Background: Balancing copper homeostasis and bioavailability is critical for morphological development in Streptomyces lividans.
Results: In Streptomyces lividans, a copper-sensitive operon repressor (CsoR) protein regulates a three-locus copper homeostasis regulon.
Conclusion: Genes regulating copper homeostasis are more extensive than the regulon under direct control of CsoR.
Significance: CsoR is part of a regulatory network controlling copper homeostasis that extends beyond the CsoR regulon.
Keywords: Bacteria, Copper, Metal Homeostasis, Transcriptomics, X-ray Crystallography
Abstract
A copper-sensitive operon repressor protein (CsoR) has been identified in Streptomyces lividans (CsoRSl) and found to regulate copper homeostasis with attomolar affinity for Cu(I). Solution studies reveal apo- and CuI-CsoRSl to be a tetramer assembly, and a 1.7-Å resolution crystal structure of apo-CsoRSl reveals that a significant conformational change is necessary to enable Cu(I) binding. In silico prediction of the CsoR regulon was confirmed in vitro (EMSA) and in vivo (RNA-seq), which highlighted that next to the csoR gene itself, the regulon consists of two Cu(I) efflux systems involving a CopZ-like copper metallochaperone protein and a CopA P1-type ATPase. Although deletion of csoR has only minor effects on S. lividans development when grown under high copper concentrations, mutations of the Cu(I) ligands decrease tolerance to copper as a result of the Cu(I)-CsoR mutants failing to disengage from the DNA targets, thus inhibiting the derepression of the regulon. RNA-seq experiments carried out on samples incubated with exogenous copper and a ΔcsoR strain showed that the set of genes responding to copper stress is much wider than anticipated and largely extends beyond genes targeted by CsoR. This suggests more control levels are operating and directing other regulons in copper homeostasis beside the CsoR regulon.
Introduction
For many bacterial organisms, maintaining cellular metal ion homeostasis is an essential requirement for viability. Sophisticated cellular machinery has evolved and consists of an extensive network of specialized proteins and transporters that respond to either metal ion deprivation or overload (1–3). Copper is an essential transition metal ion in biology and is required by many proteins and enzymes that function in electron-transfer chemistry and in the catalytic oxidation of substrates. The ability of copper to donate or accept single electrons is also detrimental as “free” cellular copper ions are complicit in a Fenton-like reaction, catalyzing the production of damaging hydroxyl radicals (4). This deleterious behavior leads to bacteria avoiding copper toxicity through buffering copper in the cytosol very tightly (2, 3, 5). Once the buffering capacity for cytosolic copper is exceeded, this is sensed by transcriptional regulatory proteins termed copper sensors that trigger the expression of genes encoding for efflux systems such as copper-exporting P1-type ATPases and copper metallochaperones that act in tandem to return the cytosol to a buffered copper state (3, 6, 7).
The filamentous Gram-positive soil-dwelling bacterium Streptomyces is important to both ecological and human welfare and is also one of the model systems for bacterial morphological and physiological development. The morphological differentiation of Streptomyces has three characteristic development stages as follows: 1) formation of branched vegetative mycelium, 2) formation of aerial hyphae, and 3) the production of spores. Concomitant or just before the morphological switch from vegetative to aerial mycelium, the metabolic development is initiated by the production of secondary metabolites. These compounds comprise a large percentage of the clinically useful antibiotics, anti-fungals, and even some of the anti-tumor agents used to date. The bioavailability of copper ions has been shown to be of crucial importance for morphological development in certain streptomycetes (8, 9). Copper dependence is restricted to the reproductive growth phase (aerial mycelium and spores), whereas vegetative growth proceeds under strongly copper -limiting conditions. Evidence from genetic knock-out studies has suggested that secreted cuproproteins or cuproenzymes are needed for the development switch from vegetative to aerial mycelium to occur, but their identification and role in the development switch have not so far been elucidated (10, 11).
The copper proteome of Streptomyces coelicolor has been determined using bioinformatics approaches, revealing a rich assortment of putative extracellular cuproenzymes, redox proteins, and copper metallochaperone-like proteins (10, 12). An exciting yet unexpected finding was the identification of a cuproenzyme along with a dedicated copper metallochaperone in the cytosol (10, 13). A metabolic requirement for copper in the bacterial cytosol has not yet been documented; however, in vitro biochemical evidence indicates that this cytosolic tyrosinase-like enzyme (MelD2) is active in the oxidation of mono- and di-phenols (13). The cytosol of Streptomyces coelicolor also contains proteins putatively involved in copper detoxification systems (10). Two operons have been identified that contain genes encoding for a CopZ-like copper metallochaperone and a CopA-like P1-type ATPase transporter. In certain bacteria, the copA and copZ resistance genes are transcriptionally regulated by a copper sensor protein belonging to the copper-sensitive operon regulator (CsoR)3 family (7). In S. coelicolor, the gene SCO4136 has been proposed to encode for a CsoR orthologue (10). However, the genomic environment of SCO4136 does not contain any known copper resistance genes and instead is located among genes encoding for proteins involved in phosphate transport (10). Therefore, if the gene product of SCO4136 functions as the cytosolic copper sensor in S. coelicolor, it is not genetically linked to copper resistance genes such as the two putative copZA-like operons.
The founding member of the CsoR family was discovered in Mycobacterium tuberculosis (CsoRMtb) (7). Under elevated copper levels, the expression of CsoRMtb was found to be strongly induced, with the binding of Cu(I) triggering the derepression of the rv0967–rv0970 operon in which rv0969 encodes for a P1-type ATPase involved in copper transport and rv0967 for CsoRMtb (7, 14). It has subsequently been found that members of the CsoR family are widespread in bacterial genomes. Examples of CsoR proteins that transcriptionally regulate copZA copper resistance operons have been identified and characterized to varying degrees in Bacillus subtilis (15), Listeria monocytogenes, and Staphylococcus aureus (16, 17). Furthermore, a recent report has identified a CsoR member that is not thought to be involved in copper homeostasis but instead appears to function in response to sulfur stress (17).
To understand the role copper plays in morphological differentiation and development, it is necessary to understand how cellular copper is handled under normal and elevated conditions. S. coelicolor and Streptomyces lividans both display a distinct dependence on copper for development with that of S. lividans being more pronounced (18). They also share a high level of sequence identity and genome organization (19, 20), and the gene numbering of the S. coelicolor genome database is used for S. lividans in this study. Sequence alignment of the S. lividans 4136 gene product with known CsoR orthologues reveals conservation of the amino acids that act as Cu(I)-binding ligands and two amino acids considered important for the Cu(I)-dependent allosteric regulation of DNA binding (Fig. 1). In this study, we have biochemically and structurally characterized the S. lividans 4136 gene product and find it to be a tetrameric CuI-CsoR (CsoRSl). In vivo studies with a genetic knock-out (ΔcsoR) and CsoRSl mutants indicate CsoRSl to be essential for the response of S. lividans to elevated copper levels. Furthermore, we have coupled bioinformatics and RNA-seq experiments to characterize the regulon under the direct control of CsoRSl. RNA-seq data suggest that the regulation of copper homeostasis is much more extensive than only the genes repressed by CsoRSl.
FIGURE 1.
ClustalW2 multiple amino acid sequence alignment of copper-sensing CsoR proteins for which biochemical or structural data have been reported. Completely and partially conserved residues are boxed in dark and light gray, respectively. The conserved Cu(I)-binding ligands are indicated (*) along with residues reported to play a role in the Cu(I)-binding allosteric regulation of DNA (+). The UniprotKB accession numbers are Q8Y646 (L. monocytogenes), A6QIT1 (S. aureus), O32222 (B. subtilis), and P71543 (M. tuberculosis).
MATERIALS AND METHODS
Streptomyces Strains, Media, and Growth Conditions
The Streptomyces strains used are as follows: S. lividans 1326 (S. lividans 66, stock number 1326 John Innes Collection), S. lividans ΔcsoR (this study), and S. coelicolor A3(2) strain M512 (21). The agar media soy flower mannitol, complex medium (R5), minimal agar medium (MM), the liquid complex medium tryptic soy broth with 10% sucrose, and the liquid defined medium NMMP were prepared according to Ref. 22. If required, glucose and/or mannitol were added to 0.5%. Agar plates were incubated at 30 °C, and liquid cultures were grown in 250-ml baffled flasks with 0.2-μm vent caps (Corning Glass) with shaking at 160 rpm. Liquid cultures were inoculated with spores to a final concentration of 2 × 107 spores/ml. Spore stocks were obtained from cultures grown on soy flower mannitol plates and stored in 20% glycerol at −20 °C. Growth was recorded by determining the dry biomass from 1.5-ml samples collected in pre-dried Eppendorf tubes, and mycelium was pelleted by centrifugation at 13,000 rpm for 10 min. After 12–16 h at 105 °C, the dry weight was determined using an analytical balance.
Generation of a CsoR Deletion Mutant (ΔcsoR) of S. lividans 1326
The 4136 gene encoding for CsoRSl was deleted in S. lividans 1326 in a two-step process using the CRE-lox system (23). First, the gene (nt +4 to 399) was replaced by homologous recombination with an apramycin resistance cassette flanked by loxP sites. For this purpose, the upstream flanking region of SL4136 (−1370 to +3) and the downstream flanking region (+399 to 1680) were amplified from genomic DNA by PCR, including EcoRI, XbaI, XbaI, and HindIII sites, respectively, for cloning purposes. These two fragments and the apramycin resistance cassette flanked by loxP sites were cloned in the delivery vector pWHM3 that is unstable in Streptomyces (24). Following protoplast transformation, recombinants that were apramycin-resistant but had lost the vector (thiostrepton resistance) were isolated. Second, an unstable plasmid encoding the Cre recombinase was introduced (23) and allowed for the excision of the apramycin resistance cassette on the lox sites. The resulting strain, ΔcsoR, has no coding sequence for CsoRSl and has only a 61-nt “scar,” including two XbaI sites left in the genome. The ΔcsoR strain was analyzed by PCR to confirm the loss of 4136, the apramycin resistance cassette, and vector sequences.
