Background: ARF and L11 can activate p53 in response to different stress signals.
Results: ARF and L11 physically and functionally interact with each other to activate p53.
Conclusion: ARF crosstalks with L11 in p53 response to stress.
Significance: Discovery of a direct link between ARF and L11 in the p53 network.
Keywords: ARF, Cell Cycle, Cell Signaling, p53, Tumor Suppressor Gene, L11, MDM2, Protein-Protein Interactions, Ribosomal Proteins, Ribosomal Stress
Abstract
The ARF tumor suppressor protein activates p53 in response to oncogenic stress, whereas ribosomal protein L11 induces p53 following ribosomal stress. Both proteins bind to central, albeit non-overlapping, regions of MDM2 and suppress MDM2 activity toward p53. However, it is not known whether the two pathways are functionally connected. Here we show that ARF directly binds to L11 in vitro and in cells, which then forms a complex with MDM2 and p53. L11 collaboratively enhances ARF-induced p53 transcriptional activity and cell cycle arrest. Supporting these results, knocking down L11 reduces ARF-mediated p53 accumulation and alleviates ARF-induced cell cycle arrest. Interestingly, overexpression of ARF increases the levels of ribosome-free L11 and enhances the interaction of L11 with MDM2 and p53. These results demonstrate that ARF activates p53, at least partly by induction of ribosomal stress, which results in L11 suppression of MDM2, and suggest that the ARF-MDM2-p53 and the L11-MDM2-p53 pathways are functionally connected.
Introduction
The tumor suppressor protein p53 plays a pivotal role in preventing cells from uncontrolled growth and tumorigenesis, and is frequently altered in human cancers (1–3). Because of its inhibitory effect on cell growth, low levels of p53 protein are tightly controlled primarily by an oncoprotein called MDM2 (4). MDM2 is both a negative regulator and a transcriptional target of p53, thus MDM2 participates in an auto-regulatory feedback loop (5–7). Various stress signals invoke distinct mechanisms to stabilize and activate p53 by blocking this loop. For example, γ-irradiation inhibits the feedback mechanism by interfering with the MDM2-p53 interaction by modifying both MDM2 and p53 (8–11), while other stresses inhibit the regulatory loop via protein-protein interactions. Examples of the latter mechanism include the oncogenic stress-induced ARF (Alternative Reading Frame,2 p14ARF in human, p19ARF in mouse)-MDM2-p53 pathway (12–15) and the ribosomal stress-induced Ribosomal Protein (RP)-MDM2-p53 pathways (16).
Oncogenic stress induced by the overexpression of oncoproteins, such as Ras (15), c-Myc (12), E2F (17), E1A (18), and β-catenin (19), induces the expression of ARF (12, 15). ARF binds to MDM2 and inhibits its ubiquitin ligase (E3) activity toward p53 (20). ARF also sequesters MDM2 in the nucleolus (21, 22) and inhibits nuclear export of both MDM2 and p53 (23). These ARF functions consequently activate p53 and induce cell cycle arrest, apoptosis, or senescence (13, 24).
Recent studies revealed that ribosomal stress can trigger a RP-MDM2-p53 signaling pathway, leading to p53 activation. Ribosomal stress, also called nucleolus stress, can be induced by perturbations of ribosomal biogenesis, such as inhibition of rRNA synthesis by treatment with a low dose of actinomycin D and mycophenolic acid, inhibition of rRNA processing by treatment with 5-fluorouricil, as well as depletion of ribosomal biogenesis factors (25–33). This type of stress induces the association of MDM2 with a group of RPs including L5, L11, L23, L26, S7, S27, and S27a (25, 26, 28, 34–42), leading to a suppression of MDM2 and consequent activation of p53. Similar to ARF, the RPs form a complex with MDM2 and p53 (26, 28, 41). They also exist in the nucleolus and are destined to be assembled onto the ribosome for protein synthesis during non-stressed conditions (28, 36, 43, 44). Thus, ARF and RPs display similar characteristics in terms of modulating the p53-MDM2 regulatory loop.
Currently, it is unknown whether ARF and the RP proteins interplay with each other in p53-MDM2 regulation. To begin to address this issue, we previously observed that L11 resembled ARF in terms of regulating MDM2 activity. Like ARF (45), L11-regulated MDM2 levels through a post-ubiquitination mechanism (46). The similarities between L11 and ARF regulatory mechanisms prompted us to further examine their functional connection in regulating the p53-MDM2 pathway. In this study, we show that L11 is directly bound to ARF and cooperatively enhanced ARF-induced p53-dependent transcription and cell cycle arrest. Supporting these ideas, ablation of L11 reduced ARF-mediated p53 accumulation. Overexpression of ARF increased the levels of ribosome-free L11 and enhanced MDM2-L11 interaction. These results suggest that L11 may play an important role in ARF-mediated activation of p53 in response to oncogenic stress.
EXPERIMENTAL PROCEDURES
Cell Lines
Human embryonic kidney epithelial 293, human p53-null lung non-small cell carcinoma H1299 and human p53-proficient osteosarcoma U2OS cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 units/ml penicillin, and 0.1 mg/ml streptomycin at 37 °C in a 5% CO2 humidified atmosphere. A human p53-null osteosarcoma Saos-2 cell line was stably transfected with the wild-type E2F1 expression vector under control of tetracycline-regulated promoters (Saos2-Tet-E2F1, a gift from Dr. Karen Vousden, Beatson Institute, UK). The stably transfected cells were maintained in 10% certified tetracycline-free FBS (Clontech) as described (17, 47). To establish Tet-inducible expression of ARF in U2OS cells, T-Rex-U2OS cells were transfected with pcDNA4-TO-V5-p14ARF. The cells were then split into selection medium containing 50 μg/ml of hygromycin and 100 μg/ml of Zeocin and selection was continued for 2 weeks. Single colonies were isolated, expanded, and screened by immunoblot (IB) analysis for Dox-induced expression of V5-ARF.
Plasmids and Antibodies
The Flag-tagged and GFP fusion human L11 expression plasmids pcDNA3-Flag-L11 and pEGFP-C1-L11 were described (46). L11 deletion mutants were generated using PCR and cloned into the pcDNA3–2Flag vector. The full-length ARF coding region was amplified using PCR and inserted into the pcDNA3–2Flag or pcDNA3-V5 vector to generate pcDNA3–2Flag-ARF or pcDNA3-V5-ARF, respectively. The primers used in the PCR were 5′-CGCGGATTCATGGTGCGCAGGTTCTTGGTG-3′ and 5′-CCGGAATTCTCAGCCAGGTCCACGGGCAGAC-3′. The Flag-ARF deletion mutants were generated by PCR. The full-length ARF coding region was subcloned into pEGFP-C1 to generate pEGFP-C1-ARF. The full-length ARF ORF, and its deletion mutants, were also cloned into the pGEX.4T.1 vector (Pharmacia Biotech) to express GST-fusion ARF proteins. The HA-MDM2 and p53 expression vectors were described (28). His-tagged L11 was expressed and purified from bacteria as previously described (26). Rabbit polyclonal anti-L11 antibodies were described (46). Anti-Flag (Sigma), anti-p21 (NeoMarkers), anti-ARF (NeoMarkers), anti-GST (GenScript), anti-V5 (Invitrogen), and anti-p53 (DO-1, Santa Cruz Biotechnology) were purchased. Anti-MDM2 (2A10) and anti-HA (12CA5) were previously described (28).
