Background: Gli1 was thought to be dispensable for normal embryogenesis.
Results: Gli1 physiologically acts as an osteogenesis-related Gli activator and mediates the functions of Gli2 and Gli3 in osteogenesis.
Conclusion: Gli1 collectively functions with Gli2 and Gli3 in osteogenesis.
Significance: This study provides insight into the elaborate molecular framework of a context-dependent divergence of functions of Hedgehog-Gli signaling in development.
Keywords: Bone, Cell Biology, Development, Hedgehog, Osteoblasts, Signal Transduction
Abstract
With regard to Hedgehog signaling in mammalian development, the majority of research has focused on Gli2 and Gli3 rather than Gli1. This is because Gli1−/− mice do not show any gross abnormalities in adulthood, and no detailed analyses of fetal Gli1−/− mice are available. In this study, we investigated the physiological role of Gli1 in osteogenesis. Histological analyses revealed that bone formation was impaired in Gli1−/− fetuses compared with WT fetuses. Gli1−/− perichondrial cells expressed neither runt-related transcription factor 2 (Runx2) nor osterix, master regulators of osteogenesis, in contrast to WT cells. In vitro analyses showed that overexpression of Gli1 up-regulated early osteogenesis-related genes in both WT and Runx2−/− perichondrial cells, and Gli1 activated transcription of those genes via its association with their 5′-regulatory regions, underlying the function of Gli1 in the perichondrium. Moreover, Gli1−/−;Gli2−/− mice showed more severe phenotypes of impaired bone formation than either Gli1−/− or Gli2−/− mice, and osteoblast differentiation was impaired in Gli1−/−;Gli3−/− perichondrial cells compared with Gli3−/− cells in vitro. These data suggest that Gli1 itself can induce early osteoblast differentiation, at least to some extent, in a Runx2-independent manner. It also plays a redundant role with Gli2 and is involved in the repressor function of Gli3 in osteogenesis. On the basis of these findings, we propose that upon Hedgehog input, Gli1 functions collectively with Gli2 and Gli3 in osteogenesis.
Introduction
Hedgehog (Hh)3 signaling is a highly conserved pathway known to play essential roles in various developmental processes. Gli1, Gli2, and Gli3 are thought to mediate transcriptional responses to Hh input in a context-dependent manner (1). Gli2 is suggested to function primarily as a transcriptional activator, and Gli3 as a transcriptional repressor (1). Gli2 and Gli3 have been demonstrated to play major roles in Hh signaling in mouse development (2, 3). In contrast, Gli1, a transcriptional activator, is thought to be dispensable for normal development (4, 5). However, several lines of evidence have indicated that Gli1 plays an important role in pathological conditions (3, 6).
Endochondral ossification is among the notable events regulated by Indian hedgehog (Ihh) (7). During this process, perichondrial cells adjacent to the mineralized hypertrophic chondrocytes turn into osteoblast precursors expressing alkaline phosphatase (Alp), bone sialoprotein (Bsp), and the type I collagen α1 chain (Col1a1). They calcify, forming the “bone collar,” and ultimately differentiate into mature osteoblasts expressing osteocalcin (Oc) (8, 9). Among transcription factors regulating the process, Runt-related transcription factor 2 (Runx2) and osterix (Osx) are master regulators of osteoblast differentiation; Osx is genetically downstream of Runx2 (8–10).
We and others have clarified essential roles of Hh signaling in endochondral ossification. Analyses of Ihh−/−, Col2a1-cre;Smo−/−, and Smo−/− chimeric mice indicate that Hh signaling specifies the osteoblast lineage in the perichondrium (7, 11). The control of osteoblast differentiation by Ihh requires both the activator function of Gli2 and derepression of the Gli3 repressor (12–14). However, the involvement of Gli1 in endochondral ossification remains poorly understood. We show here that Gli1 is involved in osteoblast differentiation.
EXPERIMENTAL PROCEDURES
Animals
Ihhtm1Amc (Ihh+/−) mice were obtained from The Jackson Laboratory, WT C57BL/6J mice were from Charles River Japan, and Gli1+/− and Gli2+/− mice were generated as described previously (4, 15). Runx2+/− mice were a gift from T. Komori (Nagasaki University, Nagasaki, Japan) (9). All experiments were performed in accordance with the protocol approved by the Animal Care and Use Committee of The University of Tokyo.
Reagents and Vectors
SAG (Smoothened agonist) was purchased from Calbiochem, and pEGFP-C1 was from Clontech. Plasmids expressing GLI1 were generated as described previously (12). The shRNA expression vector for Gli1 was constructed using the piGENEmU6 vector (iGENE Therapeutics). The target sequences were as follows: Gli1-1, AACTCCACAGGCACACAGGAT; and Gli1-2, GCCTTACATGTGTGAGCAAGA. For luciferase analysis, reporter genes including the human ALP and BSP regulatory regions were obtained by PCR using human genomic DNA as template. Deletion and mutation constructs were created by the PCR technique as described previously (16).
Cell Culture
C3H10T1/2 cells were obtained from the RIKEN Cell Bank. Perichondrial cells were isolated as described previously (17). These cells were cultured in high-glucose DMEM containing 10% FBS and 1% penicillin/streptomycin. Plasmid transfection was performed using FuGENE HD (Roche Applied Science). ALP staining was performed as described previously (16).
