Background: There are multiple interacting clathrin adaptors at the trans-Golgi network and endosomes in yeast.
Results: Autoregulation of one adaptor, Gga2, alters the temporal delay between recruitment of Gga2 and a second adaptor, Ent5, to organelles.
Conclusion: The interaction between Gga2 and Ent5 is regulated by an autoregulatory sequence.
Significance: This autoregulatory mechanism may ensure accurate membrane traffic in vivo.
Keywords: Adaptor Proteins, Cell Biology, Endosomes, Membrane Trafficking, Protein Sorting, Protein-Protein Interactions, Ent5, Gga2, Autoregulation, Clathrin
Abstract
Membrane traffic is an essential process that allows protein and lipid exchange between the endocytic, lysosomal, and secretory compartments. Clathrin-mediated traffic between the trans-Golgi network and endosomes mediates responses to the environment through the sorting of biosynthetic and endocytic protein cargo. Traffic through this pathway is initiated by the controlled assembly of a clathrin-adaptor protein coat on the cytosolic surface of the originating organelle. In this process, clathrin is recruited by different adaptor proteins that act as a bridge between clathrin and the transmembrane cargo proteins to be transported. Interactions between adaptors and clathrin and between different types of adaptors lead to the formation of a densely packed protein network within the coat. A key unresolved issue is how the highly complex adaptor-clathrin interaction and adaptor-adaptor interaction landscape lead to the correct spatiotemporal assembly of the clathrin coat. Here we report the discovery of a new autoregulatory motif within the clathrin adaptor Gga2 that drives synergistic binding of Gga2 to clathrin and the adaptor Ent5. This autoregulation influences the temporal and/or spatial location of the Gga2-Ent5 interaction. We propose that this synergistic binding provides built-in regulation to ensure the correct assembly of clathrin coats.
Introduction
Clathrin acts in many processes, including endocytosis and transport between the trans-Golgi network (TGN)3 and endosomes (reviewed in Ref. 1). Clathrin is a multimeric protein complex that forms a polyhedral lattice on the outer surface of some transport vesicles (2). Formation of the clathrin lattices in vivo is controlled by a class of proteins called adaptors. In both endocytosis and transport between TGN and endosomes, adaptors perform three general functions: membrane association, clathrin binding and assembly, and cargo collection. The adaptors that function in endocytosis and at the TGN are encoded by different genes; however, the adaptors play the key role in directing formation of clathrin-coated vesicles in both of the processes.
Clathrin-dependent traffic requires seemingly redundant adaptors (3–5). At least three different classes of adaptors act at the TGN and endosomes, the heterotetrameric AP-1 complex; the GGA proteins Gga1 and Gga2 in yeast and GGA1, GGA2, and GGA3 in mammals; and the epsin-like proteins Ent3 and Ent5 in yeast and EpsinR in mammals (6–14). Multiple adaptors can be recruited to the same transport event (Ref. 15 and reviewed in Ref. 16). The involvement of multiple adaptors is a hallmark of clathrin-dependent traffic, although the functional significance of this complexity remains unclear. Initially it was proposed that adaptor redundancy allows the transport of distinct subsets of cargo. Recent work in endocytosis also suggests that potentially redundant adaptors play different mechanical roles within a single endocytic event (17).
In addition to shared function, different adaptors utilize the same molecular interfaces to interact with clathrin. One common mechanism relies on an interaction with the globular domain at the N terminus of clathrin known as the terminal domain (reviewed in Ref. 18). Many adaptors contain “clathrin box motifs,” which bind to a pocket in the terminal domain of clathrin. Clathrin box sequences are characterized by the consensus LφXφ(D/E) (where X is any amino acid, and φ is a hydrophobic amino acid). Some adaptors contain an additional motif with the consensus sequence (D/E)LL. This DLL-type motif was first characterized in the endocytic adaptors, where, in multiple copies, it mediates the interaction of adaptors with the clathrin triskelion and clathrin cages (19). Importantly, DLL-type motifs interact with cages formed of triskelia that lack the terminal domain, suggesting that the DLL-type motifs can bind to a different region of clathrin than the clathrin box (20). The presence of different adaptors, each capable of binding to clathrin at the same sites, adds to the complexity of clathrin coats.
Supporting the possibility that different adaptors cooperate in function, adaptors interact with one another. At the TGN and endosomes, the three different classes of adaptors interact with one another. Ggas and AP-1 share a C-terminal homologous domain termed the γ-ear (5, 6, 21–23). The γ-ears mediate interactions with a motif in Epsin-like adaptors with the consensus DFXXφ (Ref. 24 and reviewed in Ref. 18). Additionally, some isoforms of Gga proteins interact directly with the γ-ear of AP-1 via a motif within a large flexible central domain of Gga known as the hinge (25, 26). The functional significance of these adaptor-adaptor interactions at the TGN and endosomes is largely unknown.
The physical interactions between TGN/endosome adaptors raises the possibility that adaptors all act coincidently; however, genetic results in yeast suggest that they function in separate events. In particular, analysis of deletion mutants provides evidence that, although Ent5 cooperates with both AP-1 and Gga proteins in vivo, AP-1 and Gga proteins seem to play roles in distinct transport functions (27). Only in the case of Ent3 has an adaptor-adaptor interaction been shown to influence function. In this case, Ent3 requires Gga proteins for recruitment to clathrin-rich structures. Thus, although evolutionary conservation implies that adaptor-adaptor interactions are important in TGN/endosome traffic in yeast, in most cases the functional significance of such interactions has not been addressed.
In the present study, we describe a previously unrecognized autoregulatory sequence within yeast Gga2 that controls interaction with Ent5. This sequence encodes overlapping motifs that can bind to clathrin or to the Gga2 γ-ear. We find that in vivo the autoregulatory sequence modulates interactions with both clathrin and Ent5 and is important for establishing a temporal delay between recruitment of Gga2 and Ent5 to clathrin-rich structures. These findings reveal a highly specialized mechanism that regulates the location/timing of the interaction of Gga2 with Ent5.
