Background: Thyroid hormone (T3) regulates skeletal development by unknown mechanisms.
Results: Canonical Wnt signaling was inhibited by T3 in osteoblastic cells, but activated in ThrbPV/PV mutant mice that display advanced bone formation.
Conclusion: The mutant thyroid hormone receptor (TRβPV) activates skeletal Wnt signaling in vivo.
Significance: T3 and Wnt signaling pathways interact during bone development.
Keywords: Beta-Catenin, Bone, Osteoblasts, Thyroid Hormone, Wnt Signaling, TR-beta-PV, Thyroid Hormone Receptor
Abstract
Thyroid hormone (T3) acts in chondrocytes and bone-forming osteoblasts to control bone development and maintenance, but the signaling pathways mediating these effects are poorly understood. ThrbPV/PV mice have a severely impaired pituitary-thyroid axis and elevated thyroid hormone levels due to a dominant-negative mutant T3 receptor (TRβPV) that cannot bind T3 and interferes with the actions of wild-type TR. ThrbPV/PV mice have accelerated skeletal development due to unknown mechanisms. We performed microarray studies in primary osteoblasts from wild-type mice and ThrbPV/PV mice. Activation of the canonical Wnt signaling in ThrbPV/PV mice was confirmed by in situ hybridization analysis of Wnt target gene expression in bone during postnatal growth. By contrast, T3 treatment inhibited Wnt signaling in osteoblastic cells, suggesting that T3 inhibits the Wnt pathway by facilitating proteasomal degradation of β-catenin and preventing its accumulation in the nucleus. Activation of the Wnt pathway in ThrbPV/PV mice, however, results from a gain of function for TRβPV that stabilizes β-catenin despite the presence of increased thyroid hormone levels. These studies demonstrate novel interactions between T3 and Wnt signaling pathways in the regulation of skeletal development and bone formation.
Introduction
Thyroid hormones regulate skeletal development and adult bone maintenance acting directly in chondrocytes and bone-forming osteoblasts and via less well defined indirect pathways (1, 2). Thyroid hormone action is mediated by the nuclear thyroid hormone receptors, TRα3 and TRβ, which function as 3,5,3′-l-triiodothyronine (T3)-inducible transcription factors that regulate expression of T3-responsive target genes (3).
In childhood, thyroid hormone deficiency causes growth retardation and delayed bone age with epiphyseal dysgenesis due to abnormal development of the growth plates resulting in short stature (2). Treatment of hypothyroid children with thyroxine induces a period of rapid catch-up growth and bone maturation (4). By contrast, thyrotoxicosis in children accelerates growth and skeletal maturation but also causes short stature due to early fusion of the growth plates. Premature fusion of the sutures of the skull is also seen in severely affected young children and may lead to craniosynostosis (5). These observations demonstrate sensitivity of the skeleton to disturbances of thyroid status during postnatal growth and indicate the importance of thyroid hormones during bone development.
In adults, thyroid hormone excess causes high bone turnover with uncoupling of the activities of bone-forming osteoblasts and bone-resorbing osteoclasts (6). This results in a net loss of bone in hyperthyroidism leading to osteoporosis and an increased susceptibility to fragility fractures (7–10). In hypothyroidism, there is reduced bone turnover with increased bone mineralization. Population studies indicate that hypothyroid patients also have an increased risk of fracture (7, 8, 10), although the underlying reasons for this have not been established. We recently showed that variation in normal thyroid status in healthy euthyroid postmenopausal women is associated with changes in bone mineral density and fracture risk (11), further demonstrating the sensitivity of the adult skeleton to thyroid status.
The mechanisms of T3 action in the skeleton have been investigated in Thra and Thrb knock-out mice lacking TRα and TRβ (12–14) and in mice with targeted mutations affecting Thra1 and Thrb (15–17). These studies demonstrated that T3 action in bone is mediated primarily by TRα and that thyroid hormones stimulate bone formation and mineralization during skeletal growth but increase bone turnover and bone loss in adulthood (2, 13). Analyses of TR mutant mice harboring a PV mutation targeted to either Thra or Thrb have been particularly instructive because of their clear skeletal phenotypes (16–18).
The mutation was originally described in a patient affected with severe resistance to thyroid hormone (RTH) (19). The PV mutation is a C-insertion at codon 448 in the TRHB gene resulting in a frameshift of the carboxyl-terminal 14 amino acids and generating a mutant TRβ protein that cannot bind T3 or activate target gene transcription and that acts as a dominant-negative antagonist of wild-type TRs (19, 20). Homozygous ThrbPV/PV mutant mice have a severely impaired pituitary-thyroid axis with markedly elevated circulating thyroid hormone levels (20), whereas Thra1PV/+ heterozygous mice have mild thyroid failure but are systemically euthyroid (21).
ThrbPV/PV mice have advanced endochondral and intramembranous ossification with increased bone mineral deposition during growth, whereas Thra1PV/+ mice have grossly delayed endochondral and intramembranous ossification, reduced bone mineral deposition, and severe growth retardation (16–18). In situ hybridization analysis of skeletal T3 target gene expression indicated evidence of increased T3 signaling in ThrbPV/PV bone but impaired T3 action in the Thra1PV/+ skeleton. Thus, we hypothesized that elevated thyroid hormones in ThrbPV/PV mice drive a phenotype of skeletal thyroid hormone excess via increased stimulation of TRα1 in bone, whereas the phenotype of skeletal thyroid hormone deficiency in Thra1PV/+ mice results from impaired TRα1 activity despite normal circulating thyroid status (18).
Although these studies have resulted in significant advances in our understanding of the molecular mechanisms of T3 action in bone, the downstream signaling pathways that mediate T3 responses in the skeleton are poorly understood. To investigate further, we performed microarray studies comparing gene expression in primary osteoblasts from wild-type mice and ThrbPV/PV mice with evidence of increased T3 signaling in bone. In ThrbPV/PV osteoblasts and in ThrbPV/PV mice in vivo, there was increased activation of canonical Wnt signaling, a pathway that is essential for both skeletal development and the maintenance of adult bone mass (22).
EXPERIMENTAL PROCEDURES
ThrbPV Mice
Animal studies were conducted in accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals and were approved by the NCI, National Institutes of Health, Animal Care and Use Committee. Wild-type and homozygous ThrbPV/PV mutant mice were bred and genotyped as described (20).
Primary Osteoblast Culture
Primary osteoblasts were prepared from calvaria of 3–5-day-old mice. Calvaria were dissected free of soft tissue and washed in PBS followed by Hanks' balanced salt solution containing penicillin/streptomycin/neomycin (PSN) (50 μg/ml, 50 μg/ml, and 100 μg/ml, respectively) and amphotericin B (1.5 μg/ml). Washed calvaria were cut into 2–3-mm strips and digested with 0.5% trypsin in PBS containing 4 mm EDTA, PSN, and fungizone. Osteoblasts were released by sequential 30-min digestions with type II collagenase (Worthington Biochemical Corp.). Fibroblasts released from the first two digestions were discarded, and osteoblasts from digestions 3–5 were pooled, centrifuged, and resuspended in α-minimum essential medium supplemented with 10% heat-inactivated FBS, PSN, and amphotericin B. After 4–5 h, the medium was replaced, and adherent cells were cultured for 7 days in differentiation-permissive α-minimum essential medium containing 10% heat-inactivated FBS or 10% thyroid hormone-deprived heat-inactivated FBS supplemented with PSN, amphotericin B, ascorbic acid (50 μg/ml), and α-glycerophosphate (5 mm) in the absence or presence of T3 (100 nm).
