Background: DNA polymerase (Pol) δ is involved in UV light-induced mutagenesis by an unknown mechanism.
Results: The C terminus of DNA Pol ζ interacts with accessory subunits of DNA Pol δ, which is required for UV light-induced mutagenesis.
Conclusion: When replication is stalled, accessory subunits of DNA Pol δ participate in recruitment of translesion DNA Pol ζ.
Significance: This finding provides a novel mechanism of DNA lesion bypass in eukaryotes.
Keywords: DNA Damage Response, DNA Polymerase, DNA Replication, Iron-Sulfur Protein, Protein-Protein Interactions, DNA Polymerase δ, DNA Polymerase ζ, Translesion DNA Synthesis
Abstract
Translesion DNA synthesis is an important branch of the DNA damage tolerance pathway that assures genomic integrity of living organisms. The mechanisms of DNA polymerase (Pol) switches during lesion bypass are not known. Here, we show that the C-terminal domain of the Pol ζ catalytic subunit interacts with accessory subunits of replicative DNA Pol δ. We also show that, unlike other members of the human B-family of DNA polymerases, the highly conserved and similar C-terminal domains of Pol δ and Pol ζ contain a [4Fe-4S] cluster coordinated by four cysteines. Amino acid changes in Pol ζ that prevent the assembly of the [4Fe-4S] cluster abrogate Pol ζ function in UV mutagenesis. On the basis of these data, we propose that Pol switches at replication-blocking lesions occur by the exchange of the Pol δ and Pol ζ catalytic subunits on a preassembled complex of accessory proteins retained on DNA during translesion DNA synthesis.
Introduction
Eukaryotes possess four B-family DNA polymerases: α, δ, ϵ, and ζ (1). Polymerase (Pol)2 α functions in initiation and early elongation steps of replication by extending RNA primers laid by a tightly associated primase. Pol δ plays an indispensable role in DNA replication and DNA repair in eukaryotic cells (2). Pol ϵ is involved in the initiation of replication at origins and in leading-strand synthesis in the vicinity of the origins (3). Its role in bulk replication is less clear because, in yeast, the N-terminal part of the protein responsible for catalytic functions is not essential for replication (4, 5). Pol ζ can bypass some lesions and, importantly, is ultimately involved in the extension of non-canonical primer-template combinations that result in mutation fixation (6, 7). Disruption of the gene encoding the catalytic subunit (REV3) results in a severe decrease in spontaneous and damage-induced mutagenesis, embryonic lethality in mice, and chromosomal instability (8). These properties put Pol ζ in a central position in the cellular machinery regulating the outcomes of DNA damage, a process that triggers many diseases, including cancer.
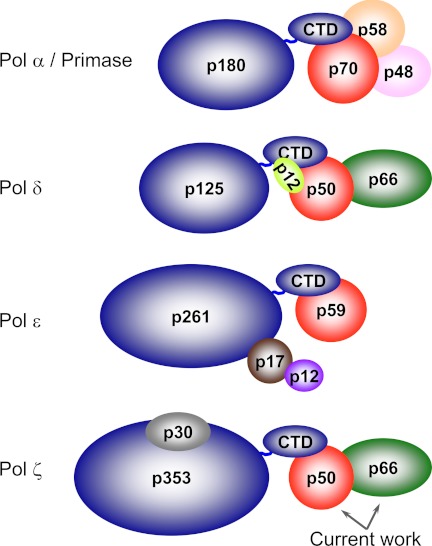
The four eukaryotic B-family DNA polymerases are multisubunit complexes composed of catalytic and regulatory subunits (Fig. 1) (9–11). DNA polymerases α, δ, and ϵ invariably have orthologous essential B-subunits bound to the C-terminal domain (CTD) of the catalytic subunit. A corresponding subunit was not found for Pol ζ; however, its CTD harbors two metal-binding sites (MBS1 and MBS2), each composed of four conserved cysteines, as in the other members of B-family DNA polymerases (11–13). The crystal structure of the yeast Pol α CTD and B-subunit complex revealed that MBS2 is directly involved in intersubunit interaction (13). Combined with electron microscopy studies of Pol α and Pol ϵ and small angle x-ray scattering studies of Pol δ, these data indicate that the CTD is organized as a separate domain connected to the catalytic core by a flexible linker (13–15). This indicates that CTD is a universal tether between the catalytic core and accessory subunits in replicative DNA polymerases. Unlike Pol α, Pol δ, and Pol ϵ, the structural role for the Pol ζ CTD was not established.