EXPERIMENTAL PROCEDURES
Yeast Strains
Yeast strains are listed in Table 1 (28, 29). Replacement of the genomic alleles used a full gene replacement strategy, in which a full gene deletion was replaced by a DNA fragment excised from a plasmid carrying the desired allele. Clones were then screened by PCR followed by restriction digestion to confirm integration of the desired alleles. Fluorescent tags were added using a PCR-based strategy with either the pFA6a-S65TGFP-HIS3Mx plasmid or pKS390 (pFA6a-mCherry-KanMx) as described previously (30, 31).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| SEY6210 | MATα ura3-52 leu2-3112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 gal2 mel | Ref. 29 |
| TVY614 | SEY6210 with pep4Δ::LEU2 prb1Δ::HISG prc1Δ::HIS3 | Ref. 28 |
| MDY250 | SEY6210 with ent5Δ::TRP1 | Ref. 12 |
| GPY178-10A | SEY6210 with apl2Δ::TRP1 | Ref. 32 |
| MDY624 | SEY6210 with GGA2::URA3 | This study |
| MDY627 | SEY6210 with gga2–355DF → AA::URA3 | This study |
| MDY632 | SEY6210 with gga2–353LI → AA::URA3 | This study |
| MDY667 | SEY6210 with gga2–358DL → AA::URA3 | This study |
| MDY655 | SEY6210 with GGA2-GFP-His3Mx::URA3 | This study |
| MDY657 | SEY6210 with gga2–355DF → AA-GFP-HIS3Mx::URA3 | This study |
| MDY659 | SEY6210 with gga2–353LI → AA-GFP-HIS3Mx::URA3 | This study |
| MDY670 | SEY6210 with gga2–358DL → AA-GFP-HIS3Mx::URA3 | This study |
| MDY726 | SEY6210 with GGA2-GFP-HIS3Mx::URA3 ENT5-mCHERRY-KanMx | This study |
| MDY728 | SEY6210 with gga2–355DF → AA-GFP-HIS3Mx::URA3 ENT5-mCHERRY-KanMx | This study |
| MDY736 | SEY6210 with gga2–353LI → AA-GFP-HIS3Mx::URA3 ENT5-mCHERRY-KanMx | This study |
| MDY730 | SEY6210 with gga2–358DL → AA-GFP-HIS3Mx::URA3 ENT5-mCHERRY-KanMx | This study |
Plasmids
The plasmids used in this study are described in Table 2. Point mutations were generated with the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions.
TABLE 2.
Plasmids used in this study
| Name | Backbone | Insert |
|---|---|---|
| Yep24-Chc-Clc | Yep24 | Genomic regions of both Clc1 and Chc1 |
| pGex-GGA2(1051) | pGEX-Kg | Gga2 aa 350-end |
| pGEX-KG-GGA2 (1171) | pGEX-Kg | Gga2 390-end |
| pMD209 | pGEX-4T | Gga2 460-end |
| pBKS+URA-GGA2 | Pbluescript KS II (+) | Gga2-genomic region linked to URA3 marker |
| pMD208 | pBKS + URA-GGA2 | Integratable GGA2 355DF to AA |
| pMD230 | pBKS + URA-GGA2 | Integratable GGA2 353LI to AA |
| pMD220 | pBKS + URA-GGA2 | Integratable GGA2 358DL to AA |
| pMD212 | pGEX5x | GST-GGA2 340-end |
| pMD213 | pGEX5x | GST-GGA2 340-end 355DF to AA |
| pMD228 | pGEX5x | GST-GGA2 340-end 353LI to AA |
| pMD222 | pGEX5x | GST-GGA2 340-end 358DL to AA |
| pMD211 | pGEX5x | GST-GGA2 340–400 |
| pMD210 | pGEX5x | GST-GGA2 340–400 355DF to AA |
| pMD224 | pGEX5x | GST-GGA2 340–400 353LI to AA |
| pMD225 | pGEX5x | GST-GGA2 340–400 358DL to AA |
| pMD312 | pet21a+ | 6-HIS-GGA2 340–400 |
| pMD297 | pet21a+ | 6-HIS-GGA2 340–400 355DF to AA |
Antibodies
Polyclonal antibodies against clathrin light chain (Clc1) and monoclonal antibodies against clathrin heavy chain (Chc1) are described elsewhere (32–34). Polyclonal antibodies were generated in rabbits against full-length GST-tagged Ent3, Ent5, Gga2, and GFP in rabbits. Gga2, Ent3, Ent5, and Clc1 antibodies were affinity-purified by passing serum first over 2 ml of Affi-Gel-10 (Bio-Rad) cross-linked to purified GST to deplete GST signal. Depleted serum was bound to 2 ml of Affi-Gel-10 cross-linked to purified GST-tagged full-length protein and eluted from the matrix with glycine according to the manufacturer's instructions.
Protein Purification
Protein expression was induced in BL21-DE2 pLysS (Promega) in mid-log phase at 30 °C in LB-Amp (ISC Bioexpress) for 4 h with 0.1 mm isopropyl β-d-thiogalactopyranoside. The cells were pelleted and resuspended in minimal water and frozen. The pellets were quick thawed in 1× PBS with 1:100 protease inhibitor mixture (Sigma) and sonicated in 0.5-s pulses for 1 min on ice. The lysates were incubated with 1% Triton X-100 for 30 min at 4 °C and pelleted at 12,000 rpm for 20 min in a ss34 rotor at 4 °C. For GST-tagged proteins, supernatants were incubated with glutathione-Sepharose (GE Life Sciences) for 1 h at 4 °C, washed with PBS and eluted with HSE (100 mm Tris, pH 9, 200 mm NaCl, 5 mm DTT, 20 mm reduced glutathione) or cleaved in place with Factor X (New England Biolabs) in 50 mm Tris, pH 7.5, 150 mm NaCl, 1 mm CaCl. Factor X was removed with p-aminobenzamidine agarose (MP Biomedicals). For His6-tagged proteins, supernatants were incubated with Talon resin (Clontech) and eluted with 150 mm imidazole in PBS with a final pH of 7.0. For light scattering experiments, cleaved proteins were further purified by passage over tandem Hightrap Q and SP (GE Life Sciences) 1-ml columns. Buffer exchange was performed by several rounds of concentration in Ultracel 10 concentration device (Millipore) at room temperature or several rounds of dialysis with Spectra/Por dialysis tubing MWCO 3500 (Spectrumlabs).
Clathrin triskelia were purified from TVY614 transformed with a Yep24-CHC-CLC, a high copy plasmid carrying both clathrin heavy and light chain genes. Six liters of cells were grown to mid-log phase in selective media, pelleted, resuspended in minimal water, and frozen in small pellets by pouring a slow stream into a liquid nitrogen bath. The lysates were generated by blending pellets in a prechilled metal canister on a warring blender until a light powder was formed. Powder was thawed with equal volume to weight in 2× Triskelia buffer (100 mm Tris, pH 7.5, 100 mm NaCl, 2 mm EDTA with protease inhibitors). High speed supernatants were generated at 4 °C in a Ti 70 at 60Kprm for 30 min. 20% weight to volume ammonium sulfate was added, and high speed pellets were generated by centrifugation at 60,000 rpm for 15 min in a Ti70 rotor. The pellets were resuspended in 1× Triskelia buffer (50 mm Tris, pH 7.5, 50 mm NaCl, 1 mm EDTA) and dialyzed against two buffer changes of Triskelia buffer. Medium speed supernatants were generated by spinning at 13,000 rpm for 10 min and were fractionated on a Superose 12 column pre-equilibrated with 10 mm KPO4, pH 7.5, 1 mm EDTA. Clathrin fractions were identified by SDS-PAGE analysis followed by Coomassie Blue staining and found to be free of adaptors (Ent5, Ent3, Gga2, AP-2, and AP-1) by immunoblotting.