Cotransfection, IB and Co-immunoprecipitation (co-IP) Analyses
Cells were transfected with plasmids as indicated in figure legends using TransIT®-LT1 transfection reagent (Mirus, Madison, WI) following the manufacturer's protocol. Cells were harvested at 48 h post-transfection and lysed in Lysis buffer solution containing 50 mm Tris/HCl (pH 8.0), 0.5% Nonidet P-40, 1 mm EDTA, 150 mm NaCl, and 1 mm phenylmethylsulfonyl fluoride (PMSF). Equal amounts of clarified cell lysates were used for IB analysis, as described (28). Co-IPs were conducted as described previously (28). Sequential co-IPs were carried out as described (48). Bound proteins in the co-IPs were detected by IB using the indicated antibodies.
Luciferase Assays
U2OS cells were transfected with a luciferase reporter plasmid driven by two copies of the p53RE motif derived from the MDM2 promoter (28). The pCMV-β-galactoside reporter plasmid was transfected into the cultured cells in combination with the indicated plasmids, with 1 μg/well of total plasmid DNA used for each transfection. Luciferase activity was determined as described previously and normalized to β-gal activity, by co-transfection with the pCMV-β-gal plasmids, as previously described (28).
Cell Cycle Analysis
U2OS cells were transfected with GFP, GFP-L11, or GFP-ARF-encoding plasmids alone or in combination with GFP-L11 and GFP-ARF expressing plasmids. Thirty-two hours after transfection, cells were treated with 200 ng/ml of nocodazole for an additional 16 h. Cells were fixed, permeabilized, and stained in 500 μl of a solution containing 50 μg/ml propidium iodide (PI, Sigma), 200 μg/ml RNase A, 0.1% Triton X-100, 0.38 m NaCl, pH 7.2 at 37 °C for 30 min, and then analyzed for DNA content using a Becton Dickinson FACScan flow cytometer (28). GFP-positive cells were gated for cell cycle analysis. Data were collected using the ModFit software program.
GST Fusion Protein Association Assays
Protein-protein interaction assays were conducted as previously described using GST fusion protein immobilized to glutathione beads (28). Purified His-tagged L11 proteins were incubated with the glutathione-Sepharose 4B beads (Sigma) containing 200 ng of GST, GST-ARF, or GST-fused deletion mutants of ARF, for 30 min at room temperature. After washing, bound proteins were analyzed by electrophoresis in a 15% SDS gel and detected by IB using anti-L11 antibody.
Polysome Profiling Assays
Cytosolic extractions, sucrose gradient sedimentation of polysomes, and analysis of the polysomes/mRNPs distribution of proteins were carried out as previously described (28). Briefly, cells were incubated with 100 μg/ml of cycloheximide (CHX) for 15 min prior to harvest. The cells were homogenized in polysome lysis buffer solution containing 30 mm Tris-HCl (pH 7.4), 10 mm MgCl2, 100 mm KCl, 0.3% Nonidet P-40, 50 μg/ml CHX, 30 units/ml RNasin inhibitor, 1 mm dithiothreitol (DTT), 1 mm PMSF, 1 mm pepstatin, and 1 mm leupeptin. After incubation on ice for 10 min, the lysates were subjected to centrifugation at 12,000 × g at 4 °C for 8 min. The resulting supernatants were subjected to sedimentation centrifugation in a 15–47% sucrose gradient solution containing 30 mm Tris-HCl (pH 7.4), 5 mm MgCl2, 100 mm KCl, and 50 μg/ml CHX in a Beckman SW41 rotor at 37,000 rpm for 2 h. Fourteen fractions were collected and assayed by IB. Similar procedures were also conducted using a Biocomp Gradient Station, and absorbance of RNA at 254 nm was recorded using an in-line UV monitor to analyze the distribution of polysomes and monosomes as described (65).
RNA Interference
RNAi-mediated ablation of endogenous L11 was performed as previously described (28). The 21-nucleotide siRNA duplexes with a 3′-dTdT overhang were purchased from Dharmacon. The target sequences for L11 and control scrambled RNA were previously described (48). These siRNA duplexes (0.2 μm) were introduced into U2OS cells using Silentfectin (Bio-Rad) following the manufacturer's protocol. Cells were then harvested 48 h after transfection for IB and cell cycle analyses.
Adenoviruses and Lentiviruses
Adenoviruses encoding human ARF were kindly provided by Dr. Yanping Zhang (49). To generate lentiviral expression of L11 shRNA, oligonucleotides containing the same L11 mRNA targeting sequence as siRNA were synthesized and annealed and ligated into H1 lentivirus vector as described (50). The resulting vector was co-transfected with gag- and env-expressing plasmids into 293FT cells using calcium chloride (Promega). The resulting viruses were then used to infect cells in the presence of polybrene (6 μg/ml). The cells were harvested at 72 h post-transduction for IB analysis.
RESULTS
Overexpression of L11 Enhances ARF-induced p53 Activation
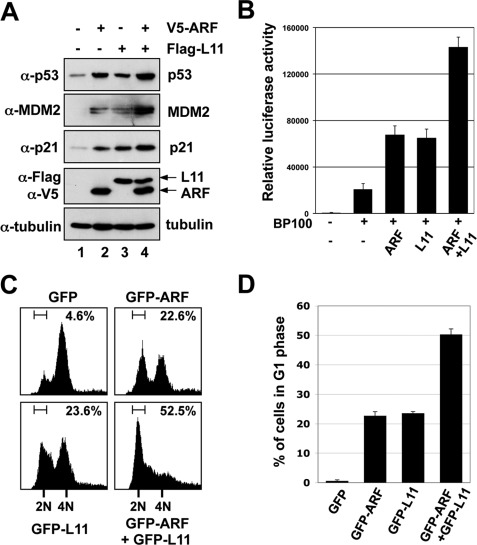
To test whether L11 interplays with ARF in the regulation of the MDM2-p53 loop, we first performed transient transfection-IB analyses in p53-proficient U2OS cells. As expected (25, 36, 41, 45), overexpression of either ARF or L11 increased p53 level compared with control transfected cells (Fig. 1A, upper panel). As a result, MDM2 and p21, two p53 targets, were also induced (second and third panels). Interestingly, coexpression of both ARF and L11 further induced the levels of p53, p21, and MDM2 (Fig. 1A, lane 4). To determine if the induction of p53 and its target genes is due to the augment of p53 transactivational activity by ARF and L11, we performed reporter assays using a plasmid encoding the firefly luciferase reporter plasmid expressed from a promoter containing the p53 response element (RE) derived from the MDM2 promoter (5, 28). As shown in Fig. 1B, ectopic expression of either ARF or L11 markedly enhanced the p53RE-driven luciferase activity. The luciferase activity was further enhanced by coexpression of ARF and L11. These results suggest that L11 and ARF can potentiate each other to induce the level and activity of p53 in cells.