FIGURE 1.
Schematic representation of multisubunit organization for human B-family DNA polymerases. Subunits with similar functions are color-coded as follows: catalytic subunits are dark blue, B-subunits are red, and the C-subunit of Pol δ (p66) is green. For quick reference: in budding yeast, the catalytic subunits of Pol α, Pol δ, Pol ϵ, and Pol ζ are Pol1, Pol3, Pol2, and Rev3, respectively; the B-subunits of Pol α, Pol δ, and Pol ϵ are Pol12, Pol31, and Dpb2, respectively; and the C-subunit of Pol δ is Pol32.
Intriguingly, mutations affecting the CTD or accessory subunits of Pol δ abolish induced mutagenesis to the same extent as the absence of Pol ζ (16–18), indicating that Pol δ participates in the regulation of error-prone translesion DNA synthesis (TLS). However, the reasons for the dependence of Pol ζ function on Pol δ and the mechanism of the key event in TLS, the Pol δ ↔ Pol ζ switch, remained a mystery. Here, we present an explanation for this by demonstrating that Pol ζ and the catalytic subunit of Pol δ share the accessory B- and C-subunits of Pol δ.
EXPERIMENTAL PROCEDURES
Construction of REV3 Mutants in Yeast and Analysis of Their Expression and in Vivo Effects
Mutations in the regions encoding the yeast Rev3 active site and CTD were first introduced into yeast integrative plasmid pRevLCav2 (19). This plasmid contains the Saccharomyces cerevisiae REV3 ORF region coding for 647 C-terminal amino acids and 335 bp of downstream noncoding sequence and the URA3 gene. We used a multiple-site plasmid mutagenesis protocol (20) to create alleles coding for changes of cysteines 1398, 1401, 1414, and 1417 at MBS1 and cysteines 1446, 1449, 1468, and 1473 at MBS2 to alanines. The resulting plasmids encoding mutant REV3 ORF ends were digested with SnaBI prior to transformation into yeast strain 8C-YUNI101 (MATa his7-2 leu2-3,112 ura3Δ bik1::ura3-29RL trp1-1UAG ade2-1UAA) (19). Transformants were selected on synthetic medium without uracil and colony-purified. In the next step, we selected clones that lost both the URA3 marker and the duplicated part of REV3 but acquired the desired mutation on medium containing 5-fluoroorotic acid, which selects against URA3+ cells (21). The resulting strains were used for studies of UV light-induced lethality and mutagenesis as described previously (19). All data points are an average of three independent trials, and error bars are S.D.
For the analysis of the protein levels of Rev3 variants, we constructed expression vectors encoding a fusion of full-length Rev3 with GST. The basic plasmid with a galactose-inducible promoter and a LEU2 marker was constructed by N. Sharma and P. Shcherbakova.3 We used an in vivo gap repair method to transfer the mutant alleles of REV3 from pRevLCav2 to the expression plasmid (22). A yeast strain with a deletion of the entire REV3 gene was transformed by a mixture of two PCR fragments: the part of the expression plasmid without the region corresponding to the C-terminal part of REV3 and the part of the REV3 gene corresponding to the mutated region from pRevLCav2. Leu+ transformants were selected, and the presence of anticipated alleles of REV3 was determined by PCR and DNA sequencing. Correct constructs were reamplified in Escherichia coli and used for transformation in the protease-deficient strain BJ2168. Induction was done as described (23). All plasmid and genomic constructs used in this study were verified by full-length sequencing.4
Analysis of Protein Interaction by Nickel-Iminodiacetic Acid (Ni-IDA) Pulldown Assay
