Background: Cells lacking both cardiolipin and mitochondrial phosphatidylethanolamine are inviable, suggesting that these lipids have overlapping functions.
Results: The loss of both lipids leads to decreased mitochondrial fusion and fragmented mitochondria.
Conclusion: One overlapping function of these lipids is in mitochondrial fusion.
Significance: Decreased mitochondrial fusion may partly explain the variation in clinical presentation observed in Barth syndrome.
Keywords: Cardiolipin, Membrane Lipids, Mitochondria, Phosphatidylethanolamine, Yeast, Barth Syndrome, Mitochondrial Fusion
Abstract
The two non-bilayer forming mitochondrial phospholipids cardiolipin (CL) and phosphatidylethanolamine (PE) play crucial roles in maintaining mitochondrial morphology. We have shown previously that CL and PE have overlapping functions, and the loss of both is synthetically lethal. Because the lack of CL does not lead to defects in the mitochondrial network in Saccharomyces cerevisiae, we hypothesized that PE may compensate for CL in the maintenance of mitochondrial tubular morphology and fusion. To test this hypothesis, we constructed a conditional mutant crd1Δpsd1Δ containing null alleles of CRD1 (CL synthase) and PSD1 (mitochondrial phosphatidylserine decarboxylase), in which the wild type CRD1 gene is expressed on a plasmid under control of the TETOFF promoter. In the presence of tetracycline, the mutant exhibited highly fragmented mitochondria, loss of mitochondrial DNA, and reduced membrane potential, characteristic of fusion mutants. Deletion of DNM1, required for mitochondrial fission, restored the tubular mitochondrial morphology. Loss of CL and mitochondrial PE led to reduced levels of small and large isoforms of the fusion protein Mgm1p, possibly accounting for the fusion defect. Taken together, these data demonstrate for the first time in vivo that CL and mitochondrial PE are required to maintain tubular mitochondrial morphology and have overlapping functions in mitochondrial fusion.
Introduction
Mitochondria exist as dynamic, double membrane-bound organelles. Mitochondrial membranes are enriched in phospholipids and proteins that are required for mitochondrial biogenesis and for maintenance of mitochondrial morphology and the tubular network (1). CL3 and PE are non-bilayer forming phospholipids in the mitochondrial membranes (2, 3) that play an essential role in mitochondrial function. Although cells lacking CL or mitochondrial PE are viable, the loss of both phospholipids is lethal, suggesting that these lipids have overlapping functions that are essential (4). Several recent studies have implicated the involvement of CL and mitochondrial PE in the maintenance of mitochondrial morphology (5–7). CL and PE are fusogenic phospholipids that form hexagonal phases in the presence of divalent cations, which confer negative curvature to the mitochondrial membrane (8, 9). In the current study, we investigated the role of CL and PE in mitochondrial fusion.
Highly conserved protein machinery strictly regulates the process of mitochondrial fusion, and recent studies suggest that phospholipids also play a vital role in this process. Mitochondrial fusion in the yeast Saccharomyces cerevisiae primarily requires three proteins. These include the outer membrane GTPase, Fzo1p (Mfn1 and Mfn2 in mammals) (10, 11), the inner membrane GTPase, Mgm1p (Opa1 in mammals) (12, 13), and the outer membrane protein Ugo1p, which links the two GTPases to form a functional complex (14–16). In S. cerevisiae, Mgm1p exists as long (l-Mgm1p) and short isoforms (s-Mgm1p), both of which are required for mitochondrial fusion (17, 18). In vitro studies demonstrated that CL stimulates the GTPase activity of the s-Mgm1p (19, 20). Moreover, it was shown in vitro that s-Mgm1p and l-Mgm1p assemble in a CL-dependent manner (20). We hypothesized that the mitochondrial phospholipids CL and PE have overlapping functions in mitochondrial fusion in vivo. Consistent with this hypothesis, we demonstrate that cells lacking both CL and mitochondrial PE have reduced levels of both Mgm1p isoforms and exhibit excessive fragmentation of mitochondria and defects in mitochondrial fusion.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Growth Media
The S. cerevisiae strains used in this study, listed in Table 1, are isogenic to BY4741 and BY4742. The single mutants were obtained from the MATa yeast deletion collection obtained from Dr. John Lopes. Double and triple mutants used in this study were obtained by tetrad dissection. Synthetic complete media contained standard concentration of amino acids, all the essential components of Difco vitamin-free yeast nitrogen base, 0.2% ammonium sulfate, and glucose (2%). Synthetic drop-out media contained all of the aforementioned ingredients except the amino acid was used as a selectable marker. Complex media contained yeast extract (1%), peptone (2%), with glucose (2%) (YPD) or galactose (2%) (YP-galactose) as a carbon source. All of the plasmids were amplified and extracted using standard protocols. The plasmids were transformed into yeast strains using a one-step transformation protocol (21). The v5 epitope-tagged CRD1 gene was cloned into the pCM189 plasmid (ATCC), in which, the TETOFF promoter regulates the expression of cloned gene, using the BamHI and NotI restriction sites. The existing URA3 marker of the plasmid was replaced by HIS3 using EcoRV and ClaI restriction sites. Bacterial transformations were performed using dam− E. coli to avoid Dam methylase sensitivity to the ClaI restriction enzyme.
TABLE 1.