Yeast Cell Lysates and Binding Studies
Cell lysates for GST-tagged protein interaction studies were generated by culturing TVY614 in yeast-rich media (10 g/liter yeast extract, 20 g/liter peptone, 2 g/liter glucose, 20 mg/liter uracil, adenine sulfate, and l-tryptophan) followed by liquid nitrogen blending as described for clathrin purification. Powder was thawed with an equal volume to weight in Buffer A (100 mm MES, pH 6.5, 0.5 mm MgCl2, 1 mm EGTA). The lysate was clarified by centrifugation at 13,000 rpm for 10 min, and the resulting supernatants were used for binding studies. For binding studies, GST-tagged proteins were prebound to glutathione-Sepharose in 1 ml of PBS for 1 h at room temperature, washed twice with Buffer A, and resuspended in lysate. Lysate binding was allowed to proceed for 1 h at 4 °C, and beads were washed twice with Buffer A and once with Triskelia buffer. The beads were resuspended in HSE and incubated for 20 min at room temperature. The supernatants were taken as the bound fraction.
Cell lysates for immunoprecipitations were generated by a combined spheroplast, bead lysis protocol. Spheroplasts were generated by first reducing the cell wall in 100 mm Tris, pH 9.4, 2% glucose, and 5 mm DTT. The cells were then resuspended in digestion buffer (yeast-rich media without additional amino acids or glucose and 0.5% glucose, 10 mm Tris, 1.2 m sorbitol, and 120 units of lyticase) and gently agitated for 30 min at 30 °C. The cells were washed in 1.2 m sorbitol and lysed with the addition of Buffer A with protease inhibitors followed by glass bead lysis and the addition of 1% Triton X-100. The lysates were clarified by centrifugation at 13,000 rpm for 10 min. The lysates were incubated overnight with 100 μl of 20% protein A-agarose slurry and 3 μl of clc1 antibody or 0.5 μl of GFP antisera. The lysates were washed three times with Buffer A, and the bound proteins were eluted in SDS sample buffer.
For cross-linking studies, the reactions were performed at room temperature in Buffer B (100 mm HEPES, 50 mm NaCl, pH 7.5) (14). GST-Gga2 460-end was added to 3.78 μm, wild-type His6 hinge fragments to 55.42 μm ΔEB fragment to 57.64 μm, and 3,3′-Dithiobis(sulfosuccinimidylpropionate) (Thermo Scientific) was added to 1 mm. After 30 min, cross-linking was quenched by the addition of 37 mm final concentration of Tris, pH 7.5. 14% of each reaction was loaded for immunoblot analysis and for Coomassie staining.
Microscopy
For fixed images, cells grown to mid-log phase were fixed by rapid mixing with equal volume of fixative (100 mm KPO4, pH 6.5, 2 mm MgCl2, 8% formaldehyde) and incubating for 1 h in the dark. The cells were washed twice with PBS. Acid-washed 22 × 22-mm coverslips were treated with 5 mg/ml concanavalin A and allowed to dry. Six microliters of fixed cells were spotted onto a treated coverslip, placed on a clean glass slide and immediately sealed with VALAP (equal parts Vaseline, lanolin, and paraffin).
The images were acquired on a spinning disk confocal microscope described by Maddox et al. (35). Well separated cells were first selected using bright field microscopy. Twenty-one Z-stack images were collected at 0.2-μm intervals through each field of cells. Integration times for the GFP and mCherry channels (1200 ms of integration/image for each mCherry image and 400 ms of integration/EGFP image in that order) were set so that a relatively high and similar signal to noise ratio was maintained for both channels.
All nonoverlapping, centrally located cells within a field were analyzed on a custom graphical user interface written in MatLAB R2009b (Natick, MD). We then conducted morphometry analysis on this down-sampled stack using functions from the Image Processing Toolbox in MatLAB. Briefly, a single cell from a Z-stack was selected manually by drawing a region of interest of arbitrary dimensions. This cropped area was then used for further analysis for the selected cell. First, a 5 × 5-pixel background region was manually selected from the maximum intensity projection image of all 21 planes in the cropped region to ensure that the dimmest region inside the cell (usually the vacuole) was specified as the background region. Using a nominal Z-depth of the confocal point spread function as 600 nm, we projected the maximum intensity pixels from a set of three successive image stacks into a single image (a stack of 21 images thus gets compressed into a stack of seven images). The coordinates of the background region were transferred to each of the seven images in the down-sampled stack to obtain background pixels for each plane. The background was estimated by using the relation: background = (mean pixel value) + (6 × standard deviation of pixel values). If one assumes that the background pixel values are normally distributed, this background estimate rejects 99% of the background pixels in our analysis. This background value was used as the threshold to define features within each plane of the cell. For the calculation of colocalization coefficients, we further discarded any patches smaller than the XY point spread function of the microscope (less than 3 pixels, where 1 pixel = ∼66 nm). Colocalization coefficient calculation was based on the method of Manders et al. (22, 38). In the analysis, we broadly categorized the cells as large budded (late mitosis) and small budded (G1 or S). The data reported in the figure provides measurements from large budded cells, although analysis of small budded cells obtained similar results.
For live cell imaging, the fixation and PBS washing steps were omitted. A single central plane was imaged with a 2-s interval. Analysis was performed in ImageJ. First the threshold command was applied to all images uniformly to generate a mask. Structures smaller than 2 squared pixels or that persisted less than five frames were omitted from analysis. Using the threshold masked Gga2 images, nonoverlapping Gga2 structures were identified. The first frame in which a structure was visible was considered T = 0. Preceding and subsequent frames from the Ent5 masked images were analyzed to identify whether and when Ent5 showed above threshold fluorescence within the defined Gga2 mask. The data reported in the figure are from >70 individual events from at least 10 cells.
Gel Filtration and Light Scattering
Purified Gga2 fragments were separated in Buffer A on a WTC-030s5 column (Wyatt) and subjected to multiangle light scattering and quasielastic light scattering on a DAWN EOS light scattering instrument (Wyatt) interfaced to an Akta FPLC (GE Life Sciences). Weighted molar mass was calculated with Astra software. The data shown are intensity from detector at position 11.
RESULTS
Binding of Gga2 to Clathrin Requires Ent5
We and others previously demonstrated that Ent5 can bind both AP-1 and Gga2 and all three bind to clathrin (12, 36). Our previous studies did not investigate whether adaptors compete for or cooperate in clathrin binding. Ent5, Gga2, and AP-1 all contain clathrin box motifs and in principle can compete for the same binding pocket on a single clathrin terminal domain. However, clathrin coats have multiple terminal domains, allowing simultaneous binding to clathrin box motifs in different adaptors. In addition, Gga2 and AP-1 contain multiple DLL-type clathrin interaction surfaces that do not bind to the terminal domain. To determine whether one adaptor influences clathrin binding to other adaptors, we performed clathrin immunoprecipitations from nondenatured lysates of different deletion mutants.