Microarray
The beaded mouse arrays contained 30,336 cDNAs. Hybridization, scanning, and image analysis were performed as described (www.nhgri.nih.gov/DIR/microarray) (23–26). Fluorescence-labeled cDNA was synthesized from 20 μg of pooled calvarial osteoblast total RNA obtained from six wild-type and seven ThrbPV/PV mice or from individual mice. Labeled cDNA was synthesized by oligo(dT)-primed polymerization in the presence of aminoallyl-dUTP (Amersham Biosciences) and coupled to either Cy-3 or Cy-5. Image analysis was performed using the DeArray software (Signal Analytics, Vienna, VA) (23, 26). The red and green channel fluorescent images from one array constituted the raw data from which differential gene expression ratios were calculated. Raw data values were thresholded to have values equal to the largest non-negative value and log-transformed. Red-to-green intensity ratios for all genes were determined, and values within each array were normalized to a median value of 1. Ratios associated with quality factors between 0.5 and 1.0 were included for further analysis, and only genes with ratios of ≥2 or <0.5 in osteoblasts from ThrbPV/PV mice relative to wild type were considered to be significantly up- or down-regulated.
qRT-PCR
Total RNA was extracted from primary osteoblasts using TRIzol (Invitrogen) according to the manufacturer's instructions. Quantitative real-time RT-PCR was performed using a SYBR Green quantitative RT-PCR kit (Sigma-Aldrich) or QuantiTect quantitative RT-PCR kit (Qiagen) according to the manufacturers' instructions and a LightCycler thermal cycler (Roche Applied Science, Mannheim, Germany). Briefly, 2.5 μl of forward primer (2 μm) and 2 μl of reverse primer (2 μm) were added to 15 μl of SYBR Green enzyme reaction mix or QuantiTect RT reaction mix. The cycles were: 55 °C for 30 min; 95 °C for 30 s; 95 °C for 15 s; 58 °C for 30 s; and 72 °C for 30 s; 65–95 °C with a heating rate of 0.1 °C/s; and a cooling step to 40 °C. Primer sequences used were: procollagen type I, αI (Col1a1) 5′-GGATTCCCGTTCGAGTACGGAA-3′ (sense) and 5′-TGCCAGTCTGCTGGTCCATGTA-3′ (antisense); integrin, α11 (Itga11) 5′-AGCTTCTACCTGGTGGGAAAC-3′ (sense) and 5′-GAGATCTCAAATGTGACCTGGC-3′ (antisense); mouse-related RAS viral oncogene homolog 2 (Rras2) 5′-GCCCGGCTGGACATTTTGGATA-3′ (sense) and 5′-CGGTCCTTTACTCTGAGAATC-3′ (antisense); cyclin D1 (Ccnd1) 5′-CTGCAAATGGAACTGCTTCTGG-3′ (sense) and 5′-TTCACATCTGTGGCACAGAGG-3′ (antisense); β-catenin (Ctnnb1) 5′-AGCCGAGATGGCCCAGAAT-3′ (sense) and 5′-AAGGGCAAGGTTTCGAATCAA-3′ (antisense); Lrp5 5′-AAGGTTGTCGGAACCAACCCATGT-3′ (sense) and 5′-TGATCGTCTTGAGGCTGACATCAGT-3′ (antisense); Lrp6 5′-GTGATCTCTCAGGTGCCAACA-3′ (sense) and 5′-GCACAAGGGTGCTGTCTGTACTC-3′ (antisense); glyceraldehyde-3-phosphatase (Gapdh) 5′-ACATCATCCCTGCATCCACT-3′ (sense) and 5′-GTCCTCAGTGTAGCCCAAG-3′ (antisense).
Wnt Signaling Pathway PCR Array
A Wnt signaling pathway RT2 ProfilerTM PCR array was used according to the manufacturer's instructions (SABiosciences, Frederick, MD). Briefly, primary osteoblast total RNA obtained from two wild-type and two homozygous TRβPV/PV mutant littermate mice was extracted using TRIzol, and 1 μg of RNA was used to synthesize wild-type and ThrbPV/PV cDNA using SuperScript II reverse transcriptase. cDNA was used as a template for Wnt signaling pathway PCR arrays and experiments were performed in duplicate. Arrays contained five reference housekeeping genes for normalization of gene expression. qRT-PCR was performed using a Bio-Rad IQ5 thermal cycler. The cycles were: 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s, 55 °C for 35 s, and 72 °C for 30 s.
In Situ Hybridization
Expression of Rankl, Runx2, and Wnt4 mRNAs in growth plate chondrocytes and osteoblasts was determined by in situ hybridization of tissue sections obtained from postnatal days 0 and 14 mice. A bacterial neomycin resistance gene cRNA probe (Roche Applied Science, Lewes, Sussex, UK) was used as a negative control for all hybridizations. Mouse Rankl (nucleotides 695–1110; GenBankTM accession number NM_011613.3), Runx2 (nucleotides 1350–1781; GenBank accession number NM_009820.2), and Wnt4 (nucleotides 161–562; GenBank accession number NM_009523.2) partial cDNAs were isolated by RT-PCR from chondrogenic ATDC5 cells (27) using the following primers: Rankl, forward, 5′-GTCACTCTGTCCTCTTGGTA-3′, reverse, 5′-GAGTCTCAGTCTATGTCCTG-3′; Runx2, forward, 5′-GTTCCCAAGCATTTCATCCC-3′, reverse, 5′-CGCCAAACAGACTCATCCAT-3′; Wnt4, forward, 5′-AAGAGGAGACGTGCGAGAAA-3′, reverse, 5′-GGACGTCCACAAAGGACTGT-3′. PCR products were subcloned into pGEM-T Easy vector (Promega, Southampton, Hampshire, UK) and sequenced. Rankl and Wnt4 constructs were linearized with SpeI, and the Runx2 construct was linearized with NcoI before digoxigenin-labeled cRNA probes were synthesized using T7 and SP6 RNA polymerases, respectively (Roche Applied Science). In situ hybridizations using alkaline phosphatase-labeled probes were performed on 3-μm deparaffinized sections as described in detail (17, 28, 29).
Transient Transfection and Adenovirus Infection
MC3T3 and UMR106 cells (1.5 × 105 cells/well of a 6-well plate) were plated 18–24 h before transfection in either α-minimum essential medium (MC3T3) or DMEM (UMR106) supplemented with 10% thyroid hormone-deprived FBS and PSN. Cells were transfected with a β-catenin-TCF4-responsive TOP-Flash reporter plasmid (TCF4; 1 μg) or thyroid hormone response element reporter plasmid (PAL-Luc; 1 μg) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. At the same time, adenovirus encoding TRα1 or TRβ1 was infected into cells at a multiplicity of infection of 10. After a 3-h incubation, medium was replaced by fresh 10% thyroid hormone-deprived medium with or without T3 (100 nm). Cells were lysed after 24 h with 3× cell lysis buffer (Pharmingen), and luciferase activity was determined according to the manufacturer's protocol using a Victor3 multilabel counter with dual-injection capability (PerkinElmer Life Sciences). Luciferase values were standardized to protein concentration.
Western Blotting
Western blot analysis of β-catenin and phospho-β-catenin was performed as described (30, 31). UMR106 cells were seeded in 6-mm wells (5 × 105 cells/well) in DMEM supplemented with 10% thyroid hormone-deprived FBS. After 24 h, the medium was changed to Opti-MEM (Invitrogen) prior to adenovirus infection. Cells were infected at a multiplicity of infection of 10 with adenovirus encoding FLAG-tagged TRβ1 or TRβ1PV. After 3 h, T3 (100 nm) was added, and cells were lysed 6 or 24 h later in 1× lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA) containing proteinase inhibitor (Complete Mini EDTA-free; Roche Applied Science) and protein phosphatase inhibitor cocktails (Thermo Scientific). Protein concentrations were determined by Bradford assay (Pierce), and 20 μg of total protein was used for Western blotting with the following antibodies: anti-β-catenin (1:1000 dilution, antibody 9562, Cell Signaling), anti-phospho-β-catenin (Ser552) (1:1000 dilution, antibody 9566, Cell Signaling), anti-GAPDH (1:1000 dilution, antibody 2118, Cell Signaling), and anti-TRβ (J53) (32). Protein expression was detected by enhanced chemiluminescence and quantified using ImageJ (rsb.info.nih.gov/ij/index.html) (30).
