Background: Rbx1/ROC1 is an E3 ligase adaptor protein that functions with the E2 enzyme CDC34.
Results: NMR and biochemical data show that Rbx1/ROC1 binds CDC34∼ubiquitin 50-fold tighter than CDC34.
Conclusion: Rbx1/ROC1 selectively recruits E2∼ubiquitin and releases the E2 after ubiquitin transfer.
Significance: Direct evidence is shown for preferential recognition of an E2∼ubiquitin complex by an E3 ligase.
Keywords: E3 Ubiquitin Ligase, Enzyme Mechanisms, Protein Complexes, Structural Biology, Ubiquitin Conjugating Enzyme (Ubc), Ubiquitination
Abstract
RING E3 ligases are proteins that must selectively recruit an E2-conjugating enzyme and facilitate ubiquitin transfer to a substrate. It is not clear how a RING E3 ligase differentiates a naked E2 enzyme from the E2∼ubiquitin-conjugated form or how this is altered upon ubiquitin transfer. RING-box protein 1 (Rbx1/ROC1) is a key protein found in the Skp1/Cullin-1/F-box (SCF) E3 ubiquitin ligase complex that functions with the E2 ubiquitin conjugating enzyme CDC34. The solution structure of Rbx1/ROC1 revealed a globular RING domain (residues 40–108) stabilized by three structural zinc ions (root mean square deviation 0.30 ± 0.04 Å) along with a disordered N terminus (residues 12–39). Titration data showed that Rbx1/ROC1 preferentially recruits CDC34 in its ubiquitin-conjugated form and favors this interaction by 50-fold compared with unconjugated CDC34. Furthermore, NMR and biochemical assays identified residues in helix α2 of Rbx1/ROC1 that are essential for binding and activating CDC34∼ubiquitin for ubiquitylation. Taken together, this work provides the first direct structural and biochemical evidence showing that polyubiquitylation by the RING E3 ligase Rbx1/ROC1 requires the preferential recruitment of an E2∼ubiquitin complex and subsequent release of the unconjugated E2 protein upon ubiquitin transfer to a substrate or ubiquitin chain.
Introduction
The intracellular destiny of many proteins in the cell is governed by their post-translational modification with the covalent attachment of ubiquitin. This process, known as ubiquitylation, plays a pivotal role in numerous cellular processes including protein turnover, cell cycle progression, transcriptional regulation, DNA repair, and signal transduction (1). Ubiquitylation involves the sequential transfer of a ubiquitin molecule through an enzyme cascade consisting of a ubiquitin activating enzyme (E1), a ubiquitin conjugating enzyme (E2), and a ubiquitin ligase (E3) until it forms an isopeptide bond between the C terminus of ubiquitin and the ϵ-amino group of a lysine on a substrate protein. The E2-E3 combination governs the specificity of the target protein for modification and the site of attachment to the substrate protein as well as the chain length and type of linkage between the ubiquitin molecules attached (2).
The really interesting new gene (RING) domain-containing E3 ligases function primarily as scaffolds that orient the E2∼ubiquitin thiol ester complex as well as the substrate protein for efficient ubiquitin transfer. To date, >650 different human RING E3 ligases have been identified (2). The cullin-RING-ligases (CRLs)3 comprise the largest superfamily of these enzymes and consist of a cullin scaffold (CUL1, CUL2, CUL3, CUL4A, CUL4B, CUL5, or CUL7) with a RING-containing catalytic protein, either RING-box protein 1 (Rbx1, also referred to as ROC1 and Hrt1), or RING-box protein 2 (Rbx2, aka ROC2/Hrt2) bound at its C-terminal catalytic core (3–5). The Rbx1/ROC1 or Rbx2/ROC2 proteins are responsible for recognizing and recruiting the E2 conjugating enzyme during ubiquitin transfer to a target protein. Crystal structures of multiple CRLs demonstrate they form elongated structures with a long stalk-like N-terminal domain that binds to an adaptor (such as Skp1; CUL3 is the only exception to this rule) and various substrate recognition factors (such as F-box proteins) to recognize different substrate proteins (6–9), whereas the C terminus forms a globular α/β domain that binds to the Rbx/ROC protein through an intermolecular β-sheet (7). Recent structural and biochemical studies show that the covalent attachment of the ubiquitin-like modifier protein NEDD8 to a conserved lysine residue near the C terminus of the cullin scaffold protein causes a conformational change in the C-terminal domain of CUL1 liberating the RING-domain of Rbx1/ROC1 (9). The increased mobility of Rbx1/ROC1 is believed to enhance its rate of ubiquitin transfer from the E2 to the substrate protein (10, 11). This modulation of Rbx1/ROC1-based E3 ligases by cullin and NEDD8 is unique among the RING E3 ligases and represents one of the most intriguing and important regulation steps in the ubiquitination process.
The best characterized CRL E3 complex to date both biochemically and structurally is the Skp1/Cullin 1/F-box (SCF) ligase complex, which consists of Rbx1/ROC1 bound to the CUL1 catalytic C-terminal domain. UbcH5 and CDC34 (cell division cycle protein 34) are the two E2 enzymes known to directly interact and function with the SCF in humans (12) with each being responsible for different facets in ubiquitylation on a target protein; UbcH5 attaches the initial ubiquitin to the substrate, whereas CDC34 is responsible for the successive addition of ubiquitin molecules to the substrate during polyubiquitin chain formation (13). A recent structural and functional study demonstrated that CDC34 can be allosterically inhibited by a small molecule that impedes the release of its ubiquitin cargo at the E2-E3 step (14), demonstrating that inhibition of CDC34-dependent polyubiquitylation might represent a possible step for therapeutic intervention. The acidic C terminus of CDC34 has recently been shown to interact with ubiquitin (15) as well as the C-terminal domain of CUL1 (16, 17). Although these observations suggest that the C terminus of CDC34 is important for bringing CDC34 and the SCF into closer proximity, the structural basis for the functional interaction between the catalytic domain of CDC34 and Rbx1/ROC1 remains unclear. Determination of the structural rationale for the enhanced activity of CDC34 bound to the RING E3 adaptor protein Rbx1/ROC1 would help to further clarify the mechanism of processive ubiquitination for this E2-E3 complex.
In this study the solution structure of Rbx1/ROC1 in the absence of its cullin scaffold was determined. The structure shows that the N terminus of Rbx1/ROC1 undergoes a large structural rearrangement from a disordered conformation to a structured β-strand in the CRL complex. Furthermore, we used NMR spectroscopy to show that Rbx1/ROC1 displays poor affinity for CDC34 alone that is increased by more than 50-fold for CDC34 in its ubiquitin-loaded form. Using biochemical reconstitution assays, we demonstrate that the cullin-free Rbx1/ROC1 is capable of activating the catalytic core of CDC34. We also identify key residues in helix α2 of Rbx1/ROC1 that are important for binding and activating CDC34∼ubiquitin. This study provides the first structural details showing that the CDC34∼ubiquitin complex is recruited by Rbx1/ROC1 and functions within the sequential model for polyubiquitylation (18, 19).
EXPERIMENTAL PROCEDURES
Cloning of Human sRbx112–108
The open reading frame of Rbx1/ROC1 was PCR-amplified from pcDNA3-Myc3-ROC1 (Addgene, Cambridge MA) with flanking NdeI and HindIII restriction sites. Primers used for cloning full-length Rbx1/ROC1 were Rbx1_fr (5′-AATCTAGACATATGGCGGCAGCGATGGATGTGGATACCC-3′) and Rbx1_rv (5′-AACTCGAGAAGCTTTTATTACTAGTGCCCATACTTTTGG-3′). The resulting PCR product was then cloned into pET28a that contained an N-terminal 6-histidine tag followed by a TEV cleavage site (MGSSHHHHHHSSGGRENLYFQ) to facilitate the removal of the tag during purification. This initial construct was then shortened from residues 1–108 to 12–108 and 36–108 using site-directed mutagenesis based upon the observed residues in the crystal structure of CUL1-Rbx1/ROC1, PDB code 1LDJ (7). The designed Rbx1/ROC112–108 and Rbx1/ROC136–108 constructs also had three extra glycine residues added between the TEV cleavage site and G12 to improve flexibility for TEV cleavage. The primers used to make Rbx1 12–108 were Rbx1d12fr (5′-AACTTGTATTTCCAGGGCGGCGGCGGCACCAACAGCGGCG-3′) and Rbx1d12rv (5′-CGCCGCTGTTGGTGCCGCCGCCGCCCTGGAAATACAAGTT-3′. The primers used to make Rbx1/ROC136–108 were Rbx1d36fr (5′-AACTTGTATTTCCAGGGCGGCGGCGGCGATATTGTGGTTGATAACTG-3′) and Rbx1d36rv (5′-CAGTTATCAACCACAATATCGCCGCCGCCGCCCTGGAAATACAAGTT3′). To improve the solubility of Rbx1/ROC112–108, four mutations (W27S, V30S, L32Q, and W33S) were incorporated into the N-terminal region of Rbx1/ROC1 that was predicted to have aggregation propensity by the online algorithm TANGO (20–22). These mutations were made using the primers Rbx1SSQSfr (5′-CGCTTTGAAGTGAAAAAGAGTAATGCAAGTGCTCAGAGTGCCTGGGATATTGTGG-3′) and Rbx1SSQSrv (5′-CCACAATATCCCAGGCACTCTGAGCACTTGCATTACTCTTTTTCACTTCAAAGCG-3′).The resulting vectors, pET28a-HisTEV-sRbx112–108 (where “sRbx1” stands for solubilized Rbx1) and pET28a-HisTEV-Rbx136–108, were verified by DNA sequencing (Robarts Research Institute, University of Western Ontario).