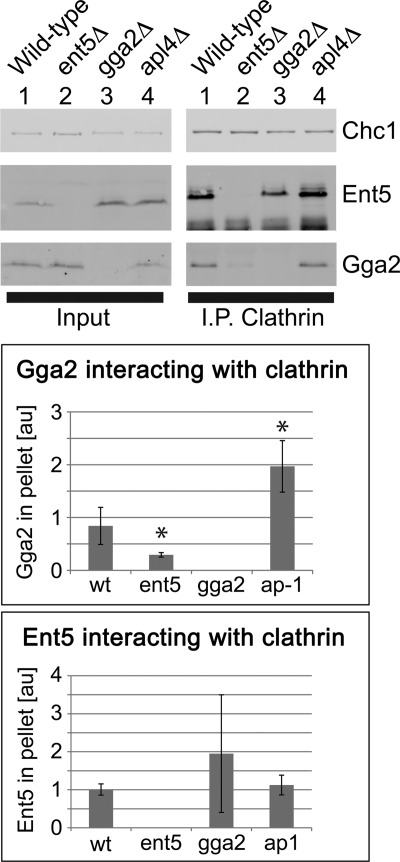
In wild-type cells, both Gga2 and Ent5 coimmunoprecipitated with native clathrin (Fig. 1, top panel). In cells lacking Ent5, substantially less Gga2 immunoprecipitated with clathrin (Fig. 1). In contrast, deletion of the γ subunit of AP-1 enhanced the amount of Gga2 associated with clathrin. We observed a minor effect of GGA2 deletion on the interaction of Ent5 with clathrin in some immunoprecipitation reactions. However, in other reactions, deletion of GGA2 did not produce an effect, suggesting that the effect of Gga2 on Ent5 binding to clathrin is at most minor (Fig. 1). These results suggest that maximal interaction of Gga2 with clathrin requires Ent5 and also that AP-1 may compete with Gga2 for clathrin. Because of the strong influence of other adaptors on Gga2 binding, we investigated clathrin binding of Gga2 in more detail.
FIGURE 1.
Gga2 interaction with clathrin is altered in adaptor deletion strains. Clathrin was immunoprecipitated from the indicated strains with a polyclonal clathrin light chain antibody, and interacting Ent5 and Gga2 were detected by immunoblot analysis. The top panel shows a representative immunoprecipitation (I.P.) reaction. The bottom panel shows data from four independent immunoprecipitation experiments. Intensity measurements of adaptors were normalized to intensity in wild-type (wt) sample of the same experiment and then normalized to the intensity of clathrin heavy chain bands (Chc1) within the same sample. *, p value <0.05 as determined by a two-tailed Student's t test.
Clathrin Box Motif in Gga2 Is Major Contributor to Its Interaction with Clathrin
Gga2 contains a single canonical clathrin box motif in the hinge region that is the major site of clathrin interaction (36). We generated three mutations, each converting different pairs of residues to alanine within or near the clathrin box motif (Fig. 2a). We first tested the effects of these mutations on interactions of the hinge fragment of Gga2 with purified clathrin. Mutation of the first two residues (ΔCB) of the motif reduced the interaction with purified clathrin to undetectable levels (Fig. 2b, lane 5). Mutation of the third and fourth residues (Δcore) also severely reduced the interaction (Fig. 2b, lane 3). Mutation of the two residues just after the canonical clathrin box (ΔEB) had no effect on the binding to purified clathrin (Fig. 2b, lane 4). In cell lysates, the ΔCB and Δcore mutations ablated the hinge interaction with clathrin, whereas the ΔEB mutations reduced but did not eliminate clathrin binding (Fig. 2c). These results confirm previous studies demonstrating that the clathrin box residues are required for direct interactions with clathrin (36). Furthermore, these results demonstrate that residues just after the clathrin box are not required for clathrin binding but that these residues contribute to clathrin affinity in the context of a complex cytosol.
FIGURE 2.
Mutations of the clathrin box region alter interaction of Gga2 with clathrin in vitro. a, top panel, schematic of Gga2 and interacting partners. Black dotted lines indicate previously defined interactions. The green dotted line indicates an intramolecular interaction defined in this study. Middle panel, schematic of fragments used in in vitro assays. Bottom panel, schematic of mutation generated. b, indicated mutations were generated in GST fusion constructs containing 60 amino acids of the Gga2 hinge, purified, and incubated with purified clathrin. Interacting clathrin was detected by immunoblot analysis. c, indicated fusion proteins were incubated with cell lysates, and interacting clathrin was detected by immunoblot analysis. d, indicated mutations were generated in GST fusion constructs containing the Gga2 hinge and ear, purified, and incubated with cell lysates. Interacting clathrin was detected by immunoblot analysis. wt, wild type.
We next investigated the effect of the mutations in the context of a larger Gga2 C-terminal fragment that contains the hinge and ear of Gga2. The Δcore and ΔCB mutations prevented interaction with clathrin, whereas the ΔEB mutation had no effect on clathrin interaction. Thus, in cell lysates, only the canonical clathrin box residues are required for clathrin binding (Fig. 2d). Within the hinge and ear fragment are three DLL-type motifs in addition to the clathrin box motif. The inability of this fragment to bind clathrin when the clathrin box motif is nonfunctional confirms that stable interaction between Gga2 and clathrin requires the clathrin box.
Clathrin Box Motif in Gga2 Regulates Ent5-Clathrin Interaction
To investigate the effects of the mutations on interactions of full-length Gga2 and Ent5 with clathrin, we replaced the endogenous GGA2 locus with mutant alleles encoding each of the three mutations. We then immunoprecipitated clathrin from cell lysates and probed for associated Ent5 and Gga2. The ΔCB mutation, which alters the first two residues of the clathrin box, dramatically reduced the interaction of Gga2 with clathrin (Fig. 3a, lane 8). At the same time, less Ent5 coimmunoprecipitated with clathrin (Fig. 3a). Surprisingly, the ΔEB mutation of residues adjacent to the clathrin box enhanced the amount of Gga2 coimmunoprecipitating with clathrin but did not alter the levels of clathrin-associated Ent5 (Fig. 3a, lane 7). The Δcore mutation did not affect either Gga2 or Ent5 levels coimmunoprecipitating with clathrin (Fig. 3a, lane 6). Together these results provide evidence that sequence elements in the clathrin box region of Gga2 influence clathrin interaction with both Ent5 and Gga2.
FIGURE 3.