FIGURE 1.
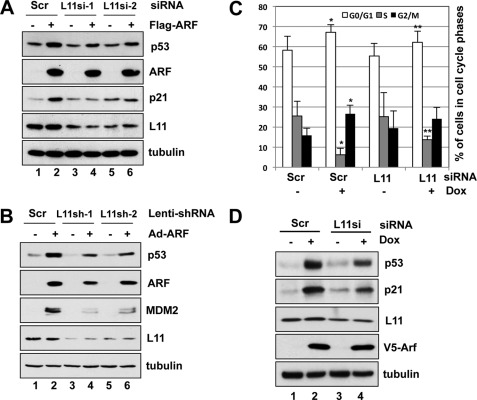
ARF and L11 cooperatively induce p53 level and activation. A, coexpression of ARF and L11 cooperatively induce p53. U2OS cells were transfected with V5-ARF and Flag-L11-expressing plasmids, individually or together as indicated. Cells were harvested at 48 h post-transfection and subjected to IB using the indicated antibodies. B, coexpression of ARF and L11 further increased p53RE-dependent luciferase activity. U2OS cells were transfected with V5-ARF and Flag-L11 plasmids, individually or together as indicated, in the presence of a luciferase reporter plasmid driven by p53RE (BP100) and pCMV-β-gal plasmids. Luciferase activity was normalized with β-galactoside activity and presented in arbitrary units. C and D, ARF and L11 collaboratively increased G1 phase cell cycle arrest. U2OS cells were transfected with GFP, GFP-ARF, GFP-L11, or GFP-ARF plus GFP-L11 plasmids and treated with nocodazole as described under “Experimental Procedures.” GFP-positive cells were gated for cell cycle analysis. The histograms of PI staining from one representative experiment are shown in C. Mean percentage of cells arrested in G1 phase summarized from three independent experiments is shown in D.
L11 Cooperates with ARF to Induce G1 Cell Cycle Arrest
Both ARF (14, 51) and L11 (25, 36, 41) induce p21 and lead to G1 cell cycle arrest. Therefore, we next determined whether coexpression of ARF and L11 could further enhance the induction of cell cycle arrest. To do so, U2OS cells were transiently transfected with the GFP-ARF or GFP-L11 plasmids, individually or together. Before the FACS analyses, cells were treated with nocodazole, an inhibitor of microtubule assembly that arrests cells at the G2/M phase, allowing us to visualize any G1 cell cycle arrest. GFP-positive cells were then gated for cell cycle analysis. As shown in a representative result in Fig. 1C, 22.6% of GFP-ARF and 23.6% of GFP-L11 expressing cells were arrested in the G1 phase, while only 4.6% of GFP-expressing cells were detected in the G1 phase. When GFP-L11 and GFP-ARF were coexpressed, 52.5% of the GFP-positive cells were arrested in the G1 phase (Fig. 1C). This result was reproduced in multiple experiments, as summarized in Fig. 1D. Since neither L11 nor ARF can induce G1 arrest in p53-deficient cells (41), these results indicate that co-expression of ARF and L11 can potentiate each other in inducing the p53-dependent G1 cell cycle arrest.
L11 Interacts with ARF in Cells and in Vitro
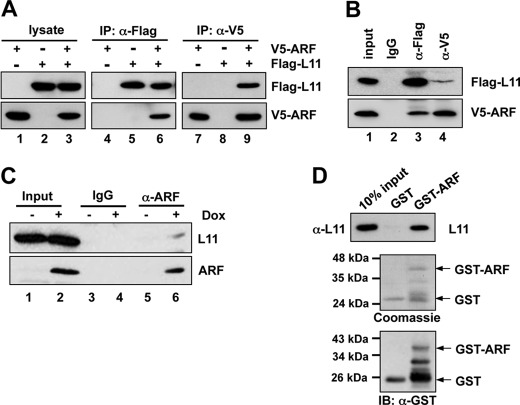
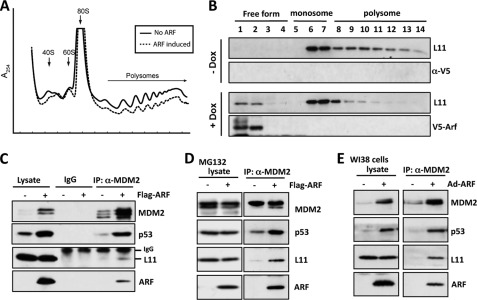
Because both ARF and L11 bind to MDM2, and both proteins cooperate with each other to regulate p53 activity (Fig. 1), we asked whether ARF and L11 also physically interact with each other. To test this idea, Flag-L11 and V5-ARF were expressed alone or together in p53-deficient H1299 cells, followed by co-IP using the anti-Flag or anti-V5 antibody. As shown in Fig. 2A, V5-ARF was specifically co-immunoprecipitated with Flag-L11 using the anti-Flag antibody when both plasmids were coexpressed in the H1299 cells (lane 6). Furthermore, Flag-L11 was specifically co-immunoprecipitated with V5-ARF by the anti-V5 antibody in cells that expressed both Flag-L11 and V5-ARF (lane 9). The interaction between ARF and L11 was specific as both proteins were not co-immunoprecipitated with control IgG (Fig. 2B, lane 2). To determine if endogenous L11 and ARF interact with each other, Soas2-Tet-E2F1 cells stably expressing the Tet-inducible E2F1 gene were analyzed. The expression of endogenous ARF was stimulated by doxycycline (Dox)-induced expression of E2F1 (17, 52). Cells cultured in the presence or absence of Dox were then subjected to co-IP using the anti-ARF antibody, or control IgG, followed by IB with the anti-L11 antibody. As shown in Fig. 2C, endogenous L11 was specifically immunoprecipitated by the anti-ARF antibody in Dox-treated cells (lane 6), but not in untreated cells (lane 5). To test whether the binding between L11 and ARF is direct, we performed GST-fusion protein-protein association assays. As shown in Fig. 2D, purified His-L11 was bound by purified GST-ARF protein, but not GST alone in vitro. These results demonstrate that L11 directly interacts with ARF.
FIGURE 2.
ARF interacts with L11 in cells and in vitro. A, ectopically expressed ARF interacts with ectopic L11 in cells. H1299 cells transfected with V5-ARF, Flag-L11, or both plasmids were subjected to co-IP with anti-Flag (lanes 4–6), or anti-V5 (lanes 7–9) antibodies followed by IB with anti-V5 or anti-Flag antibodies. B, specific interaction between L11 and ARF. H1299 cells transfected with V5-ARF and Flag-L11 were subjected to co-IP with anti-Flag, anti-V5, or control mouse IgG followed by IB with anti-Flag or anti-V5 antibodies. C, endogenous ARF interacts with endogenous L11 in cells. Saos2-Tet-E2F1 cells were cultured either in the presence or absence of doxycycline (Dox) for 48 h. The cleared cell lysates were immunoprecipitated with monoclonal anti-ARF antibody or control mouse IgG, followed by IB with polyclonal anti-ARF and anti-L11 antibodies. D, L11 directly interacts with ARF in vitro. Bacterially purified GST or GST-ARF immobilized on glutathione beads was incubated with recombinant His-L11 protein. Bound L11 was detected using IB with anti-L11 antibody (top panel). The purified GST and GST-ARF proteins were shown in the bottom panels by Coomassie Blue staining and IB with anti-GST antibodies, respectively.