RNA Isolation and Transcriptome Analysis by RNA-seq
Total RNA was isolated with Kirby mix according to standard procedures from mycelium in early log phase grown on NMMP supplemented with 0.5% glucose and mannitol (22). Cultures were induced with 400 μm Cu(II) for 2 h followed by total RNA isolation. RNA integrity was confirmed by agarose gel electrophoresis, and the absence of genomic DNA was checked by PCR. For the removal of ribosomal RNA, an Ambion kit was used. Samples were sent to BaseClear, an independent and accredited service laboratory for DNA-based research, and transcriptome analysis by RNA-seq was carried out. The sequences obtained on an Illumina sequencer were filtered for noncoding RNAs and analyzed with CLCbio bioinformatics software packages using the annotated S. coelicolor genome as reference. Expression values were expressed as reads/kb of exon model/million mapped reads (25), i.e. dividing the total number of exon reads (in this case one exon per reference sequence) by the number of mapped reads (in Millions) times the exon length (in this case the length of the reference sequence).
Promoter Probing
The DNA sequence (−300 to +3) upstream of SL4136 was obtained from genomic DNA by PCR introducing a flanking EcoRI and BamHI site to facilitate cloning in pIJ2585 digested with the same enzymes (26). The resulting plasmid, p4136-I, was introduced in strain M512 by protoplast transformation. cultures were grown in triplicate in NMMP medium supplemented with 0.5% glucose, 0.5% mannitol, and 50 μg/ml apramycin. Strain M512 transformed with the empty vector was grown under the same conditions to obtain background readings. Following extraction of mycelium with methanol, the concentration of undecylprodigiosin was quantified from the absorbance at 530 nm and the extinction coefficient of 100,500 liters/mol−1 cm−1 (27).
Determination of Tyrosinase and Cytochrome c Oxidase Activity
The wild type, S. lividans 1326, and the mutant strain ΔcsoR were each transformed with pIJ703 (plasmid harboring the Streptomyces antibioticus melC operon under control of its own promoter). Four independent transformants of each strain were grown in liquid tryptic soy broth with 10% sucrose medium supplemented with 25 μm Cu(II). Tyrosinase activity was determined in spent medium with 10 mm 3,4-dihydroxy-l-phenylalanine in 100 mm phosphate buffer, pH 6.8, as substrate (28). Cytochrome c oxidase activity was visualized with N,N,N′,N′-tetramethyl-p-phenylenediamine as substrate, essentially according to Refs. 29, 30.
Cloning, Overexpression, and Purification of CsoRSl from Escherichia coli
The 4136 gene was cloned from S. lividans 1326 genomic DNA (supplemental “Materials and Methods”) and overexpressed in Escherichia coli using a pET28a (Kanr) vector (Novagen). This construct, designated pET4136, was transformed to E. coli BL21(DE3) cells, and single colonies were transferred to 2× YT medium (Melford) with kanamycin (50 μg/liter) (Melford) at 37 °C. Overexpression of the N-terminal His-tagged CsoRSl was induced by 1 m isopropyl β-d-1-thiogalactopyranoside (Melford) to a final concentration of 1 mm, and the temperature was decreased to 25 °C for overnight incubation. Cultures were harvested by centrifugation at 4000 rpm for 20 min at 4 °C, and the cell pellet was resuspended in 50 mm Tris/HCl, 500 mm NaCl (Fisher) and 20 mm imidazole (Sigma) at pH 7.5 (Buffer A). The resuspended cell suspension was lysed using an EmulsiFlex-C5 cell disrupter (Avestin) followed by centrifugation at 18,000 rpm for 20 min at 4 °C. The clarified supernatant was loaded onto a 5-ml nickel-nitrilotriacetic acid-Sepharose column (GE Healthcare) equilibrated with Buffer A and eluted by a linear imidazole gradient using Buffer B (Buffer A with 500 mm imidazole). A single peak at ∼30% Buffer B was eluted from the column, and fractions were pooled and dialyzed overnight at 4 °C against 10 mm MES, pH 6.5, 150 mm NaCl, 2 mm dithiothreitol (DTT) (Melford), and 4 mm EDTA (Sigma) (Buffer C). Following dialysis, the N-terminal His tag was removed by incubating the protein at room temperature overnight with 125 units of thrombin (Sigma). The protein/thrombin mixture was reapplied to the nickel-nitrilotriacetic acid-Sepharose column (GE Healthcare), and the flow-through was collected and concentrated using a Centricon (VivaSpin) with a 5-kDa cutoff at 4 °C for application to a G-75 Sephadex column (GE Healthcare) equilibrated with Buffer C. Fractions eluting from the major peak of the G-75 column were analyzed by SDS-PAGE, and those deemed of good purity were concentrated and stored at −20 °C until required.
Site-directed Mutagenesis of CsoRSI and Cloning for in Vivo Studies
The QuikChange site-directed mutagenesis method (Stratagene) was used to create the C75A and H100A mutants of CsoRSl. Forward and reverse primers were designed with the respective nucleotide change(s) to create the desired mutation (supplemental “Materials and Methods”), and the pET4136 plasmid was used as template. The respective mutations were confirmed by DNA sequencing. For expression in S. lividans, the wild type and mutant CsoRSl open reading frames (ORF) were cloned under control of the 4136 promoter and the constitutive ermE promoter. For this purpose, the 4136 promoter region was obtained by PCR from S. lividans 1326 genomic DNA introducing an NdeI site on the ATG start codon of the ORF, and the ermE promoter was obtained from plasmid pHM10a (31). The wild type, C75A, and the H100A ORFs were cloned downstream of the promoters in the low copy vector pHJL401 (32).
UV-visible and Circular Dichroism Spectroscopies
A Varian Cary 50 UV-visible spectrophotometer and an Applied Photophysics Chirascan circular dichroism (CD) spectrophotometer (Leatherhead, UK) both equipped with a thermostatic cell holder controlled with a Peltier system were routinely used. An extinction coefficient (ϵ) at 280 nm of 3105 m−1 cm−1 was calculated for the CsoRSl monomer. This value was used throughout to determine the concentration of apo-CsoRSl samples. Far-UV CD spectra at 20 °C with 20 μm CsoRSl in 10 mm potassium phosphate, 50 mm potassium fluoride, pH 7.0, were acquired in the range 260 to 190 nm.
Electrospray Ionization Mass Spectrometry (ESI-MS) and Atomic Absorption Spectroscopy (AAS)
CsoRSl samples for mass spectrometry were diluted 1:20 with a 50% methanol and 1% formic acid solution. Spectra were acquired on a Micomass Quattro Ultima triple quadrupole instrument using the following experimental parameters: capillary voltage 1.7 kV, cone voltage 80–120 V, and cone gas 100 liter/h. Acquisition and processing were carried out using MassLynx software (Waters, Manchester, UK). Metal content was determined with a Unicam 939/959 atomic absorption graphite furnace spectrometer.
Cu(I) Titrations and Determination of Binding Affinity
Apo-CsoRSl samples for experiments with Cu(I) were prepared in an anaerobic chamber (DW Scientific [O2] <2 ppm) by first incubating for 2–3 h with 2 mm DTT followed by desalting using a PD-10 column (GE Healthcare) equilibrated with either 10 mm MOPS, pH 7.5, 150 mm NaCl, or 10 mm MES, pH 6.5, 150 mm NaCl. Free thiol content was determined by the reduction of 5,5′-dithiobis(2-nitrobenzoic acid) monitored at 412 nm (ϵ = 13,500 m−1 cm−1) (33). Cu(I)Cl (Sigma) was dissolved under anaerobic conditions in 10 mm HCl and 500 mm NaCl and diluted with either MES or MOPS buffer. Cu(I) concentration was determined spectrophotometrically by stepwise addition using a gastight syringe (Hamilton) to a known concentration of the Cu(I)-specific bidentate chelator bicinchoninic acid (BCA) using an extinction coefficient at 562 nm of ϵ = 7900 m−1 cm−1 for [CuI(BCA)2]3 (34). Apo-CsoRSl proteins (30–50 μm) were sealed in an anaerobic quartz cuvette (Hellma), and the absorbance change at 240 nm was monitored upon titrating in the Cu(I) solution. Competition assays were set up anaerobically with either BCA or bathocuproine disulfonate (BCS) (Sigma). Increasing protein concentrations (0–90 μm) were added to solutions of [CuIL2]3− of defined molar ratio L:Cu(I) ≥3 (to ensure the presence of the 1:2 complex [CuIL2]3− with negligible contribution from the 1:1 complex [CuIL2]−) creating a series of individual solutions with constant [CuI] and [L] and varying [apo-CsoRSl]. Samples were left for between 1 and 4 h, and the transfer of Cu(I) from the [CuIL2]3− complex to apo-CsoRSl was determined by measuring the absorbance of the [CuIL2]3− complex spectrophotometrically for L = BCA at 562 nm (ϵ = 7900 m−1 cm−1) and L = BCS at 483 nm (13,000 m−1 cm−1) (34, 35). By interchanging L, assays favoring competitive or noncompetitive Cu(I) binding could be set up, which for the latter led to an estimate of the binding stoichiometry. The dissociation constant for Cu(I) (KD(CuI)) was determined from competitive assays by assuming Reaction 1 (1),
and by using Equation 1,
where [L] is the total ligand concentration (BCA or BCS), and the overall formation constant (β2) is 1017.2 m−2 for [copper (BCA)2]3− and 1019.8 m−2 for [copper (BCS)2]3− (34, 36). Assays were performed in duplicate, and the KD(CuI) value for a series was initially calculated for each individual solution and then averaged. By using the average KD(CuI) value, a simulated curve was plotted using Equation 1.
Bioinformatic Identification of CsoRSl Operator Targets
The computational prediction of CsoRSl cis-acting elements was performed as described previously (37). Experimentally validated binding sites of CsoR orthologues in B. subtilis (TAATACCCTACGGGGGTATGG) (15, 38), S. aureus (ATATACCTATAGGGGGTACAT) (17), Geobacillus thermodenitrificans (TTATACCCGAAGGGGGTATAT) (17), and M. tuberculosis (RicR.1, ATATACCACCCGGGGGTATAG; RicR.2, ATATACCCTATAGGGGTAGG; RicR.3, ATATACCCTATACGGGTATCT; RicR.4, TTGTACCCCAGCGGGGTATCG) (39) were used to generate via the PREDetector program (40) a first weight matrix to identify similar putative cis-acting elements within S. coelicolor (SCO), Streptomyces griseus (SGR), Streptomyces scabies (SCAB), and Streptomyces avermitilis (SAV) genomes. This first round of prediction allowed the identification of highly reliable putative cis-acting sequences in terms of (i) scores, (ii) interspecies conservation, and (iii) physiological meaning (hits related to copper sensitivity). Hits conserved among the selected four streptomycetes and identified upstream of copper utilization-related genes, i.e. upstream of copZ (copper chaperone) and upstream of the CsoRStrep orthologues, were use to generate a new weight matrix (named “CsoR streptomycetes”), more specific for predictions in Streptomyces species. Sequences used to generate the CsoR streptomycetes weight matrix were copZSCO, copZSGR, copZSCAB, and copZSAV, and csoRSCO, csoRSGR, csoRSCAB, and csoRSAV and are presented in supplemental Fig. S6, and reliable CsoR-binding sites identified in these four Streptomyces species are presented in supplemental Tables S2–S5. Putative CsoR-like sequences predicted in SCO were used to identify cis-acting elements in the closely related strain S. lividans.