Strains used in this study
| Strains | Genotype | Reference |
|---|---|---|
| BY4741 | MATa, his 301, leu 200, met 1500, ura 300 | Invitrogen |
| BY4742 | MATα, his 301, leu 200, lys 200, ura 300 | Invitrogen |
| VGY1 | MATα, his 301, leu 200, lys 200, ura 300, crd1Δ::URA3 | Ref. 4 |
| crd1Δ | MATa, his 301, leu 200, met 1500, ura 300, crd1Δ::KanMX4 | Invitrogen |
| psd1Δ | MATa, his 301, leu 200, met 1500, ura 300, psd1Δ::KanMX4 | Invitrogen |
| psd1Δ | MATα, his 301, leu 200, lys 200, ura 300, psd1Δ::KanMX4 | This study |
| dnm1Δ | MATa, his 301, leu 200, met 1500, ura 300, dnm1Δ::KanMX4 | Invitrogen |
| fis1Δ | MATa, his 301, leu 200, met 1500, ura 300, fis1Δ::KanMX4 | Invitrogen |
| crd1Δpsd1Δ | MATα, his 301, leu 200, lys 200, ura 300, crd1Δ::URA3, psd1Δ::KanMX4, pCM189-CRD1 | This study |
| crd1Δpsd1Δ | MATa, his 301, leu 200, lys 200, met 1500, ura 300, crd1Δ::URA3, psd1Δ::KanMX4, pCM189-CRD1 | This study |
| crd1Δpsd1Δfis1Δ | MATa, his 301, leu 200, lys 200, ura 300, crd1Δ::URA3, psd1Δ::KanMX4, fis1Δ::KanMX4, pCM189-CRD1 | This study |
| crd1Δpsd1Δfis1Δ | MATα, his 301, leu 200, lys 200, ura 300, crd1Δ::URA3, psd1Δ::KanMX4, fis1Δ::KanMX4, pCM189-CRD1 | This study |
| crd1Δpsd1Δdnm1Δ | MATa, his 301, leu 200, ura 300, crd1Δ::URA3, psd1Δ::KanMX4, dnm1Δ::KanMX4, pCM189-CRD1 | This study |
| crd1Δpsd1Δdnm1Δ | MATα, his 301, leu 200, lys 200, met 1500, ura 300, crd1Δ::URA3, psd1Δ::KanMX4, dnm1Δ::KanMX4, pCM189-CRD1 | This study |
Fluorescence Microscopy
Fluorescence microscopy was performed using an Olympus BX41 epifluorescence microscope. Images were acquired using an Olympus Q-Color3 digitally charge-coupled device camera operated by QCapture2 software. All pictures were taken at 1000×. To stain mitochondrial DNA, yeast cells were cultured to the mid-log phase, fixed in 70% ethanol at room temperature for 30 min, washed two times with distilled water, and stained with 1 μg/ml DAPI (Sigma) for 5 min. Mitochondria were visualized by transforming the cells with either plasmid pYX142 or pYX122 expressing GFP fused to the mitochondrial presequence, pre-Su9 (22) (provided by Dr. Benedikt Westermann) or pYX142-mtRFP-expressing mitochondria-targeted RFP (provided by Dr. Janet Shaw). Cells were harvested in the appropriate medium and viewed under fluorescence microscopy.
Electron Microscopy
Cells were grown in 100 ml of YPD to an A550 of 0.5. After harvesting, cells were prepared for EM using the osmium thiocarbohydrazide osmium fixation method (23).
In Vivo Fusion Assay
The mitochondrial in vivo fusion assay was performed as described (13, 24). MATα cells of WT, crd1Δ, and psd1Δ, were transformed with pYX122-mtGFP and MATa cells were transformed with pYX142-mtRFP. MATa cells of the conditional mutant crd1Δpsd1Δ were transformed with pYX142-mtRFP, and MATα cells were transformed with pYX142-mtGFP. MATa cells of the conditional mutant crd1Δpsd1Δfis1Δ were transformed with pYX142-mtGFP, and MATα with pYX142-mtRFP. MATα cells of crd1psd1dnm1Δ were transformed with pYX142-mtGFP and MATa with pYX142-mtRFP. Cells were grown in 5 ml of selective media to an A550 of 0.5. After centrifugation, cells of opposite mating type were mixed and spotted on an YPD plate. After 3.5 h of incubation at 30° C, cells were observed for mitochondrial fusion. The images were merged and analyzed using Image J software.
Extraction, Separation, and Analysis of Yeast Total Phospholipids
Yeast cells were grown in the presence of 32Pi (10 μCi/ml) in the indicated growth conditions. Total phospholipids were extracted and analyzed by TLC as described (25). The developed chromatograms were analyzed by phosphorimaging, and the phospholipids were quantified using Image Quant software.
Flow Cytometry
Mitochondrial membrane potential was measured using whole cells as described (26). Cells were grown in YP-galactose media to the mid-logarithmic phase. Actively growing cells (5 × 104 cells) were incubated at 30° C with the dye tetramethyl rhodamine methyl ester (TMRM) (50 nm) for 30 min. To induce a decrease in membrane potential, control cells were treated with sodium azide (20 mm). Fluorescence was measured using a flow cytometer. The results were analyzed using WinMDI2.9 software.
SDS-PAGE and Western Blot Analysis
Proteins were extracted from cells grown to an A550 of 0.5, separated by 8% SDS-PAGE, transferred to PVDF membrane, and analyzed using primary antibodies to Fzo1p (1:1000), Ugo1p (1:1000), Mgm1p (1:500) (provided by Dr. Jodi Nunnari), and α-tubulin (1:1000) (Santa Cruz Biotechnology). Proteins were visualized using appropriate secondary antibody conjugated with horseradish peroxidase (1:3000) followed by detection using the ECL chemiluminescence system (GE Healthcare).
RESULTS
Maintenance of Mitochondrial Network and Mitochondrial Fusion Is Defective in Absence of CL and Mitochondrial PE
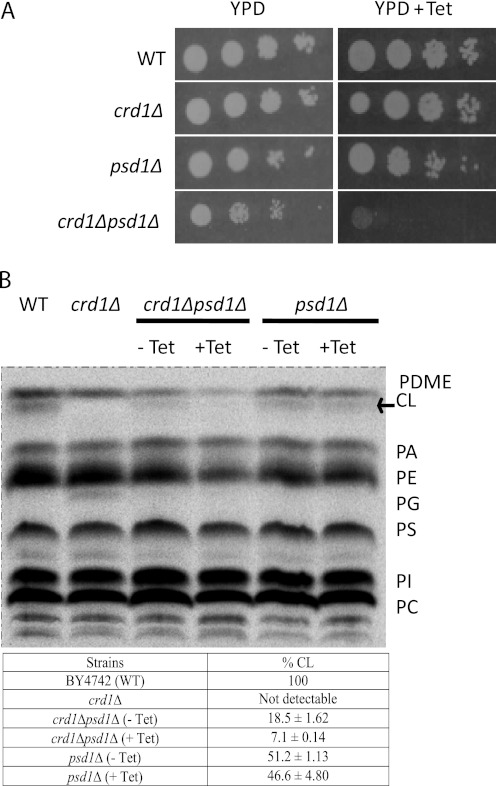
Previous studies have shown that loss of CL (crd1Δ) is lethal in combination with loss of mitochondrial PE (psd1Δ), but not cytosolic PE (psd2Δ) (4). To gain insight into the overlapping roles of these mitochondrial lipids, we constructed a conditional mutant, crd1Δpsd1Δ, in which CRD1 is expressed from a plasmid under the control of the TETOFF promoter. This mutant lacks mitochondrial PE and CL in the presence of tetracycline but contains CL in the absence of tetracycline. We used this conditional mutant as a tool to identify functions of these phospholipids in mitochondrial morphology and mitochondrial fusion. The conditional double mutant grew normally on YPD. The addition of tetracycline (200 μg/ml), which shut off CRD1 expression, inhibited growth of the double mutant but did not affect growth of WT, crd1Δ, or psd1Δ cells (Fig. 1A). To determine whether tetracycline did indeed regulate CRD1 expression, we measured the levels of CL in crd1Δpsd1Δ cells. In psd1Δ, CL was synthesized, although levels were reduced compared with those of WT, consistent with previous studies (4). In crd1Δpsd1Δ grown in the absence of tetracycline, CL levels were 40% of those of psd1Δ, indicating that CL levels from plasmid CRD1 are less than CL levels obtained from genomic CRD1. In the presence of tetracycline, CL was greatly diminished to only 14% of the levels in psd1Δ, indicating that expression from the TETOFF promoter was greatly (but not completely) repressed. Tetracycline itself did not affect CL levels in cells lacking the plasmid, which were similar in psd1Δ cells grown in the presence and absence of the drug (Fig. 1B).