Mutations of the clathrin box region alter interaction of Gga2 with clathrin in cell lysates. a, effect of Gga2 mutations on clathrin binding to Gga2 and Ent5 in lysates. The indicated mutations were generated at the endogenous Gga2 locus. Clathrin was immunoprecipitated from indicated strains with a polyclonal clathrin light chain antibody, and interacting Gga2 and Ent5 were detected by immunoblot analysis. The left panel shows representative immunoprecipitation (I.P.) results. The right panels show intensity analysis as described in Fig. 1. b, effect of Gga2 mutations on Gga2 binding to clathrin and Ent5 in lysates Indicated mutations were generated at the endogenous Gga2 locus and GFP-tagged at the C terminus. Gga2-GFP alleles were immunoprecipitated with a monoclonal GFP antibody, and interacting clathrin and Ent5 were detected by immunoblot analysis.
We also investigated the effects of the mutations on Gga2 interactions detected by coimmunoprecipitations of GFP-tagged alleles of Gga2. Similar to the results from the clathrin immunoprecipitations, gga2-ΔCB-GFP exhibited a severe reduction in interaction with clathrin; however, Ent5 binding to Gga2 was not significantly changed (Fig. 3b, lane 9). Also similar to the clathrin immunoprecipitations, the ΔEB mutation augmented interaction of Gga2p with both clathrin and Ent5 (Fig. 3b, lane 8). gga2-Δcore-GFP displayed a possible increase in clathrin binding and a clear elevation in Ent5 binding. Together, our results provide evidence for a network of pairwise interactions between Gga2, Ent5, and clathrin that can influence assembly of the adaptors with clathrin. In particular, the evidence for increased binding of Ent5 and clathrin to ΔEB and Ent5 to Δcore suggests that the clathrin box region is involved in an inhibitory interaction.
Autoregulatory Motif in Gga2 Modulates Binding to Both Clathrin and Ent5
In light of these findings, we reviewed residues in and around the clathrin box. The canonical clathrin box motif partially overlaps with a sequence similar to the characterized γ-ear binding motif from Ent5 and other related proteins, DFXXφ (Fig. 2a). The residues targeted in the ΔCB mutation are specific for the clathrin-binding motif, the ΔEB residues are specific for the γ-ear binding motif, and the Δcore residues are shared by the two motifs. Importantly, because both binding motifs share the central residues, γ-ear interaction could prevent binding to clathrin and vice versa.
To determine whether the γ-ear binding motif in the clathrin box region of Gga2 can interact with the Gga2 γ-ear domain, we performed chemical cross-linking with a GST-tagged Gga2 ear fragment and a His6 fragment of the Gga2 hinge containing the clathrin box motif. Upon cross-linking, a slow migrating species was detected by antibodies against Gga2 (Fig. 4a, lane 8). This species was not observed in reactions containing only the Gga2 ear fragment or only the Gga2 hinge fragment or in reactions lacking the cross-linker. Furthermore, when reactions were performed with a hinge fragment encoding the Δcore mutation, the level of the specific cross-linked product was substantially reduced compared with wild-type reactions (Fig. 4a, lane 10). These results provide evidence that the clathrin box region of Gga2 also encodes a sequence that can interact with the Gga2 γ-ear domain.
FIGURE 4.
The clathrin box region of Gga2 interacts with the Gga2 γ-ear. a, the clathrin box region binds to the γ-ear. A His6-tagged 60-amino acid fragment containing the wild-type (CAB-wt) or a Δcore (CAB-Δcore) clathrin box region incubated with a GST-tagged fragment encoding the γ-ear (γ-ear) were incubated with or without cross-linker. Identical samples were processed for Western blot analysis and Coomassie staining. Note that un-cross-linked CAB-wt does not adhere to membrane in the immunoblot but is present in the Coomassie-stained gel. The arrowhead indicates migration of a band present only in reactions containing the CAB, γ-ear, and cross-linker. b, a fragment containing the Δcore mutant adopts an extended conformation. Elution profiles of Gga2 amino acid 340 to the end and Gga2 Δcore mutant amino acid 340 to the end in gel filtration are shown. Weight-averaged solute molar mass for the wild-type fragment was 25.18 kDa, and for the mutant fragment was 26.02 kDa with an error of 1% for both mass values.
Next, we tested whether the clathrin box region in Gga2 mediates an intramolecular interaction. Gga2 fragments encompassing the hinge and ear with or without the Δcore mutation were subjected to gel filtration followed by quasielastic and multiangle light scattering. Both fragments eluted as a single peak; however, the mutant form eluted earlier than the wild type, indicating a slightly larger size (Fig. 4b). The molecular mass of both the wild-type and mutant fragments were determined to be that of a monomer by light scattering. These results are consistent with a model in which the core residues mediate an intramolecular interaction that compacts the structure of the hinge-ear fragment. Without this intramolecular interaction, the Δcore protein assumes a more extended form.
Together, our results demonstrate that the clathrin box region of Gga2 can bind to both clathrin and the Gga2 γ-ear. The γ-ear binding can occur as an intramolecular interaction. Mutation of the residues important for γ-ear binding enhances the interaction of full-length Gga2 with both Ent5 and clathrin in coimmunoprecipitation reactions. This suggests that the clathrin box region acts as an autoregulatory motif that, through binding to the γ-ear, modulates interaction with both clathrin and Ent5. Because of its dual roles, we will refer to the larger sequence encompassing both the clathrin box and γ-ear binding motif as the clathrin and adaptor binding motif (CAB).
Mutation of the Autoregulatory Motif in Gga2 Alters Ent5 Recruitment in Vivo
We tagged Gga2 alleles with S65T-GFP and Ent5 with mCherry at the endogenous loci to assess the function of the Gga2 CAB in cells. Ent5 and Gga2 colocalize on a subset of structures in wild-type cells (Fig. 5a). Because both Ent5 and Gga2 structures are highly motile within the cell, we fixed cells and analyzed the area of colocalization in >100 individual cells for each genotype to obtain a quantitative measure of the effect of mutations on colocalization (Fig. 5b). In the ΔCB mutant, in which Gga2 shows a reduced interaction with both clathrin and Ent5, colocalization of Ent5 and Gga2 is unaffected. In contrast, in the Δcore mutant, in which the CAB does not bind to the γ-ear or to clathrin and Ent5 binding is enhanced, there was a statistically significant increase in the colocalization of Ent5 with Gga2. Furthermore, in the ΔEB mutant, in which the CAB retains some ability to bind to clathrin and interactions with both clathrin and Ent5 are elevated, the extent of colocalization between Ent5 and Gga2 was even greater. Indeed, very few structures could be identified that did not contain both Ent5 and Gga2. Thus, when Gga2 has enhanced binding to Ent5, the two proteins almost always colocalize. These results suggest that the autoregulatory motif of Gga2 limits colocalization of Ent5 and Gga2 in vivo, consistent with the inhibitory role detected by biochemical assays.
FIGURE 5.