Electrophoretic Mobility Shift Assays of CsoRSl Targets
Intergenic DNA fragments (208, 232, and 240 bp) containing the predicted target sequence for CsoRSl identified by PREDetector were amplified from genomic S. lividans 1326 DNA as described in the supplemental “Materials and Methods”. DNA oligomers (Sigma) for use in EMSA studies were between 35 and 36 bp in length. Complementary pairs were annealed by heating at 96 °C in a water bath for 5 min and left to cool to room temperature overnight. 0.5 μm of a DNA oligomer target was incubated with concentrations of apo-CsoRSl monomer ranging between 4 and 30 μm in 10 mm HEPES, pH 7.5, 150 mm NaCl, 1 mm DTT. Cu(I)-CsoRSl samples were either prepared by pre-loading apo-CsoRSl with a stoichiometric amount of Cu(I) in an anaerobic chamber before mixing with DNA samples or added directly to the DNA-protein complex under anaerobic conditions. All samples were incubated at room temperature for 30 min and then loaded (20 μl) to a pre-run 6% Tris/borate EDTA (TBE) polyacrylamide gel. Gels were stained for 30 min in an ethidium bromide solution followed by imaging.
Crystallization and Structure Determination of Apo-CsoRSl
Crystals of apo-CsoRSl were grown using the hanging drop vapor diffusion method at 20 °C. 1 μl of protein solution at a concentration of 15 mg/ml was mixed with an equal volume of reservoir solution containing 1.26 m ammonium sulfate, 0.1 m sodium citrate, pH 4. Crystals of dimensions ∼0.2 × 0.2 × 0.2 mm grew within 1 week. A single crystal was transferred to a cryoprotectant solution containing 1.3 m ammonium sulfate, 0.1 m sodium citrate, pH 4, and 15% glycerol, prior to flash-cooling to 100 K by plunging into liquid nitrogen. Crystallographic data were measured to 1.7 Å resolution at Diamond Light Source beamline I04 using an ADSC Q315r CCD detector and an x-ray wavelength of 0.9795 Å. Data were indexed using iMosflm (41) and scaled and merged using Scala (42) in the CCP4i suite. The structure was solved by molecular replacement in BALBES (43). The initial model was built into the electron density map using Buccaneer (44) and refined using Refmac5 (45). Riding hydrogen atoms were added when refinement of the protein atoms had converged. Models were rebuilt between refinement cycles in Coot (46), and the final structure was validated using the MolProbity server (47) and Coot. Coordinates and structure factors were deposited in the RCSB Protein Data Bank with accession number 4adz. A summary of data and refinement statistics and the quality indicators for the structure are given in Table 1.
TABLE 1.
Crystallographic data collection and processing statistics for apo-CsoRSl
Values in parentheses refer to the outermost resolution shell (1.79 to 1.70 Å).
| Wavelength | 0.9795 Å |
| Resolution | 45.9 to 1.70 Å |
| Space group | P21221 |
| Unit cell | a = 41.63 Å, b = 54.55 Å, c = 91.75 Å |
| Unique reflections | 22,220 (2403) |
| Completeness | 93.8% (71.7%) |
| Rmerge | 0.058% (0.283%) |
| I/σ(I) | 12.1 (2.1) |
| Rcryst | 0.185 |
| Rfree | 0.229 |
| ESU based on ML | 0.071 Å |
| Root mean square deviation bond lengths | 0.017 Å |
| Root mean square deviation bond angles | 1.66° |
| Ramachandran favored | 100% |
| Wilson B-factor | 22.4 Å2 |
| Protein Data Bank accession code | 4adz |
RESULTS
Mass, Metal Content, Secondary Structure, and Assembly State of Purified CsoRSl
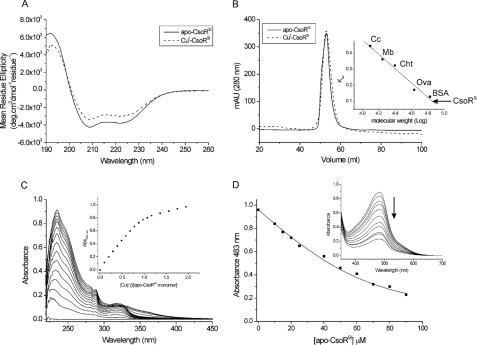
Purified CsoRSl ran as a single band on an SDS-polyacrylamide gel (supplemental Fig. S1) and denaturing ESI-MS gave a single component spectrum with a species of molecular mass 14,739 Da, in excellent agreement with the predicted mass (supplemental Table S1). AAS revealed negligible copper or nickel ion content, and thus an apo-form of CsoRSl was purified. The isolated protein reduced 5,5′-dithiobis(2-nitrobenzoic acid) to give a protein/thiol ratio of 1:1.8, suggesting that the thiol groups of Cys-75 and Cys-104, putatively involved in Cu(I) binding, remain reduced under the purification conditions (see below). The CD spectrum of apo-CsoRSl is shown in Fig. 2A with the two negative minima at 222 and 208 nm being typical signatures for α-helical secondary structure. Using this CD spectrum, a secondary structure content of 51% α-helix, 8% β-strand, 13% loops, and 27% unordered was predicted using Dichroweb (48, 49). Size-exclusion chromatography using a calibrated G-75 column revealed a major peak eluting at a mass of ∼74 kDa, suggesting that under native conditions apo-CsoRSl exists as a higher order assembly (Fig. 2B). This was confirmed from analytical ultracentrifugation (AUC) experiments (supplemental “Materials and Methods”) where sedimentation equilibrium scans were fitted to a single component model to give an average single species molecular mass of 59.3 kDa (supplemental Fig. S2A). Based on the mass determined using ESI-MS, the mass obtained from AUC is consistent with apo-CsoRSl existing as a tetrameric assembly (expected mass 58,955.6 Da).
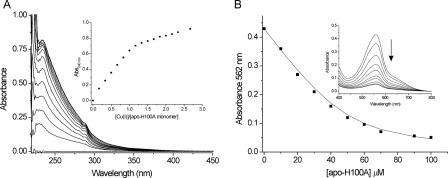
FIGURE 2.
CsoRSl exists in solution as a tetramer and binds Cu(I). A, far-UV CD spectra of CsoRSl at pH 7 and 20 °C. B, G-75 size-exclusion chromatography profile of CsoRSl. The gel phase distribution coefficients (Kav) versus log molecular weight of the following standards run individually on the column; cytochrome c (cc; 12.5 kDa), myoglobin (Mb; 17 kDa), chymotrypsin (Cht; 25 kDa), ovalbumin (ova; 43 kDa), and bovine serum albumin (BSA; 67 kDa) are plotted (inset) to estimate the molecular weight of the eluted CsoRSl. C, changes in the UV region of the apo-CsoRSl base-lined spectrum upon addition of Cu(I)Cl at pH 7.5, 20 °C. The inset shows the increase in absorbance at 240 nm plotted as a function of [Cu(I)]/[monomer-CsoRSl]. D, determination of the KD(CuI) for apo-CsoRSl at pH 7.5 under copper -limiting conditions imposed by [CuI(BCS)2]3− as a competitive probe. The absorbance at 483 nm in the visible spectrum of [CuI(BCS)2]3− decreases upon addition of apo-CsoRSl (inset) and can be used to determine the KD(CuI) using Equation 1. The line represents a best fit to the data using an average KD(CuI) 2.6 × 10−18 m. Conditions used are as follows: 10–90 μm [apo-CsoRSl], 74 μm [CuI]total, and 300 μm [BCS].
Apo-CsoRSl Binds Cu(I) with Attomolar Affinity
An anaerobic titration of Cu(I) into apo-CsoRSl gave rise to absorbance changes in the UV region of the spectrum (Fig. 2C). The absorbance increase at 240 nm is indicative of the formation of Cys-thiolate copper coordination bonds, whereas the 320 nm increase has not previously been reported for other CsoR orthologues (7, 38). The absorbance change at 240 nm saturates at ∼1 mol eq of Cu(I) per monomer of CsoRSl (Fig. 2C, inset). Beyond this point, a shallow increase in absorbance occurs upon further addition of Cu(I). The end point sample from the titration was passed through a desalting column to remove excess copper, and AAS gave a 1:1 copper :CsoRSl monomer stoichiometry. Cu(I)-CsoRSl samples were characterized by CD spectroscopy, gel filtration (Fig. 2, A and B), and AUC (supplemental Fig. S2B). The secondary structure content remains essentially unchanged compared with apo-CsoRSl and gel filtration (Fig. 2B), and AUC results are consistent with Cu(I)-CsoRSl maintaining a tetramer assembly.
Accurate determination of the KD(CuI) for CsoRSl necessitates the use of competition assays employing the specific Cu(I) bidentate chelators BCA and BCS (36, 50). At pH 7.5, addition of apo-CsoRSl into [CuI(BCA)2]3− leads to the extraction of 1 eq of Cu(I) from the [CuI(BCA)2]3− complex (supplemental Fig. S3). This indicates that under the concentrations employed, BCA cannot compete with CsoRSl for Cu(I), and that 1 eq of copper is bound per CsoRSl monomer, corroborating the results from the UV titration and AAS experiments. Competition for Cu(I) between apo-CsoRSl and [CuI(BCS)2]3− was observed (Fig. 2D), and analysis of the data set shown in Fig. 2D using Equation 1 and a β2 of 1019.8 m−2 for [CuI(BCS)2]3− gave a KD(CuI) of 2.6 × 10−18 m. This KD(CuI) was used to simulate a fit of the data, as shown by the solid line in Fig. 2D. Duplicate data sets were obtained with varying [CuI]total or [BCS], and the average KD(CuI) for CsoRSl at pH 7.5 was 6.7 × 10−18 m. Competition between CsoRSl and [CuI(BCS)2]3− was also observed at pH 6.5, and an average KD(CuI) of 3.2 × 10−17 m was determined.