Statistics
Data were expressed as mean ± S.E. The differences between groups were examined for statistical significance using one-way analysis of variance followed by Tukey's multiple comparison post hoc test or by two-tailed Student's t test as appropriate. p values <0.05 were considered significant.
RESULTS
Microarray Analysis
Microarrays comprising 30,336 cDNAs were used to compare gene expression profiles between wild-type and ThrbPV/PV mice. Only genes up-regulated at least 2-fold or down-regulated at least 50% in mutants as compared with wild-type mice were considered to be of potential biological significance. Of 192 differentially expressed genes, 85 (44%) were up-regulated and 107 (56%) were down-regulated in ThrbPV/PV mice (supplemental Fig. 1). Empirical Bayes analysis indicated >97% likelihood that differences in gene expression resulted from true biological variation rather than by chance. To obtain deeper insight into the signaling pathways in osteoblasts associated with advanced skeletal development in ThrbPV/PV mice (16, 17), we classified differentially expressed genes according to function using controlled vocabulary gene ontology terms with the Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatics database (33) and Expression Analysis Systemic Explorer (EASE) software (david.niaid.nih.gov/david/ease.htm) (34). Using this method, seven functional categories were identified in which to classify the 192 differentially expressed genes: focal adhesion pathway; signal transduction; enzyme regulation; development; transcriptional regulation; regulation of biological processes; and structural molecules (supplemental Tables 1–7).
Functional Pathway Analysis and Validation of Microarray Data
Investigation of classification categories using the Kyoto Encyclopedia of Genes and Genomes (KEGG) Resource revealed that nine differentially expressed genes act within the focal adhesion pathway (supplemental Table 1). This network converges at contact points between the cell membrane and extracellular matrix where actin filaments anchor to integrin receptors via junctional protein complexes. These focal adhesion contact points contain both structural and signaling proteins that mediate intracellular responses to extracellular signals and regulate processes such as cell shape and motility, cell proliferation, differentiation and survival, and gene expression. Various extracellular signals that include mechanical stress, kinases and phosphatases, matrix ligands, hormones, and growth factors all impact on focal adhesion networks and demonstrate the fundamental significance of the pathway (35, 36).
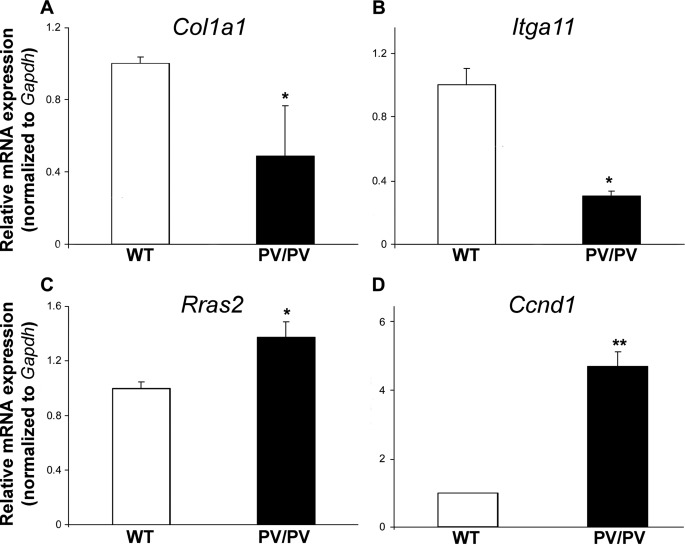
To substantiate results from microarray experiments, the expression of four focal adhesion pathway genes, Col1a1, Itga11, Rras2, and Ccnd1, was investigated by qRT-PCR in independent cultures of wild-type and ThrbPV/PV primary osteoblasts. In these studies, Col1a1 mRNA expression was reduced by 50% in ThrbPV/PV osteoblasts as compared with wild type, and Itga11 expression was decreased by 75%, whereas Rras2 expression increased 1.4-fold and Ccnd1 mRNA increased 4.7-fold (Fig. 1). These changes were consistent with the 80 and 90% reductions in Col1a1 and Itga11 expression and 2.8- and 2.5-fold increases in Rras2 and Ccnd1 expression determined in microarray studies. Furthermore, overexpression of Ccnd1 has been reported in human thyroid cancer (37–39) and in thyroid and pituitary glands of ThrbPV/PV mice, which are susceptible to both thyroid cancer and pituitary thyrotroph tumors (24, 40). Taken together, these studies validate the microarray studies and are consistent with previous studies in different tissues from ThrbPV/PV mice.
FIGURE 1.
qRT-PCR analysis of Col1a1 (A), Itga11 (B), Rras2 (C), and Ccnd1 (D) mRNA expression in primary osteoblasts isolated from wild-type (WT) and ThrbPV/PV (PV/PV) littermate mice. mRNA expression was normalized to Gapdh. Data are expressed as mean ± S.E. in ThrbPV/PV mice relative to WT and analyzed by two-tailed Student's t test (n = 3–6, *, p < 0.05, **, p < 0.01).
Expression of Wnt/β-Catenin Pathway Genes in TRβPV/PV Osteoblasts
Increased expression of Ccnd1 (2.5-fold) and the Drosophila homolog Frizzled 7 (Fzd7) (2.35-fold) in ThrbPV/PV osteoblasts (supplemental Table 4) implicates Wnt/β-catenin signaling, a key pathway that regulates bone mass (41), as a target for thyroid hormones in osteoblasts. Ccnd1 encodes the cell cycle regulator cyclin D1 and is a downstream target gene of Wnt/β-catenin signaling (42), and Fzd7 encodes a seven-transmembrane G-protein-coupled receptor that binds Wnt and mediates activation of the Wnt/β-catenin signaling cascade (43). Thus, we used a mouse Wnt signaling pathway PCR array to investigate expression of 84 genes related to Wnt-mediated signal transduction in ThrbPV/PV primary osteoblasts. 34 Wnt pathway genes were differentially expressed in ThrbPV/PV primary osteoblasts as compared with wild type (Table 1). Ccnd1 expression was increased 3.66-fold and Fzd7 expression was increased 1.83-fold in ThrbPV/PV osteoblasts, demonstrating consistency with microarray analyses. Overall, expression of 12 Wnt pathway genes was increased in ThrbPV/PV osteoblasts, whereas 22 genes were down-regulated.
TABLE 1.