FIGURE 1.
Tetracycline-dependent growth of the conditional mutant crd1Δpsd1Δ. A, 10-fold serial dilutions of cell suspensions were spotted on YPD plates supplemented with 200 μg/ml tetracycline (Tet) where indicated and incubated at 30° C. B, Cells were grown in YPD for 12 h in the presence or absence of tetracycline. Steady state labeling, phospholipid extraction, one-dimensional TLC, phosphorimaging, and quantification were carried out as described under “Experimental Procedures.” CL levels are quantified as percent of total phospholipids. Mean values ± S.D. of two independent experiments are shown. PC, phosphatidylcholine; PI, phosphatidylinositol; PS, phosphatidylserine; PG, phosphatidylglycerol; PA, phosphatidic acid.
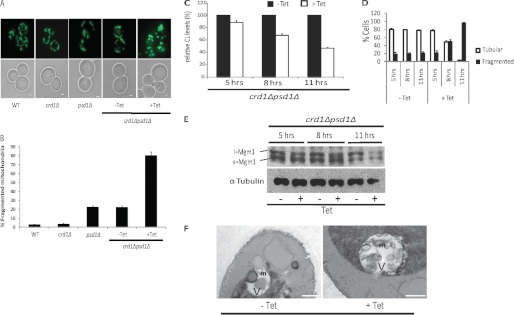
To determine whether CL and mitochondrial PE play a role in the maintenance of mitochondrial morphology, we compared the mitochondrial network in WT, crd1Δ, psd1Δ, and crd1Δpsd1Δ cells transformed with plasmids expressing mitochondria targeted GFP (mtGFP) (Fig. 2A) (22). At least 500 cells of each strain were observed for each biological replicate (Fig. 2B). Cells exhibited a normal tubular mitochondrial network in crd1Δ, consistent with earlier findings (27), indicating that the lack of CL by itself does not affect the mitochondrial network. The lack of mitochondrial PE had a small but significant effect on the mitochondrial network, as ∼23% of psd1Δ cells exhibited fragmented mitochondria. Unlike the WT-like tubular mitochondrial network, the majority of psd1Δ cells had short tubular mitochondria consistent with a mitochondrial morphology defect in these cells. The morphology of crd1Δpsd1Δ cells grown in the absence of tetracycline was similar to that of psd1Δ cells. However, the addition of tetracycline severely affected the mitochondrial network, leading to excessive mitochondrial fragmentation similar to that observed in fusion mutants (Fig. 2, A and B). These findings suggested that loss of both CL and mitochondrial PE leads to a defect in mitochondrial fusion. Tetracycline by itself did not affect the mitochondrial network in WT, crd1Δ, and psd1Δ cells (data not shown). To determine whether the increase in mitochondrial fragmentation correlated with the loss of CL, a time course experiment was performed in which crd1Δpsd1Δ cells were grown in YPD containing 32Pi, in the presence or absence of tetracycline. Total phospholipids and mitochondrial morphology were examined at 5, 8, and 11 h. Total CL decreased by ∼11, ∼31, and ∼55%, whereas the percentage of mitochondrial fragmentation increased during this time to ∼20, ∼45, and ∼96% at 5, 8, and 11 h, respectively (Fig. 2, C and D). These findings indicate that the increase in mitochondrial fragmentation corresponded with a decrease in CL in the crd1Δpsd1Δ cells.
FIGURE 2.
Mitochondrial fragmentation observed in crd1Δpsd1Δ cells. A, mitochondria were visualized using mtGFP. Cells were grown at 30° C to log phase in synthetic leucine deficient medium with or without 200 μg/ml tetracycline (Tet) and examined by fluorescence microscopy. Bars, 1 μm. B, quantitation of cells containing fragmented mitochondria. Values are mean ± S.E. (n = 3). At least 500 cells were visualized in each experiment. C–F, the crd1Δpsd1Δ mutant cells were grown at 30° C in the presence or absence of 200 μg/ml tetracycline and harvested at the indicated times. C, CL levels were analyzed by one-dimensional TLC as described under “Experimental Procedures,” and relative levels of CL are indicated. Values are mean ± S.E. (n = 3). D, cells containing fragmented and tubular mitochondrial morphology were quantified. Values are mean ± S.E. (n = 3). E, total cell proteins were extracted and analyzed by SDS-PAGE followed by Western blot. F, aliquots of crd1Δpsd1Δ cells were fixed as described under “Experimental Procedures,” and thin sections were examined by electron microscopy. Labels m and V indicate mitochondria and vacuole (white area), respectively. Bars, 500 nm.
Electron microscopic examination of the mutants revealed that crd1Δ mitochondria were somewhat smaller than those of WT but relatively unremarkable (data not shown). Mitochondria in psd1Δ cells and in crd1Δpsd1Δ cells grown in the absence of tetracycline also appeared smaller than WT. This phenotype was more significant in crd1Δpsd1Δ cells grown in the presence of tetracycline. Thus, the loss of both CL and mitochondrial PE led to highly fragmented mitochondria, consistent with defective fusion (Fig. 2F).
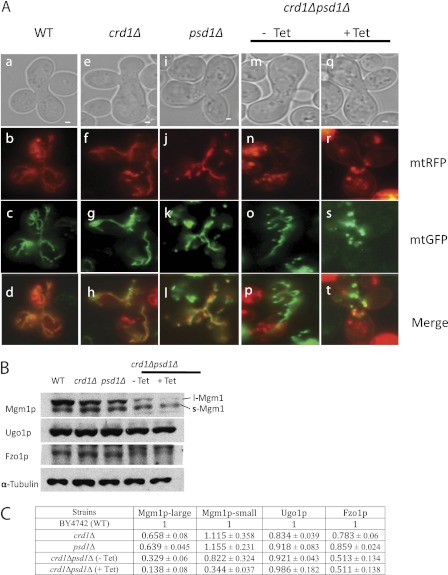
To determine the role of CL and mitochondrial PE in mitochondrial fusion, we performed an in vivo fusion assay (13, 24) as described under “Experimental Procedures.” In this assay, we examined mitochondrial fusion events in zygotes acquired by mating haploids of opposite mating types of WT, crd1Δ, psd1Δ, and crd1Δpsd1Δ cells transformed with either mtGFP or mitochondria-tagged RFP (mtRFP). As expected, crd1Δ cells exhibited complete mixing of mitochondrial content, indicating that the lack of CL alone does not affect mitochondrial fusion (Fig. 3A). Fusion occurred but was decreased in psd1Δ cells, suggesting that the lack of PE causes somewhat reduced fusion even when CL is present. As expected, the fusion phenotype of crd1Δpsd1Δ cells grown in the absence of tetracycline was similar to that of psd1Δ cells. However, in the presence of tetracycline, a complete block of mitochondrial fusion was observed in all the crd1Δpsd1Δ zygotes examined, consistent with the defective mitochondrial network observed in the absence of both CL and mitochondrial PE (Fig. 3A). These results indicate that when both CL and mitochondrial PE are deficient, mitochondrial fusion does not occur.
