11, 1081–1092 [DOI] [PubMed] [Google Scholar]
- 6. Munksgaard Persson M., Johansson M. E., Monsef N., Planck M., Beckman S., Seckl M. J., Rönnstrand L., Påhlman S., Pettersson H. M. (2012) HIF-2α expression is suppressed in SCLC cells, which survive in moderate and severe hypoxia when HIF-1α is repressed. Am. J. Pathol. 180, 494–504 [DOI] [PubMed] [Google Scholar]
- 7. Ehlén A., Brennan D. J., Nodin B., O'Connor D. P., Eberhard J., Alvarado-Kristensson M., Jeffrey I. B., Manjer J., Brändstedt J., Uhlén M., Pontén F., Jirström K. (2010) Expression of the RNA-binding protein RBM3 is associated with a favourable prognosis and cisplatin sensitivity in epithelial ovarian cancer. J. Transl. Med. 8, 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alvarado-Kristensson M., Melander F., Leandersson K., Rönnstrand L., Wernstedt C., Andersson T. (2004) p38-MAPK signals survival by phosphorylation of caspase-8 and caspase-3 in human neutrophils. J. Exp. Med. 199, 449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. De Preter K., Speleman F., Combaret V., Lunec J., Laureys G., Eussen B. H., Francotte N., Board J., Pearson A. D., De Paepe A., Van Roy N., Vandesompele J. (2002) Quantification of MYCN, DDX1, and NAG gene copy number in neuroblastoma using a real-time quantitative PCR assay. Mod. Pathol. 15, 159–166 [DOI] [PubMed] [Google Scholar]
- 10. Stransky N., Vallot C., Reyal F., Bernard-Pierrot I., de Medina S. G., Segraves R., de Rycke Y., Elvin P., Cassidy A., Spraggon C., Graham A., Southgate J., Asselain B., Allory Y., Abbou C. C., Albertson D. G., Thiery J. P., Chopin D. K., Pinkel D., Radvanyi F. (2006) Regional copy number-independent deregulation of transcription in cancer. Nat. Genet. 38, 1386–1396 [DOI] [PubMed] [Google Scholar]
- 11. Mizuarai S., Machida T., Kobayashi T., Komatani H., Itadani H., Kotani H. (2011) Expression ratio of CCND1 to CDKN2A mRNA predicts RB1 status of cultured cancer cell lines and clinical tumor samples. Mol. Cancer 10, 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elkabets M., Gifford A. M., Scheel C., Nilsson B., Reinhardt F., Bray M. A., Carpenter A. E., Jirström K., Magnusson K., Ebert B. L., Pontén F., Weinberg R. A., McAllister S. S. (2011) Human tumors instigate granulin-expressing hematopoietic cells that promote malignancy by activating stromal fibroblasts in mice. J. Clin. Invest. 121, 784–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gaber A., Johansson M., Stenman U. H., Hotakainen K., Pontén F., Glimelius B., Bjartell A., Jirström K., Birgisson H. (2009) High expression of tumour-associated trypsin inhibitor correlates with liver metastasis and poor prognosis in colorectal cancer. Br. J. Cancer 100, 1540–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Svensson K. J., Christianson H. C., Kucharzewska P., Fagerström V., Lundstedt L., Borgquist S., Jirström K., Belting M. (2011) Chondroitin sulfate expression predicts poor outcome in breast cancer. Int. J. Oncol. 39, 1421–1428 [DOI] [PubMed] [Google Scholar]
- 15. Kononen J., Bubendorf L., Kallioniemi A., Bärlund M., Schraml P., Leighton S., Torhorst J., Mihatsch M. J., Sauter G., Kallioniemi O. P. (1998) Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 4, 844–847 [DOI] [PubMed] [Google Scholar]
- 16. Chen H. Z., Tsai S. Y., Leone G. (2009) Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer 9, 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harbour J. W., Dean D. C. (2000) The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14, 2393–2409 [DOI] [PubMed] [Google Scholar]
- 18. Lee E. Y., Bookstein R., Young L. J., Lin C. J., Rosenfeld M. G., Lee W. H. (1988) Molecular mechanism of retinoblastoma gene inactivation in retinoblastoma cell line Y79. Proc. Natl. Acad. Sci. U.S.A. 85, 6017–6021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nygren P., Larsson R., Gruber A., Peterson C., Bergh J. (1991) Doxorubicin selected multidrug-resistant small cell lung cancer cell lines characterised by elevated cytoplasmic Ca2+ and resistance modulation by verapamil in absence of P-glycoprotein overexpression. Br. J. Cancer 64, 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adams P. D. (2001) Regulation of the retinoblastoma tumor suppressor protein by cyclin/cdks. Biochim. Biophys. Acta 1471, M123–133 [DOI] [PubMed] [Google Scholar]
- 21. Qin X. Q., Livingston D. M., Kaelin W. G., Jr., Adams P. D. (1994) Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc. Natl. Acad. Sci. U.S.A. 91, 10918–10922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Biswas A. K., Johnson D. G. (2012) Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res. 72, 13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Molenaar J. J., Ebus M. E., Koster J., van Sluis P., van Noesel C. J., Versteeg R., Caron H. N. (2008) Cyclin D1 and CDK4 activity contribute to the undifferentiated phenotype in neuroblastoma. Cancer Res. 68, 2599–2609 [DOI] [PubMed] [Google Scholar]
- 24. Zhang J., Mullighan C. G., Harvey R. C., Wu G., Chen X., Edmonson M., Buetow K. H., Carroll W. L., Chen I. M., Devidas M., Gerhard D. S., Loh M. L., Reaman G. H., Relling M. V., Camitta B. M., Bowman W. P., Smith M. A., Willman C. L., Downing J. R., Hunger S. P. (2011) Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children's Oncology Group. Blood 118, 3080–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]