Analysis of RT2 ProfilerTM PCR array mouse Wnt signaling pathway
| GenBank accession number | Symbol | Gene description | Fold change |
|---|---|---|---|
| Frizzled signaling pathway | |||
| NM_019725 | Tle2 | Transducin-like enhancer of split 2, homolog of Drosophila E(spl) | −1.88 |
| Frizzled-2 signaling pathway | |||
| NM_023653 | Wnt2 | Wingless-related MMTV integration site 2 | 2.22 |
| NM_009520 | Wnt2b | Wingless-related MMTV integration site 2b | −2.30 |
| NM_009525 | Wnt5b | Wingless-related MMTV integration site 5b | −2.22 |
| NM_009527 | Wnt7a | Wingless-related MMTV integration site 7a | 8.57 |
| NM_011720 | Wnt8b | Wingless-related MMTV integration site 8b | 2.97 |
| NM_009519 | Wnt11 | Wingless-related MMTV integration site 11 | 2.81 |
| NM_053116 | Wnt16 | Wingless-related MMTV integration site 16 | −6.54 |
| Negative regulation of the Wnt receptor | |||
| NM_010051 | Dkk1 | Dickkopf homolog 1 (Xenopus laevis) | 7.11 |
| Wnt binding antagonists | |||
| NM_013834 | sFRP1 | Secreted frizzled-related sequence protein 1 | −39.95 |
| NM_009144 | sFRP2 | Secreted frizzled-related sequence protein 2 | −390.72 |
| NM_016687 | sFRP4 | Secreted frizzled-related sequence protein 4 | −5.43 |
| Regulation of growth | |||
| NM_018865 | Wisp1 | WNT1-inducible signaling pathway protein 1 | −2.19 |
| Regulation of cell proliferation | |||
| NM_007614 | Ctnnb1 | Catenin (cadherin-associated protein), β 1 | −1.84 |
| NM_011098 | Pitx2 | Paired-like homeodomain transcription factor 2 | −2.38 |
| NM_009331 | Tcf7 | Transcription factor 7, T-cell specific | −2.07 |
| Other genes related to growth and proliferation | |||
| NM_008043 | Frat1 | Frequently rearranged in advanced T-cell lymphomas | −3.68 |
| Regulation of the cell cycle | |||
| NM_007631 | Ccnd1 | Cyclin D1 | 3.66 |
| NM_009829 | Ccnd2 | Cyclin D2 | 2.64 |
| Transcription factors | |||
| NM_008238 | Foxn1 | Forkhead box N1 | 4.14 |
| NM_007614 | Ctnnb1 | Catenin (cadherin-associated protein), β 1 | −1.84 |
| NM_011098 | Pitx2 | Paired-like homeodomain transcription factor 2 | −2.38 |
| NM_009331 | Tcf7 | Transcription factor 7, T-cell specific | −2.07 |
| Transcription regulators | |||
| NM_010235 | Fosl1 | Fos-like antigen 1 | 3.34 |
| NM_019725 | Tle2 | Transducin-like enhancer of split 2, homolog of Drosophila E(spl) | −1.88 |
| Protein kinase activity | |||
| NM_007631 | Ccnd1 | Cyclin D1 | 3.66 |
| Other Wnt signaling pathway-related genes | |||
| NM_008045 | Fshb | Follicle-stimulating hormone beta | 2.69 |
| NM_021457 | Fzd1 | Frizzled homolog 1 (Drosophila) | −3.41 |
| NM_008055 | Fzd4 | Frizzled homolog 4 (Drosophila) | −2.58 |
| NM_008056 | Fzd6 | Frizzled homolog 6 (Drosophila) | −2.11 |
| NM_008057 | Fzd7 | Frizzled homolog 7 (Drosophila) | 1.83 |
| NM_027280 | Nkd1 | Naked cuticle homolog (Drosophila) | −6.68 |
| NM_023638 | Porcn | Porcupine homolog (Drosophila) | −5.86 |
| NM_011915 | Wif1 | Wnt inhibitory factor 1 | −6.15 |
The canonical Wnt/β-catenin pathway is complex. Its activity is regulated by an interplay involving at least 19 Wnt ligands, 10 Frizzled receptors, lipoprotein-related receptors 5 and 6 (LRP5, LRP6), and various soluble inhibitors that act at different sites in the pathway to regulate its activity (22, 44, 45). Transduction of the Wnt signal following activation of the LRP/Frizzled co-receptor complex involves several intracellular proteins and enzymes that ultimately lead to dephosphorylation of the transcription factor β-catenin. This prevents sequestration and degradation of β-catenin in the cytoplasm, resulting in its translocation to the nucleus, where it acts as a specific co-activator of Wnt-dependent target gene expression by associating with transcription factors of the T-cell factor/lymphoid enhancer-binding factor (Tcf/Lef) family (22, 44, 45). In this context, therefore, it is not possible to predict the overall functional response of the Wnt pathway to external regulating factors such as thyroid hormones simply by analyzing changes in the expression patterns of mRNAs encoding the various Wnt pathway components.
Wnt/β-Catenin Pathway Activity in Skeleton in ThrbPV/PV Mice
Nevertheless, the increased expression of Fzd7 and Ccnd1 in ThrbPV/PV primary osteoblasts seen in independent microarray, RT-PCR, and Wnt PCR array studies suggests the Wnt/β-catenin pathway may be activated in bone in ThrbPV/PV mice. To investigate this possibility, we determined expression of the Rankl (receptor activator of NFκB ligand) and Runx2 (Runt-related transcription factor-2) Wnt target genes in bones obtained from neonatal and 2-week-old ThrbPV/PV mice by in situ hybridization. Expression of Rankl is reduced following activation of Wnt signaling (46), whereas expression of Runx2 is increased (47).
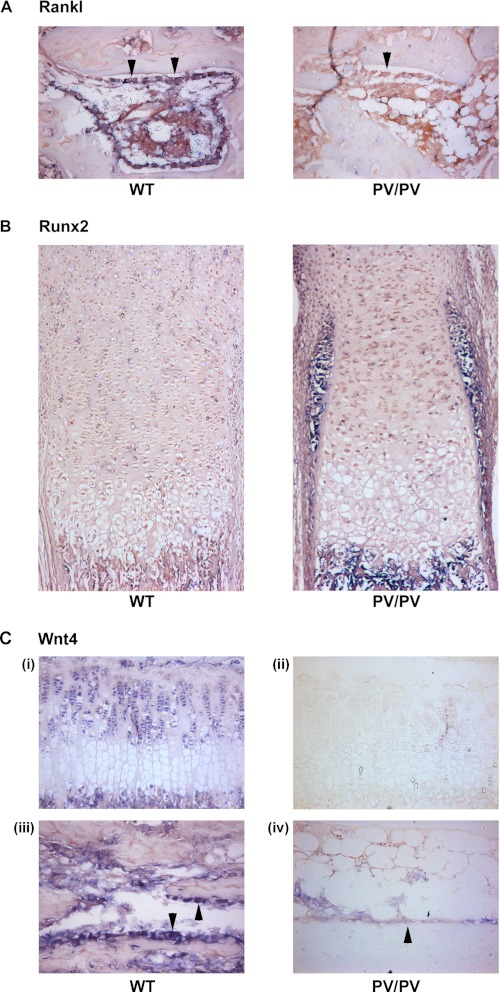
In 2-week-old ThrbPV/PV mice, Rankl expression was markedly reduced or absent in osteoblasts lining trabecular bone surfaces as compared with expression that was readily detectable in wild-type littermates (Fig. 2A). In neonatal mice, Runx2 expression was increased markedly in perichondrial cells surrounding the developing growth plate in ThrbPV/PV mice as compared with wild-type littermates (Fig. 2B). These findings are consistent with increased activation of the Wnt/β-catenin pathway in bone in ThrbPV/PV mice during endochondral ossification and postnatal growth.
FIGURE 2.
In situ hybridization analysis. A–C, in situ hybridization analysis of Rankl mRNA expression in osteoblasts (arrowheads) lining trabecular bone surfaces (400× magnification) (A), Runx2 mRNA expression in perichondrial cells adjacent to the tibial growth plate (100× magnification) (B), and Wnt4 mRNA expression (C) in tibial growth plate chondrocytes (200× magnification) (panels i and ii) and osteoblasts (arrowheads) lining trabecular bone surfaces (400× magnification) (panels iii and iv) in 2-week-old (A and C) and neonatal (B) wild-type (WT) and ThrbPV/PV (PV/PV) littermate mice.