Site-directed mutagenesis was used to substitute Trp-87, Lys-89, and Lys-91 to alanines using the following primers: for W87A, Rbx1W87Afr (5′-CCACTGCATCTCTCGGGCACTCAAAACACGACAGG-3′) and Rbx1W87Arv (5′-CCTGTCGTGTTTTGAGTGCCCGAGAGATGCAGTGG-3′); for K89A, Rbx1K89Afr (5′-GCATCTCTCGCTGGCTCGCAACACGACAGGTGTGTCC-3′) and Rbx1K89Arv (5′-GGACACACCTGTCGTGTTGCGAGCCAGCGAGAGATGC-3′); for K89A/R91A, Rbx1K89R91fr (5′-CATCTCTCGCTGGCTCGCAACCGCACAGGTGTGTCCATTGG-3′) and Rbx1K89R91rv (5′-CCAATGGACACACCTGTGCGGTTGCGAGCCAGCGAGAGATG-3′); for R91A, Rbx1R91Afr (5′-CTCGCTGGCTCAAAACAGCACAGGTGTGTCCATTGG-3′)and Rbx1R91Arv (5′-CCAATGGACACACCTGTGCTGTTTTGAGCCAGCGAG-3′). The resulting vectors coding for sRbx112–108 W87A, K89A, K89A/R91A, and R91A were verified by DNA sequencing.
Expression and Purification sRbx12–108, CDC34, and CDC34-Ubiquitin Complexes
To express unlabeled sRbx112–108 protein (where “s” stands for soluble Rbx112–108 with solubilizing mutations), a 10-ml overnight culture of BL21(DE3)-RIL transformed with pET28a-HisTEV-sRbx112–108 was used to inoculate 1 liter of prewarmed LB media supplemented with 30 μg/ml kanamycin and 34 μg/ml chloramphenicol. To prepare 13C,15N-labeled sRbx112–108, cells were grown in 1 liter of M9 minimal media supplemented with 15NH4Cl (1 g/liter) alone or with [13C6]glucose (2 g/liter) in the presence of 30 μg/ml kanamycin and 34 μg/ml chloramphenicol. The cultures were grown at 37 °C with shaking at 200 rpm. At an A600 of 0.5, the unlabeled and 15N- or 13C,15N-labeled growth was supplemented with 300 μm ZnCl2. Once the cells reached an A600 of 0.7, the cultures were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside at 16 °C for 20 h. Cells were then harvested by centrifugation at 6000 × g for 15 min at 4 °C.
The cell pellets for unlabeled and 13C,15N-labeled sRbx112–108 were resuspended in 10 ml of wash buffer (50 mm Na2HPO4, 300 mm NaCl, 10 mm imidazole (pH 8.0)) with 1 mm PMSF and an EDTA-free protease inhibitor tablet (Roche Applied Science). The resuspended cells were then sonicated and centrifuged at 38,000 rpm for 1 h at 4 °C. The supernatant was then syringe-filtered through a 0.45-mm filter (Millipore, Mississauga, ON, Canada) and loaded onto a 5-ml HisTrap column (GE Healthcare) pre-equilibrated with wash buffer at a flow rate of 0.5 ml/min using an AKTA FPLC (GE Healthcare). After the column was washed with wash buffer containing 30 mm imidazole for 15 column volumes at 3 ml/min, the protein was eluted with elution buffer (50 mm Na2HPO4, 300 mm NaCl, 250 mm imidazole, pH 8.0) at a flow rate of 2 ml/min. Fractions containing eluted sRbx112–108 protein were pooled, TEV protease was added to cleave the His tag, and the protein was dialyzed against wash buffer overnight at 4 °C. The TEV-cleaved sRbx112–108 protein was then reloaded onto the HisTrap column at a flow rate of 1 ml/min. The flow-through containing purified sRbx112–108 was pooled and dialyzed against Rbx1 dialysis buffer (20 mm Na2HPO4, 100 mm NaCl, 1 mm DTT, pH 7.5, or 6.0) in preparation for NMR experiments. The resulting sRbx112–108 contained an additional GGG at its N termini as a result of its cloning and TEV cleavage. After purification, the concentration of sRbx112–108 was determined by the better Bradford method (Bio-Rad). Purified sRbx112–108 was also submitted for ICP-AES Zn2+ analysis (Laboratory for Geochemical Analysis, University of Western Ontario) to confirm that three zinc ions were bound per sRbx112–108 molecule, similar to a previous study (23).
Unlabeled full-length CDC34 with C191S/C223S substitutions and ubiquitin K48R/G76C (UbCys) were expressed and purified as previously described (15). The CDC34-UbCys thiol ester mimetic complex was formed using full-length CDC34 disulfide linked to UbCys, as previously described (15, 24, 25). After purification, CDC34 and CDC34-UbCys were dialyzed against 20 mm Na2HPO4, 100 mm NaCl, pH 7.5, in preparation for interaction studies with Rbx1/ROC1.
NMR Spectroscopy and Structure Calculations for sRbx112–108
NMR samples for assignment and structure calculation of sRbx112–108 were prepared in 20 mm Na2HPO4, 100 mm NaCl, 1 mm DTT, 10% D2O, 90% H2O at pH 7.5. 13C,15N-labeled sRbx112–108 was concentrated to 700 μm by ultrafiltration (Millipore) in a volume of 300 μl and transferred into a Shigemi tube.
All NMR data were collected at 25 °C using a Varian Inova 600-MHz NMR spectrometer equipped with a triple resonance probe and z-field gradients. Backbone and side chain assignments for sRbx112–108 were determined from the following experiments collected using the standard pulse sequences from the Varian Biopack: 1H,15N HSQC (26), HNCO, HNCA, HNHA, HNCACB (27), CBCA(CO)NH (28), HCC(CO)NH, C(CO)NH, and HCCH-TOCSY spectroscopy (29). The 15N-NOESY-HSQC experiment was collected with a mixing time of 150 ms. After exchanging the sample into 100% D2O, 13C-NOESY-HSQC aliphatic and aromatic experiments were collected using a mixing time of 100 ms. All of the three-dimensional experiments were collected with 16–32 transients.
Heteronuclear 15N{1H} NOEs for sRbx112–108 were measured in 20 mm Na2HPO4, 100 mm NaCl, 1 mm DTT, 10% D2O, 90% H2O at pH 6.0 as previously described (15, 30, 31) using a 3-s irradiation period and 2-s relaxation delay (NOE) or a 5-s relaxation delay (no NOE).
Data processing was performed using NMRPipe and NMRDraw (32). NMRViewJ (33) was used for manual spectral analysis and resonance assignment. 2,2-Dimethyl-2-silapentane-5-sulfonate sodium salt (DSS) was used as the internal standard with 1H chemical shifts referenced at 0 ppm, whereas the 13C and 15N chemical shifts were indirectly referenced to DSS, as previously described (34). The NMR assignments for sRbx112–108 have been deposited in the BioMagResBank and assigned the identifier BMRB 17824.
Structures were determined using a combination of manually assigned NOEs and automatic NOE assignment with the program CYANA (35). Seven iterations of refinement of 100 structures per cycle were completed by using distance calibrations and parameters as previously described (36). After each cycle of refinement, the 20 structures with the lowest calculated target function were used to seed the subsequent rounds of structure calculations and were used in automated NOE assignment by CYANA. Dihedral angular restraints (ϕ,ψ) were determined by inputting the Hα, Cα, Cβ, and C′ chemical shift assignments of sRbx112–108 into TALOS+ (37). Cysteine residues in sRbx112–108 involved in Zn2+ coordination were identified using their Cα and Cβ chemical shifts (38). After the initial fold of the protein was determined, a modified CYANA amino acid library that included Zn2+-ligated cysteine residues with S-Zn2+ distance constraints was used to incorporate the three Zn2+ ions into the subsequent sRbx112–108 structure calculations, as previously described (39). The final 20 structures were subjected to water refinement using the RECOORD protocols for Xplor-NIH (40, 41) and were chosen as representative of the calculation. The atomic coordinates for sRbx112–108 were deposited in the Protein Data Bank and assigned the identifier PDB code 2LGV.
IκBα-Ubiquitin Elongation Assay
The elongation assay required the assembly of two reaction mixtures that contained CDC34∼ubiquitin and Nedd8-SCFβTrCP-IκBα-ubiquitin, respectively. The CDC34∼ubiquitin charge mix (5 μl) contained 50 mm Tris-HCl (pH 7.4), 5 mm MgCl2, 0.5 mm DTT, 2 mm ATP, 0.1 mg/ml of BSA, 2 mm NaF, 10 nm okadaic acid, 60 μm ubiquitin, 0.1 μm E1 enzyme, and CDC34 in concentrations as indicated. The mixture was incubated for 5 min at 37 °C before mixing with the Nedd8-SCFβTrCP-IκBα-ubiquitin complex.