Growth and Development of S. lividans 1326 Versus That of ΔcsoR Strain
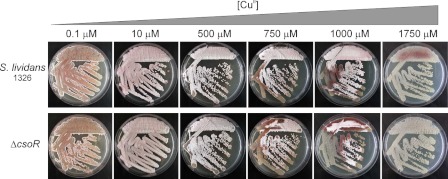
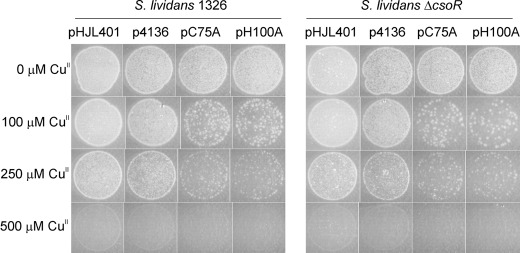
The attomolar affinity of CsoRSl for Cu(I) was consistent with a role in sensing, buffering, and handling copper in the cytosol under all growth conditions, including stress elicited by elevated copper concentrations. To assess the effect CsoRSl has on growth and morphological development of S. lividans 1326 under normal conditions and when copper stress is applied, a deletion strain (ΔcsoR) was constructed. Fig. 3 shows an example of growth on R5 agar media of the wild type S. lividans 1326 and the ΔcsoR strain at increasing copper concentrations. At low concentrations of copper, no significant differences between the two strains were observed regarding vegetative, aerial growth, and sporulation (Fig. 3). This was also the case on various other solid media and in the presence of copper chelators (supplemental Fig. S4A). However, above 750 μm Cu(II), the ΔcsoR strain appears to be slightly more affected in morphological development and in the production of the red pigment (undecylprodigiosin) than the wild type (Fig. 3). Copper therefore has a triphasic effect on development. Under conditions where all copper in the medium is bound by the chelator bathocuproine disulfonic acid, development is completely blocked (supplemental Fig. S4A). Stimulation of aerial hyphae production and spores occurs when the bioavailability of copper concentrations increase to 2–5 μm, and development is inhibited again at concentrations above 500 μm. This effect is not observed on MM or soy flower mannitol solid media, but on MM development is also severely retarded above 500 μm Cu(II) and inhibited completely at 1000 μm (supplemental Fig. S4B). Growth rate determinations in liquid defined media (NMMP) supplemented with Cu(II) in the range from 0 to 1000 μm corroborated the observations on the different solid media. Essentially no differences were seen between the growth rates of the wild type and the ΔcsoR strain under all conditions (Table 2 and supplemental Fig. S5).
FIGURE 3.
Effect of copper on morphological development of S. lividans 1326. Spores of wild type S. lividans 1326 and the ΔcsoR mutant were streaked on R5 agar plates in the presence of increasing concentrations of exogenous Cu(II) and incubated at 30 °C. Photographs were taken after 90 h.
TABLE 2.
Growth rates (doubling/h) of the wild type S. lividans 1326 and the ΔcsoR mutant in NMMP medium supplemented with the indicated [Cu(II)]
Standard deviation of the growth rates was 0.01.
| Concentration of Cu(II) | |||||
|---|---|---|---|---|---|
| μm | |||||
| Strain | 0 | 100 | 400 | 750 | 1000 |
| S. lividans 1326 | 0.27 | 0.28 | 0.28 | 0.27 | 0.28 |
| ΔcsoR | 0.28 | 0.29 | 0.27 | 0.26 | 0.26 |
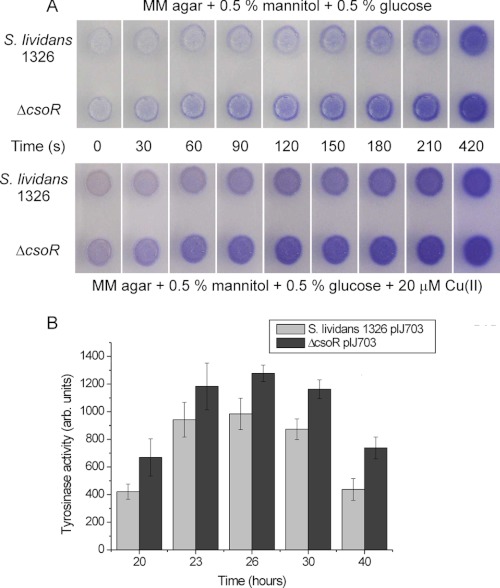
Challenging the wild type and the ΔcsoR strain with stresses that could affect copper homeostasis, such as diamide (redox stress) and hydrogen peroxide, showed a similar response in both strains (data not shown), as is the case for the addition of the iron chelator bathophenanthroline disulfonic acid to the medium (supplemental Fig. S4A). However, when the in vivo maturation of two cuproenzymes was measured, a small but consistent difference between wild type and the ΔcsoR strain was observed. Both the activity of cytochrome c oxidase and the secreted heterologous tyrosinase (MelC2) are significantly higher in the ΔcsoR strain (Fig. 4). Because deletion of the csoR gene is not likely to affect the expression of the endogenous cox genes (as confirmed by the RNA-seq data, see supplemental Table S6A) or the heterologous melC operon, these data would suggest that maturation/incorporation of the copper cofactor is more efficient in the ΔcsoR strain.
FIGURE 4.
Effect of ΔcsoR on cytochrome c oxidase and tyrosinase activity in S. lividans 1326. A,1000 spores were spotted on MM agar plates and incubated for 50 h at 30 °C. The in vivo activity of cytochrome c oxidase was determined using N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) by monitoring the production of the blue compound. The ΔcsoR mutant displays a quicker production of the blue compound than the wild type both on plates with and without added Cu(II). B, tyrosinase activity in the spent medium was determined with 3,4-dihydroxy-l-phenylalanine as substrate at the indicated time points and normalized for the biomass. The average tyrosinase activity of four transformants is plotted.
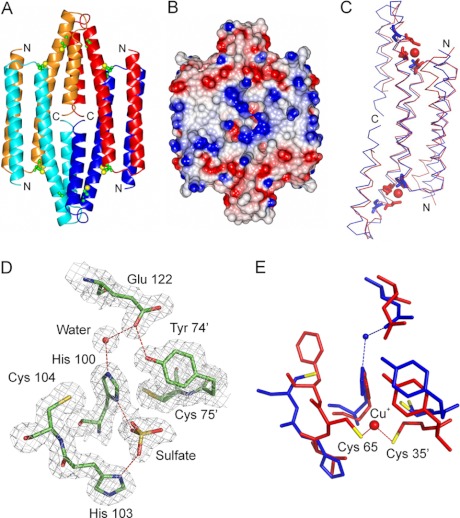
X-ray Crystal Structure of Apo-CsoRSl
The crystal structure of apo-CsoRSl was determined to 1.7 Å resolution. Two protomers (chains A and B) were found in the crystallographic asymmetric unit of a crystal of CsoRSl with well defined electron density visible for residues 44–133 in each protomer. The overall protomer fold (residues 44–133) consists of three α-helices of varying lengths. No electron density was visible for residues 1–43 suggesting that these residues are disordered. By applying crystallographic symmetry, a tetramer assembly, consistent with AUC, was generated with chain A packing against chain C and chain B packing against chain D (Fig. 5A). A surface representation of the tetramer is shown in Fig. 5B indicating that CsoRSl has a “closed” tetrameric assembly as opposed to a donut-like structure reported for a CsoR-like protein from Thermus thermophilus and CsoRMtb (7, 51). The overall fold of CsoRSl and the packing of the three α-helices of each protomer within the tetramer assembly are similar to that in the homodimer structure of CuI-CsoRMtb (Fig. 5C) (7). Superposition of the two structures by secondary structure matching gave a root mean square deviation in Cα positions of 1.98 Å. An extended α-helix 3 in CsoRSl appears to be the reason for the closed assembly such that in apo-CsoRSl the C termini of each pair of protomers pack together. Further differences between these structures are observed in the hairpin loop connecting α-helices 2 and 3, and at the N terminus of α-helix 1 (Fig. 5C). The largest differences are in the vicinity of the copper -binding sites, with residues 104–106 having deviations of >4 Å. The electrostatic potential of the CsoRSl tetramer reveals a central region of strong positive potential at the interface of two homodimers that extends out toward helix 1 (Fig. 5B). Negative potential is located at the start and end of helix-2 and flanks the central positive potential in the tetramer assembly (Fig. 5B).
FIGURE 5.
Structure of apo-CsoRSl. A, physiologically relevant tetramer assembly, with protomers colored red (A), orange (B), blue (C), and cyan (D). The close packing of the C termini of all chains is apparent. The Cys residues predicted to be involved in copper binding are shown as sticks with the Sγ atom colored yellow. B, electrostatic surface representation of the CsoRSl tetramer. Positive charges are indicated in blue and negative charges in red. C, superposition (calculated in the program Superpose, part of the CCP4 suite), based on secondary structure matching, of protomers A and C of the CsoRSl structure (blue) with the dimer of CsoRMtb (red). The copper -binding Cys residues from CsoRMtb and the corresponding residues in CsoRSl are shown as sticks. D, 2Fo − Fc electron density map, contoured at 1σ for the putative copper -binding region in the structure of CsoRSl. Hydrogen bonds are shown as dashed red lines. The proposed copper binding residues Cys-75 and Cys-104 lie some 7.6 Å apart and are separated by the imidazole side chain of His-100, which forms a hydrogen bond to a well ordered water molecule. A sulfate anion is present at the protein surface, forming bonds to His-100 and His-103. E, comparison of the copper -binding region in CsoRSl and CsoRMtb, based on the superposition shown in C. A and B prepared using CCP4MG (56) and C–E prepared using PyMOL.