FIGURE 3.
crd1Δpsd1Δ cells exhibit defective mitochondrial fusion. A, cells of opposite mating types were transformed with either mtGFP or mtRFP. Mitochondrial fusion was examined by observing merged images of mtGFP and mtRFP in WT (a–d panels), crd1Δ (e–h panels), psd1Δ (i–l panels), and crd1Δpsd1Δ cells grown without (m–p panels) or with (q–t panels) tetracycline (Tet). Bars, 1 μm. B, total cellular proteins were analyzed by SDS-PAGE followed by Western blot. Steady state levels of Mgm1p, Fzo1p, and Ugo1p were measured. α-Tubulin was used as a loading control. C, quantitation of fusion proteins. Values are mean ± S.E. (n = 3).
Loss of Mitochondrial DNA and Reduced Mitochondrial Membrane Potential in Cells Lacking CL and Mitochondrial PE
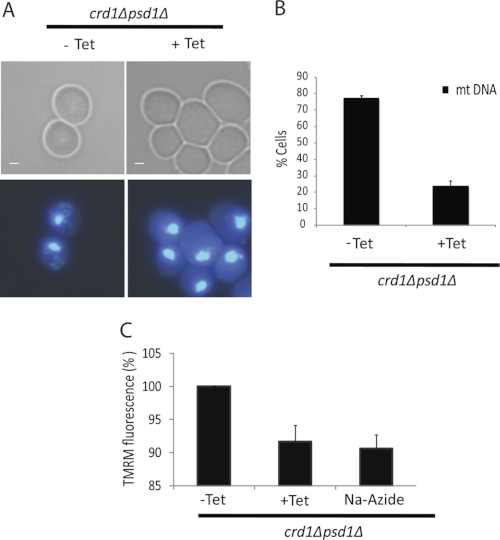
Several studies have reported that cells defective in mitochondrial fusion lose mitochondrial DNA (mtDNA) (10, 18, 24, 28, 29). Therefore, we hypothesized that crd1Δpsd1Δ cells would exhibit mtDNA loss. To address this possibility, WT, crd1Δ, psd1Δ, and crd1Δpsd1Δ cells were grown with or without tetracycline to the mid-logarithmic growth phase at 30° C. Cells were observed under the fluorescence microscope after DAPI staining for the presence of mtDNA (Fig. 4A). As expected, the majority of crd1Δpsd1Δ cells (∼80%) grown in the absence of tetracycline at the permissive temperature of 30° C retained mtDNA. This was consistent with our previous study showing that crd1Δ cells retained mtDNA at 30° C but exhibited mtDNA loss only at elevated temperatures (30). However, in the presence of tetracycline, only ∼20% of crd1Δpsd1Δ cells had mtDNA (Fig. 4B).
FIGURE 4.
crd1Δpsd1Δ cells exhibit loss of mitochondrial DNA and reduced membrane potential. A, cells were grown in YP-gal to log phase at 30° C with or without 200 μg/ml tetracycline (Tet) and stained with DAPI. Bars, 1 μm. B, quantitation of cells containing mtDNA. Values are mean ± S.E. (n = 3). At least 500 cells were visualized in each experiment. C, dissipation of the mitochondrial membrane potential demonstrated as TMRM fluorescence (%) in crd1Δpsd1Δ cells grown to log phase in YP-gal with or without tetracycline and stained with TMRM. Cells were also treated with sodium azide as a control.
Mitochondrial fusion as determined by in vitro assay involves distinct steps of outer and inner membrane fusion (31). In addition to functional protein complexes, fusion of the outer membrane requires low GTP levels and a proton gradient, whereas inner membrane fusion requires large amounts of GTP and an inner membrane potential. It is therefore possible that a decreased membrane potential led to the fusion defect in crd1Δpsd1Δ cells. To test this possibility, we used a flow cytometry assay to measure mitochondrial membrane potential (ΔΨm) in intact WT, crd1Δ, psd1Δ, and crd1Δpsd1Δ cells grown with or without tetracycline (26) in YP-galactose rather than YP-glucose to ensure actively respiring mitochondria. Cells were grown at 30° C to the mid-logarithmic growth phase and then incubated with the voltage-dependent probe TMRM (50 nm) for 30 min. The accumulation of TMRM in mitochondria is driven by the ΔΨm, which is determined by the difference in yellow fluorescence and forward scatter in the form of fluorescence peaks (26). Values were calculated relative to the control, i.e. crd1Δpsd1Δ cells grown in the absence of tetracycline. As seen in Fig. 4C, crd1Δpsd1Δ cells in the presence of tetracycline exhibited a decrease in membrane potential similar to that observed in these cells in the presence of sodium azide, a cytochrome c oxidase inhibitor that reduces the ΔΨm (26). These observations were consistent with a reduced membrane potential in cells lacking both CL and mitochondrial PE. It was recently demonstrated that mitochondrial fusion in mammalian cells requires high ΔΨm levels and is prevented by depolarization (32). Thus, the observed decrease of ΔΨm could be one explanation for the fusion defects in the crd1Δpsd1Δ mutant cells.
