Background: The effect of PS1 mutations on the active site of γ-secretase is unknown.
Results: PS1 mutations reduce photoprobe interaction with the S2 subsite of γ-secretase and Notch1 cleavage.
Conclusion: Certain PS mutations specifically alter the S2 subsite of γ-secretase active site that leads to changes in APP and Notch1 cleavage.
Significance: It provides structural insights into the mechanism of PS1 mutations in regards to γ-secretase regulation.
Keywords: Alzheimer's Disease, Notch, Photoaffinity Labeling, Presenilin, Secretases, Activity-based Probes, Notch Assay, Photophore Walking
Abstract
Presenilin-1 (PS1) is the catalytic subunit of γ-secretase, and mutations in this protein cause familial Alzheimer Disease (FAD). However, little is known about how these mutations affect the active site of γ-secretase. Here, we show that PS1 mutations alter the S2 subsite within the active site of γ-secretase using a multiple photoaffinity probe approach called “photophore walking.“ Moreover, we developed a unique in vitro assay with a biotinylated recombinant Notch1 substrate and demonstrated that PS1 FAD mutations directly and significantly reduced γ-secretase activity for Notch1 cleavage. Substitution of the Notch Cys-1752 residue, which interacts with the S2 subsite, with Val, Met, or Ile has little effect on wild-type PS1 but leads to more efficient substrates for mutant PS1s. This study indicates that alteration of this S2 subsite plays an important role in determining the activity and specificity of γ-secretase for APP and Notch1 processing, which provides structural basis and insights on how certain PS1 FAD mutations lead to AD pathogenesis.
Introduction
Presenilin-1 (PS1)2, a multipass membrane protein, is the most commonly mutated protein in early-onset familial Alzheimer disease (FAD) (1, 2). Approximately 180 PS1 mutations have been reported to be linked with FAD (2). Mutations in presenilin2 (PS2), a homolog of PS1, are also associated with FAD (3). Although the precise mechanism of how these mutations cause AD is unknown, multiple theories have arisen to explain the role of PS1 and PS2 mutations on AD pathogenesis. These mutations lead to abnormal function of γ-secretase, the β-catenin pathway, calcium homoeostasis, and the lysosomal/autophagy pathway as well as chaperones (4–10).
Among these hypotheses, the effect of PS mutations on γ-secretase has been investigated extensively. γ-Secretase is composed of at least four subunits: PS, nicastrin, Aph1, and Pen2, with a total of 19 putative transmembrane domains (11). γ-Secretase cleaves multiple substrates, including the amyloid precursor protein (APP), Notch, and other type I membrane proteins (12). Genetic and biochemical studies have demonstrated that PS is the catalytic subunit of γ-secretase (13–16). PS mutations cause aberrant processing of APP and, thus, augment the ratio of Aβ42/Aβ40, which could contribute to neurodegeneration (5). Little is known about the molecular mechanism of these mutations in altering the specificity of γ-secretase. A previous study showed that expression of PS1 FAD mutants in PS1- and PS2-null cells led to the formation of γ-secretase complexes with different molecular weights (17), whereas another study showed that the overexpression of PS1 FAD mutants can also alter the equilibrium of wild-type PS1 and PS2 in γ-secretase complexes (18). Furthermore, we have demonstrated that proteoliposomes containing PS1 mutants alone are sufficient to alter the specificity of γ-secretase for the production of Aβ40 and Aβ42 (16). Fluorescence life-time imaging microscopy (19) and the substituted cysteine accessibility method (20) have been used to investigate the conformation of γ-secretase and its catalytic core. However, both methods rely on the generation of artificial PS1 containing fluorescent epitopes or substituted cysteine residues. Moreover, these studies can only detect overall conformational change but not the active site specifically. Because γ-secretase is a membrane-bound macromolecular complex and catalyzes the scissile bond hydrolysis within the lipid bilayer, elucidation of the conformational changes and structural features of the active site of γ-secretase has been a formidable challenge that requires the development of novel approaches.
In this study, we have developed and applied a chemical biology approach to detect conformational changes within the active site of γ-secretase caused by PS1 FAD mutations. We demonstrated using small molecular probes that PS1 mutations directly influence the S2 subsite (S2) of the γ-secretase active site. To further investigate the structure and activity relationship of this S2 subsite, we developed a robust in vitro γ-secretase assay using a biotinylated recombinant Notch1 substrate and assessed the interaction of the enzyme and substrate at the S2 subsite. This study demonstrates that PS1 FAD mutations directly alter the active site of γ-secretase that leads to altered processing of Notch1. This finding offers a molecular basis for γ-secretase specificity and FAD-mediated AD pathogenesis. Moreover, this chemical biology approach can be broadly used to characterize the modulation of γ-secretase and other transmembrane enzymes and receptors.
EXPERIMENTAL PROCEDURES
Peptide Synthesis and SM320 Production
The Notch peptide VLLSRKRRRC was synthesized with an automated solid-phase peptide synthesizer (ProteinTech) using Fmoc chemistry. This peptide was purified and confirmed by LC-MS. This neoepitope peptide antigen was conjugated to maleimide-functionalized keyhole limpet hemocyanin (KLH) according to the instructions of the manufacturer (Pierce). The conjugated antigen was used to immunize rabbits for antibody production (Covance). After initial characterization of two sera, SM320 and SM321, neoepitope antibodies were purified by affinity chromatography with peptide-immobilized resin. SM320 was used for this study.
Cell-based Notch Cleavage Assay
HEK 293 cells were seeded in a 24-well plate and transfected with the Notch1-ΔE construct (N1-ΔE) (a gift from Dr. Raphael Kopan) or the empty vector using FuGENE6 transfection reagents (Roche). DMSO or 1 μm compound E was added to the transfected cells at a final DMSO concentration of 1% (v/v). After 48 h of treatment, the cells were washed with PBS and lysed with 1× radioimmune precipitation assay buffer (50 mm Tris (pH8.0), 150 mm NaCl, 0.1% (w/v) SDS, 1% (v/v) Nonidet P-40, and 0.5% (w/v) deoxycholic acid). Cell lysates were centrifuged at 13,000 × g at 4 °C to remove cell debris. The cleared supernatant was collected and resolved by SDS-PAGE. The proteins were transferred to a PVDF membrane (Millipore) using semidry transfer unit (Bio-Rad) and analyzed by Western blot analysis with anti-myc antibody (9E10) and SM320 antibody.