From the sequence alignment in Fig. 1, Cys-75, His-100, and Cys-104 are predicted to be the copper ligands in CsoRSl. In the structure of the tetrameric assembly, His-100 and Cys-104 are located toward the end of helix 2 in each protomer, and Cys-75′ is located on a loop connecting α-helices 1 and 2 of an opposite protomer creating a putative intersubunit binding site, as found in CsoRMtb. The side chain of Cys-75 has been modeled in two slightly different conformations with occupancies of 0.7:0.3 (Fig. 5D), with the Sγ atom pointing either toward or away from the His-100 side chain. It is apparent that, in the absence of a copper ion, the His-100 imidazolate in apo-CsoRSl intersects the two Cys ligands, which are some 7.6 (monomer A)/7.3 (monomer B) Å apart such that no disulfide bond between the two Cys residues is formed (Fig. 5D). The Nϵ2 atom of His-100 forms an H-bond (2.7/2.6 Å) with a well ordered water molecule (Wat1), which further hydrogen bonds (2.7/2.8 Å) to the side chain Oϵ1 atom of Glu-122 in α-helix 3 (Fig. 5D). A further H-bond interaction (2.6/2.7 Å) involving the Oϵ1 atom of Glu-122 and the OH atom of Tyr-74′ of an adjacent protomer is also observed (Fig. 5D). A number of sulfate ion-binding sites are present in the structure, consistent with the ammonium sulfate component of the crystallization solution. One of these sites is located in the vicinity of each of the putative copper -binding sites. This sulfate anion is clearly positioned to be within H-bond distance of the Nδ1 atom of His-100 (2.6/2.6 Å) and 2.9/3.1 Å from Nδ1 of His-103 (Fig. 5D). It is interesting to note that the position of the sulfate anion is close to that of the modeled copper ion in the 2.55 Å resolution CsoRMtb structure (7).
In Silico Identification of Putative CsoRSl Operator Targets
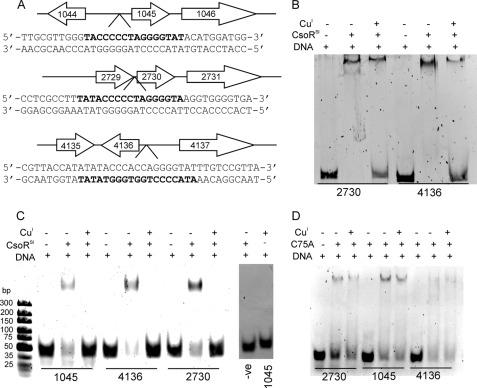
The structural similarity of the 4136 gene product to CuI-CsoRMtb adds further credence to a role as repressor in S. lividans. Based on experimentally validated cis-acting sequences bound by B. subtilis, S. aureus, and M. tuberculosis CsoR orthologues (7, 15, 17, 38), we bioinformatically predicted putative CsoRSl operator sequences in S. lividans. Regulons tend to be highly conserved in distantly related species, and therefore to ensure the identification of highly reliable CsoRSl putative target genes, the computational prediction was performed in four Streptomyces species, S. coelicolor, S. scabies, S. avermitilis, and S. griseus (supplemental Tables S2–S5), so as to increase the reliability of cis-acting element predictions. Predictions from S. coelicolor were used to identify putative CsoR operator sequences in S. lividans. Three target sequences were selected from a stringent prediction procedure for further in vitro analyses. The highest score was 21.69, and the target sequence (AAATACCCCTGGTGGGTATAT) was located −42 nucleotides upstream from the start codon of csoR and −183 nucleotides upstream from the start codon of 4137 encoding a putative phosphate transport regulator (Fig. 6A). The identification of the most highly reliable operator sequence upstream of csoR itself strongly supports the validity of the prediction procedure and suggests autoregulation of CsoRSl expression as reported for CsoR orthologues in other bacteria (7, 16, 17). The other two targets had scores of 19.37 (TTATACCCCCTAGGGGTAAGG) and 14.15 (GGGTACCCCCTAGGGGTATAC) and were found to be located −25 nucleotides upstream of the gene 2730 and −83 nucleotides upstream of the gene 1045 (Fig. 6A). Both of these genes are part of a copZA-like operon, predicted to encode a CopZ-like copper metallochaperone protein and a CopA P1-type ATPase, which in tandem operate as a specific Cu(I) efflux system in many bacterial systems. Based on our computational predictions, the deduced consensus binding sequence of streptomycetes CsoR orthologues corresponds to the 21-nt palindromic sequence ATATACCCCTNAGGGGTATAT, where positions 3–8 and 14–19 (underlined) appear to be the most conserved and thus probably the more crucial for CsoRSl recognition (supplemental Figs. S6 and S7).
FIGURE 6.
CsoRSl operator DNA targets and EMSA studies. A, three-locus regulon and the DNA sequences identified by PREDetector (bold) to be target operator binding sites for CsoRSl. The position of the DNA target sequence in the oligomers used in EMSA is highlighted in bold. B, EMSA of CsoRSl in the presence of the intergenic DNA fragments containing the target sequences for 2730 (232 bp) and 4136 (208 bp). C, EMSA of CsoRSl in the presence of the individual DNA oligomers containing the target sequences shown in A. The gel image to the right is a control EMSA showing the effect on binding CsoRSl to a randomly generated double-stranded DNA oligomer with the sequence 5′-GACGAGGACGTCTACGCCATCGACATACTG-3′ and the mobility of the 1045 DNA oligomer in the presence of only Cu(I). D, EMSA of the C75A mutant of CsoRSl in the presence of the individual DNA oligomers containing the target sequences shown in A. The components present in each lane of the gel image are indicated with 0.5 μm [DNA] and 5 μm [apo-CsoRSl monomer] and [CuI-CsoRSl monomer] used.
EMSA Analysis of CsoRSl Operator Targets and Assembly State of DNA-CsoRSl Complex
To test whether the three operator sequences identified with PREDetector would bind CsoRSl in vitro, EMSAs were carried out. These were initially performed with intergenic regions consisting of 240 bp (1044/1045), 232 bp (2729/2730), and 208 bp (4136/4137) and with smaller 35/36-bp DNA oligomers, containing the CsoRSl operator sequence flanked by 10 or 11 bp (Fig. 6A). Incubation of the intergenic regions or oligomers with apo-CsoRSl resulted in the formation of a low mobility CsoRSl-DNA complex visualized by the retardation of the DNA in the EMSA (Fig. 6, B and C). No shift in mobility of a random DNA sequence was observed suggesting that binding of the targets to CsoRSl is specific (Fig. 6C). Incubation of the DNA oligomers with CuI-CsoRSl samples or anaerobic addition of Cu(I) to preincubated apo-CsoRSl-DNA complexes resulted in the absence of a band shift, and only the high mobility band corresponding to the free DNA target was observed (Fig. 6C). This strongly suggests that once Cu(I) is bound to CsoRSl, the affinity for these small DNA fragments is significantly reduced such that a complex is not detected by EMSAs. For the intergenic regions, similar behavior is observed, but it is noted that some low mobility complex remains (Fig. 6B). DNA incubated with only Cu(I) was not affected in mobility (Fig. 6C). To determine the DNA:CsoRSl binding stoichiometry, size-exclusion chromatography was used (supplemental “Materials and Methods”). This revealed a 1:8 ratio (DNA:CsoRSl monomer) or two tetramers of CsoRSl are required to bind the DNA operator (supplemental Fig. S8).
Effect of Mutating Residues Cys-75 and His-100 in CsoRSl
Size-exclusion chromatography and AUC measurements verified that the mutants do not disrupt the tetramer assembly state, and AAS indicated the absence of stoichiometric copper in the purified proteins. Both mutants showed an absorbance increase at 240 nm upon addition of Cu(I) under anaerobic conditions, saturating at ∼1 mol eq of Cu(I) (Fig. 7A and supplemental Fig. S9), and for the H100A mutant, the absorbance increase at ∼320 nm that was observed in the wild type protein upon addition of Cu(I) was absent (Fig. 7A). The KD(CuI) values for the mutants were determined using BCA as a competitive probe (Fig. 7B). Duplicate data sets at pH 7.5 were acquired for both mutants with varying [CuI]total or [BCA], and the data averaged to give a KD(CuI) of 1.3 × 10−14 m and 9.8 × 10−16 m for the C75A and H100A variants, respectively. Both mutants therefore display an increased KD(CuI) compared with wild type CsoRSl.
FIGURE 7.
H100A mutant binds Cu(I) with a reduced affinity. A, changes in the UV region of the apo-H100A mutant base-lined spectrum upon addition of Cu(I)Cl at pH 7.5 and 20 °C. The inset shows the increase in absorbance at 240 nm plotted as a function of [Cu(I)]/[apo-H100A monomer]. B, determination of the KD(CuI) for apo-H100A under copper -limiting conditions imposed by [CuI (BCA)2]3− as a competitive probe. The inset shows the decrease in absorbance at 562 nm in the visible region of the spectrum upon increasing [apo-H100A], and Equation 1 was used to determine the KD(CuI). The line represents a best fit to the data using KD(CuI) 6.3 × 10−16 m. Conditions used are as follows: 10–100 μm [apo-H100A], 54 μm [CuI]total, and 260 μm [BCA].
The mutants were each able to bind to the three DNA operators as indicated by EMSA (Fig. 6D). Addition of Cu(I) to the mutant-DNA operator complexes did not result in the disappearance of the low mobility band or a substantial increase in the intensity of the high mobility (free) DNA band (Fig. 6D). This suggests that Cu(I) binding in the mutants does not alter the affinity for the DNA operator to the extent observed for the wild type and suggests that the allosteric mechanism of operator release or exposure is affected. This apparent inability of the CsoR mutants C75A and H100A to disengage in vitro from the DNA operator following Cu(I) binding raises the question of what the effect on growth is when these mutant proteins are expressed in the mycelium. The wild type and mutated CsoRSl genes were cloned on a low copy plasmid under control of their own promoter. These constructs did not show any effect on growth. Therefore, the CsoRSl wild type gene and the mutant genes encoding the C75A and H100A variants were cloned under control of a strong constitutive promoter, and the wild type and ΔcsoR strain were transformed with these constructs. Fig. 8 clearly shows that the mutant forms of CsoRSl have a strong inhibitory growth effect in both strains starting from 100 μm Cu(II). The effect of mutant CsoR expression is slightly more pronounced in the ΔcsoR mutant than in the wild type because the latter also produces CsoR from the genomic gene copy resulting in a mix of plasmid and genome encoded proteins. Upon prolonged incubation, the control transformants harboring the empty vector or the wild type CsoRSl gene produce confluent growth with 250 and 500 μm Cu(II), although those expressing the mutant CsoRs do not. These observations support the in silico data indicating that CsoRSl has control over the expression of genes that encode proteins involved in exporting excess copper ions and its own expression under copper stress conditions.
FIGURE 8.