Deletion of DNM1 in crd1Δpsd1Δ Cells Restores Normal Mitochondrial Tubular Network
We wished to determine whether the mitochondrial fragmentation observed in crd1Δpsd1Δ cells could be explained by increased fission rather than decreased fusion. Fusion and fission regulate mitochondrial morphology in an antagonistic manner (33). Previous studies have shown that three major proteins regulate mitochondrial fission, Dnm1p (34–36), Fis1p (36), and Mdv1p (37, 38). Abolishing mitochondrial fission by deletion of any of these genes leads to net-like mitochondria. In contrast, eliminating fusion by deletion of MGM1, FZO1, or UGO1 leads to fragmentation, which can be restored to normal tubular morphology by deletion of the fission gene DNM1 (13, 35). If mitochondrial fragmentation in crd1Δpsd1Δ cells results from a defect in fusion and not increased fission, then disruption of mitochondrial fission would restore mitochondria to the normal tubular morphology. Therefore, we examined whether the fragmented mitochondrial morphology of crd1Δpsd1Δ cells could be rescued to normal tubular mitochondrial morphology by deletion of the fission gene DNM1. To do so, we constructed a crd1Δpsd1Δdnm1Δ conditional mutant containing the plasmid with the TETOFF-regulated CRD1 expression plasmid, as well as a plasmid expressing mtGFP (Fig. 5A). In the absence of tetracycline, when CRD1 is expressed, the crd1Δpsd1Δdnm1Δ cells would be expected to exhibit net-like mitochondria characteristic of a fission defect. However, in the presence of tetracycline, the cells would be predicted to lack both fission and fusion and, hence, would exhibit WT tubular mitochondrial morphology.
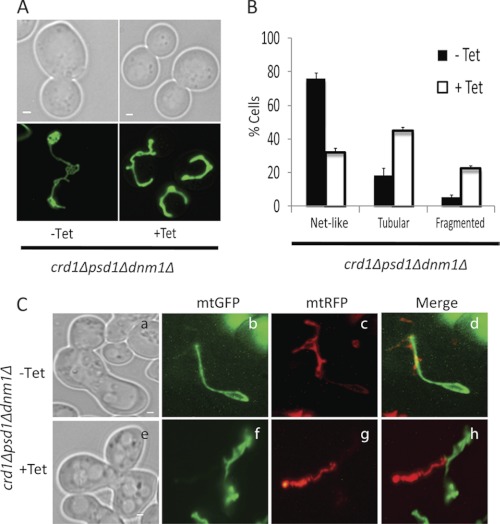
FIGURE 5.
crd1Δpsd1Δdnm1Δ cells are defective in mitochondrial fusion. A, mitochondria were visualized in the crd1Δpsd1Δdnm1Δ mutant using mtGFP. Cells were grown at 30° C to log phase in synthetic deficient glucose medium with 200 μg/ml tetracycline (Tet) where indicated and examined by fluorescence microscopy. Bars, 1 μm. B, cells containing tubular, fragmented, and net-like mitochondria were quantified. Values are mean ± S.E. (n = 3). At least 100 cells were visualized in each experiment. C, crd1Δpsd1Δdnm1Δ cells of opposite mating types were transformed with either mtGFP or mtRFP. Mitochondrial fusion was examined by observing merged images of mtGFP and mtRFP in zygotes of crd1Δpsd1Δdnm1Δ grown without (a–d panels) or with (e–h panels) tetracycline. Bars, 1 μm.
As seen in Fig. 5B, ∼76% of crd1Δpsd1Δdnm1Δ cells grown in the absence of tetracycline exhibited net-like mitochondria, the predicted phenotype. The remaining cells (∼18%) exhibited tubular mitochondria, most likely because fusion was decreased in these cells due to the low level of expression of CRD1 (Fig. 1). In the presence of tetracycline, only ∼32% of cells exhibited net-like mitochondria, whereas the majority (∼45%) exhibited tubular mitochondria, as predicted. These findings suggest that both fission and fusion were defective in these cells and that the fragmented mitochondrial morphology in crd1Δpsd1Δ cells was rescued by deletion of the fission gene DNM1 (Fig. 5, A and B). Tetracycline itself did not affect the mitochondrial morphology in dnm1Δ (data not shown). Interestingly, a significant number of crd1Δpsd1Δdnm1Δ cells (∼22%) grown in the presence of tetracycline, had fragmented mitochondria, as the network exhibited the appearance of a string of beads (data not shown). This morphology suggested the presence of a persistent fusion defect in the absence of CL and mitochondrial PE.
To further investigate the block in fusion, we performed an in vivo mitochondrial fusion experiment by mating crd1Δpsd1Δdnm1Δ cells of opposite mating types, in which one mating type contained mtGFP and the other mating type contained mtRFP. We observed decreased fusion in cells grown without tetracycline, and a complete block in mitochondrial fusion in cells grown with tetracycline (Fig. 5C). Cells grown without tetracycline that exhibited net-like structures had no defect in mitochondrial fusion (Fig. 5C). Cells grown in the presence of tetracycline displayed a complete block of mitochondrial fusion. Similar observations were made in the conditional mutant crd1Δpsd1Δfis1Δ (supplemental Fig. S1). These experiments suggest that crd1Δpsd1Δdnm1Δ cells exhibited a fusion defect due to loss of CRD1 and PSD1. Taken together, these studies indicate that mitochondrial fragmentation observed in crd1Δpsd1Δ cells is a result of defective fusion and not due to increased fission.
To determine whether deletion of the fission gene FIS1 or DNM1 could rescue the lethality of the double mutant, we crossed crd1Δdnm1Δ and crd1Δfis1Δ with psd1Δ and carried out meiotic tetrad analysis to identify viable triple mutants. However, triple mutants were not detected in 72 tetrads of the diploid crd1Δfis1ΔPSD1/CRD1FIS1psd1Δ or 75 tetrads of the diploid crd1Δdnm1ΔPSD1/CRD1DNM1psd1Δ. Therefore, although CL and mitochondrial PE have overlapping functions in mitochondrial fusion, rescue of the fusion defect could not rescue the synthetic lethality.
Reduced Steady State Levels of l-Mgm1 and s-Mgm1 Isoforms in Cells Lacking CL and Mitochondrial PE
The current study suggests that one common function of CL and PE is mitochondrial fusion. It has been reported that the lack of CL destabilizes the anchoring, assembly, and GTPase activity of fusion protein Mgm1p in vitro (19, 20, 39). To test whether mitochondrial PE compensates for the loss of CL and stabilizes the fusion proteins in vivo, we determined the steady state levels of fusion proteins Fzo1p, Ugo1p, l-Mgm1p, and s-Mgm1p in WT, crd1Δ, psd1Δ, and crd1Δpsd1Δ cells. The crd1Δpsd1Δ cells exhibited significantly diminished levels of l-Mgm1p and s-Mgm1p (Fig. 3, B and C). Fzo1p levels were slightly decreased and Ugo1p was not affected (Fig. 3, B and C). To determine whether the loss of Mgm1p isoforms correlated with the loss CL in crd1Δpsd1Δ, cells were grown in the presence or absence of tetracycline, proteins were extracted from cells harvested at 5, 8, and 11 h, and the levels of Mgm1p isoforms were determined by Western blot (Fig. 2E). The isoform levels were severely diminished at 11 h, which correlated with increased mitochondrial fragmentation as seen in Fig. 2C. These data indicate that the defect in mitochondrial fusion in crd1Δpsd1Δ can be attributed at least in part to the reduced levels of s-Mgm1p and 1-Mgm1p.