Immunostaining
HEK 293 cells overexpressing N1-ΔE were treated with DMSO or compound E. The cells were then collected through centrifugation and embedded in paraffin. The cell pellets were sliced and mounted on glass slides. The immunodetection of the Notch1 intracellular domain (NICD) was performed at the Molecular Cytology Core Facility of the Memorial Sloan Kettering Cancer Center using a Discovery XT processor (Ventana Medical Systems). The cell sections were blocked for 30 min in 10% normal goat serum in 0.2% BSA/PBS, followed by incubation for 5 h with 0.5 μg/ml of SM320 and incubation for 60 min with biotinylated goat anti-rabbit IgG (Vector Laboratories, 1:200 dilution). The detection was performed with a DAB-MAP kit (Ventana Medical Systems). The slides were scanned and digitized using the Mirax scanner (Carl Zeiss Microsystems).
Production of the Recombinant Notch1 Substrate N1-Sb1
We constructed a recombinant human Notch 1 substrate, N1-Sb1. First, we subcloned a fragment of human Notch1 (1733–1812) into a vector that contains an AviTag. This Notch1-AviTag construct was further subcloned into the pIAD16 vector that contains a maltose binding protein (MBP) to facilitate protein purification. The MBP-N1-Sb1 plasmid was cotransformed into the BL21 (DE3) Escherichia coli strain with the pACYC 184 plasmid (BirA) that encodes for biotin ligase. isopropyl 1-thio-β-d-galactopyranoside (0.1 mm) and biotin (50 μm) were added to the cell culture during induction. Cells were centrifuged at 8000 × g for 30 min, and the pelleted cells were lysed by French press (Spectronic Instruments). The cell homogenate was centrifuged at 17,000 × g for 30 min. The supernatant was loaded onto on an amylose resin column and purified using the AKTA Prime Plus (GE Healthcare). The purified MBP-N1-Sb1 was digested with thrombin and analyzed by LC-MS. The P2-substituted N1-Sb1 substrates were generated with a Stratagene site-directed mutagenesis kit, expressed, and purified as described for wild-type N1-Sb1.
Cellular Membrane Preparation
HeLa membrane fraction was isolated from HeLa-S3 cells (National Cell Culture Center) as described previously (15, 21). N2a cells overexpressing wild-type, M146L, E280A, or H163R PS1 (from Dr. Sangram Sisodia) were cultured in 50% Dulbecco's modified Eagle's medium high-glucose, 50% Opti-MEM reduced-serum media and 10% fetal bovine serum. Cultured cells were pelleted by centrifugation and resuspended in hypotonic buffer (40 mm Tris (pH 7.4), 10 mm NaCl, 1 mm EDTA, and 0.5 mm DTT) for 20 min before homogenization. Cell debris from cells was pelleted at 800 × g for 30 min. The resulting supernatants were ultracentrifuged at 100,000 × g for 1 h. The ensuing pellet was used as membrane fractions for γ-secretase studies. Mouse brains expressing PS1 WT or PS1 M146V knock-in (gift from Dr. Hui Zheng) were homogenized, and the same procedures were used for membrane preparation. Protein concentration was determined with the DC protein assay kit according to the instructions of the manufacturer instructions (Bio-Rad).
In Vitro γ-Secretase Assay with N1-Sb1
The N1-Sb1 substrate (0.4 μm) was incubated with cell membrane (40 μg/ml) in the presence of 0.25% CHAPSO and PIPES buffer (50 mm PIPES (pH 7.0), 150 mm KCl, 5 mm CaCl2, 5 mm MgCl2) at 37 °C for 2 h. SM320, protein A-conjugated acceptor beads, and streptavidin-conjugated donor beads (PerkinElmer Life Sciences) were added to the reaction at a final concentration of 0.1 μg/ml, 1.25 μg/ml, and 5 μg/ml, respectively, to detect NICD. The reaction was incubated in the dark at room temperature overnight, and the AlphaLISA signal was detected using an Envision plate reader (PerkinElmer Life Sciences). The AlphaLISA signal was used as an arbitrary unit of γ-secretase activity. The specific activity was calculated on the basis of the amount of membrane protein and reaction time and is expressed as units/μg/min.
Photolabeling Probes and Photolabeling
Total cell membrane was preincubated in the presence of DMSO or 1 μm L458 in PIPES buffer containing 0.25% CHAPSO at 37 °C for 30 min. Then, 10 nm of photolabeling probes (JC8, L646, or GY4) (15, 22, 23) were added and incubated for an additional 1 h at 37 °C. The reaction mixtures were irradiated at 350 nm for 45 min and solubilized with radioimmune precipitation assay buffer. Biotinylated proteins in the soluble fraction were captured by streptavidin resin (Pierce) overnight at 4 °C. Bound proteins were eluted by boiling with SDS sample buffer and analyzed by Western blotting with anti-PS1 NTF antibodies.
RESULTS
PS1 Mutations Alter the S2 Subsite of γ-Secretase
Activity-based probes have been widely used to identify and profile various classes of enzymes (24). L-685,458 (L458), an aspartyl protease transition state mimic that interacts with the active site of γ-secretase (Fig. 1A), has been a valuable tool to study γ-secretase (15, 25). Therefore, we have generated various potent photoactivatable probes through “photophore walking,” in which the photoactivatable benzophenone is separately incorporated into different positions along the peptidomimetic core structure of L458. These positions are also known as the P or P′ positions according to the Schecter and Berger nomenclature (26). We intended to apply these L458-based probes to sense the subsite (S and S′) conformational change within the active site of γ-secretase caused by PS1 FAD mutations. The rationale of this strategy is that the efficiency of photolabeling by these photoactivatable probes depends on the orientation of the probes and their proximity to residues within the active site. Conformational changes resulting from PS1 mutations within the active site alter the orientation or distance of contact regions with the probes and could lead to different cross-linking efficiencies.