Effect of C75A and H100A CsoRSl mutants on growth. S. lividans 1326 and the ΔcsoR mutant were transformed with pHJL401 (empty vector) or the same vector harboring the csoR gene (p4136) or the mutants C75A (pC75A) or H100A (pH100A) under control of the constitutive ermE promoter. Spores (1000) were spotted on MM agar plates containing varying [Cu(II)] and incubated at 30 °C. After 50–60 h of incubation, a confluent lawn was produced. Images were taken with a Leica M80 stereomicroscope equipped with a DFC295 camera. Note the higher toxicity of exogenous copper supply due to mutations preventing the copper -dependent modulation of CsoRSl DNA binding ability.
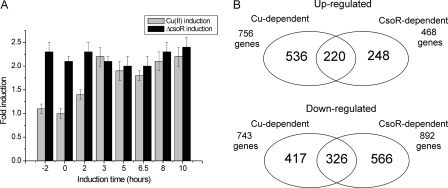
Mapping the Global Response of Wild Type S. lividans to Elevated Copper Levels and the ΔcsoR Gene by RNA-seq
RNA-seq is a second generation high resolution sequencing technique that enables gene expression profiling in an organism to be analyzed in response to a mutation or an external stimulus (52–54). We have applied RNA-seq to further our insight into the CsoR regulon in S. lividans by analysis of the transcriptome of liquid-grown cultures. The transcriptomes of wild type S. lividans strain 1326 grown without and with a 2-h exposure to 400 μm Cu(II) were compared with that of the ΔcsoR strain. From analysis of the RNA-seq data, both CopZ genes (gene numbers 2730 and 1045) and the 1044 gene encoding for an uncharacterized secreted protein are all induced by exogenous copper and in the ΔcsoR strain (Table 3). These data corroborate the in silico approach and identify these genes as bona fide CsoRSl targets. The CopA-like ATPases (2731 and 1046) are also up-regulated under elevated copper conditions (Table 3) but less so in the ΔcsoR strain. Nevertheless, this suggests regulation by the same promoter as the cognate CopZ. It is noticeable that the RNA-seq data do not provide clear support for the copper induction of gene 4137 or the csoR gene, which was predicted from PREDetector analysis (Table 3 and supplemental Table S2). Therefore, the promoter of csoR was analyzed in a promoter-probing experiment. The data clearly show that transcription originating from the csoR promoter is both copper -inducible and CsoR-dependent (Fig. 9A). The low induction by a 2-h exposure to 400 μm Cu(II) seen in the RNA-seq analysis (1.3-fold, Table 3) is confirmed by the promoter probing experiment (Fig. 9A). It also shows that upon longer exposure to Cu(II) the transcription is induced to around 2-fold, a similar level as observed in the ΔcsoR strain.
TABLE 3.
Expression levels obtained by RNA-seq of the genes predicted by PREDetector to be under control of CsoRSl along with other putative copper proteins up-regulated
| Annotation | Gene | PREDetector score | S. lividans 1326 RPKMa | S. lividans 1326 copper-induced (RPKM)a | -Fold increase | ΔcsoR (RPKM)a | -Fold increase |
|---|---|---|---|---|---|---|---|
| CsoR | 4136 | 21.7 | 169.8 | 217.5 | 1.3 | ||
| Pit accessory protein | 4137 | 21.7 | 418.2 | 470.5 | 1.1 | 496.3 | 1.2 |
| Membrane protein | 3280 | 21.3 | 21.7 | 112.8 | 5.2 | 20.0 | 0.9 |
| CopZ | 2730 | 19.4 | 86.7 | 436.0 | 5.0 | 220.9 | 2.5 |
| Secreted protein | 1044 | 14.2 | 4.0 | 17.1 | 4.2 | 7.2 | 1.8 |
| CopZ | 1045 | 14.2 | 0.0 | 9.8 | ≫ | 5.8 | ≫ |
| CopA-like P1-type ATPaseb | 2731 | 32.4 | 65.5 | 2.0 | 25.5 | 0.8 | |
| CopA-like P1-type ATPaseb | 1046 | 3.4 | 9.2 | 2.7 | 4.8 | 1.4 | |
| Uncharacterized protein with CusF domainc | 7265 | 3.2 | 23.0 | 7.3 | 18.0 | 5.7 | |
| Uncharacterized protein with a putative type 1 copper site | 6710 | 0.0 | 6.3 | ≫ | 2.1 | ≫ |
a The selected expression measure is the RKPM. It is defined as the reads/kb of exon model/Million mapped reads (25).
b Genes 2731 and 1046 are in the same operon as 2730 and 1045, respectively.
c Gene 7265 is the only putative copper-trafficking linked gene that is induced both by copper stress and by csoR deletion (see Table S6A for all putative copper proteins).
FIGURE 9.
Induction of csoR by promoter probing and global differential gene expression. A, promoter activity in response to the addition of 400 μm Cu(II) in the wild type and the ΔcsoR strain are expressed as fold change relative to the wild type strain without addition of Cu(II). Note the copper -induced Red production in the S. lividans wild type background and the constant and copper -insensitive Red production in the S. lividans ΔcsoR background. B, Venn diagrams depicting a global overview of the up- and down-regulated genes (>2-fold reads/kb of exon model/million) upon Cu(II) induction of wild type S. lividans and in the ΔcsoR strain. The supplemental Table S6, A–G, reports the lists of genes affected.
Finally, the transcriptome response to either copper or deletion of the csoR gene is not limited to the genes reported in Table 3. The global response can be appreciated visually by the use of Venn diagrams (Fig. 9B). A large number of genes are up- and down-regulated in response to copper and deletion of the csoR, with a significantly greater proportion of genes being down-regulated in the ΔcsoR strain. A considerable overlap is present between copper induction and csoR deletion (Fig. 9B and supplemental Table S6, B–G). Together, these data indicate that the response to changes in copper homeostasis in S. lividans is much more extensive than only the direct CsoRSl regulon reported in Table 3. Aside from the genes up-regulated in Table 3, there is no obvious enrichment of genes that encode proteins known to be directly related to copper homeostasis. However, a gene for a putative copper transporter (3964), part of an operon encoding for two putative copper chaperones, is down-regulated in copper -induced S. lividans but not in the csoR deletion (supplemental Table S6, E–G).
DISCUSSION
At the molecular level, our understanding of proteins involved in copper detoxification, storage, and trafficking has advanced considerably in recent times (1, 3, 4). The next challenge is to interface our molecular and mechanistic understanding of copper homeostasis with the global response of an organism to copper overload. In this study, we have extensively characterized a Cu(I)-CsoR metalloregulator from S. lividans and uncovered the direct and extended CsoR regulon.
Cu(I) Affinity for CsoR Is Unified but How DNA Is Engaged Remains Unresolved
Recent reports have highlighted the need for a unifying approach to accurately determine Cu(I) binding affinities in homeostatic cuproproteins (36, 50). The use of the Cu(I)-specific chelators BCA and BCS has been strongly advocated, and these have been used in this work. Accurate determination of metal affinities of metal sensors is particularly important as these values are considered the thresholds for homeostasis (2). By using BCS, an upper limit KD(CuI) of 1 × 10−20 m was initially reported for CsoRMtb (7) and later revised to 1 × 10−19 m (55). For CsoRBs, an upper limit of 1 × 10−21 m has been reported using BCS (38) and most recently a KD(CuI) of 7.9 × 10−19 m for CsoRSa (17). The KD(CuI) value determined for CsoRSl at pH 7.5 is 6.7 × 10−18 m, an order of magnitude higher than CsoRSa and CsoRMtb, and thus a comparatively lower affinity for Cu(I). These studies clearly highlight the consistency in using BCS as a probe to determine Cu(I) binding affinity in the attomolar range, enabling for cross-species comparisons. The KD(CuI) values determined using BCS are all consistent with the CsoR sensors being triggered by [copper] ∼9 orders of magnitude higher than the minimal intracellular copper concentration, consistent with the bacterial cytosol having no free atoms of copper (5). Furthermore, this clearly indicates the lengths to which cells go to avoid the potentially deleterious effects of uncomplexed copper ions.
At 1.7 Å resolution, the crystal structure of apo-CsoRSl is the highest resolution structure to date for a member of the CsoR family. The primary structure of CsoRSl is unique compared with other characterized CsoR proteins in that it possesses an extended (43 amino acids) N-terminal tail (Fig. 1). Structural organization of this region was not revealed from the crystal structure as electron density was only observed from residue 44 onward. Likewise, the C-terminal extension in CsoRMtb (7) was not structurally observed, and in the absence of proteolytic cleavage, dynamics outside the “core” structure are likely to be relevant. The core structure of CsoRSl is very similar to that of CuI-CsoRMtb (7) and has no recognizable DNA structural binding motif, such as a helix-turn-helix motif. Despite this, apo-CsoRSl has high specificity and affinity for operator DNA targets (Fig. 6). The absence of a recognizable DNA-binding motif leaves the question of how CsoR orthologues engage with their operator target unanswered. From our structure of CsoRSl, we observe that the tetramer assembly creates a large continuous surface area with strong electropositive charge centered in the middle of each face of the tetramer, which may be of significance for DNA binding (Fig. 5B). An alternative possibility is that the absence of a DNA-binding motif is the reason why two tetramers associate with the DNA, as also reported for CsoRBs (38), so as to enhance binding, specificity, or both.
Apo-CsoRSl Structure Provides Insights into the Mechanism of Allosteric Regulation
An hypothesis for the mechanism of allosteric Cu(I) regulation in the CsoR family has been put forward based on the crystal structure of CuI-CsoRMtb (7) and tested experimentally using unnatural amino acid substitutions of the copper -coordinating His residue (55). These unnatural amino acids were designed to abolish the H-bonding capability of the Nϵ2 atom of the imidazole ring so that, on binding Cu(I) to the Nδ1 atom, a second coordination sphere H-bond network centered on the Nϵ2 atom could not form and trigger release of the DNA (Fig. 5E). Cu(I) binding to the non-native His-substituted CsoRMtb did not significantly affect the DNA binding affinity, but the allosteric coupling free energy (ΔGc) was determined to be close to zero (55). This observation was taken to indicate that allosteric switching is initiated upon binding Cu(I) to the Nδ1 atom of the His ligand, triggering the formation of a H-bond network to the Nϵ2 atom that results in dissociation from the operator DNA sequence. However, this H-bond network is also present in the apo-CsoRSl structure and suggests that copper binding is not an essential requirement for its formation (Fig. 5E). One difference in the network found in CsoRSl is the presence of a bridging water molecule between His-100 and Glu-122 (Fig. 5D). In the CuI-CsoRMtb structure, no such water molecule is observed, and the H-bond length between the Cu(I) coordinating His and the Glu is >4 Å (7). This is indicative of a very weak interaction, which may lead to a destabilization of the DNA-bound structure. Whether or not this water molecule plays a role in the mechanism of allosteric Cu(I) regulation or in DNA binding is presently unclear. From Fig. 5E, it is clear that His-100 and Cys-75′ in the apo-CsoRSl structure are closely aligned to the corresponding residues in the CuI-CsoRMtb structure. However, Cys-104 in apo-CsoRSl must undergo a significant movement to complete the Cu(I) coordination sphere (Fig. 5E). It is conceivable that the movement associated with the positioning of the Cys-104 side chain to enable Cu(I) coordination may be of significance in allosterically regulating the dissociation from the DNA operator.