ARF Binds to the C-terminal Domain of L11 in Cells
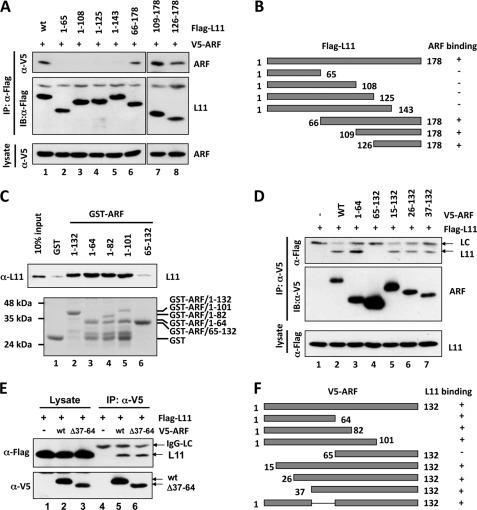
To define which portions of L11 bind ARF, we generated a panel of Flag-tagged L11 deletion mutants. H1299 cells were cotransfected with V5-ARF together with Flag-L11 or its deletion mutants. Cell lysates were then subjected to co-IP assays using anti-Flag antibodies. As shown in Fig. 3A, V5-ARF was co-immunoprecipitated with N-terminal deletion mutants L1166–178 (lane 6), L11109–178 (lane 7), and L11126–178 (lane 8), but not C-terminal deletion mutants L111–143, L111–125, L111–108, and L111–65 (lanes 2–5). These results, as summarized in Fig. 3B, indicate that the C-terminal residues of L11 (144–178) are essential for ARF-binding. It is noted that this C-terminal region of L11 is distinct from the central MDM2-binding domain of L11 (residues 65 to 125) (41), suggesting that ARF and MDM2 could bind simultaneously to L11.
FIGURE 3.
Mapping the binding domains between ARF and L11. A, ARF binds to the C-terminal domain of L11 in cells. H1299 cells were transfected with V5-ARF and Flag-tagged wild-type (wt) L11 or its deletion mutants as indicated. The cell lysates were immunoprecipitated with the anti-Flag antibody followed by IB with anti-V5 or anti-Flag antibodies (top panels). B, diagram of the Flag-L11 and its deletion mutants. The co-IP results determined in panel A are shown on the right. “+” indicates binding and “−” indicates lack of binding. C, L11 binds to the N-terminal domain of ARF in vitro. Bacterially purified GST, GST-ARF, or their deletion mutants immobilized on glutathione beads, were incubated with bacterially expressed His-L11. Bound L11 was detected using IB with anti-L11 antibody (top panel). The purified GST and GST-ARF proteins were shown in the bottom panel by Coomassie Blue staining. D and E, L11 interacts with the N-terminal domain of ARF in cells. H1299 cells were transfected with Flag-L11 together with V5-ARF, or its deletion mutants, as indicated. The cell lysates were immunoprecipitated with the anti-V5 antibody followed by IB with anti-V5 or anti-Flag antibodies. F, diagram of V5-ARF and its deletion mutants used in D and E. The co-IP results determined in panels D and E are shown on the right. “+” indicates binding and “−” indicates lack of binding.
L11 Binds to the N-terminal Domain of ARF in Vitro
To map the L11-binding domain in ARF, we first constructed a set of deletion mutants of ARF fused to GST. GST-ARF fusion proteins were then incubated with His-L11 purified from bacteria. As shown in Fig. 3C, GST-ARF mutants retaining the N-terminal 64 amino acid residues (lanes 3–5) bound L11 as efficiently as wild-type GST-ARF (lane 2). However, deletion of the N terminus of ARF (GST-ARF65–132) abolished its binding to L11 (lane 6), suggesting that L11 associates with the N-terminal domain of ARF. To further map the L11 binding sites at ARF, we constructed a panel of ARF mutants deleted with short fragments of the N terminus. H1299 cells were cotransfected with Flag-L11 and V5-ARF, or its deletion mutants, followed by co-IP assays using anti-V5 antibodies. As shown in Fig. 3D, Flag-L11 was co-immunoprecipitated with N-terminal deletion mutants ARF15–132 (lane 5), L1126–132 (lane 6), and ARF37–132 (lane 7), but not ARF65–132 (lane 4). Furthermore, deletion of amino acid residues 37–64 (ARFΔ37–64) did not abolish its association with L11 (Fig. 3E). These results, as summarized in Fig. 3F, indicate that at least two regions in the ARF N terminus (amino acid residues 1–36 and 37–64) bind to L11.
ARF and L11 Together Form a Ternary Complex with MDM2 in Cells
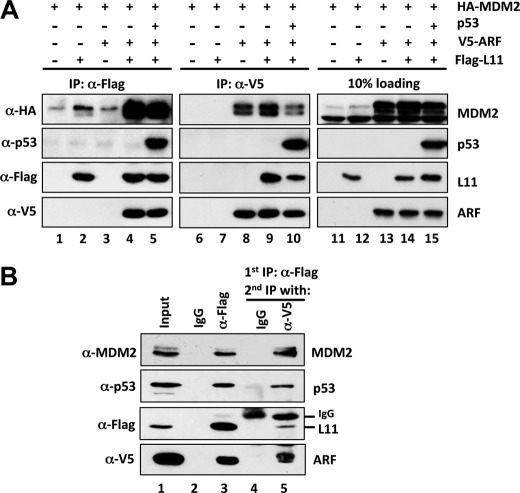
ARF (53) or L11 (36, 41) have been shown to suppress MDM2 function by forming a complex with MDM2 and p53. To determine whether ARF and L11 can simultaneously associate with MDM2 and p53, we performed a series of co-IP/IB assays using H1299 cells transfected with different combinations of plasmids encoding V5-ARF, Flag-L11, HA-MDM2, and p53. As shown in Fig. 4A, MDM2 associated with ARF or L11 individually, as expected (lanes 2 and 8). Coexpression of these three proteins resulted in a complex containing all of them as detected by reverse co-IP with antibodies against V5 or Flag (lanes 4 and 9). However the level of MDM2 in this ternary complex was remarkably increased in cells expressing all three proteins even though the levels of ARF or L11 were unchanged (compare lanes 2 and 8 with lanes 4 and 9). This result suggests that the presence of both L11 and ARF in the complex can enhance their association with MDM2. Furthermore, when p53 was coexpressed with ARF, L11, and MDM2, it formed a complex with L11, ARF, and MDM2 as determined by co-IP using anti-Flag or anti-V5 antibodies (lanes 5 and 10).