CAB mutations alter colocalization of Ent5 and Gga2. a, representative images of cells analyzed. The indicated mutations were generated at the endogenous Gga2 locus and GFP-tagged at the C terminus, and Ent5 tagged at the C terminus with mCherry was introduced into these strains through crosses. The cells were imaged as described under “Experimental Procedures.” b, colocalization analysis of Gga2 alleles with Ent5. The box plot diagrams show the Manders' coefficients for analyzed data. On each box, the central mark is the median, the edges of the box are the 25th and 75th percentiles, the whiskers extend to the 95th percentile, and the outliers are plotted individually. The outliers are indicated as circles. p values reflect a Mann-Whitney test with Null hypothesis (h = 0) that the two samples have the identical median with 95% confidence interval. *, p value greater than 1 × 10−3 for samples compared with wild type.
To better understand the effects of Gga2 autoregulation on Ent5 and Gga2 localization in vivo, we monitored adaptor recruitment dynamics in live cells. Similar to previous reports, Gga2 and Ent5 exhibit a stereotypical recruitment order in wild-type cells (Fig. 6, a and b) (37). When monitored by single-plane confocal imaging, numerous events were observed where structures rich in Gga2 initially lack detectable Ent5. Over time, many of these Gga2-rich structures acquired significant Ent5 fluorescence. In contrast, very few events were observed where structures rich in Ent5 but lacking Gga2 then acquired Gga2. In wild-type cells, Ent5 and Gga2 colocalized at some point in ∼60% of events. Events were observed where Gga2 disappeared without ever recruiting Ent5 and vice versa. To describe the relationship between Ent5 and Gga2 in live cells, we determined the time differential between recruitment of Gga2 and Ent5. In events where Gga2 was recruited first, the value is positive, and where Gga2 was recruited second, the value is negative. Gga2-rich structures that did not recruit Ent5 were omitted from this analysis and made up 23% of all events. In wild-type cells, the median recruitment differential is 8 s with a broad standard deviation (Fig. 6b).
FIGURE 6.
Clathrin box region mutations alters the recruitment differential between Gga2 and Ent5. a, representative images of events analyzed. The green arrow indicates the time point utilized for t = 0. The red arrow indicates the time point used to calculate the differential between Gga2 and Ent5 recruitment. The black arrow indicates the time point used to calculate event life span. Frames of time lapse are 2 s apart for each channel. Approximately 800 ms separates channels at a single time point. b, box plot diagrams of analyzed recruitment differential data. c, box plot diagrams of analyzed life span data. On each box, the central mark is the median, the edges of the box are the 25th and 75th percentiles, the whiskers extend to the 95th percentile, and the outliers are plotted individually. The outliers are indicated as circles. p values reflect a Mann-Whitney test with Null hypothesis (h = 0) that the two samples have the identical median with 95% confidence interval. wt, wild type.
In contrast to wild-type cells, in the gga2-ΔEB-GFP cells Gga2 was rarely recruited before Ent5. Instead the majority of events observed were coincident appearance of Gga2 and Ent5. Only 7% of Gga2 events failed to recruit Ent5 in gga2-ΔEB-GFP cells. The median differential recruitment interval was 0 with low standard deviation. However, we cannot distinguish coincident recruitment from migration of a structure with both Gga2 and Ent5 into the plane of focus. Consequently, it was possible that the apparent decrease in differential recruitment time could result from enhanced stability of Gga2/Ent5 structures in the gga2-ΔEB-GFP cells rather than an actual change in the time between Gga2 and Ent5 recruitment. To investigate whether Gga2 and Ent5 structures are more stable in the gga2-ΔEB-GFP cells, we analyzed the life span of Gga2 foci observed by single plane imaging. In wild-type and gga2-ΔEB-GFP mutant cells, the median life spans of a Gga2 structure were 20 and 18 s, respectively, with a large variability (Fig. 6c). The loss of Gga2-first events together with no change in event life-time suggests that the absence of Gga2 autoregulation leads to a loss in sequential recruitment of Gga2 and Ent5. Thus, the autoregulatory motif of Gga2 ensures a temporal delay of Ent5 recruitment to Gga2 rich structures.
DISCUSSION
Our study reveals a complex interplay of interactions between Ent5, Gga2, and clathrin. In particular, we have identified a novel bifunctional motif in Gga2p, the CAB, which consists of overlapping sequences for clathrin and γ-ear binding. The evidence supports formation of an intramolecular interaction between CAB and the Gga2 γ-ear that inhibits binding of Gga2p to clathrin and Ent5. Based on these results, we propose that CAB acts as an autoinhibitory element to restrict Gga2 interaction with Ent5 and impose a temporal delay in recruitment of Ent5 relative to Gga2 during formation of clathrin-coated vesicles.
Most, but not all, of the effects of CAB mutations can be explained by the finding that the motif binds to the γ-ear of Gga2. The two CAB mutations that prevent interaction with the γ-ear, ΔEB and Δcore, enhance binding of Gga2 to Ent5. Furthermore, the ΔEB mutation, which leaves the clathrin box intact, also stimulates binding to clathrin. Binding of CAB to the γ-ear domain would occlude both the CAB clathrin box and the DFXXφ binding pocket in the γ-ear. The ΔEB mutation prevents interaction between the CAB and γ-ear, which allows clathrin unrestricted access to the clathrin box motif in CAB and frees the γ-ear to interact with Ent5, thereby enhancing the interaction of Gga2 with both proteins. In a similar fashion, the Δcore mutation releases the γ-ear for Ent5 binding, but clathrin binding would not be enhanced because of the change in the clathrin box. It is not evident why this mutant retains clathrin binding; perhaps the enhanced binding to Ent5 allows an increase in indirect binding to clathrin that compensates for the loss of direct binding or supplements the weak interactions provided by the DLL-type interaction surfaces on Gga2.
Some effects of the ΔCB mutation can be explained by interaction of the CAB with the γ-ear. The ΔCB mutation reduces Gga2 and Ent5 binding to clathrin. The ΔCB mutation inactivates the clathrin box, accounting for the significant decrease in clathrin binding. Because the ΔCB retains residues required for γ-ear binding, interaction of the CAB with the γ-ear occludes interaction with Ent5, thus reducing interaction with Ent5 as well. However, the effects of the ΔCB mutation on the interactions of Ent5 with clathrin and Gga2 are not as easily explained by a simple three-protein interaction scheme. This suggests that a more complicated network involving clathrin, Ent5, and Gga2 may contribute to the core interactions defined in our studies.
Our findings strongly suggest that the Gga2-CAB is a key motif that limits the interaction of Ent5 and Gga2 in vitro and in vivo. However, deletion of Gga2 or Ent5 does not influence recruitment of the other protein to clathrin-rich structures in vivo (20), indicating that most Gga2 and most Ent5 are recruited independently of one another in wild-type cells. Nevertheless, loss of regulation caused by CAB mutation resulted in coordinate assembly and increased colocalization of Ent5 with Gga2. This finding suggests that interaction of Gga2 with Ent5 can either promote Ent5 recruitment to Gga2-containing structures or stabilize an Ent5-Gga2 complex once it forms.