Real-time RT-PCR
Total RNA extraction, reverse transcription, and real-time PCR were performed as described previously (12). All reactions were run in triplicate. The primer sequences were as follows: β-actin, AGATGTGGATCAGCAAGCAG (forward) and GCGCAAGTTAGGTTTTGTCA (reverse); Alp, GCTGATCATTCCCACGTTTT (forward) and CTGGGCCTGGTAGTTGTTGT (reverse); Bsp, CAGAGGAGGCAAGCGTCACT (forward) and CTGTCTGGGTGCCAACACTG (reverse); Oc, AAGCAGGAGGGCAATAAGGT (forward) and TTTGTAGGCGGTCTTCAAGC (reverse); Gli1, GCACCACATCAACAGTGAGC (forward) and GCGTCTTGAGGTTTTCAAGG (reverse); and Runx2, CCGCACGACAACCGCACCAT (forward) and CGCTCCGGCCCACAAATCTC (reverse).
Histology
The procedures for von Kossa staining and in situ hybridization have been described previously (18). Immunohistochemistry was performed using anti-Runx2 (1:50 dilution; sc-10758, Santa Cruz Biotechnology, Inc.) and anti-osterix (1:5000 dilution; ab22552, Abcam) antibodies as described previously (16). For immunohistochemistry of the Gli1−/− fetus, frozen sections were prepared at 10 μm. Alexa Fluor 488 (1:500 dilution; A-11034, Invitrogen) was used for detection.
Microscopy
Images were taken using an Axio Imager A1 or Axiovert 200 microscope (Carl Zeiss) and processed using AxioVision (Carl Zeiss).
Luciferase Assay
Cells were plated onto 24-well plates and transfected with 0.4 μg of DNA mixture containing the test reporter plasmids, control reporter plasmids encoding Renilla luciferase, and effector plasmids. A Dual-Luciferase assay was performed as described previously (12).
EMSA
Nuclear extracts were obtained as described previously (19). Digoxigenin-labeled probes were constructed using a DIG gel shift kit (Roche Applied Science). A total of 1 μg of nuclear extracts of GFP- and GLI1-transfected C3H10T1/2 cells was used. The binding mixture contained 50 mm KCl, 25 mm HEPES-KOH (pH 7.9), 5 mm MgCl2, 10 mm ZnSO4, 1 mm DTT, 12% glycerol, 0.1% Nonidet P-40, 0.25 μg of poly(dI-dC), 0.1 μg of poly-l-lysine, and 30 fmol of the digoxigenin-labeled probe. The binding reaction was performed for 30 min at room temperature, and a 50-fold molar excess of unlabeled probes was used for the competition. When antibodies were used, nuclear extracts were incubated with 1 μg of anti-GLI1 antibody (sc-20687X, Santa Cruz Biotechnology, Inc.) or non-immune IgG (sc-2027, Santa Cruz Biotechnology, Inc.) for 30 min at room temperature prior to the binding reaction. Electrophoresis was performed with a 6% Tris borate/EDTA gel (Invitrogen) in 0.25× Tris borate/EDTA at 150 V for 60 min, and samples were subjected to blotting on nylon membranes and detection for digoxigenin-labeled probes.
Oligonucleotide probes containing the Gli-G/C-containing region in the human ALP region and the Gli motif-containing region in the human BSP region were used, and their mutations were as follows: Gli-G/C-containing region in the WT human ALP region, 5′-GGGCTCGGGCCGGGGGCGGGGCCGGG-3′; Gli-G/C-containing region in the human ALP region mutant, 5′-GGGCTCGGGCCGGAAGCGGGGCCGGG-3′; Gli motif-containing region in the WT human BSP region, 5′-TGACAGTGATTGGCTGTTGGAAGGC-3′; and Gli motif-containing region in the human BSP region mutant, 5′-TGACAGTGATTAACTGTTGGAAGGC-3′.
ChIP
ChIP was performed with a OneDay ChIP kit (Diagenode) using 5 μg of anti-GLI1 antibody, anti-GFP antibody (ab290, Abcam), non-immune IgG, and anti-acetyl-histone H3 antibody (06-599, Upstate) following the manufacturer's instructions. To shear genomic DNA, a shearing ChIP kit (Diagenode) was used according to the manufacturer's instructions. PCR was performed after purification of DNA. The primer sequences were as follows: −220 to −21 in the Alp region, CAGGATGAGCCGCAGGGAAAGAG (forward) and ATGAAGGCGGCCTGGCCGAG (reverse); −1000 to −801 in the Alp region, ACCCCCCTCCTTGCATCAAAAGTGGT (forward) and CTACACTTTCCCTGTTGTTGGTCTCTG (reverse); −240 to −41 in the Bsp region, GAACTCTAACTACCATCTTCTCCTCACCCT (forward) and CCTTCTCCGCCTCTCAACAGCCAAT (reverse); and −2000 to −1801 in the Bsp region, AGGCGTAACTCATGTCCTTGGATGGC (forward) and TGTCAGAGACCATCCCGTGAGAAAGTG (reverse).
Statistical Analysis
The means of groups were compared by analysis of variance, and the significance of differences was determined by post hoc testing using Tukey's method.