FIGURE 4.
ARF and L11 form a complex with MDM2 and p53. A, H1299 cells were transfected with various combinations of plasmids encoding Flag-L11, V5-ARF, HA-MDM2, and p53 as indicated. Cell lysates were immunoprecipitated with anti-Flag or anti-V5 antibody, followed by IB with antibodies as indicated. B, sequential co-IP of L11 and ARF with MDM2 and p53. H1299 cells transfected with Flag-L11, V5-ARF, HA-MDM2, and p53 plasmids were subjected to IPs with control mouse IgG (lane 2) or anti-Flag (M2) antibody (lane 3), followed by elution with Flag peptide. 10% of the elution was loaded (lanes 2 and 3, respectively). The remaining 90% of the anti-Flag elution was immunoprecipitated with anti-V5 antibody (lane 5) or control IgG (lane 4), followed by IB with the indicated antibodies.
To further examine whether L11, ARF, MDM2, and p53 form a four-part complex in cells, we performed sequential co-IP assays. H1299 cells transfected with Flag-L11, V5-ARF, HA-MDM2, and p53 plasmids were first subjected to IP with anti-Flag antibody, followed by elution with Flag peptide (Fig. 4B, lane 3). The elution containing Flag-L11-associated proteins was then immunoprecipitated with anti-V5 antibody (Fig. 4B, lane 5). Both MDM2 and p53 were co-immunoprecipitated in the first anti-Flag IP, as well as in the secondary anti-V5 IP, demonstrating that L11, ARF, MDM2, and p53 form a complex in cells. This is in agreement with the non-overlapping binding domains of these proteins (Fig. 3), as ARF binds to the acidic domain of MDM2 (13) whereas L11 associates with the zinc finger domain of MDM2 (41), MDM2 binds to the central domain (residues 66–125) of L11 (41), and ARF interacts with the C-terminal domain (residues 144–178) of L11 (Fig. 4).
Knockdown of Endogenous L11 Reduces ARF-mediated p53 Activation
It was reported that ARF is not required for L11-mediated p53 activation (54). To determine the role of L11 in ARF-mediated p53 activation, we addressed whether ablation of L11 by siRNA affects ARF-induced p53 level and activity in U2OS cells. As shown in Fig. 5A, overexpression of ARF induced the levels of p53 and its target gene p21. Knocking down endogenous L11 by either of the two different siRNAs reduced the ARF-induced levels of p53 and p21 (compare lanes 4 and 6 to lane 2). This effect is less likely an off-target effect, as similar results were observed using two different siRNAs against L11. Similarly, knockdown of L11 by lentiviral-based shRNAs also alleviated p53 induction by adenoviral-mediated expression of ARF in human normal fibroblast WI38 cells (Fig. 5B).
FIGURE 5.
Endogenous L11 contributes to ARF-responsive p53 activation. A, knockdown of L11 by siRNA reduced ARF-induced levels of p53. U2OS cells were transfected with Flag-ARF, or empty vector, followed by transfection with L11 siRNA or scrambled RNAs. Cells were harvested after 48 h and assayed for protein expression by IB with the indicated antibodies. B, ablation of endogenous L11 by siRNA reduced ARF-induced levels of p53 in WI38 cells. Cells were infected with control or adenoviruses encoding ARF, followed by infection with scrambled or L11 shRNA lentiviruses. Cells were harvested after 72 h and assayed for protein expression by IB with indicated antibodies. C and D, knockdown of L11 alleviated ARF-mediated p53 induction and G1 cell cycle arrest. U2OS-tet-V5-ARF cells were transfected with scrambled or L11 siRNA and cultured in the presence or absence of Dox for 48 h, followed by cell cycle analyses using PI staining. The mean percentage of cells in G0/G1, S, and G2/M cell cycle phases obtained from seven independent experiments (n = 7) is shown in C. *, p < 0.01, compared with cells transfected with scrambled RNA; **, p < 0.05, compared with cells transfected with scrambled RNA in the presence of Dox induction. The representative protein expression is shown in D.
To test whether knockdown of L11 also affects ARF-mediated cell cycle arrest, we generated a U2OS cell line with Tet-inducible expression of ARF (U2OS-Tet-V5-ARF). Dox-induced expression of ARF markedly suppressed cell proliferation by inducing both G1 and G2/M cell cycle arrest, which was partially suppressed by knockdown of L11 (Fig. 5C). These results suggest that endogenous L11 may play a role in ARF-mediated cell cycle arrest. Consistently, knocking down L11 reduced the levels of p53 and p21 upon Dox-induced ARF expression (Fig. 5D).
ARF Induces the Ribosome-free Form of L11
Recent studies have suggested that increased ARF expression can reduce ribosomal biogenesis and protein synthesis (49, 55–59). Consistent with these ideas, Dox-induced expression of ARF in U2OS cells significantly reduced the ratio of ploysome/monosome (1.42 ± 0.06 in control versus 0.96 ± 0.09 in Dox-induced cells, p < 0.01) in U2OS-Tet-V5-ARF cells (Fig. 6A), suggesting that ARF expression can adversely affect ribosomal biogenesis and/or protein synthesis, thus causing ribosomal stress. Upon ribosomal stress, RPs including L11 can be released as a ribosome-free form from the nucleolus to bind to MDM2, leading to p53 activation (25, 36, 41). To test whether L11 contributes to ARF-induced p53 activation via the L11-MDM2-p53 pathway, we examined whether ARF induces the ribosome-free form of L11 by performing polysome profile assays. Cytoplasmic extracts prepared from U2OS-Tet-V5-ARF cultured with or without Dox were subjected to a linear sucrose gradient sedimentation centrifugation. Fourteen fractions were collected and assayed for the level of L11 and ARF by IB. As shown in Fig. 6B, induced expression of ARF (+Dox) significantly increased the levels of ribosome-free L11 (fractions 1 and 2), whereas polysomes (fractions 8 through 14) were significantly reduced by ARF expression. ARF was not associated with either polysomes or monosomes in cytosolic extracts. These results suggest that ARF suppresses ribosomal biogenesis and causes the release of the free form of L11 from the ribosome.
FIGURE 6.