In previous studies, Wang et al. (48, 49) found that T3 activates Wnt signaling in growth plate chondrocytes. They demonstrated increased expression of β-catenin (Ctnnb1) and its target gene Runx2, as well as increased expression of the activating Wnt ligand Wnt4 following T3 treatment.
To investigate further, we determined expression of Wnt4 in bones obtained from 2-week-old ThrbPV/PV mice. In ThrbPV/PV mice, Wnt4 expression was markedly reduced or absent in both growth plate chondrocytes and osteoblasts lining trabecular bone surfaces as compared with expression that was readily detectable in wild-type littermates (Fig. 2C). These data are consistent with the dominant-negative activity of mutant TRβPV protein (19, 20) and with previous studies indicating that Wnt4 expression is increased in chondrocytes in response to T3 (48, 49). Nevertheless, the findings reveal an overall discrepancy in ThrbPV/PV mice, in which increased Wnt/β-catenin activity in bone and cartilage is accompanied by reduced expression of the activating ligand Wnt4.
Regulation of β-Catenin Signaling in Osteoblasts by T3
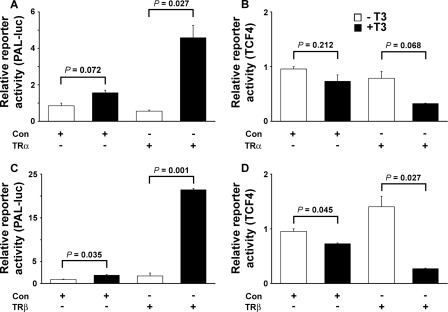
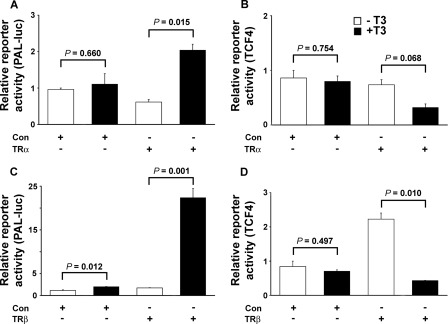
To investigate whether T3 influences activity of the Wnt/β-catenin pathway, we determined whether T3 regulated expression of a β-catenin/Tcf/Lef-responsive reporter construct (50, 51) in transfected osteoblasts infected with adenovirus expressing TRα1 or TRβ1 (Figs. 3 and 4). Transfection of rat UMR106 osteosarcoma cells with a well characterized thyroid hormone response element reporter (PAL-Luc) (52, 53) in the presence of TRα1, but not TRβ1, resulted in unliganded apoTRα-mediated repression of reporter gene activity in the absence of T3, as reported previously (53). In the presence of T3, TRα1 and TRβ1 mediated 8.2- and 12.5-fold increases in reporter gene activity, respectively (Fig. 3), thus demonstrating functional integrity of TR signaling. By contrast, T3 treatment repressed activity of the β-catenin/Tcf/Lef reporter in UMR106 cells co-transfected with TRα1 (expression reduced 2.4-fold) or TRβ1 (expression reduced 5.2-fold). Treatment with T3 in the absence of co-transfected TR did not affect β-catenin/Tcf/Lef reporter activity, and apoTR in the absence of T3 also did not affect reporter gene expression. Thus, β-catenin signaling in UMR106 cells was inhibited by T3 by a TR-dependent mechanism (Fig. 3). Experiments in mouse osteoblastic MC3T3 cells similarly resulted in repression of β-catenin/Tcf/Lef reporter gene activity by T3 by a TR-dependent mechanism, with equivalent results also obtained using adenovirus expressing either TRα1 or TRβ1 (Fig. 4). Thus, T3 inhibits activity of the Wnt/β-catenin signaling pathway in osteoblasts.
FIGURE 3.
UMR106 cells. A–D, activities of T3-responsive (PAL-Luc) (A and C) and β-catenin-TCF4-responsive TOP-Flash (TCF4) (B and D) luciferase reporter constructs in the absence or presence of TRα1 (A and B) or TRβ1 (C and D) or control adenovirus following treatment of infected UMR106 cells with vehicle or T3 (100 nm) for 24 h. Data were expressed as mean ± S.E. and analyzed by one-way analysis of variance followed by Tukey's multiple comparison post hoc test (n = 3).
FIGURE 4.
MC3T3 cells. A–D, activities of T3-responsive (PAL-Luc) (A and C) and β-catenin-TCF4-responsive TOP-Flash (TCF4) (B and D) luciferase reporter constructs in the absence or presence of TRα1 (A and B) or TRβ1 (C and D) or control adenovirus following treatment of infected MC3T3 cells with vehicle or T3 (100 nm) for 24 h. Data were expressed as mean ± S.E. and analyzed by one-way analysis of variance followed by Tukey's multiple comparison post hoc test (n = 3).
TRβPV Regulates β-Catenin stability in Osteoblasts
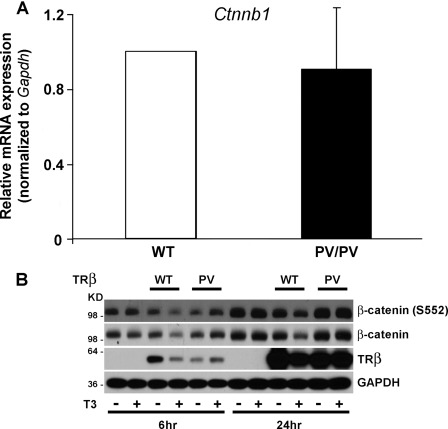
In microarray studies, β-catenin mRNA expression did not differ between ThrbPV/PV and wild-type osteoblasts (data not shown), whereas in Wnt PCR array experiments, expression of β-catenin was reduced in ThrbPV/PV osteoblasts by 2.38-fold (60%) (Table 1). However, in independent RT-PCR analyses, the level of β-catenin mRNA did not differ in ThrbPV/PV osteoblasts, suggesting that changes in β-catenin mRNA expression do not account for increased activation of the canonical Wnt pathway in ThrbPV/PV osteoblasts (Fig. 5A). Similarly, expression of Lrp5 and Lrp6 mRNAs did not differ in ThrbPV/PV osteoblasts as compared with wild-type in RT-PCR studies, suggesting that altered expression of these Wnt co-receptors also does not account for increased Wnt signaling in ThrbPV/PV osteoblasts (supplemental Fig. 2).
FIGURE 5.
A, qRT-PCR analysis of Ctnnb1 mRNA expression in primary osteoblasts isolated from wild-type (WT) and ThrbPV/PV (PV/PV) littermate mice. mRNA expression was normalized to Gapdh. Data expressed as mean ± S.E. in PV/PV mice relative to WT. B, representative Western blot analysis of β-catenin and phosphorylated β-catenin (β-catenin (S552)) expression in UMR106 cells infected with adenovirus encoding FLAG-tagged TRβ (WT) or TRβPV (PV). Cells were treated with vehicle or T3 (100 nm) for 6 or 24 h to determine endogenous β-catenin and TR protein expression. GAPDH is used as a loading control.
To investigate in further detail, we determined the effect of T3 on endogenous β-catenin and TRβ protein in UMR106 osteoblastic cells in the absence and presence of wild-type receptor or mutant TRβPV (Fig. 5B). Wild-type TRβ and β-catenin protein concentrations were decreased following T3 treatment for 6 and 24 h, consistent with the previously reported T3-induced proteasome-mediated degradation of TRβ in other cell types and the physical interaction between unliganded TRβ and β-catenin that is dissociated by T3 (54). By contrast, TRβPV levels were not affected by T3 as the mutant protein cannot bind ligand. Accordingly, there was no effect of T3 treatment on β-catenin levels in cells lacking wild-type or mutant TR. Furthermore, β-catenin was stabilized in TRβPV-expressing UMR106 cells (Fig. 5B). Taken together, these data demonstrate a gain of function for TRβPV in osteoblasts that stabilizes β-catenin in the presence of T3.