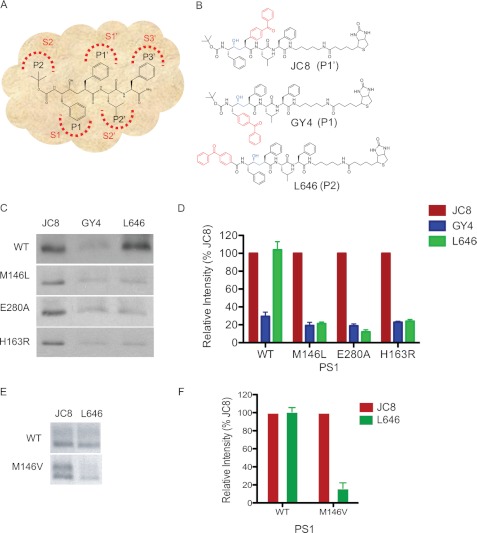
FIGURE 1.
PS1 mutations alter the S2 subsite conformation of γ-secretase. A, schematic representation of the interactions between L458 side chain residues (P/P′ positions) with corresponding γ-secretase subsites (S/S′). B, structure of JC8, GY4, and L646, in which a benzophenone was incorporated into the P1′, P1, or P2 site of L458, respectively. Biotin was linked to the probes to facilitate isolation of photolabeled proteins. C, photolabeling of PS1 NTF with JC8, GY4, and L646. Membranes isolated from WT, M146L, E280A, or H163R PS1 stable cell lines were used for this study. D, the photolabeled PS1 NTF were quantified by comparing the labeling intensities of GY4 and L646 to the labeling intensity of JC8. E, analysis of γ-secretase isolated from PS1 M146V knock-in or WT mouse brains. Photolabeled PS1 was detected with Western blot analysis using anti-PS1 NTF antibody. F, the analyses of photolabeled PS1 NTF were quantified by comparing the labeling intensities of L646 to JC8.
L646, GY4, and JC8, which photolabel the S2, S1, and S1′ subsites within the active site of PS1, respectively, were used for this study (Fig. 1B). All three probes only photolabel PS1-NTF but not PS1-CTF or other subunits (18, 22, 23, 25). Membranes isolated from N2a cells stably expressing WT, M146L, E280A, or H163R PS1 were photolabeled with JC8, GY4, or L646 in the presence of 0.25% CHAPSO. Additionally, we analyzed the level of FAD PS1 in cell membranes. The Western blot analysis showed that total amount of full-length and PS1-NTF in each cell line is virtually the same (supplemental Fig. S1). The labeling was determined to be specific because incubating the probes in the presence of excess L458 completely prevented labeling (data not shown). After UV irradiation, biotinylated proteins were isolated with streptavidin resins and analyzed by Western blotting with anti-PS1 NTF antibodies. We showed that JC8, GY4, and L646 all photolabeled WT, M146L, E280A, and H163R PS1 NTF (Fig. 1C) but not full-length PS1, which is consistent with our previous findings (15, 16). To eliminate the effect of various amounts of γ-secretase existing in different cell lines, we normalized the labeling intensity of each probe with the level of JC8-labeled PS1 NTF in the same cell lines. L646-labeled M146L, E280A, and H163R PS1 NTF with ∼80% less intensity than JC8, whereas L646 was photoinserted into WT PS1 with the same efficiency as JC8 (Fig. 1D). This result strongly indicates that the reduced labeling of PS1 mutants by L646 is not due to decreased expression of γ-secretase in the different stable cell lines but due to conformational changes within the S2 subsites of M146L, E280A, and H163R PS1. Finally, we showed that GY4-labeled WT, M146L, E280A, and H163R PS1 NTF with similar efficiencies (Fig. 1, C and D).
To further investigate the effect of PS1 mutations on the active site in an animal model, we performed the same study using PS1 M146V knock-in and PS1 WT mouse brains. The PS1 M146V knock-in mice have been well characterized for memory formation and Aβ production (18, 27, 28). Membranes isolated from the M146V knock-in and PS1 WT mouse brain were labeled with the photoprobes JC8 and L646. Similarly, we found that JC8 and L646 labeled WT PS1 with similar intensities (Fig. 1, E and F). However, L646 labeled M146V PS1 with 84% less efficiency than JC8 (Fig. 1, E and F). This data also showed that M146V PS1 leads to a similar conformation change at the S2 subsite. Taken together, these studies indicate that PS1 FAD mutations can directly influence the shape of the active site within the PS1 γ-secretase complex. However, it is still unclear how this change in the S2 subsite of the γ-secretase active site affects the interaction and catalysis of substrates such as Notch1.
Development of a Robust in Vitro Notch1 γ-Secretase Assay
To further elucidate how PS1 FAD mutations induce S2 subsite changes, we turned to mutate substrates in which the corresponding interacting residue (P2) is substituted with different amino acids. However, our current assays with APP substrates rely on the P2 residues for antibody recognition of Aβ40 and Aβ42 fragments and, thus, mutating the P2 residue within APP destroys the antibody-binding epitopes. Therefore, we attempted to use Notch1 substrate for this strategy. Because Notch signaling plays an important role in neurogenesis and has been implicated in AD pathogenesis (29–31), biochemical examination of the PS1 FAD mutation on Notch1 cleavage is necessary to understand the overall effect of PS1 mutations on substrate cleavage. First, we generated a polyclonal antibody against the N-terminal fragment of the NICD, designated as SM320. After affinity purification, the specificity of the SM320 antibody was determined. HEK293 cells were transfected with a Myc-tagged Notch1-ΔE construct (N1-ΔE) encoding a γ-secretase substrate that does not require ligand activation (32). The transfected cells were treated with either DMSO or compound E, a potent γ-secretase inhibitor (33). Anti-Myc analysis indicates that the N1-ΔE protein was expressed in the transfected cells, but not in the mock control (Fig. 2A, top panel). The lesser amount of N1-ΔE substrate in DMSO-treated cells than the Compound E-treated ones indicates an inhibition of N1-ΔE cleavage. The SM320 antibody only detected a band in the DMSO-treated cells, but not in the compound E and mock control (Fig. 2A, bottom panel). These analyses established that the SM320 antibody specifically recognizes the γ-secretase-cleaved product, NICD, but not the N1-ΔE substrate. We also used SM320 antibody for immunostaining. HEK293 cells expressing N1-ΔE substrate were treated with DMSO or compound E. The cells were pelleted and embedded in paraffin for SM320 staining. The embedded cell pellets were sectioned and mounted on glass slides and stained with SM320. SM320 clearly stained NICD in cells treated with DMSO but not with compound E, which further indicates that SM320 can detect NICD specifically (Fig. 2B). Finally, we showed that SM320 detects NICD within the nucleus using confocal microscopy in which the SM320 signal colocalizes with DAPI and anti-myc staining (supplemental Fig. S2A, panel 4). However, in the presence of compound E, we were able to detect only full-length N1-ΔE substrate (anti-myc) but no NICD (SM320) signal (supplemental Fig. S2B)
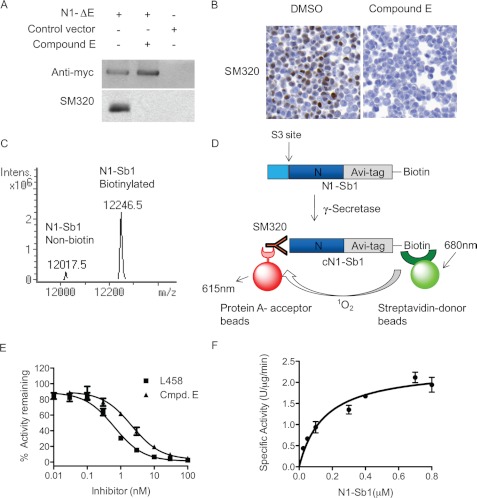
FIGURE 2.