RESULTS
Gli1 Contributes to Hh Signaling-mediated Osteoblast Differentiation
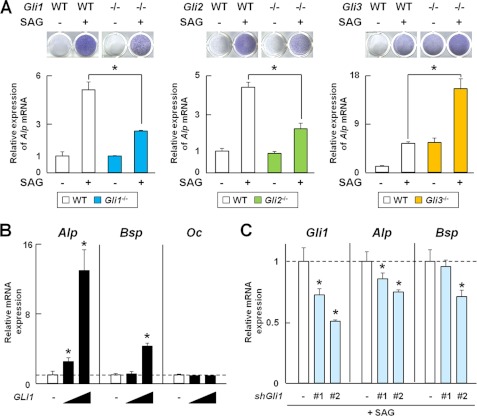
To examine the contribution of Gli1 to Hh signaling-mediated osteoblast differentiation compared with Gli2 and Gli3, we examined the induction of osteoblast differentiation by SAG, a Smoothened agonist, in perichondrial cells isolated from Gli knock-out or WT mice. SAG-induced osteoblast differentiation was impaired in Gli1−/− and Gli2−/− cells relative to that in WT cells as shown by a decrease in ALP staining intensity and Alp mRNA expression, whereas it was enhanced in Gli3−/− cells (Fig. 1A). In addition, overexpression of Gli1 alone induced mRNA expression of Alp and Bsp, early and middle markers of osteoblast differentiation. Overexpression of Gli1 alone did not affect Oc mRNA (Fig. 1B), a late maker in the C3H10T1/2 cell line, which has been reported to be able to differentiate into osteoblastic cells upon Hh signaling (20). shRNA for Gli1 inhibited the SAG-induced expression of Gli1, Alp, and Bsp in C3H10T1/2 cells (Fig. 1C). These data suggest that in addition to Gli2 and Gli3, Gli1 contributes to Hh-mediated osteoblast differentiation, mainly in its early phase.
FIGURE 1.
Involvement of Gli1 in Hh-mediated osteoblast differentiation. A, ALP staining and Alp mRNA expression determined by real-time RT-PCR in perichondrial cells from WT, Gli1−/−, Gli2−/−, and Gli3−/− mouse embryos (E17.5). Cells were obtained form three pairs of WT and mutant mice. Cells were treated with SAG for 7 days for ALP staining and for 3 days for real-time RT-PCR analysis. *, p < 0.05. B, mRNA expression of osteoblast marker genes determined by real-time RT-PCR in C3H10T1/2 cells. Cells were transfected with the GFP (1 μg) or GLI1 (0.1 or 1 μg) plasmid and cultured for 48 h. C, mRNA expression of osteoblast marker genes determined by real-time RT-PCR in C3H10T1/2 cells. After being transfected with shRNA for GFP or Gli1, the cells were cultured for 12 h, followed by exposure to SAG (1 μm) for 48 h. shGli1, Gli1 shRNA. *, p < 0.05. For A–C, data are means ± S.D. of triplicate wells, and representative data of independent experiments are shown.
Gli1 Is Involved in Hh Signaling-mediated Specification of Osteoblast Lineage
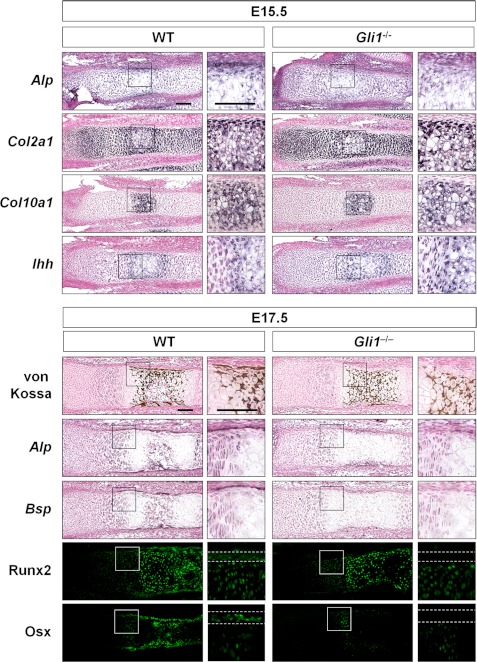
Although in vitro data have suggested that Gli1 contributes to osteoblast differentiation, Gli1−/− mice are viable and appear normal in adulthood (4), and no detailed analyses in fetal Gli1−/− mice are available. Therefore, we investigated the physiological roles of Gli1 in endochondral ossification by analyzing the skeletal phenotypes of fetal Gli1−/− mice. At embryonic day (E) 15.5, the Gli1−/− perichondrium, unlike the WT perichondrium, did not express Alp. There was no difference in the expression pattern of chondrogenic markers, including Col2a1, type X collagen α1 chain (Col10a1), and Ihh, between WT and Gli1−/− mice (Fig. 2). At E17.5, no bone collar was observed in Gli1−/− mice, even though mineralized chondrocytes were already present. The Gli1−/− perichondrium showed a decreased region of Alp-expressing cells compared with the WT perichondrium, and it lacked Bsp expression (Fig. 2). Expression of Runx2 or Osx was not discernible in the Gli1−/− perichondrial cells at this stage (Fig. 2), indicating that osteogenesis in the Gli1−/− perichondrial cells was delayed.
FIGURE 2.
Involvement of Gli1 in endochondral ossification. Shown is the in situ hybridization (Alp, Bsp, Col2a1, and Col10a1), von Kossa staining, and immunohistochemistry (Runx2 and Osx) of the representative sections of metatarsals obtained from three pairs of WT and Gli1−/− mouse embryos (E15.5 and E17.5). The region between the dashed white lines indicates the perichondrium. Scale bars = 100 μm.
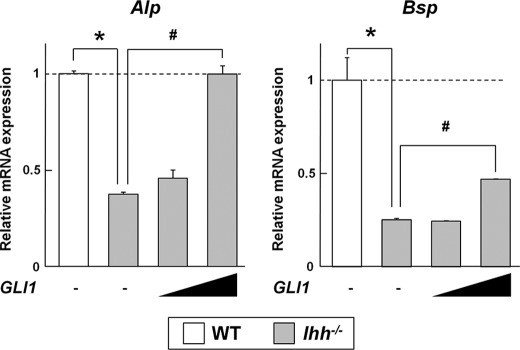
Because the expression of Runx2 and Osx is absent in the Ihh−/− perichondrium (7, 13), this suggests that the Gli1−/− fetus partially phenocopies Ihh−/− in osteogenesis. We then tested whether Gli1 overexpression could rescue the phenotype of the Ihh−/− perichondrium in vitro. Perichondrial cells of Ihh−/− embryos showed decreased mRNA expression of Alp and Bsp compared with WT cells (Fig. 3). Gli1 overexpression rescued the expression of Alp completely and that of Bsp partially (Fig. 3). These data suggest that, at least in part, Gli1 is likely to contribute to the specification of the osteoblast lineage downstream of Ihh.