The Gga2 autoregulatory mechanism described here provides insight into the basis of sequential clathrin adaptor recruitment to the TGN described in a recent report (37). In agreement with our results, this study presented evidence that Gga2 and Ent5 are recruited in sequence, with Ent5 recruitment peaking together with AP-1. Notably, apparent colocalization of Gga2 and Ent5 (or AP-1) in wild-type cells observed by wide field fluorescence microscopy was resolved in many cases into separate closely spaced structures visualized by super-resolution methods. Thus, the delay in Ent5 recruitment relative to Gga2 can be attributed to Ent5 assembly into a later forming class of AP-1-containing clathrin coats distinct from Gga2-enriched coats (Fig. 7a). However, ∼20% of Ent5 was recruited concurrently with Gga2, and some colocalization could be observed by super-resolution microscopy (37). The coassembly and colocalization in small structures suggests that Ent5 and Gga2 act together in some instances in vivo. Indeed a functional role of Ent5 in Gga2 mediated traffic was previously indicated by genetic analysis (27). Based on these findings and the results presented here, the role of CAB-mediated autoregulation is likely to be 2-fold: first, to prevent premature interaction between Gga2 and Ent5 in the cytoplasm prior to assembly into clathrin coats; second, to limit levels of Ent5 recruited to Gga2-enriched clathrin coats at the TGN so that most Ent5 is free to assemble independently into later forming AP-1-containing coats.
FIGURE 7.
Schematic of possible adaptor recruitment models. a, different types of adaptor vesicle form in a temporal order on an organelle. Event type 1, which utilizes only Gga2 (orange) forms early on an organelle. Later in the life span of the same organelle, event type 2 vesicles form, which utilizes both Gga2 and Ent5 (purple). Finally in late stage organelles, only event type 3 vesicles form, which contain Ent5 and AP-1 (blue). Mutations of the autoregulatory domain of Gga2 may cause the loss of event type 1, resulting in the observed increase in Ent5 and Gga2 colocalization and the loss of temporal order of recruitment between Gga2 and Ent5. b, model of possible role of CAB in Ent5 recruitment in event type 2 vesicles. Gga2 in its autoregulated form becomes concentrated at a cellular membrane. DLL-type motifs of Gga2 loosely recruit clathrin. Now in local high concentration, clathrin can bind to the CAB, thus promoting the open form of Gga2. In the open form, Gga2 helps to stabilize Ent5 in the clathrin coat.
In this model, the intramolecular interaction between Gga2-CAB and Gga2 γ-ear domain limits Gga2 interaction with Ent5 outside of the context of assembling coats. Once Gga2 associates with the membrane to initiate coat formation, clathrin can be recruited through interactions with DLL motifs and limited interaction with the CAB clathrin box. In this nascent coat, higher concentrations of clathrin can compete with the γ-ear for binding to the CAB. Binding of clathrin to the clathrin box would release the γ-ear from the CAB, allowing Ent5 to be recruited to Gga2 structures through cooperative binding to the Gga2 γ-ear and clathrin. Ent5 recruitment would stabilize the coat assembly, both by binding directly to clathrin and by promoting the “open” Gga2 conformation with the exposed clathrin box (Fig. 7b). Supporting this view, deletion of Ent5 reduced coimmunoprecipitation of Gga2 with clathrin. In addition to the interactions described here, it is also possible that interactions with other proteins and post-translational modifications contribute to the autoregulatory process.
The functional significance of the limited association of Ent5 and Gga2 in vivo is currently unclear. We found no major defect in the maturation of α factor in cells carrying any of the Gga2 mutations (data not shown). Maturation of α factor depends on clathrin-dependent TGN-endosome trafficking of the processing protease Kex2. However, this assay may not capture cargo-specific, subtle, or kinetic defects in traffic. A more general compression of the delay between Gga2 and both AP-1 and Ent5 recruitment resulted in subtle α factor maturation defects, providing evidence that the temporal regulation of assembly is important for optimal function of the TGN (37). We therefore speculate that the function of Gga2 autoregulation of Ent5 recruitment is most important for TGN function in the complex natural environment of yeast growing in the wild.
Acknowledgments
We thank Edward D. Salmon and Kerry S. Bloom for resources and advice; Ashutosh Tripathy at the Macromolecular Interactions Facility (UNC) for technical support and insightful comments; and Santiago Di Pietro (Colorado State University) for insightful comments.
This work was supported, in whole or in part, by National Institutes of Health Grants GM092741 (to M. C. D.) and GM39040 (to G. P.). This work was also supported by funds from the state of North Carolina (to M. C. D. and Q. A.).
- TGN
- trans-Golgi network
- CAB
- clathrin and adaptor binding motif.