Development of in vitro γ-secretase assay with SM320 antibody and recombinant Notch substrate (N1-Sb1). A, SM320 antibody detects NICD specifically. HEK293 cells were transfected with N1-ΔE, and cell lysates were analyzed with Western blot analysis. Anti-myc antibody detected full-length N1-ΔE in N1-ΔE-transfected cells (top panel, first and second lanes). SM320 antibody recognized NICD in DMSO-treated cells (bottom panel, first lane). This product is absent in the presence of Compound E (bottom panel, second lane). B, immunostaining of HEK293 cells overexpressing N1-ΔE. SM320 antibody detects NICD (brown) in DMSO but not in compound E-treated cells. C, mass spectrometry confirmed that the majority of N1-Sb1 was biotinylated. D, schematic representation of the in vitro γ-secretase assay with N1-Sb1 and SM320 antibody. HeLa cell membrane was incubated with N1-Sb1 in the presence of 0.25% CHAPSO. The C-terminal of cleaved N1-Sb1 (cN1-Sb1) was detected with SM320 antibody. Streptavidin-conjugated donor beads and protein A-conjugated acceptors beads were added to the reaction to detect cN1-Sb1. E, potencies of γ-secretase inhibitors L458 and compound E were determined to be 0.7 nm and 1 nm, respectively. F, the kinetic analysis of N1-Sb1 proteolysis. The specific activity was expressed as the arbitrary AlphaLISA assay unit.
Next, we designed and generated a biotinylated recombinant Notch1 substrate for development of the γ-secretase assay. We cloned a Notch1 protein fragment that contains the γ-secretase cleavage site into a vector that has an MBP tag and an Avi tag for in vivo biotinylation. This construct was designated as MBP-N1-Sb1. After coexpression of this construct with a biotin ligase, we purified this fusion protein and treated it with thrombin to remove the MBP tag. The resulting product, N1-Sb1, was analyzed by LC-MS. Two proteins, 12017.5 and 12246.5 that matched the calculated molecular masses of N1-Sb1 in non-biotinylated (12017.7) and biotinylated form (12245.7), were detected (Fig. 2C), and our data showed that the majority of N1-Sb1 exists as a biotinylated form.
Finally, we developed a γ-secretase activity assay with N1-Sb1 using AlphaLISA technology, in which interaction between donor and acceptor beads generates an amplified luminescence signal when they are brought into close proximity (34) (Fig. 2D). The N1-Sb1 substrate was incubated with a HeLa membrane, which is the source of γ-secretase, in the presence of 0.25% CHAPSO. The cleaved product (cN1-Sb1) was detected with SM320 antibody, a streptavidin-conjugated donor, and protein A-conjugated acceptor beads (Fig. 2D). We detected γ-secretase activity with a ratio of signal (in the absence of L458) to background (in the presence of L458) of more than 20 (data not shown). Furthermore, L458 and compound E inhibited γ-secretase activity with IC50 values of 0.7 nm and 1 nm in our assay, respectively (Fig. 2E), and these values are similar to the reported IC50 values of Aβ40 inhibition (25, 32, 35). In addition, we determined that γ-secretase cleaved N1-Sb1 with an apparent Km value of 0.15 ± 0.03 μm (Fig. 2F). These characteristics indicate that the newly designed in vitro γ-secretase assay is viable and can be used to further characterize PS1 FAD γ-secretase cleavage of Notch1.
PS1 Mutations Reduced γ-Secretase Activity for Notch 1 Cleavage
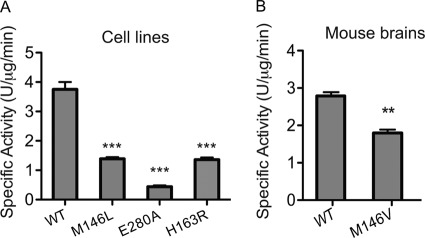
Previous data showed that both M146L, E280A, and H163R PS1 FAD mutations affect the specificity of γ-secretase processing of APP, resulting in an elevated Aβ42/Aβ40 ratio (18). How these mutations biochemically affect γ-secretase cleavage of Notch1 is unknown. Therefore, we determined the γ-secretase activity of WT, M146L, E280A, and H163R PS1 for the cleavage of N1-Sb1 using this new γ-secretase assay (Fig. 3A) and demonstrated that M146L, E280A, and H163R PS1 were 60%, 86%, and 60% less active for N1-Sb1 cleavage, respectively, compared with WT PS1 (Fig. 3A). Next, we examined whether PS1 mutants from mice have the same effects on N1-Sb1 cleavage as the cell line model. Membrane isolated from mouse brain expressing either knock-in M146V or WT PS1 (27, 36) were used to measure N1-Sb1 cleavage, and we show that M146V PS1 produced significantly less cN1-Sb1 compared with WT PS1 (Fig. 3B).
FIGURE 3.
PS1 mutants have different effects on the γ-secretase activity for Notch1 (N1-Sb1) catalysis. Membrane from N2a cells stably expressing WT, M146L, E280A, or H163R PS1 (A) or mouse brains expressing PS1 M146V knock-in or PS1 WT (B) were used to determine γ-secretase activity of N1-Sb1 catalysis. The specific activity was expressed as the arbitrary AlphaLISA assay unit. n = 3. Data are mean ± S.D. ***, p < 0.001; **, p < 0.01.