FIGURE 3.
Effects of overexpression of Gli1 on expression of osteoblast marker genes in WT and Ihh−/− perichondrial cells. mRNA expression was determined by real-time RT-PCR analysis in perichondrial cells from three pairs of WT and Ihh−/− mouse embryos (E17.5). Cells were transfected with the GFP (1 μg) or GLI1 (0.1 or 1 μg) plasmid and cultured for 48 h. Data are means ± S.D. of triplicate wells, and representative data of independent experiments are shown. *, p < 0.05; #, p < 0.05.
Gli1 Itself, as Well as Activation of Hh Signaling, Can Induce Early Osteoblast Differentiation in Runx2-independent Manner
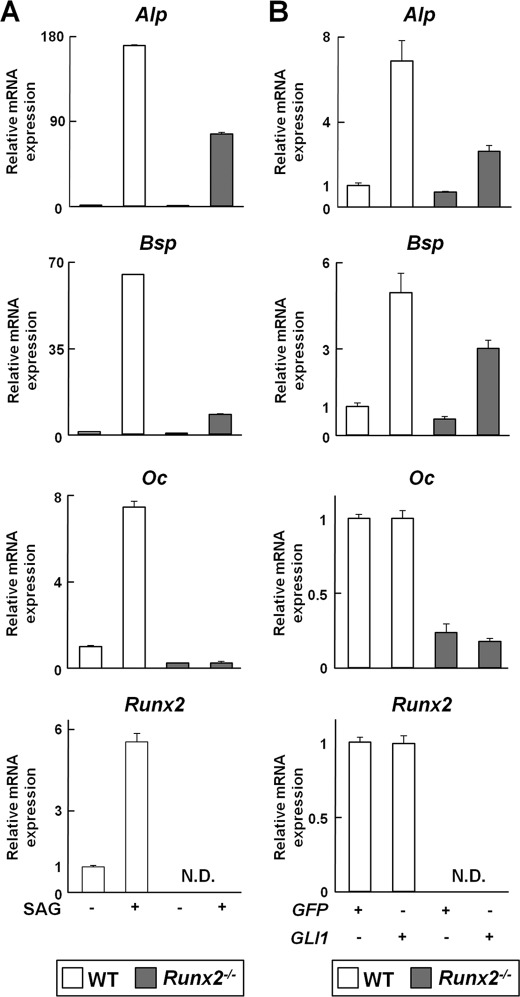
Our data up to this point led us to consider two possibilities for molecular mechanisms underlying the induction of early osteoblast differentiation by the Hh-Gli1 pathway: 1) indirect action via induction of Runx2 and 2) Gli1-mediated transcriptional regulation of osteogenesis-related genes. We initially focused on the first possibility and investigated whether activation of the Hh-Gli1 pathway could induce osteoblast differentiation in Runx2−/− perichondrial cells in vitro. Activation of Hh signaling with SAG up-regulated mRNA expression of osteogenesis-related genes, including Alp, Bsp, and Oc, as well as Runx2, in WT cells (Fig. 4A). In Runx2−/− perichondrial cells, SAG up-regulated Alp and Bsp, but not Oc (Fig. 4A). Overexpression of Gli1 up-regulated Alp and Bsp, but not Oc, in both WT and Runx2−/− cells, and it did not alter the mRNA expression of Runx2 and Oc in WT cells (Fig. 4B). The expression levels of Alp and Bsp induced by SAG or Gli1 in Runx2−/− cells were lower than those in WT cells (Fig. 4, A and B). These data indicate that activation of the Hh-Gli1 pathway can induce some markers of the early stage of osteoblast differentiation in a Runx2-independent manner.
FIGURE 4.
Effects of Hh-Gli1 signaling on expression of osteoblast marker genes in WT and Runx2−/− perichondrial cells. mRNA expression of osteoblast marker genes and Runx2 was determined by real-time RT-PCR analysis in perichondrial cells from WT or Runx2−/− mouse embryos (E17.5). A, cells were treated with SAG (1 μm) for 6 days. B, cells were transfected with the GFP (1 μg) or GLI1 (1 μg) plasmid and cultured for 48 h. N.D., none detected. Data are means ± S.D. of triplicate wells, and representative data of independent experiments are shown.