CsoR Regulon in Streptomycetes
A CsoR-responsive element prediction in four different Streptomyces species (supplemental Tables S2–S5) confirmed a very limited occurrence of highly conserved sequences that have been identified only upstream of one or two sets of orthologues of the copper chaperone copZ and the efflux ATPase copA, as well as upstream of the copper -sensing repressor csoR itself. EMSA studies (Fig. 6) revealed that CsoRSl recognizes specifically these sequences, and the effect of Cu(I) on DNA binding supports a mechanism of allosteric regulation most likely similar to other CsoR orthologues (7, 17, 38). RNA-seq data corroborate the in silico predictions, revealing clear induction of transcripts for two operons encoding copper efflux systems (2730/2731 and 1045/1046) (Table 3) and the divergent expression of 1044 (Table 3), a secreted protein of unknown function but not predicted to bind copper. RNA-seq data also indicates that CsoRSl transcript levels, under normal conditions, are relatively high compared with the two copZ genes and suggest that a significant proportion of CsoRSl is constitutively present (Table 3). The observation that expression of the mutant csoR genes (C75A and H100A) under control of their own promoter does not lead to growth inhibition at higher [copper] illustrates that CsoRSl controls its own transcription. Failure of the mutants to disengage from the DNA operator has been reported for other copper ligand variants of CsoR orthologues (7, 16, 17) and appears to lead to permanent transcription repression, resulting in too low protein expression levels to block transcription of the other genes with CsoRSl-binding sites. The induction of csoR was not observed from the RNA-seq data (Table 3) but was confirmed after >2 h of incubation with exogenous copper (Fig. 9A). This suggests the possibility that a modular system of CsoR repression may be in operation. Interestingly, the in silico analysis in S. griseus predicted four other CsoR-responsive elements suggesting a somewhat wider regulon in this strain (supplemental Table S5). The SGR3189 (cutC) gene encoding a putative copper homeostasis protein (cutC) is located upstream of a copA-like gene but is divergently transcribed. This offers the possibility that the CsoR-responsive elements could act for both copA and cutC in S. griseus. Two other CsoR-binding sites are found at position −30 nt from the SGR5260 gene, which could encode for the first member of an operon, including a putative multicopper oxidase (SGR5259) and at position −36 nt from SGR1262 (cstR) encoding for a putative non-copper sensing member of the CsoR family (17).
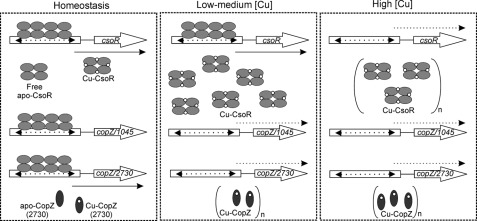
A Model of the Directly Regulated CsoR Regulon in S. lividans
Our findings clearly show that CsoRSl directly acts to regulate a three-locus regulon. Under homeostasis conditions, all operator sequences are occupied by two tetramers of CsoRSl resulting in repression of transcription with some free apo-CsoR and some copper -bound CsoR and CopZ (2730) acting as buffers (Fig. 10). Upon elevated cytosolic copper levels, we suggest, based on our data, a three-step response of the system. Low [copper] can be buffered by free apo-CsoR and CopZ (2730), and no further action is required. Medium [copper] will require a stronger response, achieved by derepressing the 2730 and 1044/1045 operons resulting in expression of CopZ and the CopA P1-type ATPases (Table 3). A further increase in [copper] will result in copper binding by the apo-CsoRSl still occupying the csoR operator. As a consequence, CsoR will be produced at a higher level and will assist in buffering copper in the cytoplasm. We assume that CsoR can also mediate the trafficking of copper by donating its copper to CopZ, which in turn will have the specificity to deliver the metal to its cognate CopA P1-type ATPase for export. As soon as copper levels are restored, apo-CsoRSl begins to occupy the operator sequences, and the system returns to its “ground state.” This model assumes that the up-regulation of CsoRSl occurs later. Quite how this may be possible is not known, but promoter probing and RNA-seq data are consistent with this phenomenon.
FIGURE 10.
A model of CsoR regulon and its response to copper stress in S. lividans. Under normal conditions, both csoR and 2730 (CopZ) are expressed at a low CsoR-independent level to ensure an immediate response to an increase in [copper] is present (left panel). Upon higher copper levels, the apo-CsoR and apo-CopZ can buffer copper at first, but as soon as copper levels increase, the free apoproteins will all become copper -bound, and the repression of 2730 and 1045 will be relieved because of Cu(I) binding to the DNA-bound CsoR resulting in production of sufficient export capacity to maintain homeostasis (middle panel). Upon further increase of [copper] the repression of csoR will be removed, and the full response becomes available (right panel).
What Can Be Learned About Copper Homeostasis in S. lividans from RNA-seq Analysis?
Although RNA-seq analysis clearly provides insight into the response of transcripts under the direct control of CsoRSl (Table 3), it is very much evident that response to exogenous copper or the deletion of the csoR gene is a complex process (Fig. 9B and supplemental Table S6, A–G). Over 400 genes show significantly higher expression in the ΔcsoR strain without presenting putative CsoRSl binding sequences in their adjacent genetic environments. Therefore, the expression of these genes must be responding in an indirect way to CsoRSl and may possibly be due to the copper -binding ability of CsoRSl rather than its DNA-binding ability. Indeed, the inactivation of csoR would somehow mimic copper overload due to the absence of free cytosolic apo-CsoRSl involved in buffering excess copper.
Of the genes annotated to encode cuproenzymes and putative copper transport proteins, the expression of some could be CsoRSl-dependent (supplemental Table S6A). For example, from the initial PREDetector analysis of the streptomycetes CsoR regulon, a fourth type of gene that could potentially be controlled by CsoRSl was identified (supplemental Table S2). Gene 3280 encodes for a membrane protein, but experimental validation of the predicted site was not included in our present EMSA studies due to the unusual location of the predicted cis-acting element at position +24 nt within the predicted 3280 coding sequence. Similar sequences identified at position +4 and −186 nt relative to the translational start of the 3280 orthologue in S. avermitilis and S. scabies are more appropriately located for controlling gene expression by a transcriptional repressor (supplemental Tables S3 and S4). The RNA-seq data highlighted a 5.2-fold higher expression of 3280 in RNA samples collected from copper -induced cultures, whereas the expression level in the ΔcsoR remains unchanged (Table 3). This suggests that 3280 is involved in copper homeostasis, and an as yet unidentified transcription control system plays a role when CsoRSl is no longer present due to the gene deletion. A closer inspection of the 3280 product predicts a protein (∼200 amino acids) with four transmembrane helices and a putative Cu(I) Cys-Xaa-Xaa-Xaa-Cys-binding motif at the start of the predicted first transmembrane helix.
The 220 genes that are induced by both exogenous copper and ΔcsoR (Table S6D) indicate that the CsoRSl and copper overload regulons only partially overlap with each other. One of these overlap genes is 7265, annotated as a membrane protein with a CusF domain and therefore implemented in copper binding. RNA-seq data reveal this gene to have the greatest fold increase of all known and predicted genes that encode for copper -binding proteins/transporters (Table 3) and may therefore have an as yet uncharacterized role in copper homeostasis. Gene 6710 encoding a protein containing a putative type I copper site is not expressed in the control culture but does show a low level of expression in the csoR mutant and upon copper induction. However, it is not clear at present how this protein could be involved in copper homeostasis. The strongest transcription increase both in response to copper overload and in the ΔcsoR mutant, by 29- and 15-fold, respectively, is a putative [2Fe-2S] thioredoxin-like protein (SCO5830). A functional reason for this is unclear as is the case for the hundreds of other genes that are induced by either copper or by the ΔcsoR. It is reasonable to assume from these data that besides CsoRSl control, one or most likely several other control systems operate in copper homeostasis, e.g. redox homeostasis. One possibility may be that the way the copper excess regulon is stimulated requires more sophisticated and efficient mechanisms than the slow and energy-consuming means involving transcriptional control by a DNA-binding protein.
Supplementary Material
Acknowledgments
We thank Professor Michael Wilson for discussions on metal competition experiments; Katie Blundell for assistance with Dichroweb; Dr. Richard Strange for assistance with data collection at Diamond beamline I04 as part of the University of Liverpool BAG MX7044, and Dr. Tom Clarke (University of East Anglia) for assistance and analysis of the AUC data. Christopher Beckwith is acknowledged for contributing to the initial stages of constructing site-directed variants of CsoRSl.
This work was supported by the University of Essex (to S. D., A. K. C., M. A. H., and J. A. R. W.), The Biochemical Society (to A. K. C.), and Leiden University and NWO (to E. V.).

This article contains “Materials and Methods,” supplemental Figs. S1–S9, and Tables S1–S6.
The atomic coordinates and structure factors (code 4adz) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CsoR
- copper-sensitive operon repressor
- AAS
- atomic absorption spectroscopy
- MM
- minimal agar medium
- BCS
- bathocuproine disulfonate
- BCA
- bicinchoninic acid
- AUC
- analytical ultracentrifugation.