Moreover, we investigated whether these PS1 mutations affect the kinetics of N1-Sb1 cleavage. Our data showed that the Km values of WT, M146L, and E280A FAD PS1 are 57 ± 10 nm, 49 ± 7 nm, and 50 ± 12 nm, respectively (supplemental Fig. S3A), which are similar, whereas the Vmax values of WT, M146L, and E280A FAD PS1 are significantly different: 17.4 ± 0.8, 9.8 ± 0.3, and 2.9 ± 0.2 units/μg/min, respectively. Consistently, the Km values of WT and M146V PS1 from mouse brain are indifferent, 60 ± 11 nm and 48 ± 6 nm, respectively, whereas the Vmax values are also significantly different (3.1 ± 0.15 and 2.1 ± 0.06 units/μg/min, respectively) (supplemental Fig. S3B). These data suggest that PS1 FAD mutations appear to reduce the turnover rate of N1-Sb1.
In addition, we showed that these PS1 mutations did not affect the potencies of two γ-secretase inhibitors, L458 and compound E, for N1-Sb1 cleavage. Compound E inhibited WT, M146L, and E280A PS1 with IC50 values of 0.78 ± 0.11, 0.76 ± 0.12, and 0.73 ± 0.03 nm, respectively (supplemental Fig. S4A), whereas L458 inhibited WT, M146L, and E280A PS1 with IC50 values of 3.16 ± 0.28, 3.21 ± 0.22, and 3.34 ± 0.81 nm, respectively (supplemental Fig. S4B).
PS1 Mutants Prefer N1-Sb1 with Larger P2 Residues
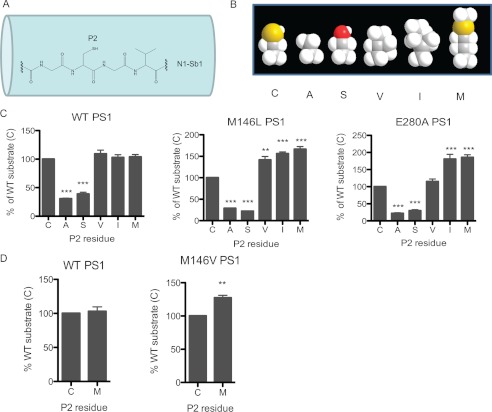
Finally, we probed the S2 subsite of the γ-secretase active site with P2-residue substituted Notch1 substrates. The rationale of this study is to investigate whether larger P2 residue of the Notch substrate can enhance the activity of PS1 FAD mutants. The wild-type P2 residue of the human Notch1 substrate is Cys-1752, which interacts with the corresponding S2 subsite within the γ-secretase active site (Fig. 4A). We substituted this Cys on N1-Sb1 with either Ala, Ser, Val, Ile, or Met using site-directed mutagenesis (Fig. 4B). After these P2-substituted N1-Sb1 substrates were purified, we determined the rate of γ-secretase cleavage from each cell membrane against these five substrates (Fig. 4C). First, the C1752A and C1752S substrates had significantly reduced reactivity for all three forms of γ-secretase (20–40% remaining) compared with the WT substrate. Secondly, the C1752V substrate had considerably increased reactivity for M146L PS1 γ-secretase (141.3 ± 7.9%,) (Fig. 4C, center panel) and had no effect on WT (left panel) and E280A PS1 (right panel). Thirdly, C1752I was a significantly better substrate for M146L (156 ± 3.8%) and E280A (180.7 ± 13.9%) PS1 (Fig. 4C, center and right panel) but has no effect on WT PS1 (left panel). Finally, the C1752M substrate had no effect on WT (Fig. 4C, left panel) but had significantly enhanced activity for M146L (166.4 ± 6.1%) (center panel) and E280A (185.5 ± 7.4%) PS1 (right panel). Similarly, we showed that the C1752M substrate also had no effect on WT PS1 (Fig. 4D, left panel) but had enhanced activity for M146V knock-in PS1 (127.4 ± 3.7%) (right panel) in our mouse model (Fig. 4D). Taken together, these data demonstrate that N1-Sb1 with the Met or Ile residue at the P2 position is a better substrate for both PS1 mutants, suggesting that Met and Ile fit the altered S2 subsites of PS1 mutants better than Cys. On the other hand, N1-Sb1 with a Val at the P2 position is a better substrate for M146L PS1 but not for WT PS1 and E280A PS1, suggesting that the S2 subsite of M146L PS1 is distinguishable from the S2 subsite of E280A PS1. These results indicate that the different PS1 mutations, such as M146L and E280A, can lead to S2 subsite variation and cause different effects on γ-secretase processing of APP and Notch1.
FIGURE 4.
PS1 mutants preferred N1-Sb1 substrates with larger P2 residues. A, schematic representation of N1-Sb1 with wild-type P2 residue cysteine. B, the relative size differences of the wild-type cysteine residue, which was substituted with either alanine, serine, valine, isoleucine, or methionine, are shown. C, M146L, E280A, and WT PS1 from stable cell lines were used to determine the catalysis of Cys or P2-substituted N1-Sb1. D, the catalysis of Cys and Met N1-Sb1 were compared between WT and M146V PS1 from mouse brain membrane. n = 3. Data are mean ± S.D. ***, p < 0.001; **, p < 0.01.
DISCUSSION
Because PS1 mutations are associated with early onset of FAD, the effects of these mutations on γ-secretase activity, protein trafficking, and processing have been well characterized in the cellular system. Recently, biochemical studies established that PS1 mutations directly affect γ-secretase specificity for Aβ42 and Aβ40 production (18). However, how these mutations influence the structure and function of PS1 has been elusive because of its transmembrane and macromolecular nature and a lack of high-resolution structure of the γ-secretase complex. Therefore, activity-based probes have been valuable tools to identify and to characterize γ-secretase. In this work, we developed a chemical biology approach, termed photophore walking, to sense the active site changes caused by PS1 FAD mutations within the lipid bilayers of the γ-secretase complex. This multiple probes approach unprecedentedly provides direct evidence that M146L, E280A, and H163R PS1 alter the conformation in the S2 subsite within the γ-secretase complex.