Gli1 Transactivates Osteogenesis-related Genes via Its Association with Their 5′-Regulatory Regions
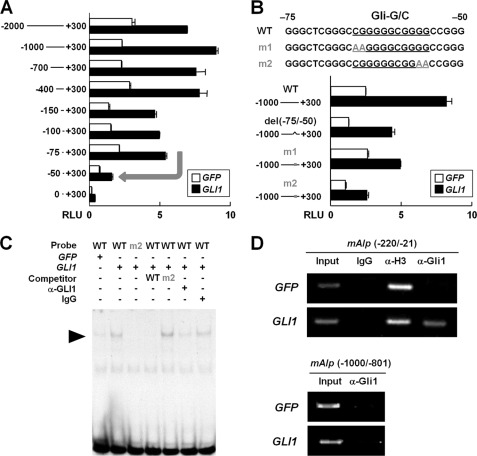
We then verified the second possible molecular mechanism underlying the induction of early osteoblast differentiation by the Hh-Gli1 pathway. We hypothesized that Gli1 regulates the transcription of Alp and Bsp via its association with the 5′-regulatory regions of those genes because Alp and Bsp were up-regulated by Gli1 overexpression in both WT and Runx2−/− cells, and their expression was attenuated in the Gli1−/− perichondrium (Figs. 2 and 4). Through deletion analyses using the 5′-regions of BSP associated with luciferase genes, we identified the core-responsive element to Gli1 between −81 and −40 of the BSP region (Fig. 5A). Within this region, we found two Gli consensus motifs (TGGGTGGTC, Gli motif), which were fully conserved between human and mouse. Transactivation by Gli1 was suppressed by deletion mutagenesis in the region (del(−81/−40)) and site-directed mutagenesis of Gli motif 2 (m2), but not by site-directed mutagenesis of Gli motif 1 (m1) (Fig. 5B). EMSA and ChIP revealed Gli1 binding to Gli motif 2 in the BSP region (Fig. 5, C and D). With regard to ALP, a core-responsive element to Gli1 was identified between −75 and −50, where we found a G/C-rich sequence (Gli-G/C) that was shown to be enriched in Gli-binding regions by a ChIP-on-chip analysis for Gli (21) (Fig. 6, A and B). EMSA and ChIP revealed Gli1 binding to the Gli-G/C region in the ALP region (Fig. 6, C and D). These data suggest that Gli1 regulates the expression of osteogenesis-related genes by associating with their transcriptional regulatory regions.
FIGURE 5.
Gli1-mediated transactivation of Bsp. A, deletion analyses using luciferase reporter constructs containing the 5′-regulatory region of BSP. C3H10T1/2 cells were transfected with a series of deletion fragments in combination with the indicated plasmids. The arrow denotes the major down-regulation of luciferase activity by deletion. RLU, relative light units. B, deletion and site-directed mutagenesis analyses for GLI1-responsive elements in BSP. del(−80/−40), deletion mutant of GLI1-responsive elements; m1 and m2, site-directed mutants of GLI1-responsive elements. C, EMSA for the specific binding of GLI1 to GLI1-responsive elements in BSP. Nuclear extracts of C3H10T1/2 cells transfected with GFP or GLI1 were used. The arrowhead indicates shifted bands of the GLI1-DNA complex. D, ChIP assay in the perichondrial cells transfected with the indicated plasmids. DNA was amplified by primer sets spanning (−240 to −41) and not spanning (−2000 to −1801) the Gli motif-containing region in the murine Bsp (mBsp) region after immunoprecipitation with anti-GLI1 antibody, IgG (negative control), or anti-acetyl histone H3 antibody (positive control), as well as input DNA. For A and B, data are means ± S.D. of triplicate wells, and representative data of independent experiments are shown.
FIGURE 6.
Gli1-mediated transactivation of Alp. A, deletion analyses using luciferase reporter constructs containing the 5′-regulatory region of ALP. C3H10T1/2 cells were transfected with a series of deletion fragments in combination with the indicated plasmids. The arrow denotes the major down-regulation of luciferase activity by deletion. RLU, relative light units. B, deletion and site-directed mutagenesis analyses for GLI1-responsive elements in ALP. del(−80/−40), deletion mutant of GLI1-responsive elements; m1 and m2, site-directed mutants of GLI1-responsive elements. C, EMSA for the specific binding of GLI1 to GLI1-responsive elements in ALP. Nuclear extracts of C3H10T1/2 cells transfected with GFP or GLI1 were used. The arrowhead indicates shifted bands of the GLI1-DNA complex. D, ChIP assay in the perichondrial cells transfected with the indicated plasmids. DNA was amplified by primer sets spanning (−220 to −21) and not spanning (−1000 to −801) the Gli motif-containing region in the murine Alp (mAlp) region after immunoprecipitation with anti-GLI1 antibody, IgG (negative control), or anti-acetyl histone H3 antibody (positive control), as well as input DNA. For A and B, data are means ± S.D. of triplicate wells, and representative data of independent experiments are shown.
Gli1 Functions Collectively with Gli2 and Gli3 in Osteogenesis
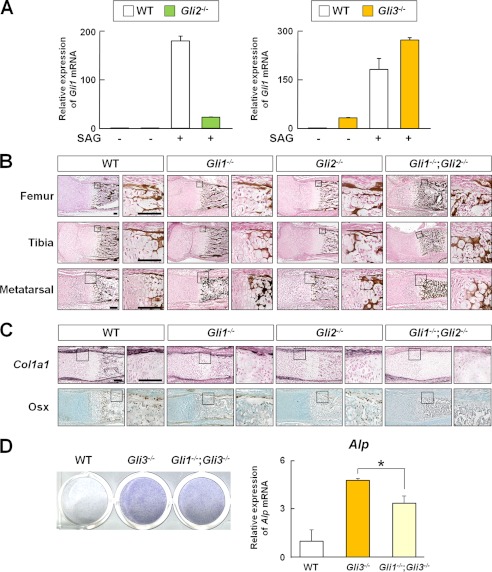
We next attempted to clarify how Gli1 functions in osteogenesis in association with Hh input, Gli2, and Gli3. First, we confirmed that Gli1 acted downstream of Gli2 and Gli3 in the perichondrium, as shown previously in other tissues (22). Gli2−/− perichondrial cells showed decreased induction of Gli1 expression by SAG compared with WT cells. This induction was increased in Gli3−/− cells (Fig. 7A). In contrast, Gli1−/− cells did not alter Gli2 or Gli3 mRNA expression, protein expression, or protein subcellular localization in vitro (data not shown).
FIGURE 7.