ARF increases non-ribosome-associated form of L11 and the interaction of L11 with MDM2 and p53 in cells. A and B, ARF suppresses ribosomal biogenesis and increases non-ribosome-associated form of L11. Cytoplasmic extracts containing polysomes from U2OS-tet-V5-ARF cells cultured in the presence or absence of Dox for 48 h were subjected to a 15 to 47% linear sucrose gradient sedimentation centrifugation. After centrifugation, the distribution of polysomes and monosomes was measured by recording the absorbance of RNA at 254 nm using an in-line UV monitor in a Biocomp Gradient Station (A). Additionally, fourteen fractions were collected, and 30 μl of each fraction was subjected to IB with anti-L11 or anti-ARF antibodies as indicated. The fractions containing polysomes, monosomes, and ribosome-free forms of L11 are indicated on the top (B). C and D, overexpression of ARF enhanced endogenous L11-MDM2-p53 interaction in U2OS cells. Cells transfected with Flag-ARF or empty vector were untreated (C) or treated with 20 μm MG132 (D) for 8 h before harvest. Cell lysates were immunoprecipitated with control IgG or anti-MDM2 (SMP14 and 4B11), followed by IB with polyclonal anti-p53, anti-MDM2 (2A10), anti-ARF, or anti-L11 antibodies, as indicated. E, overexpression of ARF enhanced endogenous L11-MDM2-p53 interaction in WI38 cells. Cells infected with Ad-ARF or control viruses were subjected to co-IP using anti-MDM2 (SMP14), followed by IB with polyclonal anti-p53, anti-MDM2 (2A10), anti-ARF, or anti-L11 antibodies.
ARF Enhances the MDM2-L11 Interaction in Cells
Because ribosomal stress induces and activates p53 via RP binding and inhibition of MDM2 (25, 26, 28, 34–42), we next addressed whether ARF may induce p53 activation, at least partially, by enhancing the association of L11 with MDM2, leading to more effective inhibition of MDM2. To test this idea, we transfected U2OS cells with the Flag-ARF plasmid and carried out co-IPs using the anti-MDM2 antibody or control IgG followed by IB. As shown in Fig. 6C, overexpression of ARF markedly increased the binding of endogenous MDM2 to endogenous L11 as well as p53. This enhanced association was not solely due to the elevated level of MDM2 that occurred with the overexpression of ARF, as the increased binding was also observed following ARF expression when the MDM2 level was equivalent following the treatment of the cells with the proteasome inhibitor MG132 (Fig. 6D). Similar enhanced interactions between L11 and MDM2 and p53 were also detected in human WI38 cells using adenovirus-mediated expression of ARF (Fig. 6E). These results suggest that ARF induces ribosomal stress and stimulates the association of L11 with MDM2.
DISCUSSION
It is well known that ARF binds to MDM2 and inhibits MDM2-mediated p53 ubiquitination and degradation, leading to p53 activation, in response to oncogenic stress (12, 15). Recently, we and others have found that a number of RPs, including L5, L11, L23, L26, S7, S27, and S27a, also suppress MDM2 function and subsequently activate p53 in response to ribosomal stress (25, 26, 28, 34–42). This study suggests that the oncogenic stress-induced ARF-MDM2-p53 and ribosomal stress-induced L11-MDM2-p53 pathways are physically and functionally connected. We show that L11 directly associates with ARF and forms a complex with MDM2-p53. Consequently, L11 collaborates with ARF in activating p53 (Fig. 1). Knockdown of L11 attenuated ARF-induced p53 stabilization and cell cycle arrest (Fig. 5). ARF is not required for L11-mediated p53 activation in response to ribosomal stress, as L11 is functional in inducing p53 in ARF-null U2OS cells (36, 41, 46, 60). Also, p19ARF is not required for the in vivo induction of p53 in response to ribosomal stress (54). However, our data indicates that L11 plays a role in ARF-induced p53 activation.
Several lines of evidence suggest that ARF suppresses ribosomal biogenesis. First, ARF inhibits ribosomal RNA processing (59) by binding to and inhibiting nucleophosmin (B23) (49). Second, ARF suppresses RNA polymerase (Pol) I activity through binding to the upstream binding factor (UBF) and inhibiting its phosphorylation (56). ARF also binds to the RNA Pol I transcription termination factor TTF1 and inhibits its nucleolar import, causing the accumulation of TTF1 in the nucleoplasm (57). Third, ARF suppresses RNA Pol III-mediated tRNA synthesis (58). Consistently, deletion of ARF increases rRNA synthesis, ribosomal nuclear export, protein translation, and consequent cell volume (55). Our data also suggest that overexpression of ARF represses ribosomal biogenesis and protein synthesis as viewed by the decrease in the ratio of polysome versus monosome (Fig. 6A).
Because inhibition of ribosomal biogenesis causes ribosomal stress, the role of L11 in ARF-mediated p53 activation could be explained by ribosomal stress-induction of the L11-MDM2-p53 pathway. Thus, ARF induces p53 activity by direct suppression of MDM2 and indirect induction of L11 alleviation of MDM2 repression of p53. Indeed, ARF expression reduced the levels of polysomes, increased the levels of ribosome-free L11 (Fig. 6A), and enhanced the interaction of L11 with MDM2 (Fig. 6, C–E). Therefore, our study provides evidence indicating that L11 contributes to oncogenic stress-induced p53 activation via responding to ARF-induced ribosomal stress (Fig. 7). Supporting our model, loss of the RP-MDM2 interaction via genetically introducing the L11- and L5-binding defective MDM2 mutant (MDM2C305F) gene significantly accelerated Eμ-Myc-induced lymphomagenesis in mice and reduces c-Myc-induced p53 induction in mouse embryonic fibroblast (MEF) cells (54).
FIGURE 7.
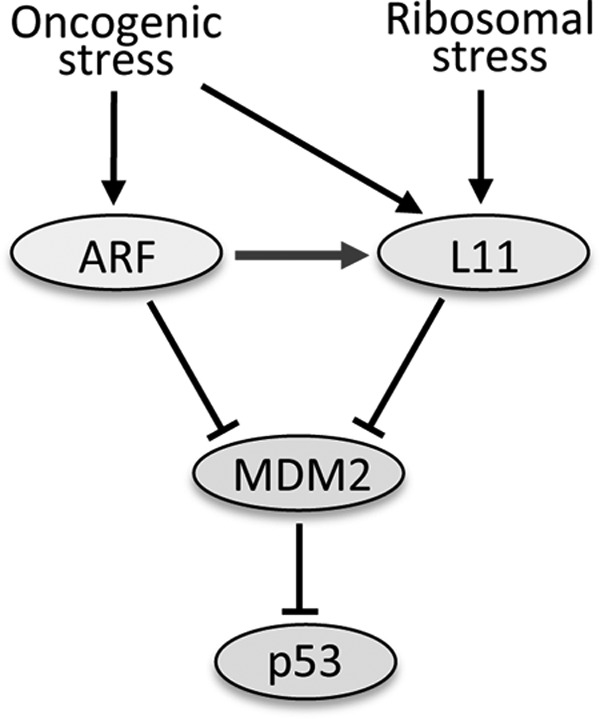
Model of the functional interaction of the L11-MDM2-p53 and the ARF-MDM2-p53 pathways in response to stress. Arrows indicate activation, whereas bars refer to inhibition in the signaling pathways regulating p53.