REFERENCES
- 1. Ma Z., Jacobsen F. E., Giedroc D. P. (2009) Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 109, 4644–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Waldron K. J., Robinson N. J. (2009) How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 7, 25–35 [DOI] [PubMed] [Google Scholar]
- 3. Waldron K. J., Rutherford J. C., Ford D., Robinson N. J. (2009) Metalloproteins and metal sensing. Nature 460, 823–830 [DOI] [PubMed] [Google Scholar]
- 4. Robinson N. J., Winge D. R. (2010) Copper metallochaperones. Annu. Rev. Biochem. 79, 537–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Changela A., Chen K., Xue Y., Holschen J., Outten C. E., O'Halloran T. V., Mondragón A. (2003) Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301, 1383–1387 [DOI] [PubMed] [Google Scholar]
- 6. Solioz M., Stoyanov J. V. (2003) Copper homeostasis in Enterococcus hirae. FEMS Microbiol. Rev. 27, 183–195 [DOI] [PubMed] [Google Scholar]
- 7. Liu T., Ramesh A., Ma Z., Ward S. K., Zhang L., George G. N., Talaat A. M., Sacchettini J. C., Giedroc D. P. (2007) CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat. Chem. Biol. 3, 60–68 [DOI] [PubMed] [Google Scholar]
- 8. Ueda K., Tomaru Y., Endoh K., Beppu T. (1997) Stimulatory effect of copper on antibiotic production and morphological differentiation in Streptomyces tanashiensis. J. Antibiot. 50, 693–695 [DOI] [PubMed] [Google Scholar]
- 9. Keijser B. J., van Wezel G. P., Canters G. W., Kieser T., Vijgenboom E. (2000) The ram-dependence of Streptomyces lividans differentiation is bypassed by copper. J. Mol. Microbiol. Biotechnol. 2, 565–574 [PubMed] [Google Scholar]
- 10. Worrall J. A., Vijgenboom E. (2010) Copper mining in Streptomyces. Enzymes, natural products, and development. Nat. Prod. Rep. 27, 742–756 [DOI] [PubMed] [Google Scholar]
- 11. Fujimoto M., Yamada A., Kurosawa J., Kawata A., Beppu T., Takano H., Ueda K. (2011) Pleiotropic role of the Sco1/SenC family copper chaperone in the physiology of Streptomyces Microbial. Biotechnol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andreini C., Banci L., Bertini I., Rosato A. (2008) Occurrence of copper proteins through the three domains of life. A bioinformatic approach. J. Proteome Res. 7, 209–216 [DOI] [PubMed] [Google Scholar]
- 13. Yang H. Y., Chen C. W. (2009) Extracellular and intracellular polyphenol oxidases cause opposite effects on sensitivity of Streptomyces to phenolics. A case of double-edged sword. PLoS One 4, e7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ward S. K., Hoye E. A., Talaat A. M. (2008) The global responses of Mycobacterium tuberculosis to physiological levels of copper. J. Bacteriol. 190, 2939–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smaldone G. T., Helmann J. D. (2007) CsoR regulates the copper efflux operon copZA in Bacillus subtilis. Microbiology 153, 4123–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Corbett D., Schuler S., Glenn S., Andrew P. W., Cavet J. S., Roberts I. S. (2011) The combined actions of the copper-responsive repressor CsoR and copper-metallochaperone CopZ modulate CopA-mediated copper efflux in the intracellular pathogen Listeria monocytogenes. Mol. Microbiol. 81, 457–472 [DOI] [PubMed] [Google Scholar]
- 17. Grossoehme N., Kehl-Fie T. E., Ma Z., Adams K. W., Cowart D. M., Scott R. A., Skaar E. P., Giedroc D. P. (2011) Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J. Biol. Chem. 286, 13522–13531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vijgenboom E. K., B. J. F. (2002) in Copper and the Morphological Development of Streptomyces, Handbook of Copper Pharmacology and Toxicology, pp. 503–525, Humana Press Inc., Totowa, NJ [Google Scholar]
- 19. Jayapal K. P., Lian W., Glod F., Sherman D. H., Hu W. S. (2007) Comparative genomic hybridizations reveal absence of large Streptomyces coelicolor genomic islands in Streptomyces lividans. BMC Genomics 8, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lewis R. A., Laing E., Allenby N., Bucca G., Brenner V., Harrison M., Kierzek A. M., Smith C. P. (2010) Metabolic and evolutionary insights into the closely related species Streptomyces coelicolor and Streptomyces lividans deduced from high resolution comparative genomic hybridization. BMC Genomics 11, 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Floriano B., Bibb M. (1996) afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 21, 385–396 [DOI] [PubMed] [Google Scholar]
- 22. Kieser T. B., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. (2000) Practical Streptomyces Genetics, pp. 1–613, John Innes Foundation, Norwich, UK [Google Scholar]
- 23. Fedoryshyn M., Welle E., Bechthold A., Luzhetskyy A. (2008) Functional expression of the Cre recombinase in actinomycetes. Appl. Microbiol. Biotechnol. 78, 1065–1070 [DOI] [PubMed] [Google Scholar]
- 24. Vara J., Lewandowska-Skarbek M., Wang Y. G., Donadio S., Hutchinson C. R. (1989) Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus). J. Bacteriol. 171, 5872–5881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 [DOI] [PubMed] [Google Scholar]
- 26. van Wezel G. P., White J., Hoogvliet G., Bibb M. J. (2000) Application of redD, the transcriptional activator gene of the undecylprodigiosin biosynthetic pathway, as a reporter for transcriptional activity in Streptomyces coelicolor A3(2) and Streptomyces lividans. J. Mol. Microbiol. Biotechnol. 2, 551–556 [PubMed] [Google Scholar]
- 27. Tsao S. W., Rudd B. A., He X. G., Chang C. J., Floss H. G. (1985) Identification of a red pigment from Streptomyces coelicolor A3(2) as a mixture of prodigiosin derivatives. J. Antibiot. 38, 128–131 [DOI] [PubMed] [Google Scholar]
- 28. Lerch K., Ettlinger L. (1972) Purification and properties of a tyrosinase from Streptomyces glaucescens. Pathol. Microbiol. 38, 23–25 [PubMed] [Google Scholar]
- 29. Green G. N., Gennis R. B. (1983) Isolation and characterization of an Escherichia coli mutant lacking cytochrome d terminal oxidase. J. Bacteriol. 154, 1269–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mueller J. P., Taber H. W. (1989) Isolation and sequence of ctaA, a gene required for cytochrome aa3 biosynthesis and sporulation in Bacillus subtilis. J. Bacteriol. 171, 4967–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Motamedi H., Shafiee A., Cai S. J. (1995) Integrative vectors for heterologous gene expression in Streptomyces spp. Gene 160, 25–31 [DOI] [PubMed] [Google Scholar]
- 32. Larson J. L., Hershberger C. L. (1986) The minimal replicon of a streptomycete plasmid produces an ultrahigh level of plasmid DNA. Plasmid 15, 199–209 [DOI] [PubMed] [Google Scholar]
- 33. Ellman G. L. (1959) Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82, 70–77 [DOI] [PubMed] [Google Scholar]
- 34. Xiao Z., Donnelly P. S., Zimmermann M., Wedd A. G. (2008) Transfer of copper between bis(thiosemicarbazone) ligands and intracellular copper-binding proteins. Insights into mechanisms of copper uptake and hypoxia selectivity. Inorg. Chem. 47, 4338–4347 [DOI] [PubMed] [Google Scholar]
- 35. Xiao Z., Loughlin F., George G. N., Howlett G. J., Wedd A. G. (2004) C-terminal domain of the membrane copper transporter Ctr1 from Saccharomyces cerevisiae binds four Cu(I) ions as a cuprous-thiolate polynuclear cluster. Sub-femtomolar Cu(I) affinity of three proteins involved in copper trafficking. J. Am. Chem. Soc. 126, 3081–3090 [DOI] [PubMed] [Google Scholar]
- 36. Xiao Z., Brose J., Schimo S., Ackland S. M., La Fontaine S., Wedd A. G. (2011) Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1, and related proteins. Detection probes and affinity standards. J. Biol. Chem. 286, 11047–11055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bertram R., Rigali S., Wood N., Lulko A. T., Kuipers O. P., Titgemeyer F. (2011) Regulon of the N-acetylglucosamine utilization regulator NagR in Bacillus subtilis. J. Bacteriol. 193, 3525–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ma Z., Cowart D. M., Scott R. A., Giedroc D. P. (2009) Molecular insights into the metal selectivity of the copper(I)-sensing repressor CsoR from Bacillus subtilis. Biochemistry 48, 3325–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Festa R. A., Jones M. B., Butler-Wu S., Sinsimer D., Gerads R., Bishai W. R., Peterson S. N., Darwin K. H. (2011) A novel copper-responsive regulon in Mycobacterium tuberculosis. Mol. Microbiol. 79, 133–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hiard S., Marée R., Colson S., Hoskisson P. A., Titgemeyer F., van Wezel G. P., Joris B., Wehenkel L., Rigali S. (2007) PREDetector. A new tool to identify regulatory elements in bacterial genomes. Biochem. Biophys. Res. Commun. 357, 861–864 [DOI] [PubMed] [Google Scholar]
- 41. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. (2011) iMOSFLM. A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 43. Long F., Vagin A. A., Young P., Murshudov G. N. (2008) BALBES. A molecular replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 64, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 45. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 46. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 47. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity. All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Whitmore L., Wallace B. A. (2004) DICHROWEB. An online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Whitmore L., Wallace B. A. (2008) Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89, 392–400 [DOI] [PubMed] [Google Scholar]
- 50. Xiao Z., Wedd A. G. (2010) The challenges of determining metal-protein affinities. Nat. Prod. Rep. 27, 768–789 [DOI] [PubMed] [Google Scholar]
- 51. Sakamoto K., Agari Y., Agari K., Kuramitsu S., Shinkai A. (2010) Structural and functional characterization of the transcriptional repressor CsoR from Thermus thermophilus HB8. Microbiology 156, 1993–2005 [DOI] [PubMed] [Google Scholar]
- 52. Croucher N. J., Thomson N. R. (2010) Studying bacterial transcriptomes using RNA-seq. Curr. Opin. Microbiol. 13, 619–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ozsolak F., Milos P. M. (2011) RNA sequencing. Advances, challenges, and opportunities. Nat. Rev. Genet. 12, 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pinto A. C., Melo-Barbosa H. P., Miyoshi A., Silva A., Azevedo V. (2011) Application of RNA-seq to reveal the transcript profile in bacteria. Genet. Mol. Res. 10, 1707–1718 [DOI] [PubMed] [Google Scholar]
- 55. Ma Z., Cowart D. M., Ward B. P., Arnold R. J., DiMarchi R. D., Zhang L., George G. N., Scott R. A., Giedroc D. P. (2009) Unnatural amino acid substitution as a probe of the allosteric coupling pathway in a mycobacterial Cu(I) sensor. J. Am. Chem. Soc. 131, 18044–18045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Developments in the CCP4 molecular graphics project. Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.