DISCUSSION
These studies demonstrate increased activity of the canonical Wnt/β-catenin pathway in the skeleton of ThrbPV/PV mice in vivo and in ThrbPV/PV osteoblasts in vitro. By contrast, T3 treatment of osteoblastic cells results in inhibition of Wnt/β-catenin signaling.
Activation of Wnt signaling leads to a postnatal increase in bone mass that is mediated by osteoblasts, whereas deletion of the Wnt co-receptor Lrp5 results in reduced bone mass (55, 56). Similarly, deletion of sclerostin, a secreted Wnt inhibitor in bone, results in a phenotype of high bone mass (57). Thus, activation of the Wnt pathway stimulates osteoblastic bone formation during growth and accrual of bone mass in adults, whereas loss-of-function mutations in the Wnt pathway lead to low bone mass due to defective osteoblast proliferation and maturation (22, 58). The finding of increased activation of the Wnt/β-catenin pathway in the skeleton in ThrbPV/PV mice in vivo and in ThrbPV/PV osteoblasts is consistent with the established role of the Wnt pathway in bone (59) and the phenotype of advanced ossification in ThrbPV/PV mice (17).
In the context of our previous hypothesis that advanced ossification in ThrbPV/PV mice results from increased activation of the predominantly expressed wild-type TRα in skeletal cells (18), the finding of increased Wnt signaling activity in ThrbPV/PV bone suggested that T3 would stimulate Wnt/β-catenin signaling in osteoblasts. However, in two different osteoblastic cell lines in the presence of either TRα or TRβ, T3 inhibited activity of a β-catenin/Tcf/Lef reporter construct in a TR-dependent manner.
In previous studies, we showed that β-catenin activity was also increased in thyroid tumors that develop in ThrbPV/PV mice with increasing age (40). In studies to investigate the molecular basis for increased β-catenin activity in ThrbPV/PV thyroid tissue, we demonstrated that unliganded wild-type TRβ and TRβPV proteins physically interact with β-catenin, leading to its stabilization and an increase in Wnt signaling (54). The association between wild-type TRβ and β-catenin was disrupted by T3, leading to targeted degradation of β-catenin by the proteasome. T3- and TRβ-dependent degradation of β-catenin resulted in repression of Wnt signaling by thyroid hormone. By contrast, the interaction between mutant TRβPV and β-catenin was not disrupted by T3, and thyroid hormone treatment had no effect on the increased activation of Wnt signaling in TRβPV-expressing cells (54). These data are consistent with the current findings of increased Wnt/β-catenin signaling in bone in ThrbPV/PV mice in vivo and the stabilization of β-catenin protein in osteoblasts expressing TRβPV. They are also consistent with the inhibition of Wnt signaling observed in osteoblasts treated with T3 and the degradation of β-catenin in osteoblasts expressing wild-type TRβ.
Nevertheless, our previous studies in mutant mice indicate that T3 action in bone is mediated primarily by TRα, although both TRα and TRβ are expressed in skeletal cells (1, 2, 13). The finding that Wnt/β-catenin signaling was inhibited equally by T3 in the presence of either TRα or TRβ (Figs. 3 and 4) suggests that, like TRβ (54), TRα is also able to associate with β-catenin. By analogy, it could be expected that Wnt/β-catenin signaling activity would be increased in the skeleton of Thra1PV/+ mice. Although this has not been investigated, the finding of an opposite phenotype of delayed ossification, reduced bone mineral deposition, and growth retardation in Thra1PV/+ mice as compared with ThrbPV/PV mice (16, 17) is not consistent with such a hypothesis. One possibility to account for the difference in skeletal phenotypes between Thra1PV/+ and ThrbPV/PV mice is that TRα1PV differs from TRβPV and might not be able to interact with and stabilize β-catenin and thus be unable to increase activity of the Wnt pathway. Alternatively, complete blockade of thyroid hormone signaling in Thra1PV/+ bone that results in severe developmental delay in the skeleton is very likely to override any effect of an increase in Wnt signaling activity that might result from stabilization of β-catenin by interaction with mutant TRα1PV protein. Studies of the Wnt/β-catenin pathway in Thra1PV/+ mice and osteoblasts will be required to determine which of these possibilities is correct.
Taken together, these considerations suggest that the skeletal phenotype of delayed ossification in Thra1PV/+ mice results solely from deficient thyroid hormone action in bone, whereas the current studies demonstrate that the advanced ossification in ThrbPV/PV mice results from activation of the Wnt signaling pathway in bone as well as from increased T3 action. Although we show that T3 normally inhibits activity of the Wnt pathway in osteoblasts, this paradoxical finding results from a gain of function of the mutant TRβPV protein, which is able to associate with and stabilize β-catenin in the presence of T3 (54). In addition to this gain-of-function activity, the reduced levels of Wnt4 expression in ThrbPV/PV chondrocytes and osteoblasts in vivo suggest that the mutant TRβPV protein also exhibits dominant-negative activity in the skeleton to inhibit previously described stimulatory effects of T3 on Wnt4 expression (48, 49).
Overall, the skeletal consequences of the TRβ PV mutation are a complex balance resulting from a combination of (i) increased T3 action mediated by the actions of elevated thyroid hormones on TRα in bone, (ii) a gain of function of the mutant TRβPV protein to stabilize β-catenin and activate Wnt signaling, and (iii) dominant-negative effects of TRβPV on the expression of certain T3 target genes such as Wnt4.
Supplementary Material
This work was authored, in whole or in part, by National Institutes of Health staff.

This article contains supplemental Figs. 1 and 2 and Tables 1–7.
- TR
- thyroid hormone receptor
- T3
- 3,5,3′-l-triiodothyronine
- PSN
- penicillin/streptomycin/neomycin
- qRT-PCR
- quantitative RT-PCR
- LRP
- lipoprotein-related receptor
- MMTV
- murine mammary tumor virus
- Tcf/Lef
- T-cell factor/lymphoid enhancer-binding factor.