DISCUSSION
In this study, we demonstrate that crd1Δpsd1Δ cells lacking both CL and mitochondrial PE have fragmented mitochondria due to a defect in mitochondrial fusion. In addition to this defect, we show that crd1Δpsd1Δ cells exhibit loss of mtDNA, decreased membrane potential, and reduced steady state levels of short and long isoforms of Mgm1p, a mitochondrial inner membrane protein essential for fusion. The fragmented mitochondrial morphology along with the fusion defect observed in crd1Δpsd1Δ cells were rescued by deletion of the fission genes DNM1 or FIS1. These data indicate that CL and mitochondrial PE are required for mitochondrial fusion in vivo.
Our previous studies have shown a synthetic lethal interaction between crd1Δ and psd1Δ mutant cells, suggesting essential overlapping roles of CL and mitochondrial PE (4). PE synthesized by the non-mitochondrial pathway (Psd2p catalyzed PE synthesis in Golgi/vacuole) (40–42) did not rescue this lethality. Externally synthesized PE is inefficiently transported to the inner mitochondrial membrane, as reduced levels of PE were observed in the inner mitochondrial membrane of the psd1Δ mutant cells (43). Taken together, these studies suggested that PE synthesized in the mitochondrial inner membrane has functions that cannot be compensated by externally synthesized PE. In the current study, we demonstrate that the loss of mitochondrial phospholipids CL and PE leads to mitochondrial fragmentation (Fig. 2, A, B, and F) and defective mitochondrial fusion (Fig. 3A). Although mitochondrial fusion is an overlapping function of CL and PE, the lack of mitochondrial fusion is probably not the cause of lethality observed in crd1Δpsd1Δ cells, as lethality was not rescued by deletion of the fission gene FIS1 or DNM1. Mitochondria are required not only for cellular bioenergetics, but also for the synthesis of essential metabolites. In addition, our previous studies have shown that CL is required for non-mitochondrial functions, including vacuolar function, the high osmolarity glycerol (44) pathway, and cell wall synthesis (45–47). Thus, it is possible that lethality in cells lacking CL and PE could be caused by deficiencies in both mitochondrial and non-mitochondrial functions. The identification of suppressors of crd1Δpsd1Δ synthetic lethality will very likely identify the essential cellular functions shared by these phospholipids. These studies are currently in progress.
How do CL and mitochondrial PE affect mitochondrial fusion? Non-bilayer lipids are known to affect the function and stability of many mitochondrial membrane proteins (48). Recent studies have proposed that scaffolding proteins such as prohibitin recruit membrane proteins to CL- and PE-rich regions, forming protein-rich lipid domains (49). The lack of CL and mitochondrial PE might influence the distribution of these domains, which in turn, would affect several mitochondrial processes, including mitochondrial fusion. Although early studies suggested that the non-bilayer forming phospholipids CL and PE play an important role in mitochondrial fusion, very little was known about the mechanism by which this could occur (8, 50, 51). In this study, we show that the lack of CL and mitochondrial PE leads to reduced steady state levels of both large and small isoforms of Mgm1p (Figs. 2E and 3B), which are required for fusion. Recent studies have shown that l-Mgm1p acts as an anchor in the inner membrane (17). Both CL and PE are synthesized and predominantly localized in the inner mitochondrial membrane, and the loss of both CL and mitochondrial PE might affect the stability of this isoform, leading to its degradation. The formation of s-Mgm1p requires functional mitochondrial protein import machinery, membrane potential and adequate ATP levels, all of which are defective in cells lacking CL (52–56). This is a first report describing overlapping roles of CL and mitochondrial PE in fusion in vivo and suggests a mechanistic role for these phospholipids in regulating mitochondrial structure and function.
How is the role of CL and PE in mitochondrial fusion relevant to human disease? The role of mitochondrial phospholipids in fusion is relevant to studies that implicate function of mitochondrial fusion in cardiac function (57). Fragmented mitochondria are associated with the loss of Opa1 (the human homolog of Mgm1p) in mitochondrial myopathies involving cardiac and skeletal muscle (53) and in ischemic cardiomyopathy (58). Overexpression of the fusion proteins Mfn1/2 (human homolog of Fzo1p) prevents cardiac cell death from ischemia (59). Elucidating the role of CL and PE in mitochondrial fusion may also shed light on defects observed in lymphoblast mitochondria from patients with Barth syndrome (BTHS), a severe genetic disorder characterized by dilated cardiomyopathy and skeletal myopathy (60, 61). BTHS is caused by mutation in the CL remodeling enzyme tafazzin, resulting in decreased CL and altered fatty acid composition of major mitochondrial phospholipids, including CL and PE (62). Defects in mitochondrial fusion may account for the observed morphological variation in BTHS mitochondria, including enlarged size, fragmentation, adhesion of opposing membranes, and deformed intercristae space observed in BTHS lymphoblasts as well as in cardiac and skeletal muscle mitochondria of the mouse model of BTHS (63, 64). Identifying the role of CL and PE in mitochondrial fusion may thus explain, in part, the wide variation in the clinical presentation observed in BTHS.
Acknowledgments
We are grateful to Jodi Nunnari and Suzanne Hoppins for discussions and useful suggestions in this study. EM images were taken at The Integrated Imaging Center (The Johns Hopkins University). We thank Vishal Gohil, Vinay Patil, and Shuliang Chen for valuable suggestions, Icksoo Lee for help with the FACS experiment, and Cunqi Ye for assistance with protein work.
This work was supported, in whole or in part, by National Institutes of Health Grant R21 HL 084218. This work was also supported by grants from The Barth Syndrome Foundation (to M. L. G.) and by Wayne State University Graduate Enhancement Research Fellowship and Graduate Enhancement Research Funds (to A. S. J.).

This article contains supplemental Fig. 1.
- CL
- cardiolipin
- PE
- phosphatidylethanolamine
- PDME
- phosphatidyldimethylethanolamine
- TMRM
- tetramethyl rhodamine methyl ester
- mtGFP
- mitochondria-targeted GFP
- RFP
- red fluorescent protein.