REFERENCES
- 1. Brodsky F. M., Chen C. Y., Knuehl C., Towler M. C., Wakeham D. E. (2001) Biological basket weaving. Formation and function of clathrin-coated vesicles. Annu. Rev. Cell Dev. Biol. 17, 517–568 [DOI] [PubMed] [Google Scholar]
- 2. Woodward M. P., Roth T. F. (1978) Coated vesicles. Characterization, selective dissociation, and reassembly. Proc. Natl. Acad. Sci. U.S.A. 75, 4394–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Copic A., Starr T. L., Schekman R. (2007) Ent3p and Ent5p exhibit cargo-specific functions in trafficking proteins between the trans-Golgi network and the endosomes in yeast. Mol. Biol. Cell 18, 1803–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chidambaram S., Müllers N., Wiederhold K., Haucke V., von Mollard G. F. (2004) Specific interaction between SNAREs and epsin N-terminal homology (ENTH) domains of epsin-related proteins in trans-Golgi network to endosome transport. J. Biol. Chem. 279, 4175–4179 [DOI] [PubMed] [Google Scholar]
- 5. Black M. W., Pelham H. R. (2000) A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J. Cell Biol. 151, 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dell'Angelica E. C., Puertollano R., Mullins C., Aguilar R. C., Vargas J. D., Hartnell L. M., Bonifacino J. S. (2000) GGAs. A family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 149, 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wasiak S., Legendre-Guillemin V., Puertollano R., Blondeau F., Girard M., de Heuvel E., Boismenu D., Bell A. W., Bonifacino J. S., McPherson P. S. (2002) Enthoprotin. A novel clathrin-associated protein identified through subcellular proteomics. J. Cell Biol. 158, 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kalthoff C., Groos S., Kohl R., Mahrhold S., Ungewickell E. J. (2002) Clint. A novel clathrin-binding ENTH-domain protein at the Golgi. Mol. Biol. Cell 13, 4060–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bonifacino J. S. (2004) The GGA proteins. Adaptors on the move. Nat. Rev. Mol. Cell Biol. 5, 23–32 [DOI] [PubMed] [Google Scholar]
- 10. Panek H. R., Stepp J. D., Engle H. M., Marks K. M., Tan P. K., Lemmon S. K., Robinson L. C. (1997) Suppressors of YCK-encoded yeast casein kinase 1 deficiency define the four subunits of a novel clathrin AP-like complex. EMBO J. 16, 4194–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rad M. R., Phan H. L., Kirchrath L., Tan P. K., Kirchhausen T., Hollenberg C. P., Payne G. S. (1995) Saccharomyces cerevisiae Apl2p, a homologue of the mammalian clathrin AP β subunit, plays a role in clathrin-dependent Golgi functions J. Cell Sci. 108, 1605–1615 [DOI] [PubMed] [Google Scholar]
- 12. Duncan M. C., Costaguta G., Payne G. S. (2003) Yeast epsin-related proteins required for Golgi-endosome traffic define a gamma-adaptin ear-binding motif. Nat. Cell Biol. 5, 77–81 [DOI] [PubMed] [Google Scholar]
- 13. Hirst J., Motley A., Harasaki K., Peak Chew S. Y., Robinson M. S. (2003) EpsinR. An ENTH domain-containing protein that interacts with AP-1. Mol. Biol. Cell 14, 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mills I. G., Praefcke G. J., Vallis Y., Peter B. J., Olesen L. E., Gallop J. L., Butler P. J., Evans P. R., McMahon H. T. (2003) EpsinR. An AP1/clathrin interacting protein involved in vesicle trafficking. J. Cell Biol. 160, 213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brady R. J., Wen Y., O'Halloran T. J. (2008) The ENTH and C-terminal domains of Dictyostelium epsin cooperate to regulate the dynamic interaction with clathrin-coated pits. J. Cell Sci. 121, 3433–3444 [DOI] [PubMed] [Google Scholar]
- 16. Boettner D. R., Chi R. J., Lemmon S. K. (2012) Lessons from yeast for clathrin-mediated endocytosis. Nat. Cell Biol. 14, 2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmid E. M., Ford M. G., Burtey A., Praefcke G. J., Peak-Chew S. Y., Mills I. G., Benmerah A., McMahon H. T. (2006) Role of the AP2 β-appendage hub in recruiting partners for clathrin-coated vesicle assembly PLoS Biol. 4, e262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Owen D. J., Collins B. M., Evans P. R. (2004) Adaptors for clathrin coats. Structure and function. Annu. Rev. Cell Dev. Biol. 20, 153–191 [DOI] [PubMed] [Google Scholar]
- 19. Morgan J. R., Prasad K., Hao W., Augustine G. J., Lafer E. M. (2000) A conserved clathrin assembly motif essential for synaptic vesicle endocytosis. J. Neurosci. 20, 8667–8676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scheele U., Alves J., Frank R., Duwel M., Kalthoff C., Ungewickell E. (2003) Molecular and functional characterization of clathrin- and AP-2-binding determinants within a disordered domain of auxilin. J. Biol. Chem. 278, 25357–25368 [DOI] [PubMed] [Google Scholar]
- 21. Boman A. L., Zhang C., Zhu X., Kahn R. A. (2000) A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol. Biol. Cell 11, 1241–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hirst J., Lui W. W., Bright N. A., Totty N., Seaman M. N., Robinson M. S. (2000) A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J. Cell Biol. 149, 67–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Costaguta G., Stefan C. J., Bensen E. S., Emr S. D., Payne G. S. (2001) Yeast Gga coat proteins function with clathrin in Golgi to endosome transport. Mol. Biol. Cell 12, 1885–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mattera R., Ritter B., Sidhu S. S., McPherson P. S., Bonifacino J. S. (2004) Definition of the consensus motif recognized by γ-adaptin ear domains. J. Biol. Chem. 279, 8018–8028 [DOI] [PubMed] [Google Scholar]
- 25. Doray B., Ghosh P., Griffith J., Geuze H. J., Kornfeld S. (2002) Cooperation of GGAs and AP-1 in packaging MPRs at the trans-Golgi network. Science 297, 1700–1703 [DOI] [PubMed] [Google Scholar]
- 26. Bai H., Doray B., Kornfeld S. (2004) GGA1 interacts with the adaptor protein AP-1 through a WNSF sequence in its hinge region. J. Biol. Chem. 279, 17411–17417 [DOI] [PubMed] [Google Scholar]
- 27. Costaguta G., Duncan M. C., Fernández G. E., Huang G. H., Payne G. S. (2006) Distinct roles for TGN/endosome epsin-like adaptors Ent3p and Ent5p. Mol. Biol. Cell 17, 3907–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vida T. A., Emr S. D. (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. (1988) Protein sorting in Saccharomyces cerevisiae. Isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell Biol. 8, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Snaith H. A., Samejima I., Sawin K. E. (2005) Multistep and multimode cortical anchoring of tea1p at cell tips in fission yeast. EMBO J. 24, 3690–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 32. Yeung B. G., Phan H. L., Payne G. S. (1999) Adaptor complex-independent clathrin function in yeast. Mol. Biol. Cell 10, 3643–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lemmon S., Lemmon V. P., Jones E. W. (1988) Characterization of yeast clathrin and anticlathrin heavy-chain monoclonal antibodies. J. Cell Biochem. 36, 329–340 [DOI] [PubMed] [Google Scholar]
- 34. Phan H. L., Finlay J. A., Chu D. S., Tan P. K., Kirchhausen T., Payne G. S. (1994) The Saccharomyces cerevisiae APS1 gene encodes a homolog of the small subunit of the mammalian clathrin AP-1 complex. Evidence for functional interaction with clathrin at the Golgi complex. EMBO J. 13, 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maddox P. S., Moree B., Canman J. C., Salmon E. D. (2003) Spinning disk confocal microscope system for rapid high-resolution, multimode, fluorescence speckle microscopy and green fluorescent protein imaging in living cells. Methods Enzymol. 360, 597–617 [DOI] [PubMed] [Google Scholar]
- 36. Mullins C., Bonifacino J. S. (2001) Structural requirements for function of yeast GGAs in vacuolar protein sorting, α-factor maturation, and interactions with clathrin. Mol. Cell Biol. 21, 7981–7994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Daboussi L., Costaguta G., Payne G. S. (2012) Phosphoinositide-mediated clathrin adaptor progression at the trans-Golgi network. Nat. Cell Biol. 19, 239–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Manders E. M. M., Verbeek F. J., Aten J. A. (1993) Measurement of colocalization of objects in dual-color confocal images. J. Microsc.-Oxf. 169, 375–382 [DOI] [PubMed] [Google Scholar]