The Nedd8-SCFβTrCP-IκBα-ubiquitin complex was assembled in sequential steps. First, Rbx1/ROC1-CUL1 was neddylated in a reaction mixture (3 μl) containing 50 mm Tris-HCl (pH 7.4), 5 mm MgCl2, 0.5 mm DTT, 2 mm ATP, 0.1 mg/ml of BSA, 2 mm NaF, 10 nm okadaic acid, 1.5 pmol of Rbx1/ROC1-CUL1, 20 μm Nedd8, 83 nm Nedd8 E1 (APPBP1/Uba3), and 15 μm Ubc12. This mixture was incubated at room temperature for 10 min. βTrCP/Skp1 (3 pmol) was then added to the mix, and the mixture was incubated at room temperature for 10 min to allow for the assembly of Nedd8-SCFβTrCP. 32P-Labeled IκBα-Ub (1 pmol), prepared as described previously (13), was added and incubated for an additional 10 min at room temperature to form the Nedd8-SCFβTrCP-IκBα-ubiquitin complex. The final volume was adjusted to 5 μl.
The ubiquitylation reaction was initiated by mixing CDC34∼ubiquitin and Nedd8-SCFβTrCP-IκBα-ubiquitin and incubating the mixture for 10 min at 37 °C. To assess the effects of CDC34-UbCys, CDC34-UbCys, in concentrations as indicated, was added to the CDC34 (1.3 μm)∼ubiquitin containing mix that had already been preincubated for 5 min at 37 °C. The final volume was adjusted to 6 μl. This mixture was then combined with Nedd8-SCFβTrCP-IκBα-Ub to a final volume of 10 μl and incubated at 37 °C for 10 min. The reaction products were separated by 4–20% SDS-PAGE, visualized by autoradiogram, and quantified by phosphor-imager analysis.
Interaction of Rbx1/ROC1 with CDC34 and CDC34-UbCys Examined by NMR Spectroscopy
Sensitivity-enhanced 1H,15N HSQC spectra (26) were recorded at 30 °C for 15N-labeled sRbx112–108 (50 μm) in the presence of unlabeled CDC34 (100 μm), in the presence of 1 mm DTT and unlabeled CDC34-UbCys complex (100 μm), and in the absence of DTT. Excess DTT (5 mm) was added to the 15N-labeled sRbx112–108-CDC34-UbCys complex to determine the effect of reducing the disulfide bond in the complex. Amide chemical shift perturbations were calculated and normalized using ΣΔδ = |Δδ(1H)| + |(0.2)Δδ(15N)|, where Δδ(1H) and Δδ(15N) represent the chemical shift differences between the free and bound states (42).
The binding affinity (Kd) for 15N-labeled sRbx112–108 with CDC34-UbCys was determined from the intermediate exchange titration data by simultaneous fitting normalized multiple traces using the global fit option in Prism 4 for Mac OSX (GraphPad Software) with shared values for Kd and the number of binding sites (n).
Di-ubiquitin Synthesis Assay
The di-ubiquitin synthesis assay proceeded with the assembly of two reaction mixtures that contained 1) CDC34∼32P-labeled ubiquitin and 2) Rbx1/ROC1 with receptor ubiquitin, respectively. The CDC34∼32P-labeled ubiquitin charge mix (5 μl) contained 50 mm Tris-HCl (pH 7.4), 5 mm MgCl2, 0.5 mm DTT, 2 mm ATP, 0.1 mg/ml of BSA, 2 mm NaF, 10 nm okadaic acid, 8.5 pmol of 32P-labeled ubiquitin (32P-PK-Ub, ubiquitin carrying a phosphorylated N-terminal protein kinase C recognition site prepared as previously described; Ref 43), 0.1 μm E1, and CDC34 in concentrations as indicated. This mixture was incubated for 5 min at 37 °C before mixing with Rbx1/ROC1 and receptor ubiquitin.
The second mix (5 μl) contained 50 mm Tris-HCl (pH 7.4), 5 mm MgCl2, 0.5 mm DTT, 2 mm ATP, 0.1 mg/ml of BSA, 2 mm NaF, 10 nm okadaic acid, ∼430 pmol of bovine ubiquitin (in 50× molar excess to 32P-labeled ubiquitin, thus acting as the receptor ubiquitin), and either the Rbx1/ROC1-CUL1 split complex (assembled in bacteria and purified as previously described; Ref. 7) or sRbx1/ROC112–108 or Rbx1/ROC136–108 in concentrations as indicated.
Ubiquitylation was initiated by combining CDC34∼32P-labeled ubiquitin with the second mixture containing Rbx1/ROC1 and receptor ubiquitin. The reaction was then incubated for 10 min at 37 °C. The reaction products were separated by 4–20% SDS-PAGE, visualized by autoradiogram, and quantified by phosphor-imager analysis.
RESULTS AND DISCUSSION
Design of Soluble Rbx1/ROC1 Protein
Rbx1/ROC1 is the E3 ligase protein found in the SCF and other CRL complexes that interacts with, and modulates the activity of CDC34 in polyubiquitin chain assembly (2–5, 10). To date, all previous functional and structural studies concerning Rbx1/ROC1 have required the co-expression of Rbx1/ROC1 with its cullin scaffold protein. To deepen our understanding of the function and regulation of Rbx1/ROC1, we sought to solve its solution structure by NMR spectroscopy in the absence of the cullin scaffold. However, our initial expression and purification trials of human Rbx1/ROC1 (Rbx112–108) with or without GST or GB1 fusion tags from Escherichia coli were unsuccessful due to protein insolubility, a problem that appears to have lingered for years in the field. Subsequent analysis using the online algorithm TANGO (20–22) identified a stretch of residues (Trp-27–Trp-33; WNAVALW) in the N-terminal extension of Rbx1/ROC1 projected to contribute to its insolubility. To improve the overall behavior of Rbx1/ROC1, four residues within this hydrophobic stretch were substituted (W27S, V30S, L32Q, W33S) providing a soluble, highly expressed protein (sRbx112–108, where “s” stands for solubilized Rbx112–108) used for solution structure determination by NMR spectroscopy and biochemical analysis.
Structure of sRbx112–108 Reveals Disordered N Terminus
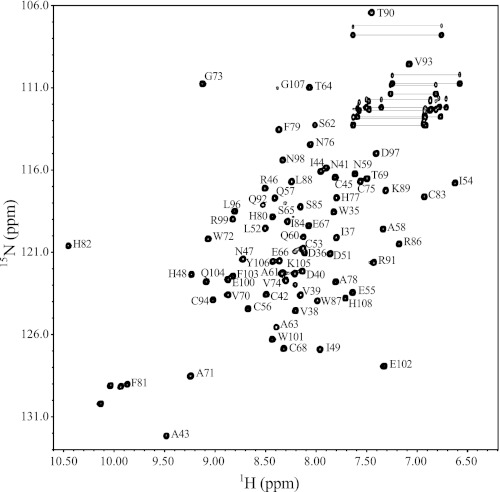
Initial 1H,15N HSQC spectra for sRbx112–108 at pH 7.5 were well dispersed and representative of a folded protein (Fig. 1). The solution structure for sRbx112–108 was determined using data collected at pH 7.5 as input to CYANA (35) and a combination of manual and automated NOE assignments (Table 1) followed by water refinement using Xplor-NIH (40, 41). The superposition of the 20 final solution structures (Fig. 2A) showed that sRbx112–108 consists of a well structured domain (root mean square deviation 0.30 ± 0.04 Å; Asp40–Ala-58, Val-70–Gly-73, His-77–Leu-96) with two well defined α-helices (Fig. 2B; α1, Glu-55–Asn-59; α2, Phe-81–Thr-90) and a two-strand β-sheet (β1, Val-70–Gly-73; β2, His-77–His-80) that accommodates three structural Zn2+ ions (Fig. 2B). Two Zn2+ ions found in site I (Cys-42, Cys-45, His-80, Cys-83) and site II (Cys-75, His-77, Cys-94, Asp-97) form the canonical RING domain that folds in a cross-brace fashion, as proposed previously (44). Using heteronuclear 15N{1H} NOE methods, these portions of the protein were among the most rigid region of the protein with an average 15N{1H} NOE of 0.75 (supplemental Fig. 1), a value expected for a globular 8-kDa protein. The structure also shows that residues Gly-12–Val-39 at the N terminus of sRbx112–108 are disordered, as indicated from a lack of inter-residue or long distance NOEs and near zero or negative 15N{1H} heteronuclear NOEs (supplemental Fig. 1). This was further supported by the absence of several resonances from residues within the N terminus of sRbx112–108 at pH 7.5 that were only observable at lower pH. In addition, a third variant Zn2+ binding site was evident (Cys-53, Cys-56, Cys-68, His-82). This site included an extended loop (Loop 2) after helix α1 that displayed poor definition reflected by lower 15N{1H} heteronuclear NOEs (0.2 minimum for Ser-65; supplemental Fig. 1). A hydrophobic groove composed of loops 1 and 3 and helix α2, previously observed in c-Cbl (45) as the E2 binding site, is maintained in the solution structure. Furthermore, the root mean square deviation for residues Asp40–His-108 of sRbx112–108 compared closely (0.85–1.17 Å) to other Rbx1/ROC1-Cullin structures (7, 9), demonstrating that a large conformational change does not occur for the ordered portion of the protein upon binding to the cullin scaffold.
FIGURE 1.