In addition, γ-secretase also cleaves Notch1, and the effects of PS1 FAD mutations on Notch1 cleavage have been investigated in a cell-based system. Although cell-based assays can reveal overall effects of these mutations on protein trafficking, localization, or concentration of Notch1, it is difficult to dissect the exact effects of PS1 FAD mutations on Notch1 cleavage. In addition, the lack of well controlled substrate concentration adds another complication to these analyses. Not surprisingly, inconsistent observations in PS1 mutations on Notch1 cleavage have been reported. Song et al. (31) reported that M146V and G384A PS1 rescued γ-secretase activity for Notch cleavage in PS1 knockout cells by 40–60% compared with WT PS1. However, Nakajima et al. (37) showed that M146V PS1 cleaves Notch with the same efficiency as WT PS1, whereas Bentahir et al. (17) demonstrated that G384A PS1 virtually lost its function in cleaving Notch. This study demonstrates that the PS1 mutants M146L, E280A, and H163R directly affect γ-secretase activity, which leads to a reduction in the rate of Notch1 cleavage. Our in vitro assay offers a unique way to characterize the effects of PS1 mutations on Notch1 and APP cleavage, as well as providing a way to address whether PS1 could contribute to AD through altering the processing of APP and Notch1.
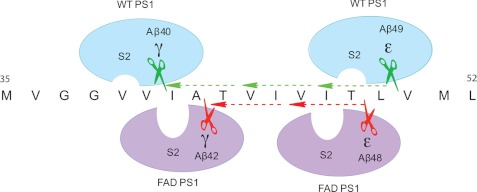
Furthermore, our P2-subsituted Notch1 substrate study demonstrated that both PS1 mutations, M146L and E280A, led to similar and yet distinguishable S2 subsite alterations. Although both PS1 mutants prefer Met and Ile P2 residues over Cys, M146L and E280A exhibited different activities for Val at the P2 position. Moreover, smaller residues at the P2 position (Ala and Ser) of the Notch1 substrate dramatically reduced its reactivity with γ-secretase. Taken together, these multiple photoaffinity labeling and substrate complement studies indicate that both M146L and E280A mutations lead to deeper and distinct S2 subsites. This conformational change in the active site could be a plausible mechanism on how PS1 mutations affect γ-secretase activity for Aβ40 and Aβ42 production. It has been shown through a sequential cleavage mechanism that Aβ40 and Aβ42 peptides are generated from Aβ49 and Aβ48 peptides, respectively (Fig. 5) (38). The P2 residues for Aβ40 and Aβ49 cleavages are Val and Thr, which are relatively smaller residues, whereas the P2 residue for both Aβ42 and Aβ48 is Ile, a larger residue. Because we showed that certain PS1 FAD mutants have deeper S2 subsites, this would favor Aβ48 cleavage, leading to an increase in Aβ42. Concurrently, these changes can affect Aβ49 and Aβ40 cleavages (Fig. 5). It has been reported that PS1 FAD mutations reduce Aβ49 production while concomitantly increasing Aβ48 levels (39), which supports our S2 subsite alteration model (Fig. 5). It is noteworthy to point out that this is a working model that was devised without knowing the high-resolution structure of γ-secretase, and the applicability of this model for PS1 FAD mutations should be further verified using an APP substrate.
FIGURE 5.
Schematic representation of WT PS1 and FAD PS1 cleavage of APP. γ-Secretase cleaves APP at several positions, such as the γ site, for Aβ40 and Aβ42 production, as well as the ϵ site to produce Aβ48 and Aβ49 peptides. The sequential cleavage model proposes that γ-secretase either first cleaves APP at the Aβ49 site followed by Aβ46, Aβ43, and Aβ40 (green dotted line) or that γ-secretase can cleave APP at the Aβ48 site followed by Aβ45 and Aβ42 (red dotted line). Because augmentation of the Aβ42/40 and Aβ48/49 ratio is associated with FAD PS1, this suggests that the deeper S2 subpocket of FAD PS1 prefers Aβ42 and Aβ48 cleavages, which have larger P2 residue (Ile), whereas WT PS1, which has a shallower S2 subsite, prefers Aβ40 and Aβ49 cleavages, which have smaller P2 residues such as Val and Thr.
In summary, we have developed a practical chemical approach to investigate the active site conformation of γ-secretase. We have demonstrated that PS1 mutations can alter the active site of γ-secretase, resulting in a potential shift in cleavage sites within substrates. These findings offer a molecular and structural foundation in elucidating the mechanism of PS1-γ-secretase mutations. Furthermore, our data suggest that the dual effect of PS1 mutations on the increased ratio of Aβ42:Aβ40 and a reduced γ-secretase activity for Notch1 cleavage could cooperatively lead to neurodegeneration in AD and, thus, contribute to our understanding of AD pathogenesis. Additionally, this unique γ-secretase assay and photophore walking approach will facilitate the characterization of γ-secretase from different tissue sources, the investigation of allosteric regulation of γ-secretase, as well as the development of selective γ-secretase inhibitors for treatment of AD and other human disorders.
Supplementary Material
Acknowledgments
We thank Dr. Min-tian Lai for the PS1 NTF antibody, Dr. Sam Sisodia for the N2a stable cell lines, Dr. Hui Zheng for the mouse brains, and Dr. Raphael Kopan for the ΔE construct. We also thank Dr. Katia Manova from the Molecular Cytology Core Facility.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-AG026660 (to Y. M. L.). This work was also supported by the American Health Assistance Foundation (to Y. M. L.), by the Alzheimer Association (Y. M. L.), by Institutional Training Grants T32 GM073546-01A1 9 (to C. J. C.) and T32 GM073546–04 (to J. C. V.), by Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, by the Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center, and by the William Randolph Hearst Fund in Experimental Therapeutics.

This article contains supplemental Figs. S1–S4.
- PS1
- presenilin 1
- FAD
- familial Alzheimer's disease
- APP
- amyloid precursor protein
- DMSO
- dimethyl sulfoxide
- NICD
- Notch1 intracellular domain
- MBP
- maltose binding protein
- AD
- Alzheimer's disease
- CHAPSO
- 3-[(3-Cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate
- NTF
- N-terminal fragment.