REFERENCES
- 1. Gohil V. M., Greenberg M. L. (2009) Mitochondrial membrane biogenesis: Phospholipids and proteins go hand in hand. J. Cell Biol. 184, 469–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gonzalvez F., Gottlieb E. (2007) Cardiolipin: Setting the beat of apoptosis. Apoptosis 12, 877–885 [DOI] [PubMed] [Google Scholar]
- 3. Ardail D., Privat J. P., Egret-Charlier M., Levrat C., Lerme F., Louisot P. (1990) Triggering of mannosyltransferase activity in inner mitochondrial membranes by dolichyl-monophosphate incorporation mediated through phospholipids or fatty acids. J. Biol. Chem. 265, 18797–18802 [DOI] [PubMed] [Google Scholar]
- 4. Gohil V. M., Thompson M. N., Greenberg M. L. (2005) Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine and cardiolipin biosynthetic pathways in Saccharomyces cerevisiae. J. Biol. Chem. 280, 35410–35416 [DOI] [PubMed] [Google Scholar]
- 5. Tamura Y., Endo T., Iijima M., Sesaki H. (2009) Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J. Cell Biol. 185, 1029–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Osman C., Haag M., Potting C., Rodenfels J., Dip P. V., Wieland F. T., Brügger B., Westermann B., Langer T. (2009) The genetic interactome of prohibitins: Coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 184, 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuroda T., Tani M., Moriguchi A., Tokunaga S., Higuchi T., Kitada S., Kuge O. (2011) FMP30 is required for the maintenance of a normal cardiolipin level and mitochondrial morphology in the absence of mitochondrial phosphatidylethanolamine synthesis. Mol. Microbiol. 80, 248–265 [DOI] [PubMed] [Google Scholar]
- 8. van den Brink-van der Laan E., Killian J. A., de Kruijff B. (2004) Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim. Biophys. Acta 1666, 275–288 [DOI] [PubMed] [Google Scholar]
- 9. Rand R. P., Sengupta S. (1972) Cardiolipin forms hexagonal structures with divalent cations. Biochim. Biophys. Acta 255, 484–492 [DOI] [PubMed] [Google Scholar]
- 10. Hermann G. J., Thatcher J. W., Mills J. P., Hales K. G., Fuller M. T., Nunnari J., Shaw J. M. (1998) Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143, 359–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rapaport D., Brunner M., Neupert W., Westermann B. (1998) Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 273, 20150–20155 [DOI] [PubMed] [Google Scholar]
- 12. Wong E. D., Wagner J. A., Gorsich S. W., McCaffery J. M., Shaw J. M., Nunnari J. (2000) The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 151, 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wong E. D., Wagner J. A., Scott S. V., Okreglak V., Holewinske T. J., Cassidy-Stone A., Nunnari J. (2003) The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion. J. Cell Biol. 160, 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sesaki H., Jensen R. E. (2004) Ugo1p links the Fzo1p and Mgm1p GTPases for mitochondrial fusion. J. Biol. Chem. 279, 28298–28303 [DOI] [PubMed] [Google Scholar]
- 15. Sesaki H., Jensen R. E. (2001) UGO1 encodes an outer membrane protein required for mitochondrial fusion. J. Cell Biol. 152, 1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoppins S., Horner J., Song C., McCaffery J. M., Nunnari J. (2009) Mitochondrial outer and inner membrane fusion requires a modified carrier protein. J. Cell Biol. 184, 569–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zick M., Duvezin-Caubet S., Schäfer A., Vogel F., Neupert W., Reichert A. S. (2009) Distinct roles of the two isoforms of the dynamin-like GTPase Mgm1 in mitochondrial fusion. FEBS Lett. 583, 2237–2243 [DOI] [PubMed] [Google Scholar]
- 18. Herlan M., Vogel F., Bornhovd C., Neupert W., Reichert A. S. (2003) Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 278, 27781–27788 [DOI] [PubMed] [Google Scholar]
- 19. Rujiviphat J., Meglei G., Rubinstein J. L., McQuibban G. A. (2009) Phospholipid association is essential for dynamin-related protein Mgm1 to function in mitochondrial membrane fusion. J. Biol. Chem. 284, 28682–28686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. DeVay R. M., Dominguez-Ramirez L., Lackner L. L., Hoppins S., Stahlberg H., Nunnari J. (2009) Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J. Cell Biol. 186, 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen D. C., Yang B. C., Kuo T. T. (1992) One-step transformation of yeast in stationary phase. Curr. Genet. 21, 83–84 [DOI] [PubMed] [Google Scholar]
- 22. Westermann B., Neupert W. (2000) Mitochondria-targeted green fluorescent proteins: Convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16, 1421–1427 [DOI] [PubMed] [Google Scholar]
- 23. Willingham M. C., Rutherford A. V. (1984) The use of osmium-thiocarbohydrazide-osmium (OTO) and ferrocyanide-reduced osmium methods to enhance membrane contrast and preservation in cultured cells. J. Histochem. Cytochem. 32, 455–460 [DOI] [PubMed] [Google Scholar]
- 24. Nunnari J., Marshall W. F., Straight A., Murray A., Sedat J. W., Walter P. (1997) Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell 8, 1233–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vaden D. L., Gohil V. M., Gu Z., Greenberg M. L. (2005) Separation of yeast phospholipids using one-dimensional thin-layer chromatography. Anal. Biochem. 338, 162–164 [DOI] [PubMed] [Google Scholar]
- 26. Ludovico P., Sansonetty F., Côrte-Real M. (2001) Assessment of mitochondrial membrane potential in yeast cell populations by flow cytometry. Microbiology 147, 3335–3343 [DOI] [PubMed] [Google Scholar]
- 27. Chen S., Liu D., Finley R. L., Jr., Greenberg M. L. (2010) Loss of mitochondrial DNA in the yeast cardiolipin synthase crd1 mutant leads to up-regulation of the protein kinase Swe1p that regulates the G2/M transition. J. Biol. Chem. 285, 10397–10407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guan K., Farh L., Marshall T. K., Deschenes R. J. (1993) Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1 gene. Curr. Genet. 24, 141–148 [DOI] [PubMed] [Google Scholar]
- 29. Chen H., Vermulst M., Wang Y. E., Chomyn A., Prolla T. A., McCaffery J. M., Chan D. C. (2010) Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 141, 280–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhong Q., Gohil V. M., Ma L., Greenberg M. L. (2004) Absence of cardiolipin results in temperature sensitivity, respiratory defects, and mitochondrial DNA instability independent of pet56. J. Biol. Chem. 279, 32294–32300 [DOI] [PubMed] [Google Scholar]
- 31. Meeusen S., McCaffery J. M., Nunnari J. (2004) Mitochondrial fusion intermediates revealed in vitro. Science 305, 1747–1752 [DOI] [PubMed] [Google Scholar]
- 32. Mitra K., Wunder C., Roysam B., Lin G., Lippincott-Schwartz J. (2009) A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc. Natl. Acad. Sci. U.S.A. 106, 11960–11965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hoppins S., Lackner L., Nunnari J. (2007) The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76, 751–780 [DOI] [PubMed] [Google Scholar]