Assigned 600 MHz 1H,15N HSQC spectrum of sRbx112–108. A sample of sRbx112–108 (0.7 mm) was prepared in 20 mm Na2HPO4, 100 mm NaCl, 1 mm DTT, 10% D2O/90% H2O, and pH 7.5, and data were collected at 25 °C on a Varian Inova NMR spectrometer. The amide resonances were assigned using standard triple resonance experiments outlined under “Experimental Procedures.” The spectrum is labeled using the one-letter amino acid code and residue number according to the human Rbx1/ROC1 sequence.
TABLE 1.
Structural statistics for 20 lowest energy sRbx112–108 structures
| Completeness of resonance assignments (residues (Asp-40–His-108) | |
| Backbone (N, CA, C)a | (202/207), 97.6% |
| Side chain (C, H) | (549/654), 83.9% |
| HN | (67/68), 98.5% |
| HA | (71/71), 100.0% |
| HB | (109/118), 92.4% |
| Experimental restraints | |
| NOE-derived distances | 1305 |
| Short (|i − j| = 1) | 647 |
| Medium (2 < |i − j| <5) | 271 |
| Long (5< |i − j|) | 387 |
| Distance restraints per residue | 19.2 |
| Zinc restraints | 26 |
| Dihedral angle restraints (ϕ/ψ)b | 51 |
| H-bond restraintsc | 20 |
| Violations (per structure) | |
| NOE violations >0.5 Å | 0.4 |
| Dihedral constraint violations >2.0 Å | 0 |
| Ramachandran statistics (%)d | |
| Most favored | 84.0% |
| Additionally allowed | 14.8% |
| Generously allowed | 1.2% |
| Disallowed | 0.0% |
| Root mean square deviations to mean structure (Å) | |
| Structured regions (41–58, 70–73, 77–96) | |
| Backbone | |
| Full (40–108) | 0.48 ± 0.08 Å |
| Residues (40–58) | 0.31 ± 0.06 Å |
| Residues (70–73) | 0.05 ± 0.03 Å |
| Residues (77–96) | 0.15 ± 0.03 Å |
| Structured regions (40–58, 70–73, 77–96) | 0.30 ± 0.04 Å |
| Heavy atom, full (40–108) | 0.94 ± 0.08 Å |
a Excluding backbone hydrogens.
b ψ/ψ dihedral restraints determined using TALOS+.
c Determined from 15N-labeled HSQC H2O/D2O exchange.
d Procheck derived Ramachandran statistics including the unstructured N-terminus of sRbx12–108.
FIGURE 2.
The solution structure of sRbx112–108. A, superposition of the 20 lowest energy structures of sRbx112–108 (residues 40–108, shown in gray) is shown. The N terminus of sRbx112–108 (residues Gly-12–Val-39, magenta) is shown for five structures to demonstrate its disordered state. B, a ribbon structure of sRbx112–108 shows helices α1 (Cys-53–Ala-58) and α2 (Phe-81–Thr-90) and β-sheets β1 (Val-70–Gly-73) and β2 (His-77–His-80). The coordinating residues for the three Zn2+ binding sites in sRbx112–108 are shown in yellow. C, shown is the crystal structure of Rbx1/ROC1 bound to the C-terminal domain of cullin1 (PDB 1LDJ) (7). In contrast to A, the N terminus of Rbx1/ROC1 forms an intermolecular β-sheet (magenta) used in the recruitment of Rbx1/ROC1 to the cullin proteins. All structures were created using PyMol (64).
Disordered-to-ordered Transition for the N Terminus of Rbx1/ROC1
Structures of Rbx1/ROC1 in complex with cullin proteins show that the N terminus of Rbx1/ROC1 forms an ordered 16-residue intermolecular β-sheet with the C-terminal domain of the cullin scaffold protein (Fig. 2C; Ref 7). However, the conformation of the N terminus of Rbx1/ROC1 in the absence of a cullin protein was unknown. Upon completion of the backbone resonance assignments for sRbx112–108, we noticed that the resonances from residues Gly-12 to Ala-34 of the N terminus of sRbx112–108 were not detected, likely due to rapid amide exchange with water. At pH 6.0, 1H,15N HSQC of sRbx112–108 showed numerous new peaks that could be assigned to the N terminus of the protein, particularly focused in the 8.0–8.5 ppm range of the 1H dimension. As shown from the structure of sRbx112–108 (Fig. 2A), we observed that the N terminus of Rbx1/ROC1 adopts a variety of positions in the absence of a cullin scaffold ranging from elongated to more compact, the trademark of a disordered protein. Finally, sRbx112–108 Gly-12–Val-39 lacked inter-residue or long distance NOEs and near zero or negative 15N{1H} heteronuclear NOEs (supplemental Fig. 1). Taken together, these data suggest that residues Gly-12–Val-39 at the N terminus of sRbx112–108 in the absence of a bound cullin molecule are disordered.
The disordered structure of the Rbx1/ROC1 N terminus in the absence of a cullin protein clarifies secondary structure predictions using algorithms such as JPred3 (46) and PredictProtein (47). Although these algorithms predict β-sheet character in the N termini of both wild-type Rbx1/ROC1 and sRbx112–108, they also display low confidence (0–2) in their assessments. The Rbx1/ROC1 N terminus interaction with the CUL1 C-terminal domain (CUL1CTD) buries several hydrophobic residues from the Rbx1/ROC1 N terminus (Trp-27, Val-30, Leu-32, Trp-33). These residues, modified in sRbx112–108 used in this work, likely give rise to its insolubility in the absence of the cullin protein but help form the β-sheet hydrophobic core with the C-terminal domain of CUL1 (7). Combining these results, we suggest that the N terminus of Rbx1/ROC1 undergoes a dramatic disordered-to-ordered conformational transition that is induced by protein-protein interactions with CUL1 (Fig. 2, A and C). This transition is reminiscent of the disordered-to-ordered β-sheet state exemplified by α-synuclein upon Lewy body formation that has been linked to the development of Parkinson disease (48). The proposed transformation of the N terminus of Rbx1/ROC1 from disordered-to-ordered conformation upon binding to CUL1 likely underscores a key step to form the catalytic Rbx1/ROC1-cullin core subcomplex that defines the CRL E3 ubiquitin ligases.
Rbx1/ROC1 Preferentially Binds to CDC34∼Ubiquitin and Releases CDC34
Rbx1/ROC1 recruits and facilitates ubiquitin transfer from the E2 enzyme CDC34 to a substrate protein or an elongating ubiquitin chain. Despite recent work that illustrates a high affinity interaction between the basic canyon of CUL1 and the acidic C terminus of CDC34 (16), it remained largely unknown how the Rbx1/ROC1 RING domain interacts with the catalytic core of CDC34, a presumed pivotal step that leads to ubiquitination. The availability of the NMR structure of cullin-free Rbx1/ROC1 RING protein permitted us to examine this step through its interaction with CDC34 or CDC34∼ubiquitin using chemical shift perturbation analysis.
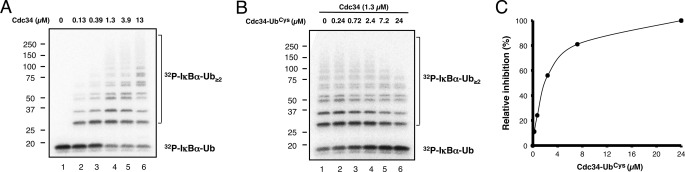
In place of the CDC34∼ubiquitin thiol ester conjugate, we used a stable E2-UbCys disulfide complex that has been shown to be a good structural mimic of E2∼ubiquitin thiol ester complexes (15, 24, 25). Because the disulfide E2-UbCys complex is catalytically inert as it lacks the thiol ester required to conjugate with another ubiquitin molecule, we reasoned that adding the disulfide complex to ubiquitination reactions would inhibit polyubiquitin assembly by directly competing for the binding site on Rbx1/ROC1 with the CDC34∼ubiquitin thiol ester. To test this possibility, we assessed the effect of CDC34-UbCys on ubiquitin chain elongation on an IκBα-ubiquitin fusion substrate, a reaction depending on the SCF loaded with the F-box protein β-TrCP (SCFβTrCP) E3 complex and CDC34 (13) (see Fig. 3A). As shown, CDC34-UbCys blocked ubiquitination, inhibiting the production of polyubiquitin chains and accumulating the IκBα-ubiquitin substrate in a dose-dependent manner (Fig. 3, B and C). This result demonstrates that the disulfide CDC34-UbCys complex disrupts the Rbx1/ROC1-dependent ubiquitination activity by out-competing CDC34∼ubiquitin for the E3-ligase binding site, thus validating the use of CDC34-UbCys as a structural and functional mimic of CDC34∼ubiquitin.
FIGURE 3.
CDC34-UbCys is an excellent functional mimic of the E2∼ubiquitin thiol ester intermediate. A, dose-dependent increases in the polyubiquitylation of 32P-labeled IκBα-ubiquitin by CDC34 are shown. B and C, CDC34-UbCys competitively inhibits CDC34/SCF-dependent ubiquitylation. All reactions contained CDC34 (1.3 μm) with increasing concentrations of CDC34-UbCys and show decreased ubiquitin conjugation to 32P-labeled IκBα-ubiquitin, demonstrating that CDC34-UbCys competes for the same site as CDC34∼ubiquitin. Reaction mixtures were prepared as described under “Experimental Procedures” visualized by autoradiogram, and the amount of remaining unreacted 32P-labeled IκBα-ubiquitin was quantified by phosphorimaging analysis.