REFERENCES
- 1. Sherrington R., Rogaev E. I., Liang Y., Rogaeva E. A., Levesque G., Ikeda M., Chi H., Lin C., Li G., Holman K., Tsuda T., Mar L., Foncin J. F., Bruni A. C., Montesi M. P., Sorbi S., Rainero I., Pinessi L., Nee L., Chumakov I., Pollen D., Brookes A., Sanseau P., Polinsky R. J., Wasco W., Da Silva H. A., Haines J. L., Perkicak-Vance M. A., Tanzi R. E., Roses A. D., Fraser P. E., Rommens J. M., St. George-Hyslop P. H. (1995) Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature 375, 754–760 [DOI] [PubMed] [Google Scholar]
- 2. Lessard C. B., Wagner S. L., Koo E. H. (2010) And four equals one. Presenilin takes the γ-secretase role by itself. Proc. Natl. Acad. Sci. U.S.A. 107, 21236–21237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Levy-Lahad E., Wasco W., Poorkaj P., Romano D. M., Oshima J., Pettingell W. H., Yu C. E., Jondro P. D., Schmidt S. D., Wang K. (1995) Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science 269, 973–977 [DOI] [PubMed] [Google Scholar]
- 4. Borchelt D. R., Ratovitski T., van Lare J., Lee M. K., Gonzales V., Jenkins N. A., Copeland N. G., Price D. L., Sisodia S. S. (1997) Accelerated amyloid deposition in the brains of transgenic mice coexpressing mutant presenilin 1 and amyloid precursor proteins. Neuron 19, 939–945 [DOI] [PubMed] [Google Scholar]
- 5. Borchelt D. R., Thinakaran G., Eckman C. B., Lee M. K., Davenport F., Ratovitsky T., Prada C. M., Kim G., Seekins S., Yager D., Slunt H. H., Wang R., Seeger M., Levey A. I., Gandy S. E., Copeland N. G., Jenkins N. A., Price D. L., Younkin S. G., Sisodia S. S. (1996) Familial Alzheimer's disease-linked presenilin 1 variants elevate Aβ1–42/1–40 ratio in vitro and in vivo. Neuron 17, 1005–1013 [DOI] [PubMed] [Google Scholar]
- 6. Citron M., Westaway D., Xia W., Carlson G., Diehl T., Levesque G., Johnson-Wood K., Lee M., Seubert P., Davis A., Kholodenko D., Motter R., Sherrington R., Perry B., Yao H., Strome R., Lieberburg I., Rommens J., Kim S., Schenk D., Fraser P., St George Hyslop P., Selkoe D. J. (1997) Mutant presenilins of Alzheimer's disease increase production of 42-residue amyloid β-protein in both transfected cells and transgenic mice. Nat. Med. 3, 67–72 [DOI] [PubMed] [Google Scholar]
- 7. Lee J. H., Yu W. H., Kumar A., Lee S., Mohan P. S., Peterhoff C. M., Wolfe D. M., Martinez-Vicente M., Massey A. C., Sovak G., Uchiyama Y., Westaway D., Cuervo A. M., Nixon R. A. (2010) Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141, 1146–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nishimura M., Yu G., Levesque G., Zhang D. M., Ruel L., Chen F., Milman P., Holmes E., Liang Y., Kawarai T., Jo E., Supala A., Rogaeva E., Xu D. M., Janus C., Levesque L., Bi Q., Duthie M., Rozmahel R., Mattila K., Lannfelt L., Westaway D., Mount H. T., Woodgett J., St. George-Hyslop P. (1999) Presenilin mutations associated with Alzheimer disease cause defective intracellular trafficking of β-catenin, a component of the presenilin protein complex. Nat. Med. 5, 164–169 [DOI] [PubMed] [Google Scholar]
- 9. Naruse S., Thinakaran G., Luo J. J., Kusiak J. W., Tomita T., Iwatsubo T., Qian X., Ginty D. D., Price D. L., Borchelt D. R., Wong P. C., Sisodia S. S. (1998) Effects of PS1 deficiency on membrane protein trafficking in neurons. Neuron 21, 1213–1221 [DOI] [PubMed] [Google Scholar]
- 10. Tu H., Nelson O., Bezprozvanny A., Wang Z., Lee S. F., Hao Y. H., Serneels L., De Strooper B., Yu G., Bezprozvanny I. (2006) Presenilins form ER Ca2+ leak channels, a function disrupted by familial Alzheimer's disease-linked mutations. Cell 126, 981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jorissen E., De Strooper B. (2010) γ-Secretase and the intramembrane proteolysis of Notch. Curr. Top. Dev. Biol. 92, 201–230 [DOI] [PubMed] [Google Scholar]
- 12. Kopan R., Ilagan M. X. (2004) γ-Secretase. Proteasome of the membrane? Nat. Rev. Mol. Cell Biol. 5, 499–504 [DOI] [PubMed] [Google Scholar]
- 13. De Strooper B., Saftig P., Craessaerts K., Vanderstichele H., Guhde G., Annaert W., Von Figura K., Van Leuven F. (1998) Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature 391, 387–390 [DOI] [PubMed] [Google Scholar]
- 14. Wolfe M. S., Xia W., Ostaszewski B. L., Diehl T. S., Kimberly W. T., Selkoe D. J. (1999) Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and γ-secretase activity. Nature 398, 513–517 [DOI] [PubMed] [Google Scholar]
- 15. Li Y. M., Xu M., Lai M. T., Huang Q., Castro J. L., DiMuzio-Mower J., Harrison T., Lellis C., Nadin A., Neduvelil J. G., Register R. B., Sardana M. K., Shearman M. S., Smith A. L., Shi X. P., Yin K. C., Shafer J. A., Gardell S. J. (2000) Photoactivated γ-secretase inhibitors directed to the active site covalently label presenilin 1. Nature 405, 689–694 [DOI] [PubMed] [Google Scholar]
- 16. Ahn K., Shelton C. C., Tian Y., Zhang X., Gilchrist M. L., Sisodia S. S., Li Y. M. (2010) Activation and intrinsic γ-secretase activity of presenilin 1. Proc. Natl. Acad. Sci. U.S.A. 107, 21435–21440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bentahir M., Nyabi O., Verhamme J., Tolia A., Horré K., Wiltfang J., Esselmann H., De Strooper B. (2006) Presenilin clinical mutations can affect γ-secretase activity by different mechanisms. J. Neurochem. 96, 732–742 [DOI] [PubMed] [Google Scholar]