Our results also suggest that ARF enhances or stabilizes the L11-MDM2 interaction by forming a multiprotein complex with L11 and MDM2 (Fig. 4). This complex formation is supported by the finding that they bind synergistically at non-overlapping domains. ARF binds to the acidic domain of MDM2 (13), whereas L11 binds to the zinc finger domain of MDM2 (61, 62). ARF binds to the C terminus (Fig. 3A), whereas MDM2 binds to the central region of L11 (46). Furthermore, while MDM2 binds to residues 1–14 and 26–37 of the N terminus of ARF (63, 64), L11 binds to the residues 37–64, in addition to 1–37, of the N terminus of ARF (Fig. 3D). This binding arrangement would allow the formation of a stable ARF-L11-MDM2 complex. Thus, it is likely that upon oncogenic stress, ARF inhibits ribosomal biogenesis, which would subsequently result in accumulation of ribosome-free L11. The accumulated L11 would then bind to ARF and MDM2, forming a ternary complex that further restricts MDM2 activity toward p53 and consequently activating p53. In the future, it would be interesting to test whether other MDM2-binding RPs, such as L5, may also participate in this regulation.
Acknowledgments
We thank the members of the Lu laboratory for active discussion. We thank Dr. Charlie D. Lopez for providing reagents.
This work was supported, in whole or in part, by NCI, National Institutes of Health Grants CA93614, CA095441, and CA079721 (to H. L.), CA127134 (to M.-S. D.), and GM049164 (to R. C. W.).
- ARF
- alternative reading frame
- GFP
- green fluorescence protein
- Co-IP
- co-immunoprecipitation
- IB
- immunoblot
- RP
- ribosomal protein.
REFERENCES
- 1. Soussi T., Dehouche K., Béroud C. (2000) p53 website and analysis of p53 gene mutations in human cancer: forging a link between epidemiology and carcinogenesis. Hum. Mutat. 15, 105–113 [DOI] [PubMed] [Google Scholar]
- 2. Vogelstein B., Lane D., Levine A. J. (2000) Surfing the p53 network. Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 3. Oren M. (2003) Decision making by p53: life, death and cancer. Cell Death Differ. 10, 431–442 [DOI] [PubMed] [Google Scholar]
- 4. Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J. (1992) The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 5. Wu X., Bayle J. H., Olson D., Levine A. J. (1993) The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 7, 1126–1132 [DOI] [PubMed] [Google Scholar]
- 6. Picksley S. M., Lane D. P. (1993) The p53-mdm2 autoregulatory feedback loop: a paradigm for the regulation of growth control by p53? Bioessays 15, 689–690 [DOI] [PubMed] [Google Scholar]
- 7. Barak Y., Juven T., Haffner R., Oren M. (1993) mdm2 expression is induced by wild type p53 activity. EMBO J. 12, 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Banin S., Moyal L., Shieh S., Taya Y., Anderson C. W., Chessa L., Smorodinsky N. I., Prives C., Reiss Y., Shiloh Y., Ziv Y. (1998) Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281, 1674–1677 [DOI] [PubMed] [Google Scholar]
- 9. Canman C. E., Lim D. S., Cimprich K. A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M. B., Siliciano J. D. (1998) Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281, 1677–1679 [DOI] [PubMed] [Google Scholar]
- 10. Maya R., Balass M., Kim S. T., Shkedy D., Leal J. F., Shifman O., Moas M., Buschmann T., Ronai Z., Shiloh Y., Kastan M. B., Katzir E., Oren M. (2001) ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 15, 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Siliciano J. D., Canman C. E., Taya Y., Sakaguchi K., Appella E., Kastan M. B. (1997) DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 11, 3471–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zindy F., Eischen C. M., Randle D. H., Kamijo T., Cleveland J. L., Sherr C. J., Roussel M. F. (1998) Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12, 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Y., Xiong Y. (2001) Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ. 12, 175–186 [PubMed] [Google Scholar]
- 14. Zhang Y., Xiong Y., Yarbrough W. G. (1998) ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92, 725–734 [DOI] [PubMed] [Google Scholar]
- 15. Palmero I., Pantoja C., Serrano M. (1998) p19ARF links the tumour suppressor p53 to Ras. Nature 395, 125–126 [DOI] [PubMed] [Google Scholar]
- 16. Zhang Y., Lu H. (2009) Signaling to p53: ribosomal proteins find their way. Cancer Cell 16, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bates S., Phillips A. C., Clark P. A., Stott F., Peters G., Ludwig R. L., Vousden K. H. (1998) p14ARF links the tumour suppressors RB and p53. Nature 395, 124–125 [DOI] [PubMed] [Google Scholar]
- 18. de Stanchina E., McCurrach M. E., Zindy F., Shieh S. Y., Ferbeyre G., Samuelson A. V., Prives C., Roussel M. F., Sherr C. J., Lowe S. W. (1998) E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 12, 2434–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Damalas A., Kahan S., Shtutman M., Ben-Ze'ev A., Oren M. (2001) Deregulated β-catenin induces a p53- and ARF-dependent growth arrest and cooperates with Ras in transformation. EMBO J. 20, 4912–4922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Honda R., Yasuda H. (1999) Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 18, 22–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weber J. D., Taylor L. J., Roussel M. F., Sherr C. J., Bar-Sagi D. (1999) Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1, 20–26 [DOI] [PubMed] [Google Scholar]
- 22. Tao W., Levine A. J. (1999) P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl. Acad. Sci. U.S.A. 96, 6937–6941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y., Xiong Y. (1999) Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol. Cell 3, 579–591 [DOI] [PubMed] [Google Scholar]
- 24. Lowe S. W., Sherr C. J. (2003) Tumor suppression by Ink4a-Arf: progress and puzzles. Curr. Opin. Genet. Dev. 13, 77–83 [DOI] [PubMed] [Google Scholar]
- 25. Bhat K. P., Itahana K., Jin A., Zhang Y. (2004) Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 23, 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dai M. S., Lu H. (2004) Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 279, 44475–44482 [DOI] [PubMed] [Google Scholar]
- 27. Dai M. S., Sun X. X., Lu H. (2008) Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol. Cell Biol. 28, 4365–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dai M. S., Zeng S. X., Jin Y., Sun X. X., David L., Lu H. (2004) Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell Biol. 24, 7654–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fumagalli S., Di Cara A., Neb-Gulati A., Natt F., Schwemberger S., Hall J., Babcock G. F., Bernardi R., Pandolfi P. P., Thomas G. (2009) Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat. Cell Biol. 11, 501–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun X. X., Dai M. S., Lu H. (2007) 5-Fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J. Biol. Chem. 282, 8052–8059 [DOI] [PubMed] [Google Scholar]