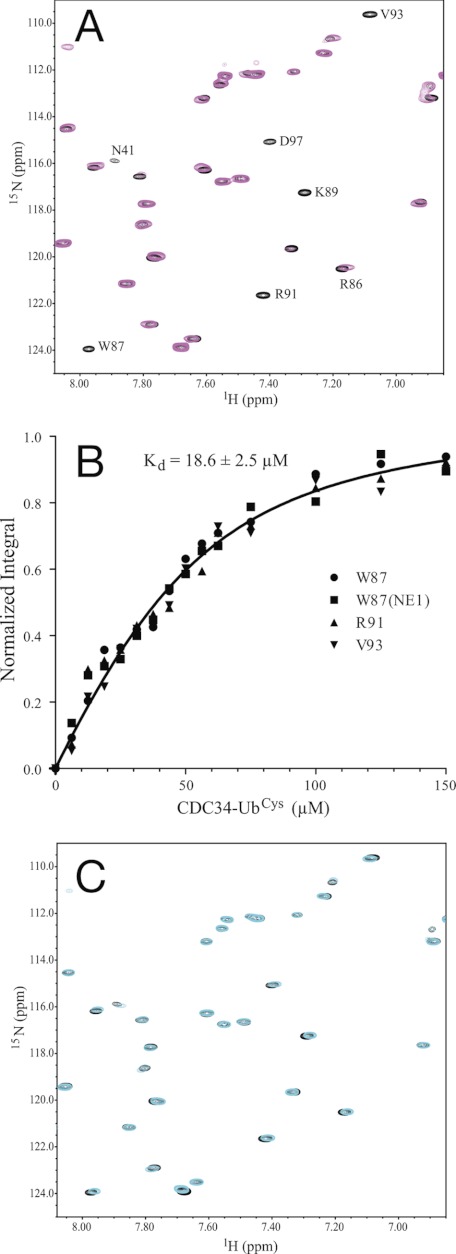
Because our ubiquitination assay indicated that the CDC34-UbCys complex accesses the Rbx1/ROC1 binding site, 1H,15N HSQC spectra of 15N-labeled sRbx112–108 in the absence and presence of unlabeled CDC34 or the thiol ester mimetic CDC34-UbCys complex were used to analyze the E2-E3 interactions. Surprisingly, in the presence of a 2-fold excess CDC34 there were no observable changes in the peak positions or intensities in sRbx112–108, suggesting an extremely poor interaction between these two proteins. Based on the concentrations of the proteins used in these experiments, the dissociation was estimated to be >1 mm. In contrast, the spectrum of 15N-labeled sRbx112–108 in the presence of the unlabeled CDC34-UbCys complex showed numerous peaks with significant chemical shift changes and broadening consistent with the overall size of Rbx1/ROC1 changing from ∼11 to ∼46 kDa (Fig. 4A) and indicative of binary protein complex formation. To determine the dissociation constant (Kd) between the two proteins, we performed a titration of 15N-labeled sRbx112–108 with unlabeled CDC34-UbCys and measured the integral of the affected signals involved in the slow-intermediate exchange process (Fig. 4B). Using a global fit for several affected residues (Trp-87, Arg-91, Val-93), a dissociation constant of 18.6 ± 2.5 μm was determined for the sRbx112–108-CDC34-UbCys interaction (Fig. 4B).
FIGURE 4.
Preferential binding of Rbx1/ROC1 to CDC34-UbCys. A, shown are selected regions of the 1H,15N HSQC spectra for 50 μm 15N-labeled sRbx112–108 in the absence (black contours) and the presence of 2 eq of unlabeled CDC34-UbCys (magenta). Labeled signals represent those that underwent chemical shift changes upon the addition of CDC34-UbCys. B, shown are normalized titration data for the interaction of 15N-labeled sRbx112–108 with CDC34-UbCys, monitored using residues Trp-87, Arg-91, and Val-93 and globally fit (Kd 18.6 ± 2.5 μm). C, shown are selected regions of the 1H,15N HSQC spectra of the identical sample used in A after the addition of 5 mm DTT (cyan). This spectrum demonstrates that the reduction of the disulfide in the CDC34-UbCys complex causes the release of the unconjugated CDC34 from Rbx1/ROC1.
The use of the CDC34-UbCys complex also allowed us to mimic the interaction with Rbx1/ROC1 upon transfer of the CDC34-bound ubiquitin during the ubiquitin-chain transfer process. We reasoned that reducing the disulfide bond within the CDC34-UbCys complex would act much like the transfer of ubiquitin to a substrate. The addition of excess DTT to the preformed 15N-labeled sRbx112–108/unlabeled CDC34-UbCys complex (Fig. 4A) resulted in the loss of shifted and broadened signals from the complex and the appearance of new, sharper signals in their original positions found for sRbx112–108 in the absence of CDC34 (Fig. 4C). Furthermore, there were no other obvious changes in peak positions or intensities. These results indicate that reduction of the disulfide bond in the CDC34-Ub complex causes the loss of interaction with sRbx112–108, indicating that the ubiquitin-unloaded form of CDC34 has a significantly reduced affinity for Rbx1/ROC1 compared with the CDC34∼Ub thiol ester. This shows that Rbx1/ROC1 favors the CDC34∼Ub conjugate by more than 50-fold compared with the E2 protein alone.
Our cumulative results clearly demonstrate an increased affinity for CDC34∼ubiquitin for Rbx1/ROC1 and suggest that ubiquitin dissociation may be the leading cause for the disassembly of the E2-RING complex. These findings suggest a critical role for ubiquitin in the dynamic cycles of the association and dissociation of the CDC34 catalytic core with Rbx1/ROC1. On one hand, the ubiquitin moiety in the CDC34∼ubiquitin thiol ester likely provides a binding surface that is critical to establish interactions with the Rbx1/ROC1 RING domain. Once ubiquitin is conjugated to a substrate or an elongating ubiquitin chain, it may trigger the dissociation of CDC34 from the RING domain, which permits the initiation of the next reaction cycle. This ubiquitin-enhanced E2 affinity to RING E3 mechanism may be “ubiquitous.” In support of this hypothesis, recent studies have shown that an unrelated bacterial E3 enzyme SspH2 has an increased affinity for ubiquitin-conjugated UbcH5 (49), the crystal structure of a UbcH5B∼Ub/HECTNEDD4L complex shows the ubiquitin simultaneously makes contacts with both the E2 and E3 enzymes (50), biochemical data show that UbcH5b∼ubiquitin binds to the bacterial effector protein OspG (51), and UbcM2∼ubiquitin binds to the nuclear import protein importin-11 (52).
Cullin-free Rbx1/ROC1 Activates CDC34 Catalytic Core for Ubiquitination
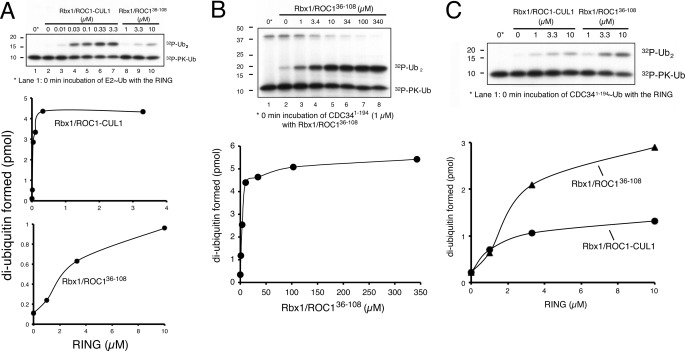
Given the above structure-based studies that underlie interactions between the Rbx1/ROC1 RING domain and the CDC34∼ubiquitin complex, we next explored biochemical approaches to determine whether these interactions are critical for ubiquitination. In keeping with previous observations (13, 53, 54), the Rbx1/ROC1-CUL1CTD complex activated CDC34 to yield di-ubiquitin products (Fig. 5A, lanes 3–7). Cullin-free Rbx1/ROC136–108 (Fig. 5A, lanes 8–10) or Rbx112–108 (not shown) stimulated CDC34 as well but were 300-fold less effective than ROC1/Rbx1-CUL1CTD (Fig. 5A). These findings demonstrate the ability of the cullin-free Rbx1/ROC1 RING protein to activate CDC34 for ubiquitination, albeit at levels significantly lower than the cullin-bound form. The drastic disparity between the cullin-bound or -free Rbx1/ROC1 molecules in CDC34-catalyzed di-ubiquitin synthesis can be best explained by recent studies from Deshaies and co-workers (16, 17), who demonstrate a high affinity interaction between the C-terminal basic canyon of CUL1 and the acidic C terminus of CDC34. Taken together, these results suggest an indispensible role for the charge-based CUL1-CDC34 C terminus interaction in recruiting CDC34 by SCF for ubiquitination.
FIGURE 5.
Rbx1/ROC1-dependent di-ubiquitin synthesis. A, full-length CDC34 (residues 1–236) with Rbx1/ROC1-CUL1 shows enhanced activity compared with cullin-free Rbx1/ROC136–108. This enhanced activity can be attributed to the interaction of the acidic C-terminal extension of CDC34 binding to the CUL1, which brings the catalytic core of CDC34 into closer proximity of Rbx1/ROC1. Ub, ubiquitin. PK-Ub, ubiquitin with an N-terminal protein kinase C recognition site. B, Rbx1/ROC136–108 can promote di-ubiquitin synthesis in the absence of CUL1 using only the catalytic core of CDC34 (residues 1–194; CDC341–194). C, cullin-free Rbx1/ROC136–108 is more efficient than Rbx1/ROC1-CUL1 in synthesizing di-ubiquitin with CDC341–194. All reaction mixtures and protein concentrations were prepared as described under “Experimental Procedures.” Gels were visualized by autoradiogram, and the amount of 32P-labeled di-ubiquitin synthesized was quantified by phosphorimaging analysis.