Relationship between Gli1, Gli2, Gli3, and Hh input in osteogenesis. A, Gli1 mRNA expression determined by real-time RT-PCR in perichondrial cells from three pairs of WT, Gli2−/−, and Gli3−/− mouse embryos (E17.5). Cells were treated with SAG (1 μm) for 3 days. B, von Kossa staining of the representative sections of femurs, tibias, and metatarsals obtained from two pairs of WT, Gli1−/−, Gli2−/−, and Gli1−/−;Gli2−/− newborn mice. Scale bars = 100 μm. C, in situ hybridization (Col1a1) and immunohistochemistry (Osx) of the representative sections of metatarsals obtained from two pairs of WT, Gli1−/−, Gli2−/−, and Gli1−/−;Gli2−/− newborn mice. Scale bars = 100 μm. D, ALP staining and Alp mRNA expression determined by real-time RT-PCR in perichondrial cells from three pairs of WT, Gli3−/−, and Gli1−/−;Gli3−/− mouse embryos (E17.5). Cells were treated with SAG for 7 days for ALP staining and for 3 days for real-time RT-PCR analysis. *, p < 0.05. For A and C, data are means ± S.D. of triplicate wells, and representative data of independent experiments are shown.
Second, given that skeletal phenotypes observed in Gli1−/− metatarsals at E17.5 were partially normalized at postnatal day 0 (Fig. 7B), we hypothesized that Gli plays a redundant role in this context. Because genetic and functional redundancies between Gli1 and Gli2 have been shown in the development of the CNS and lung lobes (4, 22, 23), we analyzed the skeletal tissues of Gli1−/−;Gli2−/− mice at postnatal day 0. The bone collar was absent in Gli1−/−;Gli2−/− metatarsals and tibias and partially impaired in Gli1−/−;Gli2−/− femurs (Fig. 7B). In Gli1−/−;Gli2−/− metatarsals, perichondrial cells did not express Col1a1 or Osx. In contrast, the WT, Gli1−/−, and Gli2−/− metatarsals appeared normal in terms of the expression pattern of osteoblast marker genes (Fig. 7C). These data suggest that Gli1 and Gli2 play redundant roles in the specification of the osteoblast lineage in the perichondrium.
Finally, regarding the interaction of Gli1 and Gli3, our data had raised the possibility that induction of osteogenesis in Gli3−/− perichondrial cells was mediated by the expression of Gli1. We found that Gli1−/−;Gli3−/− perichondrial cells showed decreased expression of Alp compared with Gli3−/− cells in vitro (Fig. 7D). These data suggest that the inhibitory actions of the Gli3 repressor on osteoblast differentiation are mediated, at least in part, by the inhibition of Gli1 expression.
DISCUSSION
This study had four major findings. First, in addition to Gli2 and Gli3, Gli1 is involved in the Hh signaling-mediated specification of the osteoblast lineage. Second, Gli1 induces an early stage of osteoblast differentiation, at least to some extent, in a Runx2-independent manner. Third, Gli1 promotes the transcription of osteogenesis-related genes via its binding to DNA, which possibly accounts for the activity of Gli1 in the perichondrium. Finally, Gli1 plays a redundant role with Gli2 and is involved in the repressor function of Gli3 in osteogenesis. On the basis of these findings, we propose that Gli1 functions collectively with Gli2 and Gli3 in osteogenesis, playing a pivotal role in the specification of the osteoblast lineage upon Hh input. In this context, Gli1 may specifically promote early osteoblast differentiation, at least to some extent, in a Runx2-independent manner.
Our findings shed new light on the physiological function of Gli1 in a context-dependent manner. We observed abnormalities in osteogenesis in the Gli1−/− perichondrium, but chondrogenesis in Gli1−/− growth plates appeared normal, even though Ihh regulates both of these processes (24, 25). These data suggest that the transient requirement of Gli1 at a certain point of differentiation appears to depend on cell types, even in skeletogenesis. In line with our findings, several groups recently reported that Gli1 affected the proliferation and differentiation of hematopoietic stem cells and thymocytes in studies of Gli1−/− mice (26, 27). Our findings support the idea that different Gli factors function in different contexts (28), and detailed analyses of Gli1 may help further reveal such diverse functions of Gli factors in various developmental processes.
In the past few years, accumulating evidence has led to determination of the transcription factors that mediate the function of Ihh signaling in osteogenesis (29). Runx2, a master regulator of osteogenesis, is thought to play a pivotal role in Hh-mediated osteogenesis because Runx2 expression is absent in the perichondrium of the Ihh−/− fetus (7, 20). However, Tu et al. (30) recently reported that recovery of Runx2 expression in skeletal cells does not restore bone formation in Ihh−/− embryos, suggesting that additional factors besides Runx2 mediate functions of Ihh signaling in the induction of osteoblast differentiation. Indeed, ALP activity and expression of early osteoblast-related genes, including Alp and Bsp, have been observed in the Runx2−/− perichondrium (9, 30), whereas none of these genes is expressed in the Ihh−/− perichondrium (7, 20). Our findings support this notion and further suggest that Runx2 is involved in certain stages of Hh signaling-mediated osteoblast differentiation. Additionally, Hh signaling is known to sequentially induce osteogenesis in both early and late differentiation (20, 31). Our data show that SAG induced both early and late osteoblast marker genes, as well as Runx2. Remarkably, SAG promoted early osteoblast differentiation, at least to some extent, in a Runx2-independent manner; however, a late marker gene was not induced by SAG in Runx2−/− cells. Overall, Runx2 does not appear to be important in the Hh signaling-mediated specification of the osteoblast lineage, but it is important in subsequent late osteoblast differentiation.