- 34. Bleazard W., McCaffery J. M., King E. J., Bale S., Mozdy A., Tieu Q., Nunnari J., Shaw J. M. (1999) The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1, 298–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sesaki H., Jensen R. E. (1999) Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147, 699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mozdy A. D., McCaffery J. M., Shaw J. M. (2000) Dnm1p GTPase-mediated mitochondrial fission is a multistep process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151, 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tieu Q., Nunnari J. (2000) Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151, 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tieu Q., Okreglak V., Naylor K., Nunnari J. (2002) The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 158, 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ban T., Heymann J. A., Song Z., Hinshaw J. E., Chan D. C. (2010) OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Hum. Mol. Genet. 19, 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Trotter P. J., Pedretti J., Voelker D. R. (1993) Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J. Biol. Chem. 268, 21416–21424 [PubMed] [Google Scholar]
- 41. Trotter P. J., Pedretti J., Yates R., Voelker D. R. (1995) Phosphatidylserine decarboxylase 2 of Saccharomyces cerevisiae. Cloning and mapping of the gene, heterologous expression, and creation of the null allele. J. Biol. Chem. 270, 6071–6080 [DOI] [PubMed] [Google Scholar]
- 42. Trotter P. J., Voelker D. R. (1995) Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270, 6062–6070 [DOI] [PubMed] [Google Scholar]
- 43. Bürgermeister M., Birner-Grünberger R., Nebauer R., Daum G. (2004) Contribution of different pathways to the supply of phosphatidylethanolamine and phosphatidylcholine to mitochondrial membranes of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1686, 161–168 [DOI] [PubMed] [Google Scholar]
- 44. Schüller C., Brewster J. L., Alexander M. R., Gustin M. C., Ruis H. (1994) The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 13, 4382–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen S., Tarsio M., Kane P. M., Greenberg M. L. (2008) Cardiolipin mediates cross-talk between mitochondria and the vacuole. Mol. Biol. Cell 19, 5047–5058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhong Q., Li G., Gvozdenovic-Jeremic J., Greenberg M. L. (2007) Up-regulation of the cell integrity pathway in Saccharomyces cerevisiae suppresses temperature sensitivity of the pgs1Δ mutant. J. Biol. Chem. 282, 15946–15953 [DOI] [PubMed] [Google Scholar]
- 47. Zhou J., Zhong Q., Li G., Greenberg M. L. (2009) Loss of cardiolipin leads to longevity defects that are alleviated by alterations in stress response signaling. J. Biol. Chem. 284, 18106–18114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schlame M., Ren M. (2009) The role of cardiolipin in the structural organization of mitochondrial membranes. Biochim. Biophys. Acta 1788, 2080–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Osman C., Voelker D. R., Langer T. (2011) Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 192, 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Furt F., Moreau P. (2009) Importance of lipid metabolism for intracellular and mitochondrial membrane fusion/fission processes. Int. J. Biochem. Cell Biol. 41, 1828–1836 [DOI] [PubMed] [Google Scholar]
- 51. Cullis P. R., de Kruijff B. (1979) Lipid polymorphism and the functional roles of lipids in biological membranes. Biochim. Biophys. Acta 559, 399–420 [DOI] [PubMed] [Google Scholar]
- 52. Jiang F., Ryan M. T., Schlame M., Zhao M., Gu Z., Klingenberg M., Pfanner N., Greenberg M. L. (2000) Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 275, 22387–22394 [DOI] [PubMed] [Google Scholar]
- 53. Duvezin-Caubet S., Jagasia R., Wagener J., Hofmann S., Trifunovic A., Hansson A., Chomyn A., Bauer M. F., Attardi G., Larsson N. G., Neupert W., Reichert A. S. (2006) Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J. Biol. Chem. 281, 37972–37979 [DOI] [PubMed] [Google Scholar]
- 54. Herlan M., Bornhövd C., Hell K., Neupert W., Reichert A. S. (2004) Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J. Cell Biol. 165, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gebert N., Joshi A. S., Kutik S., Becker T., McKenzie M., Guan X. L., Mooga V. P., Stroud D. A., Kulkarni G., Wenk M. R., Rehling P., Meisinger C., Ryan M. T., Wiedemann N., Greenberg M. L., Pfanner N. (2009) Mitochondrial cardiolipin involved in outer membrane protein biogenesis: Implications for Barth syndrome. Curr. Biol. 19, 2133–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Claypool S. M., Oktay Y., Boontheung P., Loo J. A., Koehler C. M. (2008) Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J. Cell Biol. 182, 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dorn G. W., 2nd, Clark C. F., Eschenbacher W. H., Kang M. Y., Engelhard J. T., Warner S. J., Matkovich S. J., Jowdy C. C. (2011) MARF and Opa1 control mitochondrial and cardiac function in Drosophila. Circ. Res. 108, 12–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen L., Gong Q., Stice J. P., Knowlton A. A. (2009) Mitochondrial OPA1, apoptosis, and heart failure. Cardiovasc. Res. 84, 91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ong S. B., Subrayan S., Lim S. Y., Yellon D. M., Davidson S. M., Hausenloy D. J. (2010) Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation 121, 2012–2022 [DOI] [PubMed] [Google Scholar]
- 60. Barth P. G., Van den Bogert C., Bolhuis P. A., Scholte H. R., van Gennip A. H., Schutgens R. B., Ketel A. G. (1996) X-linked cardioskeletal myopathy and neutropenia (Barth syndrome): Respiratory chain abnormalities in cultured fibroblasts. J. Inherit. Metab. Dis. 19, 157–160 [DOI] [PubMed] [Google Scholar]
- 61. Bolhuis P. A., Hensels G. W., Hulsebos T. J., Baas F., Barth P. G. (1991) Mapping of the locus for X-linked cardioskeletal myopathy with neutropenia and abnormal mitochondria (Barth syndrome) to Xq28. Am. J. Hum. Genet. 48, 481–485 [PMC free article] [PubMed] [Google Scholar]
- 62. Xu Y., Sutachan J. J., Plesken H., Kelley R. I., Schlame M. (2005) Characterization of lymphoblast mitochondria from patients with Barth syndrome. Lab. Invest. 85, 823–830 [DOI] [PubMed] [Google Scholar]
- 63. Acehan D., Xu Y., Stokes D. L., Schlame M. (2007) Comparison of lymphoblast mitochondria from normal subjects and patients with Barth syndrome using electron microscopic tomography. Lab. Invest. 87, 40–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Acehan D., Vaz F., Houtkooper R. H., James J., Moore V., Tokunaga C., Kulik W., Wansapura J., Toth M. J., Strauss A., Khuchua Z. (2011) Cardiac and skeletal muscle defects in a mouse model of human barth syndrome. J. Biol. Chem. 286, 899–908 [DOI] [PMC free article] [PubMed] [Google Scholar]