To more closely assess the functional interaction between Rbx1/ROC1 and CDC34, we employed a truncated protein comprising only the catalytic core of the E2 enzyme (CDC341–194). This E2 protein was shown previously to exhibit an ability to form thiol esters with ubiquitin at levels comparable with the wild-type E2 and showed significant defects in binding to ROC1/Rbx1-CUL1CTD with diminished activity for ubiquitin chain assembly (55). When assayed with Rbx1/ROC136–108, CDC341–194 formed di-ubiquitin products effectively (Fig. 5B). Contrary to reactions with the full-length CDC34, Rbx1/ROC136–108 appeared more active than ROC1/Rbx1-CUL1CTD in producing di-ubiquitin in the presence of CDC341–194 (Fig. 5C). These observations underscore a functional interaction between the Rbx1/ROC1 RING domain and the catalytic core of CDC34 that leads to ubiquitin transfer. It has been shown previously that the extreme C-terminal domain/C-terminal winged-helix subdomain (ECTD/WH-B) of CUL1 forms an autoinhibitory interaction with the ROC1/Rbx1 RING domain (56) and that this inhibition is liberated upon NEDD8-imposed conformational change (9). Our work is in agreement with such hypotheses, indicating that when facing a form of CDC34 that lacks the acidic C terminus in the absence of NEDD8 modification, CUL1 modulates the activity of the Rbx1/ROC1 in a negative fashion.
Trp-87 and Arg-91 in Rbx1/ROC1 Are Key Residues for Di-ubiquitin Synthesis by CDC34
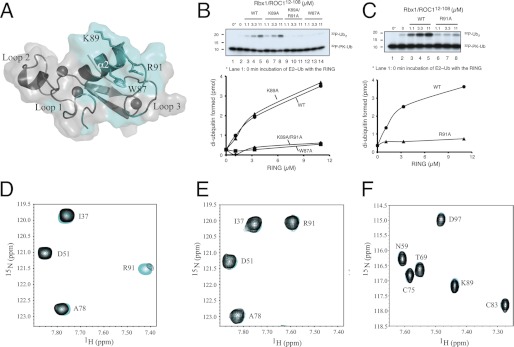
Having identified that ubiquitin-loaded CDC34 interacts preferentially with Rbx1/ROC1 to promote ubiquitin chain synthesis, we used NMR chemical shift perturbation experiments to identify the surface used by Rbx1/ROC1 to recruit the E2-ubiquitin complex. The residues most affected in 15N-labeled sRbx112–108 were monitored upon the addition of the disulfide mimetic CDC34-UbCys (supplemental Fig. 2) and mapped to the surface of sRbx112–108 (Fig. 6A). The experiments show that the largest changes in sRbx112–108 occurred near the C terminus of helix α2 (Arg-86, Trp-87, Lys-89) and the subsequent loop (Thr-90, Arg-91, Gln-92, Val-93, Leu-96, Asp-97, Asn-98). Minor changes were also observed for Asn-41, Ile-44, Asn-47, and Ile-49 (loop 1). The residues that were the most affected (particularly Ile-44, Trp-87, Pro-95, and Leu-96) were found to face toward a conserved hydrophobic cleft between loop 1 and helix α2 similar to those identified in the E3 ligases cCbl (45) and CNOT4 (57) for recruitment of their E2 enzymes. To identify the functional importance of these residues in helix α2 of Rbx1/ROC1, we examined the effects of alanine substitutions at positions Trp-87, Lys-89, and Arg-91 in Rbx1/ROC1 toward CDC34∼ubiquitin binding and di-ubiquitin chain formation (Fig. 6, B–F). These experiments revealed that the W87A and R91A proteins diminished the ability of Rbx1/ROC1 to activate CDC341–194 for di-ubiquitin synthesis (Fig. 6, B and C) and to recruit CDC34-UbCys (Fig. 6, E and F). Interestingly, although Lys-89 in Rbx1/ROC1 showed the largest chemical shift perturbation upon binding to CDC34-UbCys (supplemental Fig. 2), the K89A-substituted protein stimulated di-ubiquitin synthesis by CDC341–194 at levels similar to the wild-type Rbx1/ROC1 (Fig. 6B), indicating that Lys-89 has a secondary role in the association of CDC34∼ubiquitin. Of note, both R91A and K89A/R91A proteins abolished the activating function of Rbx1/ROC1 (Fig. 6, B and C), confirming the essential role for residue Arg-91 of Rbx1/ROC1 in forming a functional complex with CDC34∼ubiquitin. Furthermore, 1H,15N HSQC spectra for 15N-labeled sRbx112–108 W87A- and R91A-substituted proteins demonstrated little observable chemical shift changes upon the addition of CDC34-UbCys (Fig. 6, E and F), indicating these folded Rbx1/ROC1 proteins have a poor interaction with the E2∼ubiquitin complex.
FIGURE 6.
Residues in helix α2 of sRbx112–108 are essential for binding and di-ubiquitin synthesis activity of CDC34∼ubiquitin. A, the interaction surface on Rbx1 for CDC34-UbCys (cyan) was determined using chemical shift perturbation analysis (supplemental Fig. 2). Residues that exhibited large chemical shift changes are located on helix α2 (Trp-87, Lys-89, and Arg-91) and were selected for alanine substitution. This figure was created using atomic co-ordinates from Fig. 2 and visualized using PyMOL (64). Cullin-free sRbx112–108-dependent 32P-labeled di-ubiquitin synthesis for wild type, K89A, K89A/R91A, W87A (B) and R91A (C) was visualized by autoradiogram and quantified by phosphorimaging analysis. All reactions contained CDC341–194 (0.5 μm) with increasing concentrations of Rbx1/ROC1 proteins as indicated. Reaction mixtures were prepared as described under “Experimental Procedures.” Shown are 1H,15N HSQC spectra for 15N-labeled sRbx112–108 (50 μm) in the absence (cyan) and presence of unlabeled CDC34-UbCys (100 μm) for wild type (D), W87A (E) and R91A (E) sRbx112–108 .
Previous work has shown that Rbx1/ROC1 Lys-89 is in direct contact with the CUL1 WH-B domain (7) and the K89A substitution enhances the ability of Rbx1/ROC1-CUL1CTD to synthesize di-ubiquitin (56). It remains to be determined whether despite its contact with CDC34-UbCys, the side chain of Rbx1/ROC1 Lys-89 is specifically required for the proposed autoinhibitory interactions with the ECTD/WH-B of CUL1 (56). In all, these results demonstrate that residues Trp-87 and Arg-91 at the C terminus of helix α2 in Rbx1/ROC1 play essential roles in E2∼ubiquitin binding and activation.
Comparison to Other E2-E3 Interactions
The molecular and structural basis for an active E2-E3 complex and whether a ubiquitin-conjugated E2 is the driving force for ubiquitin transfer has been a point of interest for years in the ubiquitin field (2). Our observations that show a ubiquitin-charged CDC34 interacts preferentially with Rbx1/ROC1 compared with the naked E2 enzyme contrasts previous structural studies that demonstrate a RING E3 can associate with an uncharged E2 binding partner (45, 57). However, it is important to note that several E2-E3 complexes, including UbcH7 in complex with c-Cbl (58) and Bcra1/Bard1 (59), show poor activity, indicating that a tight E2-E3 complex does not necessarily translate into efficient ubiquitin transfer properties. The tight binding of CDC34∼Ub (∼18 μm) compared with CDC34 alone (>1 mm) to Rbx1/ROC1 indicates there are some important differences for CDC34 compared with other E2 enzymes involved in E2-E3 complexes. The interfaces between several E2-E3 complexes have been recently compared (60) and show some common regions in both enzymes at the protein-protein interface. Key conserved residues in the E3 enzyme include bulky hydrophobic residues in the loop regions (L1, L2) and in the α2 helix directed at the E2 enzyme. In Rbx1/ROC1 these positions all showed large chemical shift changes upon interaction with CDC34-UbCys including Ile-44 (Loop 1), Leu-96 (Loop 3 in Rbx1/ROC1, similar to L2 in other RING E3s), and Trp-87 (α2). Less conserved is Arg-91 in Rbx1/ROC1, which we demonstrate is necessary for di-ubiquitin synthesis and interaction with the ubiquitin-loaded E2 enzyme. In general, the strong agreement for important residues in Rbx1/ROC1 with other E3 enzymes found in existing E2-E3 complexes suggests the selective recruitment of a ubiquitin-loaded E2 is likely not a function of the E3 but rather is a result of differences in the E2 sequence and structure. Structures reveal that conserved E2 residues for E3 binding are found near the N terminus of helix α1 and in two connecting loops between β-sheets β3 and β4 (L4) and near the active site just before helix α2 (L7) (60). Notable differences exist in CDC34 and most other E2 enzymes in two of these regions. For example, in helix α1 a series of basic residues is found in most E2 enzymes that are not present in CDC34 (Lys-11, Ala-12, Leu-15). Furthermore, there is an acidic loop insertion in CDC34 adjacent to or possibly within the L7 recognition region. It is likely these two features make significant contributions to the increased affinity of ubiquitin-charged CDC34 for Rbx1/ROC1 compared with the unloaded form and may indicate that the ubiquitin protein makes important contributions to the E3 interaction site or influence a rearrangement of the L7 recognition site.