With regard to the contribution of Gli transcription factors in osteogenesis, disruption of Gli3 partially rescued the impairment of early osteoblast differentiation in Ihh−/− embryos, as shown by the partial recovery of Alp and Runx2, but not Bsp or Oc, in the perichondrium or impaired bone collar formation (13, 14, 32). It has previously been shown that skeletal phenotypes of Ihh−/− embryos were completely rescued in Ihh−/−;Gli3−/−;C2ΔNGli2 embryos, in which ΔNGli2 (an N-terminally truncated form of Gli2, as a constitutively active form of Gli2) was exogenously expressed in Col2a1-positive cells (14). On the basis of these findings, Joeng and Long (14) suggested that the Gli2 activator and the Gli3 repressor collectively mediate all major aspects of Ihh function in endochondral ossification. Our findings demonstrate how the function of Gli1 is combined with the model of Joeng and Long to allow specification of the osteoblast lineage in the perichondrium. Gli1 was present in Ihh−/−;Gli3−/−;C2ΔNGli2 mice, although it was not detectable in Ihh−/−;Gli3−/− mice (14). These results suggested that complete rescue of the Ihh−/− phenotype in Ihh−/−;Gli3−/−;C2ΔNGli2 mice was caused, at least in part, by the recovery of Gli1 in addition to the recovery of Runx2 and the function of the Gli2 activator itself. Furthermore, in this study, we demonstrated that Gli1, as well as Hh input, can promote early osteoblast differentiation, at least to some extent, in a Runx2-independent manner. Taken together, these results suggest that the absence of the Runx2-independent function of Gli1, in addition to the function of Gli2 and Gli3, may account for inadequate rescue of osteoblast phenotypes by the recovery of Runx2 alone in Ihh−/− embryos (30).
In late osteoblast differentiation, Gli2 and Gli3, but not Gli1, are likely to play pivotal roles. Several studies have suggested interactions of Gli2 and Gli3 with Runx2 in late osteoblast differentiation. Shimoyama et al. (33) reported that Gli2 stimulates Runx2 expression, interacts with Runx2, and enhances its activity. The repressor form of Gli3, which was derepressed by Hh input, was previously shown to inhibit Runx2 function by binding to its DNA-binding sites (12). In contrast, no functional interaction between Runx2 and Gli1 was reported so far (12). Our study further shows that expression of Runx2 and Oc was enhanced by SAG, but not by Gli1 overexpression, in WT perichondrial cells, and no expression of Runx2 was observed in the perichondrium of the Gli1−/− fetus. Collectively, these results suggest that Gli1 acts as a permissive factor for the expression of Runx2 at a certain point of development or is indirectly involved in its expression, which also underlies the osteogenic function of Gli1.
In conclusion, this study shows that Gli1 functions collectivity with Gli2 and Gli3 in the Hh-mediated specification of the osteoblast lineage. Gli1 plays a redundant role with other Gli activators and is partially involved in the actions of Gli repressors. It remains to be elucidated whether all members of the Gli family function independently of each other at different stages or whether they act collectively at a particular stage in osteogenesis. However, our study provides insight into the elaborate molecular framework of context-dependent differences in Hh-Gli signaling during development, as well as insight into potential clinical applications of Hh-Gli1 signaling for the treatment of bone defects and non-unions in fracture repair.
Acknowledgments
We thank Drs. G. Yamada and T. Komori for providing the experimental materials and Dr. F. Long and D. K. Ebner for helpful input.
This work was supported in part by grants-in-aid for scientific research from the Japan Society for the Promotion of Science (JSPS) and by the Center for NanoBio Integration, the Center for Medical System Innovation, and the Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST program).
- Hh
- Hedgehog
- Ihh
- Indian hedgehog
- Alp
- alkaline phosphatase
- Bsp
- bone sialoprotein
- Col1a1
- type I collagen α1 chain
- Oc
- osteocalcin
- Runx2
- Runt-related transcription factor 2
- Osx
- osterix
- Col2a1
- type II collagen α1 chain
- E
- embryonic day
- Col10a1
- type X collagen α1 chain.
REFERENCES
- 1. Jiang J., Hui C. (2008) Hedgehog signaling in development and cancer. Dev. Cell 15, 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jacob J., Briscoe J. (2003) Gli proteins and the control of spinal cord patterning. EMBO Rep. 4, 761–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stecca B., Ruiz I, Altaba A. (2010) Context-dependent regulation of the Gli code in cancer by Hedgehog and non-Hedgehog signals. J. Mol. Cell. Biol. 2, 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Park H. L., Bai C., Platt K. A., Matise M. P., Beeghly A., Hui C. C., Nakashima M., Joyner A. L. (2000) Mouse Gli1 mutants are viable but have defects in Shh signaling in combination with a Gli2 mutation. Development 127, 1593–1605 [DOI] [PubMed] [Google Scholar]
- 5. Bai C. B., Auerbach W., Lee J. S., Stephen D., Joyner A. L. (2002) Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129, 4753–4761 [DOI] [PubMed] [Google Scholar]
- 6. Kimura H., Stephen D., Joyner A., Curran T. (2005) Gli1 is important for medulloblastoma formation in Ptc1+/− mice. Oncogene 24, 4026–4036 [DOI] [PubMed] [Google Scholar]
- 7. St-Jacques B., Hammerschmidt M., McMahon A. P. (1999) Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 13, 2072–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J. M., Behringer R. R., de Crombrugghe B. (2002) The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108, 17–29 [DOI] [PubMed] [Google Scholar]