REFERENCES
- 1. Bassett J. H., Williams G. R. (2008) Critical role of the hypothalamic-pituitary-thyroid axis in bone. Bone 43, 418–426 [DOI] [PubMed] [Google Scholar]
- 2. Gogakos A. I., Duncan Bassett J. H., Williams G. R. (2010) Thyroid and bone. Arch. Biochem. Biophys. 503, 129–136 [DOI] [PubMed] [Google Scholar]
- 3. Cheng S. Y., Leonard J. L., Davis P. J. (2010) Molecular aspects of thyroid hormone actions. Endocr. Rev. 31, 139–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rivkees S. A., Bode H. H., Crawford J. D. (1988) Long-term growth in juvenile acquired hypothyroidism: the failure to achieve normal adult stature. N. Engl. J. Med. 318, 599–602 [DOI] [PubMed] [Google Scholar]
- 5. Segni M., Leonardi E., Mazzoncini B., Pucarelli I., Pasquino A. M. (1999) Special features of Graves' disease in early childhood. Thyroid 9, 871–877 [DOI] [PubMed] [Google Scholar]
- 6. Mosekilde L., Eriksen E. F., Charles P. (1990) Effects of thyroid hormones on bone and mineral metabolism. Endocrinol. Metab. Clin. North Am. 19, 35–63 [PubMed] [Google Scholar]
- 7. Bauer D. C., Ettinger B., Nevitt M. C., Stone K. L. (2001) Risk for fracture in women with low serum levels of thyroid-stimulating hormone. Ann. Intern. Med. 134, 561–568 [DOI] [PubMed] [Google Scholar]
- 8. Vestergaard P., Mosekilde L. (2002) Fractures in patients with hyperthyroidism and hypothyroidism: a nationwide follow-up study in 16,249 patients. Thyroid 12, 411–419 [DOI] [PubMed] [Google Scholar]
- 9. Vestergaard P., Mosekilde L. (2003) Hyperthyroidism, bone mineral, and fracture risk: a meta-analysis. Thyroid 13, 585–593 [DOI] [PubMed] [Google Scholar]
- 10. Vestergaard P., Rejnmark L., Mosekilde L. (2005) Influence of hyper- and hypothyroidism, and the effects of treatment with antithyroid drugs and levothyroxine on fracture risk. Calcif. Tissue Int. 77, 139–144 [DOI] [PubMed] [Google Scholar]
- 11. Murphy E., Glüer C. C., Reid D. M., Felsenberg D., Roux C., Eastell R., Williams G. R. (2010) Thyroid function within the upper normal range is associated with reduced bone mineral density and an increased risk of nonvertebral fractures in healthy euthyroid postmenopausal women. J. Clin. Endocrinol. Metab. 95, 3173–3181 [DOI] [PubMed] [Google Scholar]
- 12. Bassett J. H., O'Shea P. J., Sriskantharajah S., Rabier B., Boyde A., Howell P. G., Weiss R. E., Roux J. P., Malaval L., Clement-Lacroix P., Samarut J., Chassande O., Williams G. R. (2007) Thyroid hormone excess rather than thyrotropin deficiency induces osteoporosis in hyperthyroidism. Mol. Endocrinol. 21, 1095–1107 [DOI] [PubMed] [Google Scholar]
- 13. Bassett J. H., Williams G. R. (2009) The skeletal phenotypes of TRα and TRβ mutant mice. J. Mol. Endocrinol. 42, 269–282 [DOI] [PubMed] [Google Scholar]
- 14. Gauthier K., Plateroti M., Harvey C. B., Williams G. R., Weiss R. E., Refetoff S., Willott J. F., Sundin V., Roux J. P., Malaval L., Hara M., Samarut J., Chassande O. (2001) Genetic analysis reveals different functions for the products of the thyroid hormone receptor α locus. Mol. Cell. Biol. 21, 4748–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bassett J. H., Nordström K., Boyde A., Howell P. G., Kelly S., Vennström B., Williams G. R. (2007) Thyroid status during skeletal development determines adult bone structure and mineralization. Mol. Endocrinol. 21, 1893–1904 [DOI] [PubMed] [Google Scholar]
- 16. O'Shea P. J., Bassett J. H., Sriskantharajah S., Ying H., Cheng S. Y., Williams G. R. (2005) Contrasting skeletal phenotypes in mice with an identical mutation targeted to thyroid hormone receptor α1 or β. Mol. Endocrinol. 19, 3045–3059 [DOI] [PubMed] [Google Scholar]
- 17. O'Shea P. J., Harvey C. B., Suzuki H., Kaneshige M., Kaneshige K., Cheng S. Y., Williams G. R. (2003) A thyrotoxic skeletal phenotype of advanced bone formation in mice with resistance to thyroid hormone. Mol. Endocrinol. 17, 1410–1424 [DOI] [PubMed] [Google Scholar]
- 18. O'Shea P. J., Bassett J. H., Cheng S. Y., Williams G. R. (2006) Characterization of skeletal phenotypes of TRα1 and TRβ mutant mice: implications for tissue thyroid status and T3 target gene expression. Nucl. Recept. Signal 4, e011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parrilla R., Mixson A. J., McPherson J. A., McClaskey J. H., Weintraub B. D. (1991) Characterization of seven novel mutations of the c-erbA β gene in unrelated kindreds with generalized thyroid hormone resistance: evidence for two ”hot spot“ regions of the ligand binding domain. J. Clin. Invest. 88, 2123–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaneshige M., Kaneshige K., Zhu X., Dace A., Garrett L., Carter T. A., Kazlauskaite R., Pankratz D. G., Wynshaw-Boris A., Refetoff S., Weintraub B., Willingham M. C., Barlow C., Cheng S. (2000) Mice with a targeted mutation in the thyroid hormone β receptor gene exhibit impaired growth and resistance to thyroid hormone. Proc. Natl. Acad. Sci. U.S.A. 97, 13209–13214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaneshige M., Suzuki H., Kaneshige K., Cheng J., Wimbrow H., Barlow C., Willingham M. C., Cheng S. (2001) A targeted dominant-negative mutation of the thyroid hormone α1 receptor causes increased mortality, infertility, and dwarfism in mice. Proc. Natl. Acad. Sci. U.S.A. 98, 15095–15100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Clevers H. (2006) Wnt/β-Catenin signaling in development and disease. Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 23. Chen Y., Dougherty E. R., Bittner M. (1997) Ratio-based decisions and the quantitative analysis of cDNA microarray images. J. Biomed. Opt. 2, 364–374 [DOI] [PubMed] [Google Scholar]
- 24. Furumoto H., Ying H., Chandramouli G. V., Zhao L., Walker R. L., Meltzer P. S., Willingham M. C., Cheng S. Y. (2005) An unliganded thyroid hormone β receptor activates the cyclin D1/cyclin-dependent kinase/retinoblastoma/E2F pathway and induces pituitary tumorigenesis. Mol. Cell. Biol. 25, 124–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Khan J., Wei J. S., Ringnér M., Saal L. H., Ladanyi M., Westermann F., Berthold F., Schwab M., Antonescu C. R., Peterson C., Meltzer P. S. (2001) Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat. Med. 7, 673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nimmakayalu M., Henegariu O., Ward D. C., Bray-Ward P. (2000) Simple method for preparation of fluor/hapten-labeled dUTP. BioTechniques 28, 518–522 [DOI] [PubMed] [Google Scholar]
- 27. Barnard J. C., Williams A. J., Rabier B., Chassande O., Samarut J., Cheng S. Y., Bassett J. H., Williams G. R. (2005) Thyroid hormones regulate fibroblast growth factor receptor signaling during chondrogenesis. Endocrinology 146, 5568–5580 [DOI] [PubMed] [Google Scholar]
- 28. Stevens D. A., Harvey C. B., Scott A. J., O'Shea P. J., Barnard J. C., Williams A. J., Brady G., Samarut J., Chassande O., Williams G. R. (2003) Thyroid hormone activates fibroblast growth factor receptor-1 in bone. Mol. Endocrinol. 17, 1751–1766 [DOI] [PubMed] [Google Scholar]
- 29. Stevens D. A., Hasserjian R. P., Robson H., Siebler T., Shalet S. M., Williams G. R. (2000) Thyroid hormones regulate hypertrophic chondrocyte differentiation and expression of parathyroid hormone-related peptide and its receptor during endochondral bone formation. J. Bone Miner. Res. 15, 2431–2442 [DOI] [PubMed] [Google Scholar]