Model for CDC34∼Ubiquitin Control of Ubiquitin Transfer by Rbx1/ROC1
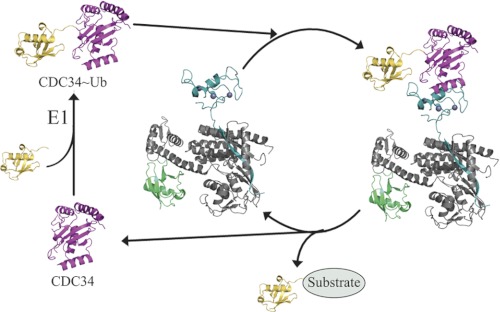
In this work we described structural and biochemical evidence for selective recognition of CDC34∼ubiquitin by the Rbx1/ROC1 protein (Figs. 3 and 4) through its helix α2 (Fig. 6, supplemental Fig. 2). Our study is in line with a hypothesis for a role of ubiquitin dissociation in promoting the discharge of CDC34 from the RING finger protein. Based on these findings and previous work, we suggest a model depicting a central role for CDC34∼ubiquitin in the control of RING-dependent ubiquitin transfer (Fig. 7). In this model, SCF and perhaps other CRL E3s as well engage CDC34 via a two-step mechanism. In the first step CUL1 engages with CDC34 through an electrostatic interaction between a basic canyon in CUL1 with the acidic C terminus of CDC34 (Kd ∼ 0.1 μm (16)). In this state little association between Rbx1/ROC1 and the catalytic core of CDC34 occurs. Upon E1-catalyzed ubiquitin loading of CDC34 in the second step, the proximity of Rbx1/ROC1 favors an interaction with the CDC34∼ubiquitin conjugate (Kd = 18 μm, this work). This interaction, mediated primarily by residues Trp-87 and Arg-91 of Rbx1/ROC1, promotes ubiquitin transfer to a substrate and returns the system to a dissociated Rbx1/ROC1 state with the now ubiquitin-unloaded CDC34 and allows for the initiation of the next ubiquitin transfer reaction cycle. It remains to be determined, however, whether CDC34 remains to be bound with SCF through the interactions between the acidic C terminus of CDC34 and the basic canyon of CUL1 (16) after each successive ubiquitin transfer.
FIGURE 7.
Rbx1/ROC1-CDC34∼ubiquitin (Ub) ternary complex controls polyubiquitylation. Structures shown represent the catalytic domain of CDC34 (pink), ubiquitin (yellow), Rbx1/ROC1 (teal), the C-terminal domain of CUL1 (gray), and NEDD8 (green). CDC34∼Ub was modeled by aligning CDC34 (PDB 2OB4) with the Ubc13∼Ub structure (PDB 2GMI) (65). The Rbx1/ROC1-CUL1-NEDD8 structure was derived from PDB 3DQV (CUL5) (9). After the charging of CDC34 with ubiquitin, CDC34∼ubiquitin binds to Rbx1/ROC1, forming the ternary Rbx1/ROC1-CDC34∼ubiquitin complex. Upon ubiquitin transfer to a substrate protein, the spent CDC34 is released from Rbx1/ROC1. This release may allow another CDC34∼ubiquitin to bind to Rbx1/ROC1 for the next round of ubiquitin transfer. Alternatively, CDC34 may remain associated with CUL1 via its extended C terminus (not shown) proposed to bind with a basic canyon on the lower side of CUL1. In either case CDC34 must be recharged with ubiquitin by the E1 enzyme before it can re-engage with Rbx1/ROC1 for subsequent ubiquitin transfer.
Our model is consistent with previous observations that a reorganization of the CDC34 active site may be required for the efficient nucleophilic attack of the thiol ester bond by the substrate protein (61) that may be mediated by its association with Rbx1/ROC1. Our findings suggest that once the CDC34∼ubiquitin transfers its cargo ubiquitin to a substrate, the uncharged CDC34 catalytic core must dissociate from the SCF to be recharged with another ubiquitin by the E1 enzyme before it can transfer another ubiquitin molecule. This is consistent with the E1-E2 and E2-E3 stages in the ubiquitination cascade being mutually exclusive as the binding sites for the E1 and E3 enzymes on an E2, such as CDC34, overlap (62). Our findings confirm that CDC34 would only be released from Rbx1/ROC1 after transfer of ubiquitin from the E2 to the substrate, as previously proposed (18), and rule out the possibility of the diffusion based hit-and-run model for CDC34-dependent ubiquitylation (63).
In summary our work shows that a large structural transition of the disordered N terminus of Rbx1/ROC1 to a structured β-sheet in the SCF complex is likely a key recruiting step. Furthermore, the selectivity of the CDC34∼ubiquitin by Rbx1/ROC1 provides a mechanism whereby ubiquitin can be transferred to a substrate mediated by Rbx1/ROC1 followed by release of the E2 protein for reactivation by an E1 enzyme necessary for subsequent rounds of ubiquitylation.
Supplementary Material
Acknowledgments
We thank Steven Beasley and Pascal Mercier for helpful expertise with CYANA and Xplor-NIH. The Biomolecular NMR Facility at the University of Western Ontario was supported by a Research Resource grant from the Canadian Institutes of Health Research.
This work was supported by Canadian Cancer Society Grant CCS-018414, Canadian Institutes of Health Research Grant MOP-14606, and by the Canada Research Chairs Program (to G. S. S.). This work was also supported, in whole or in part, by National Institutes of Health Grants GM061051 and CA095634 (to Z.-Q. P.).

This article contains supplemental Figs. 1 and 2.
The atomic coordinates and structure factors (code 2LGV) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
NMR assignments for sRbx112-108 were deposited to the BioMagResBank (BMRB 17824).
- CRL
- Cullin RING ligase
- UbCys
- K48R/G76C substituted ubiquitin
- CDC34-UbCys
- disulfide complex between CDC34 and UbCys
- Rbx1/ROC1
- RING box protein 1
- SCF
- Skp1/Cullin 1/F-box
- HSQC
- heteronuclear single quantum coherence
- TEV
- tobacco etch virus.
REFERENCES
- 1. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 3. Petroski M. D., Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 4. Zimmerman E. S., Schulman B. A., Zheng N. (2010) Structural assembly of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 20, 714–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duda D. M., Scott D. C., Calabrese M. F., Zimmerman E. S., Zheng N., Schulman B. A. (2011) Structural regulation of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 21, 257–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Angers S., Li T., Yi X., MacCoss M. J., Moon R. T., Zheng N. (2006) Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443, 590–593 [DOI] [PubMed] [Google Scholar]
- 7. Zheng N., Schulman B. A., Song L., Miller J. J., Jeffrey P. D., Wang P., Chu C., Koepp D. M., Elledge S. J., Pagano M., Conaway R. C., Conaway J. W., Harper J. W., Pavletich N. P. (2002) Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 [DOI] [PubMed] [Google Scholar]
- 8. Goldenberg S. J., Cascio T. C., Shumway S. D., Garbutt K. C., Liu J., Xiong Y., Zheng N. (2004) Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119, 517–528 [DOI] [PubMed] [Google Scholar]
- 9. Duda D. M., Borg L. A., Scott D. C., Hunt H. W., Hammel M., Schulman B. A. (2008) Structural insights into NEDD8 activation of cullin-RING ligases. Conformational control of conjugation. Cell 134, 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deshaies R. J., Emberley E. D., Saha A. (2010) Control of cullin-ring ubiquitin ligase activity by nedd8. Subcell Biochem. 54, 41–56 [DOI] [PubMed] [Google Scholar]
- 11. Boh B. K., Smith P. G., Hagen T. (2011) Neddylation-induced conformational control regulates Cullin RING ligase activity in vivo. J. Mol. Biol. 409, 136–145 [DOI] [PubMed] [Google Scholar]
- 12. Saha A., Deshaies R. J. (2008) Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu K., Kovacev J., Pan Z. Q. (2010) Priming and extending. A UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol. Cell 37, 784–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ceccarelli D. F., Tang X., Pelletier B., Orlicky S., Xie W., Plantevin V., Neculai D., Chou Y. C., Ogunjimi A., Al-Hakim A., Varelas X., Koszela J., Wasney G. A., Vedadi M., Dhe-Paganon S., Cox S., Xu S., Lopez-Girona A., Mercurio F., Wrana J., Durocher D., Meloche S., Webb D. R., Tyers M., Sicheri F. (2011) An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell 145, 1075–1087 [DOI] [PubMed] [Google Scholar]
- 15. Spratt D. E., Shaw G. S. (2011) Association of the disordered C terminus of CDC34 with a catalytically bound ubiquitin. J. Mol. Biol. 407, 425–438 [DOI] [PubMed] [Google Scholar]
- 16. Kleiger G., Saha A., Lewis S., Kuhlman B., Deshaies R. J. (2009) Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell 139, 957–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kleiger G., Hao B., Mohl D. A., Deshaies R. J. (2009) The acidic tail of the Cdc34 ubiquitin-conjugating enzyme functions in both binding to and catalysis with ubiquitin ligase SCFCdc4. J. Biol. Chem. 284, 36012–36023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petroski M. D., Kleiger G., Deshaies R. J. (2006) Evaluation of a diffusion-driven mechanism for substrate ubiquitination by the SCF-Cdc34 ubiquitin ligase complex. Mol. Cell 24, 523–534 [DOI] [PubMed] [Google Scholar]
- 19. Pierce N. W., Kleiger G., Shan S. O., Deshaies R. J. (2009) Detection of sequential polyubiquitylation on a millisecond timescale. Nature 462, 615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fernandez-Escamilla A. M., Rousseau F., Schymkowitz J., Serrano L. (2004) Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 22, 1302–1306 [DOI] [PubMed] [Google Scholar]