- 9. Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764 [DOI] [PubMed] [Google Scholar]
- 10. Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89, 747–754 [DOI] [PubMed] [Google Scholar]
- 11. Long F., Chung U. I., Ohba S., McMahon J., Kronenberg H. M., McMahon A. P. (2004) Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development 131, 1309–1318 [DOI] [PubMed] [Google Scholar]
- 12. Ohba S., Kawaguchi H., Kugimiya F., Ogasawara T., Kawamura N., Saito T., Ikeda T., Fujii K., Miyajima T., Kuramochi A., Miyashita T., Oda H., Nakamura K., Takato T., Chung U. I. (2008) Patched1 haploinsufficiency increases adult bone mass and modulates Gli3 repressor activity. Dev. Cell 14, 689–699 [DOI] [PubMed] [Google Scholar]
- 13. Hilton M. J., Tu X., Cook J., Hu H., Long F. (2005) Ihh controls cartilage development by antagonizing Gli3, but requires additional effectors to regulate osteoblast and vascular development. Development 132, 4339–4351 [DOI] [PubMed] [Google Scholar]
- 14. Joeng K. S., Long F. (2009) The Gli2 transcriptional activator is a crucial effector for Ihh signaling in osteoblast development and cartilage vascularization. Development 136, 4177–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mo R., Freer A. M., Zinyk D. L., Crackower M. A., Michaud J., Heng H. H., Chik K. W., Shi X. M., Tsui L. C., Cheng S. H., Joyner A. L., Hui C. (1997) Specific and redundant functions of Gli2 and Gli3 zinc finger genes in skeletal patterning and development. Development 124, 113–123 [DOI] [PubMed] [Google Scholar]
- 16. Saito T., Fukai A., Mabuchi A., Ikeda T., Yano F., Ohba S., Nishida N., Akune T., Yoshimura N., Nakagawa T., Nakamura K., Tokunaga K., Chung U. I., Kawaguchi H. (2010) Transcriptional regulation of endochondral ossification by HIF-2α during skeletal growth and osteoarthritis development. Nat. Med. 16, 678–686 [DOI] [PubMed] [Google Scholar]
- 17. Arai F., Ohneda O., Miyamoto T., Zhang X. Q., Suda T. (2002) Mesenchymal stem cells in perichondrium express activated leukocyte cell adhesion molecule and participate in bone marrow formation. J. Exp. Med. 195, 1549–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hojo H., Yano F., Ohba S., Igawa K., Nakajima K., Komiyama Y., Kan A., Ikeda T., Yonezawa T., Woo J. T., Takato T., Nakamura K., Kawaguchi H., Chung U. I. (2010) Identification of oxytetracycline as a chondrogenic compound using a cell-based screening system. J. Bone Miner. Metab. 28, 627–633 [DOI] [PubMed] [Google Scholar]
- 19. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu H., Hilton M. J., Tu X., Yu K., Ornitz D. M., Long F. (2005) Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development 132, 49–60 [DOI] [PubMed] [Google Scholar]
- 21. Vokes S. A., Ji H., Wong W. H., McMahon A. P. (2008) A genome-scale analysis of the cis-regulatory circuitry underlying Sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 22, 2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ingham P. W., McMahon A. P. (2001) Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 15, 3059–3087 [DOI] [PubMed] [Google Scholar]
- 23. Bai C. B., Joyner A. L. (2001) Gli1 can rescue the in vivo function of Gli2. Development 128, 5161–5172 [DOI] [PubMed] [Google Scholar]
- 24. Kronenberg H. M. (2003) Developmental regulation of the growth plate. Nature 423, 332–336 [DOI] [PubMed] [Google Scholar]
- 25. Chung U. I., Schipani E., McMahon A. P., Kronenberg H. M. (2001) Indian hedgehog couples chondrogenesis to osteogenesis in endochondral bone development. J. Clin. Invest. 107, 295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Drakopoulou E., Outram S. V., Rowbotham N. J., Ross S. E., Furmanski A. L., Saldana J. I., Hager-Theodorides A. L., Crompton T. (2010) Non-redundant role for the transcription factor Gli1 at multiple stages of thymocyte development. Cell Cycle 9, 4144–4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Merchant A., Joseph G., Wang Q., Brennan S., Matsui W. (2010) Gli1 regulates the proliferation and differentiation of HSCs and myeloid progenitors. Blood 115, 2391–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karlstrom R. O., Tyurina O. V., Kawakami A., Nishioka N., Talbot W. S., Sasaki H., Schier A. F. (2003) Genetic analysis of zebrafish gli1 and gli2 reveals divergent requirements for gli genes in vertebrate development. Development 130, 1549–1564 [DOI] [PubMed] [Google Scholar]
- 29. Long F. (2012) Building strong bones: molecular regulation of the osteoblast lineage. Nat. Rev. Mol. Cell Biol. 13, 27–38 [DOI] [PubMed] [Google Scholar]
- 30. Tu X., Joeng K. S., Long F. (2012) Indian hedgehog requires additional effectors besides Runx2 to induce osteoblast differentiation. Dev. Biol. 362, 76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mak K. K., Bi Y., Wan C., Chuang P. T., Clemens T., Young M., Yang Y. (2008) Hedgehog signaling in mature osteoblasts regulates bone formation and resorption by controlling PTHrP and RANKL expression. Dev. Cell 14, 674–688 [DOI] [PubMed] [Google Scholar]
- 32. Koziel L., Wuelling M., Schneider S., Vortkamp A. (2005) Gli3 acts as a repressor downstream of Ihh in regulating two distinct steps of chondrocyte differentiation. Development 132, 5249–5260 [DOI] [PubMed] [Google Scholar]
- 33. Shimoyama A., Wada M., Ikeda F., Hata K., Matsubara T., Nifuji A., Noda M., Amano K., Yamaguchi A., Nishimura R., Yoneda T. (2007) Ihh/Gli2 signaling promotes osteoblast differentiation by regulating Runx2 expression and function. Mol. Biol. Cell 18, 2411-2418 [DOI] [PMC free article] [PubMed] [Google Scholar]