- 31. Sun X. X., Dai M. S., Lu H. (2008) Mycophenolic acid activation of p53 requires ribosomal proteins L5 and L11. J. Biol. Chem. 283, 12387–12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun X. X., Wang Y. G., Xirodimas D. P., Dai M. S. (2010) Perturbation of 60 S ribosomal biogenesis results in ribosomal protein L5- and L11-dependent p53 activation. J. Biol. Chem. 285, 25812–25821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yuan X., Zhou Y., Casanova E., Chai M., Kiss E., Gröne H. J., Schütz G., Grummt I. (2005) Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol. Cell 19, 77–87 [DOI] [PubMed] [Google Scholar]
- 34. Chen D., Zhang Z., Li M., Wang W., Li Y., Rayburn E. R., Hill D. L., Wang H., Zhang R. (2007) Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene 26, 5029–5037 [DOI] [PubMed] [Google Scholar]
- 35. Jin A., Itahana K., O'Keefe K., Zhang Y. (2004) Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell Biol. 24, 7669–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lohrum M. A., Ludwig R. L., Kubbutat M. H., Hanlon M., Vousden K. H. (2003) Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3, 577–587 [DOI] [PubMed] [Google Scholar]
- 37. Ofir-Rosenfeld Y., Boggs K., Michael D., Kastan M. B., Oren M. (2008) Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol. Cell 32, 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sun X. X., DeVine T., Challagundla K. B., Dai M. S. (2011) Interplay between ribosomal protein S27a and MDM2 protein in p53 activation in response to ribosomal stress. J. Biol. Chem. 286, 22730–22741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xiong X., Zhao Y., He H., Sun Y. (2011) Ribosomal protein S27-like and S27 interplay with p53-MDM2 axis as a target, a substrate and a regulator. Oncogene 30, 1798–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Y., Wang J., Yuan Y., Zhang W., Guan W., Wu Z., Jin C., Chen H., Zhang L., Yang X., He F. (2010) Negative regulation of HDM2 to attenuate p53 degradation by ribosomal protein L26. Nucleic Acids Res. 38, 6544–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Y., Wolf G. W., Bhat K., Jin A., Allio T., Burkhart W. A., Xiong Y. (2003) Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell Biol. 23, 8902–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhu Y., Poyurovsky M. V., Li Y., Biderman L., Stahl J., Jacq X., Prives C. (2009) Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol. Cell 35, 316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Olson M. O., Hingorani K., Szebeni A. (2002) Conventional and nonconventional roles of the nucleolus. Int. Rev. Cytol 219, 199–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Raska I., Koberna K., Malinský J., Fidlerová H., Masata M. (2004) The nucleolus and transcription of ribosomal genes. Biol. Cell 96, 579–594 [DOI] [PubMed] [Google Scholar]
- 45. Xirodimas D., Saville M. K., Edling C., Lane D. P., Laín S. (2001) Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene 20, 4972–4983 [DOI] [PubMed] [Google Scholar]
- 46. Dai M. S., Shi D., Jin Y., Sun X. X., Zhang Y., Grossman S. R., Lu H. (2006) Regulation of the MDM2-p53 pathway by ribosomal protein L11 involves a post-ubiquitination mechanism. J. Biol. Chem. 281, 24304–24313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen D., Padiernos E., Ding F., Lossos I. S., Lopez C. D. (2005) Apoptosis-stimulating protein of p53–2 (ASPP2/53BP2L) is an E2F target gene. Cell Death Differ. 12, 358–368 [DOI] [PubMed] [Google Scholar]
- 48. Dai M. S., Arnold H., Sun X. X., Sears R., Lu H. (2007) Inhibition of c-Myc activity by ribosomal protein L11. EMBO J. 26, 3332–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Itahana K., Bhat K. P., Jin A., Itahana Y., Hawke D., Kobayashi R., Zhang Y. (2003) Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell 12, 1151–1164 [DOI] [PubMed] [Google Scholar]
- 50. He H., Sun Y. (2007) Ribosomal protein S27L is a direct p53 target that regulates apoptosis. Oncogene 26, 2707–2716 [DOI] [PubMed] [Google Scholar]
- 51. Kamijo T., Weber J. D., Zambetti G., Zindy F., Roussel M. F., Sherr C. J. (1998) Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sci. U.S.A. 95, 8292–8297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Irwin M., Marin M. C., Phillips A. C., Seelan R. S., Smith D. I., Liu W., Flores E. R., Tsai K. Y., Jacks T., Vousden K. H., Kaelin W. G., Jr. (2000) Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407, 645–648 [DOI] [PubMed] [Google Scholar]
- 53. Sherr C. J. (2001) The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell Biol. 2, 731–737 [DOI] [PubMed] [Google Scholar]
- 54. Macias E., Jin A., Deisenroth C., Bhat K., Mao H., Lindström M. S., Zhang Y. (2010) An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 interaction. Cancer Cell 18, 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Apicelli A. J., Maggi L. B., Jr., Hirbe A. C., Miceli A. P., Olanich M. E., Schulte-Winkeler C. L., Saporita A. J., Kuchenreuther M., Sanchez J., Weilbaecher K., Weber J. D. (2008) A non-tumor suppressor role for basal p19ARF in maintaining nucleolar structure and function. Mol. Cell Biol. 28, 1068–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ayrault O., Andrique L., Fauvin D., Eymin B., Gazzeri S., Séité P. (2006) Human tumor suppressor p14ARF negatively regulates rRNA transcription and inhibits UBF1 transcription factor phosphorylation. Oncogene 25, 7577–7586 [DOI] [PubMed] [Google Scholar]
- 57. Lessard F., Morin F., Ivanchuk S., Langlois F., Stefanovsky V., Rutka J., Moss T. (2010) The ARF tumor suppressor controls ribosome biogenesis by regulating the RNA polymerase I transcription factor TTF-I. Mol. Cell 38, 539–550 [DOI] [PubMed] [Google Scholar]
- 58. Morton J. P., Kantidakis T., White R. J. (2007) RNA polymerase III transcription is repressed in response to the tumour suppressor ARF. Nucleic Acids Res. 35, 3046–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sugimoto M., Kuo M. L., Roussel M. F., Sherr C. J. (2003) Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol. Cell 11, 415–424 [DOI] [PubMed] [Google Scholar]
- 60. Park Y. B., Park M. J., Kimura K., Shimizu K., Lee S. H., Yokota J. (2002) Alterations in the INK4a/ARF locus and their effects on the growth of human osteosarcoma cell lines. Cancer Genet. Cytogenet 133, 105–111 [DOI] [PubMed] [Google Scholar]
- 61. Lindström M. S., Jin A., Deisenroth C., White Wolf G., Zhang Y. (2007) Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol. Cell Biol. 27, 1056–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang Q., Xiao H., Chai S. C., Hoang Q. Q., Lu H. (2011) Hydrophilic residues are crucial for ribosomal protein L11 (RPL11) interaction with zinc finger domain of MDM2 and p53 protein activation. J. Biol. Chem. 286, 38264–38274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Clark P. A., Llanos S., Peters G. (2002) Multiple interacting domains contribute to p14ARF-mediated inhibition of MDM2. Oncogene 21, 4498–4507 [DOI] [PubMed] [Google Scholar]
- 64. Weber J. D., Kuo M. L., Bothner B., DiGiammarino E. L., Kriwacki R. W., Roussel M. F., Sherr C. J. (2000) Cooperative signals governing ARF-mdm2 interaction and nucleolar localization of the complex. Mol. Cell Biol. 20, 2517–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Palam L. R., Baird T. D., Wek R. C. (2011) Phosphorylation of eIf2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 286, 10939–10949 [DOI] [PMC free article] [PubMed] [Google Scholar]