- 18. Placanica L., Tarassishin L., Yang G., Peethumnongsin E., Kim S. H., Zheng H., Sisodia S. S., Li Y. M. (2009) Pen2 and presenilin-1 modulate the dynamic equilibrium of presenilin-1 and presenilin-2 γ-secretase complexes. J. Biol. Chem. 284, 2967–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Berezovska O., Lleo A., Herl L. D., Frosch M. P., Stern E. A., Bacskai B. J., Hyman B. T. (2005) Familial Alzheimer's disease presenilin 1 mutations cause alterations in the conformation of presenilin and interactions with amyloid precursor protein. J. Neurosci. 25, 3009–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Isoo N., Sato C., Miyashita H., Shinohara M., Takasugi N., Morohashi Y., Tsuji S., Tomita T., Iwatsubo T. (2007) Aβ42 overproduction associated with structural changes in the catalytic pore of γ-secretase. Common effects of Pen-2 N-terminal elongation and fenofibrate. J. Biol. Chem. 282, 12388–12396 [DOI] [PubMed] [Google Scholar]
- 21. Tian Y., Bassit B., Chau D., Li Y. M. (2010) An APP inhibitory domain containing the Flemish mutation residue modulates γ-secretase activity for Aβ production. Nat. Struct. Mol. Biol. 17, 151–158 [DOI] [PubMed] [Google Scholar]
- 22. Chun J., Yin Y. I., Yang G., Tarassishin L., Li Y. M. (2004) Stereoselective synthesis of photoreactive peptidomimetic γ-secretase inhibitors. J. Org. Chem. 69, 7344–7347 [DOI] [PubMed] [Google Scholar]
- 23. Yang G., Yin Y. I., Chun J., Shelton C. C., Ouerfelli O., Li Y. M. (2009) Stereo-controlled synthesis of novel photoreactive γ-secretase inhibitors. Bioorg. Med. Chem. Lett. 19, 922–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cravatt B. F., Wright A. T., Kozarich J. W. (2008) Activity-based protein profiling. From enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 77, 383–414 [DOI] [PubMed] [Google Scholar]
- 25. Shelton C. C., Zhu L., Chau D., Yang L., Wang R., Djaballah H., Zheng H., Li Y. M. (2009) Modulation of γ-secretase specificity using small molecule allosteric inhibitors. Proc. Natl. Acad. Sci. U.S.A. 106, 20228–20233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schechter I., Berger A. (1967) On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 27, 157–162 [DOI] [PubMed] [Google Scholar]
- 27. Wang R., Dineley K. T., Sweatt J. D., Zheng H. (2004) Presenilin 1 familial Alzheimer's disease mutation leads to defective associative learning and impaired adult neurogenesis. Neuroscience 126, 305–312 [DOI] [PubMed] [Google Scholar]
- 28. Wang R., Wang B., He W., Zheng H. (2006) Wild-type presenilin 1 protects against Alzheimer disease mutation-induced amyloid pathology. J. Biol. Chem. 281, 15330–15336 [DOI] [PubMed] [Google Scholar]
- 29. Louvi A., Artavanis-Tsakonas S. (2006) Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 7, 93–102 [DOI] [PubMed] [Google Scholar]
- 30. Hitoshi S., Alexson T., Tropepe V., Donoviel D., Elia A. J., Nye J. S., Conlon R. A., Mak T. W., Bernstein A., van der Kooy D. (2002) Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 16, 846–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Song W., Nadeau P., Yuan M., Yang X., Shen J., Yankner B. A. (1999) Proteolytic release and nuclear translocation of Notch-1 are induced by presenilin-1 and impaired by pathogenic presenilin-1 mutations. Proc. Natl. Acad. Sci. U.S.A. 96, 6959–6963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kopan R., Schroeter E. H., Weintraub H., Nye J. S. (1996) Signal transduction by activated mNotch. Importance of proteolytic processing and its regulation by the extracellular domain. Proc. Natl. Acad. Sci. U.S.A. 93, 1683–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Seiffert D., Bradley J. D., Rominger C. M., Rominger D. H., Yang F., Meredith J. E., Jr., Wang Q., Roach A. H., Thompson L. A., Spitz S. M., Higaki J. N., Prakash S. R., Combs A. P., Copeland R. A., Arneric S. P., Hartig P. R., Robertson D. W., Cordell B., Stern A. M., Olson R. E., Zaczek R. (2000) Presenilin-1 and -2 are molecular targets for γ-secretase inhibitors. J. Biol. Chem. 275, 34086–34091 [DOI] [PubMed] [Google Scholar]
- 34. Bielefeld-Sevigny M. (2009) αLISA immunoassay platform. The “no-wash” high-throughput alternative to ELISA. Assay Drug Dev. Technol. 7, 90–92 [DOI] [PubMed] [Google Scholar]
- 35. Li Y. M., Lai M. T., Xu M., Huang Q., DiMuzio-Mower J., Sardana M. K., Shi X. P., Yin K. C., Shafer J. A., Gardell S. J. (2000) Presenilin 1 is linked with γ-secretase activity in the detergent-solubilized state. Proc. Natl. Acad. Sci. U.S.A. 97, 6138–6143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guo Q., Sebastian L., Sopher B. L., Miller M. W., Ware C. B., Martin G. M., Mattson M. P. (1999) Increased vulnerability of hippocampal neurons from presenilin-1 mutant knockin mice to amyloid β-peptide toxicity. Central roles of superoxide production and caspase activation. J. Neurochem. 72, 1019–1029 [DOI] [PubMed] [Google Scholar]
- 37. Nakajima M., Shimizu T., Shirasawa T. (2000) Notch-1 activation by familial Alzheimer's disease (FAD)-linked mutant forms of presenilin-1. J. Neurosci. Res. 62, 311–317 [DOI] [PubMed] [Google Scholar]
- 38. Takami M., Nagashima Y., Sano Y., Ishihara S., Morishima-Kawashima M., Funamoto S., Ihara Y. (2009) γ-Secretase. Successive tripeptide and tetrapeptide release from the transmembrane domain of β-carboxyl terminal fragment. J. Neurosci. 29, 13042–13052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sato T., Dohmae N., Qi Y., Kakuda N., Misonou H., Mitsumori R., Maruyama H., Koo E. H., Haass C., Takio K., Morishima-Kawashima M., Ishiura S., Ihara Y. (2003) Potential link between amyloid β-protein 42 and C-terminal fragment γ 49–99 of β-amyloid precursor protein. J. Biol. Chem. 278, 24294–24301 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.