- 21. Linding R., Schymkowitz J., Rousseau F., Diella F., Serrano L. (2004) A comparative study of the relationship between protein structure and β-aggregation in globular and intrinsically disordered proteins. J. Mol. Biol. 342, 345–353 [DOI] [PubMed] [Google Scholar]
- 22. Rousseau F., Schymkowitz J., Serrano L. (2006) Protein aggregation and amyloidosis. Confusion of the kinds? Curr. Opin. Struct. Biol. 16, 118–126 [DOI] [PubMed] [Google Scholar]
- 23. Hristova V. A., Beasley S. A., Rylett R. J., Shaw G. S. (2009) Identification of a novel Zn2+ binding domain in the autosomal recessive juvenile Parkinson-related E3 ligase parkin. J. Biol. Chem. 284, 14978–14986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Serniwka S. A., Shaw G. S. (2009) The structure of the UbcH8-ubiquitin complex shows a unique ubiquitin interaction site. Biochemistry 48, 12169–12179 [DOI] [PubMed] [Google Scholar]
- 25. Merkley N., Barber K. R., Shaw G. S. (2005) Ubiquitin manipulation by an E2 conjugating enzyme using a novel covalent intermediate. J. Biol. Chem. 280, 31732–31738 [DOI] [PubMed] [Google Scholar]
- 26. Kay L. E., Keifer P., Saarinen T. (1992) Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114, 10663–10665 [Google Scholar]
- 27. Grzesiek S., Bax A. (1992) Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 114, 6291–6293 [Google Scholar]
- 28. Grzesiek S., Anglister J., Bax A. (1993) Correlation of backbone amide and aliphatic side-chain resonances in 13C,15N-enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Reson. Series B, 101, 114–119 [Google Scholar]
- 29. Kay L. E., Xu G., Singer A. U., Muhandirum D. R., Forman-Kay J. D. (1993) A gradient-enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlations in H2O samples of proteins. J. Magn. Reson. 101, 333–337 [Google Scholar]
- 30. Merkley N., Shaw G. S. (2004) Solution structure of the flexible class II ubiquitin-conjugating enzyme Ubc1 provides insights for polyubiquitin chain assembly. J. Biol. Chem. 279, 47139–47147 [DOI] [PubMed] [Google Scholar]
- 31. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 32. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe. A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 33. Johnson B. A., Belvins R. A. (1994) NMRView. A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 34. Wishart D. S., Bigam C. G., Yao J., Abildgaard F., Dyson H. J., Oldfield E., Markley J. L., Sykes B. D. (1995) 1H, 13C, and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 6, 135–140 [DOI] [PubMed] [Google Scholar]
- 35. Güntert P., Mumenthaler C., Wüthrich K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 36. Herrmann T., Güntert P., Wüthrich K. (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 37. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+. A hybrid method for predicting backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kornhaber G. J., Snyder D., Moseley H. N., Montelione G. T. (2006) Identification of zinc-ligated cysteine residues based on 13Cα and 13Cβ chemical shift data. J. Biomol. NMR 34, 259–269 [DOI] [PubMed] [Google Scholar]
- 39. Beasley S. A., Hristova V. A., Shaw G. S. (2007) Structure of the Parkin in-between ring domain provides insights for E3 ligase dysfunction in autosomal recessive Parkinson disease. Proc. Natl. Acad. Sci. U.S.A. 104, 3095–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) The Xplor-NIH NMR molecular structure determination package. J. Magn Reson 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 41. Schwieters C. D., Kuszewski J. J., Clore G. M. (2006) Using Xplor-NIH for NMR molecular structure determination. Prog. Nucleic Acid Res. Mol. Biol. 48, 47–62 [Google Scholar]
- 42. Shuker S. B., Hajduk P. J., Meadows R. P., Fesik S. W. (1996) Discovering high affinity ligands for proteins. SAR by NMR. Science 274, 1531–1534 [DOI] [PubMed] [Google Scholar]
- 43. Tan P., Fuchs S. Y., Chen A., Wu K., Gomez C., Ronai Z., Pan Z. Q. (1999) Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol. Cell 3, 527–533 [DOI] [PubMed] [Google Scholar]
- 44. Borden K. L. (1998) RING fingers and B-boxes. Zinc-binding protein-protein interaction domains. Biochem. Cell Biol. 76, 351–358 [DOI] [PubMed] [Google Scholar]
- 45. Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. (2000) Structure of a c-Cbl-UbcH7 complex. RING domain function in ubiquitin-protein ligases. Cell 102, 533–539 [DOI] [PubMed] [Google Scholar]
- 46. Cole C., Barber J. D., Barton G. J. (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36, W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rost B., Yachdav G., Liu J. (2004) The PredictProtein server. Nucleic Acids Res. 32, W321–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Uversky V. N., Eliezer D. (2009) Biophysics of Parkinson disease. Structure and aggregation of α-synuclein. Curr. Protein Pept. Sci. 10, 483–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Levin I., Eakin C., Blanc M. P., Klevit R. E., Miller S. I., Brzovic P. S. (2010) Identification of an unconventional E3 binding surface on the UbcH5∼Ub conjugate recognized by a pathogenic bacterial E3 ligase. Proc. Natl. Acad. Sci. U.S.A. 107, 2848–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kamadurai H. B., Souphron J., Scott D. C., Duda D. M., Miller D. J., Stringer D., Piper R. C., Schulman B. A. (2009) Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol. Cell 36, 1095–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim D. W., Lenzen G., Page A. L., Legrain P., Sansonetti P. J., Parsot C. (2005) The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. U.S.A. 102, 14046–14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Plafker S. M., Plafker K. S., Weissman A. M., Macara I. G. (2004) Ubiquitin charging of human class III ubiquitin-conjugating enzymes triggers their nuclear import. J. Cell Biol. 167, 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gazdoiu S., Yamoah K., Wu K., Escalante C. R., Tappin I., Bermudez V., Aggarwal A. K., Hurwitz J., Pan Z. Q. (2005) Proximity-induced activation of human Cdc34 through heterologous dimerization. Proc. Natl. Acad. Sci. U.S.A. 102, 15053–15058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gazdoiu S., Yamoah K., Wu K., Pan Z. Q. (2007) Human Cdc34 employs distinct sites to coordinate attachment of ubiquitin to a substrate and assembly of polyubiquitin chains. Mol. Cell Biol. 27, 7041–7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu K., Chen A., Tan P., Pan Z. Q. (2002) The Nedd8-conjugated ROC1-CUL1 core ubiquitin ligase utilizes Nedd8-charged surface residues for efficient polyubiquitin chain assembly catalyzed by Cdc34. J. Biol. Chem. 277, 516–527 [DOI] [PubMed] [Google Scholar]
- 56. Yamoah K., Oashi T., Sarikas A., Gazdoiu S., Osman R., Pan Z. Q. (2008) Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1 C-terminal tail. Proc. Natl. Acad. Sci. U.S.A. 105, 12230–12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dominguez C., Bonvin A. M., Winkler G. S., van Schaik F. M., Timmers H. T., Boelens R. (2004) Structural model of the UbcH5B-CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure 12, 633–644 [DOI] [PubMed] [Google Scholar]
- 58. Huang A., de Jong R. N., Wienk H., Winkler G. S., Timmers H. T., Boelens R. (2009) E2-c-Cbl recognition is necessary but not sufficient for ubiquitination activity. J. Mol. Biol. 385, 507–519 [DOI] [PubMed] [Google Scholar]
- 59. Christensen D. E., Brzovic P. S., Klevit R. E. (2007) E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 14, 941–948 [DOI] [PubMed] [Google Scholar]
- 60. Budhidarmo R., Nakatani Y., Day C. L. (2012) RINGs hold the key to ubiquitin transfer. Trends Biochem. Sci. 37, 58–65 [DOI] [PubMed] [Google Scholar]
- 61. Saha A., Lewis S., Kleiger G., Kuhlman B., Deshaies R. J. (2011) Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from cdc34 to substrate. Mol. Cell 42, 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Eletr Z. M., Huang D. T., Duda D. M., Schulman B. A., Kuhlman B. (2005) E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat. Struct. Mol. Biol. 12, 933–934 [DOI] [PubMed] [Google Scholar]
- 63. Deffenbaugh A. E., Scaglione K. M., Zhang L., Moore J. M., Buranda T., Sklar L. A., Skowyra D. (2003) Release of ubiquitin-charged Cdc34-S-Ub from the RING domain is essential for ubiquitination of the SCF(Cdc4)-bound substrate Sic1. Cell 114, 611–622 [DOI] [PubMed] [Google Scholar]
- 64. DeLano W. L. (2008) The Pymol Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 65. Eddins M. J., Carlile C. M., Gomez K. M., Pickart C. M., Wolberger C. (2006) Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 13, 915–920 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.