- 30. Bhat M. K., Dace A., Cheng S. Y. (1999) Tissue-specific differential repression of gene expression by a dominant-negative mutant of thyroid hormone β1 receptor. Thyroid 9, 411–418 [DOI] [PubMed] [Google Scholar]
- 31. Miller L. D., Park K. S., Guo Q. M., Alkharouf N. W., Malek R. L., Lee N. H., Liu E. T., Cheng S. Y. (2001) Silencing of Wnt signaling and activation of multiple metabolic pathways in response to thyroid hormone-stimulated cell proliferation. Mol. Cell. Biol. 21, 6626–6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin K. H., Willingham M. C., Liang C. M., Cheng S. Y. (1991) Intracellular distribution of the endogenous and transfected β form of thyroid hormone nuclear receptor visualized by the use of domain-specific monoclonal antibodies. Endocrinology 128, 2601–2609 [DOI] [PubMed] [Google Scholar]
- 33. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 34. Hosack D. A., Dennis G., Jr., Sherman B. T., Lane H. C., Lempicki R. A. (2003) Identifying biological themes within lists of genes with EASE. Genome Biol. 4, R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mitra S. K., Hanson D. A., Schlaepfer D. D. (2005) Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 6, 56–68 [DOI] [PubMed] [Google Scholar]
- 36. Turner C. E. (2000) Paxillin and focal adhesion signaling. Nat. Cell Biol. 2, E231–236 [DOI] [PubMed] [Google Scholar]
- 37. Bièche I., Franc B., Vidaud D., Vidaud M., Lidereau R. (2001) Analyses of MYC, ERBB2, and CCND1 genes in benign and malignant thyroid follicular cell tumors by real-time polymerase chain reaction. Thyroid 11, 147–152 [DOI] [PubMed] [Google Scholar]
- 38. Khoo M. L., Beasley N. J., Ezzat S., Freeman J. L., Asa S. L. (2002) Overexpression of cyclin D1 and underexpression of p27 predict lymph node metastases in papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 87, 1814–1818 [DOI] [PubMed] [Google Scholar]
- 39. Khoo M. L., Ezzat S., Freeman J. L., Asa S. L. (2002) Cyclin D1 protein expression predicts metastatic behavior in thyroid papillary microcarcinomas but is not associated with gene amplification. J. Clin. Endocrinol. Metab. 87, 1810–1813 [DOI] [PubMed] [Google Scholar]
- 40. Ying H., Suzuki H., Furumoto H., Walker R., Meltzer P., Willingham M. C., Cheng S. Y. (2003) Alterations in genomic profiles during tumor progression in a mouse model of follicular thyroid carcinoma. Carcinogenesis 24, 1467–1479 [DOI] [PubMed] [Google Scholar]
- 41. Krishnan V., Bryant H. U., Macdougald O. A. (2006) Regulation of bone mass by Wnt signaling. J. Clin. Invest. 116, 1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tetsu O., McCormick F. (1999) β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398, 422–426 [DOI] [PubMed] [Google Scholar]
- 43. Wei W., Chua M. S., Grepper S., So S. K. (2011) Soluble Frizzled-7 receptor inhibits Wnt signaling and sensitizes hepatocellular carcinoma cells toward doxorubicin. Mol. Cancer 10, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Corr M. (2008) Wnt/β-Catenin signaling in the pathogenesis of osteoarthritis. Nat. Clin. Pract. Rheumatol. 4, 550–556 [DOI] [PubMed] [Google Scholar]
- 45. Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004) WNT and β-catenin signaling: diseases and therapies. Nat. Rev. Genet. 5, 691–701 [DOI] [PubMed] [Google Scholar]
- 46. Spencer G. J., Utting J. C., Etheridge S. L., Arnett T. R., Genever P. G. (2006) Wnt signaling in osteoblasts regulates expression of the receptor activator of NFκB ligand and inhibits osteoclastogenesis in vitro. J. Cell Sci. 119, 1283–1296 [DOI] [PubMed] [Google Scholar]
- 47. Dong Y. F., Soung do Y., Schwarz E. M., O'Keefe R. J., Drissi H. (2006) Wnt induction of chondrocyte hypertrophy through the Runx2 transcription factor. J. Cell. Physiol. 208, 77–86 [DOI] [PubMed] [Google Scholar]
- 48. Wang L., Shao Y. Y., Ballock R. T. (2007) Thyroid hormone interacts with the Wnt/β-catenin signaling pathway in the terminal differentiation of growth plate chondrocytes. J. Bone Miner. Res. 22, 1988–1995 [DOI] [PubMed] [Google Scholar]
- 49. Wang L., Shao Y. Y., Ballock R. T. (2010) Thyroid hormone-mediated growth and differentiation of growth plate chondrocytes involves IGF-1 modulation of β-catenin signaling. J. Bone Miner. Res. 25, 1138–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. He T. C., Sparks A. B., Rago C., Hermeking H., Zawel L., da Costa L. T., Morin P. J., Vogelstein B., Kinzler K. W. (1998) Identification of c-MYC as a target of the APC pathway. Science 281, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 51. Korinek V., Barker N., Morin P. J., van Wichen D., de Weger R., Kinzler K. W., Vogelstein B., Clevers H. (1997) Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science 275, 1784–1787 [DOI] [PubMed] [Google Scholar]
- 52. Harvey C. B., Bassett J. H., Maruvada P., Yen P. M., Williams G. R. (2007) The rat thyroid hormone receptor (TR) Δβ3 displays cell-, TR isoform-, and thyroid hormone response element-specific actions. Endocrinology 148, 1764–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Williams G. R. (2000) Cloning and characterization of two novel thyroid hormone receptor β isoforms. Mol. Cell. Biol. 20, 8329–8342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guigon C. J., Zhao L., Lu C., Willingham M. C., Cheng S. Y. (2008) Regulation of β-catenin by a novel nongenomic action of thyroid hormone β receptor. Mol. Cell. Biol. 28, 4598–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Boyden L. M., Mao J., Belsky J., Mitzner L., Farhi A., Mitnick M. A., Wu D., Insogna K., Lifton R. P. (2002) High bone density due to a mutation in LDL receptor-related protein 5. N. Engl. J. Med. 346, 1513–1521 [DOI] [PubMed] [Google Scholar]
- 56. Gong Y., Slee R. B., Fukai N., Rawadi G., Roman-Roman S., Reginato A. M., Wang H., Cundy T., Glorieux F. H., Lev D., Zacharin M., Oexle K., Marcelino J., Suwairi W., Heeger S., Sabatakos G., Apte S., Adkins W. N., Allgrove J., Arslan-Kirchner M., Batch J. A., Beighton P., Black G. C., Boles R. G., Boon L. M., Borrone C., Brunner H. G., Carle G. F., Dallapiccola B., De Paepe A., Floege B., Halfhide M. L., Hall B., Hennekam R. C., Hirose T., Jans A., Jüppner H., Kim C. A., Keppler-Noreuil K., Kohlschuetter A., LaCombe D., Lambert M., Lemyre E., Letteboer T., Peltonen L., Ramesar R. S., Romanengo M., Somer H., Steichen-Gersdorf E., Steinmann B., Sullivan B., Superti-Furga A., Swoboda W., van den Boogaard M. J., Van Hul W., Vikkula M., Votruba M., Zabel B., Garcia T., Baron R., Olsen B. R., Warman M. L. (2001) LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 107, 513–523 [DOI] [PubMed] [Google Scholar]
- 57. Li X., Ominsky M. S., Niu Q. T., Sun N., Daugherty B., D'Agostin D., Kurahara C., Gao Y., Cao J., Gong J., Asuncion F., Barrero M., Warmington K., Dwyer D., Stolina M., Morony S., Sarosi I., Kostenuik P. J., Lacey D. L., Simonet W. S., Ke H. Z., Paszty C. (2008) Targeted deletion of the sclerostin gene in mice results in increased bone formation and bone strength. J. Bone Miner. Res. 23, 860–869 [DOI] [PubMed] [Google Scholar]
- 58. Baron R., Rawadi G. (2007) Wnt signaling and the regulation of bone mass. Curr. Osteoporos. Rep. 5, 73–80 [DOI] [PubMed] [Google Scholar]
- 59. Hartmann C. (2006) A Wnt canon orchestrating osteoblastogenesis. Trends Cell Biol. 16, 151–158 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.