Different human DNA Pol constructs cloned in pCOLADuet-1 (see details under supplemental “Experimental Procedures”) were expressed in E. coli strain BL21(DE3) at 17 °C for 16 h following induction with 1 mm isopropyl β-d-thiogalactopyranoside at A600 = 1. For constructs containing the p70 subunit of human Pol α, we used Rosetta-2(DE3) cells and the same expression conditions. Afterward, expression cells were harvested, washed with PBS, aliquoted, and kept at −80 °C. Cells were disrupted in ice water by sonication in lysis buffer containing 20 mm Tris-HCl (pH 7.9), 0.15 m NaCl, 3% glycerol, 3 mm β-mercaptoethanol, 0.4 mm PMSF, and 1 μg/ml leupeptin. After centrifugation, 0.4 ml of lysate (corresponding to a 5-ml culture volume) was incubated with 20 μl of Ni-IDA resin (Bio-Rad) for 1 h by rocking at 4 °C. The resin was washed one time with 0.4 ml of lysis buffer, two times with 0.4 ml of 0.3 m NaCl in lysis buffer, and again with 0.4 ml of lysis buffer. The bound proteins were eluted with 65 μl of 0.3 m imidazole HCl (pH 7.7). For preliminary small ubiquitin-like modifier (SUMO) tag proteolysis, 1 μg of doubly tagged UD1 (dtUD1) was added to 0.4 ml of lysate, followed by incubation 1 h at 6 °C before loading on Ni-IDA. Samples loaded on and eluted from Ni-IDA were subjected to 12% SDS-PAGE, followed by detection with Coomassie Blue staining or Western blotting using the ECL Plex system (GE Healthcare). B-subunits of different DNA polymerases were visualized using anti-His monoclonal antibody (6G2A9, GenScript); SUMO-tagged CTDs were visualized by anti-SUMO monoclonal antibody (4G11E9, GenScript).
Purification of p353C-p50-p66N, p125C-p50-p66N, p180C-p70, p261C-p59, and p50-p66N
Expression of these complexes (with SUMO-tagged CTDs and His6-tagged B-subunits) was carried out in 1–2 liters of E. coli culture in LB medium with 25 μg/ml kanamycin under the conditions described above. After cell disruption using EmulsiFlex-C5, the protein complexes were purified according to the same scheme consisting of chromatography on Ni-IDA (Bio-Rad) and Mono Q (GE Healthcare) columns. His6-tagged dtUD1 protease was added to lysate at a 1:10,000 mass ratio before loading on Ni-IDA. After SDS-PAGE analysis, the most pure fractions were combined, and their UV-visible absorbance spectra were measured using a NanoDrop 2000c spectrophotometer (Thermo Scientific) and trUView cuvettes (Bio-Rad). Protein concentrations were estimated by measuring the absorbance at 280 nm and using extinction coefficients of 61.8, 50.8, 57.2, 60.3, and 47.8 mm−1 cm−1 for complexes containing p125C, p353C, p180C, p261C, and p50-p66N only, respectively (calculated with ProtParam (24)). Protein concentrations obtained this way were 7–20% higher compared with concentrations obtained by the Bradford method using BSA as the standard.
Purification of His6-SUMO-tagged p261C and p353C
Expression was carried out for 12 h at 17 °C in E. coli BL21(DE3) cells transformed with pASHSUL encoding the N-terminally His6-SUMO-tagged Pol ϵ or Pol ζ CTD (including p353C variants with mutated MBS1 or MBS2). Cells were grown in 10 ml of LB medium to A600 = 0.7 at 37 °C and cooled down to 17 °C (∼30 min), followed by induction with 25 ng/ml anhydrotetracycline. After expression, the cells were washed with PBS and lysed, and the soluble protein fraction was precipitated by 40% ammonium sulfate and purified using Ni-IDA resin. Protein concentrations were estimated by the Bradford method using BSA as the standard.
Determination of Iron Content in Protein Samples
The concentration of non-heme iron in protein samples was determined by colorimetry with the iron chelator Ferrozine using the iron assay kit from Pointe Scientific, Inc. The original protocol was optimized to avoid the effect of protein aggregation on absorbance measurement. After the addition of iron color reagent, the reaction was incubated for 10 min at 37 °C and spun for 3 min at 13,000 rpm, and the absorbance was recorded at 560 nm against a blank solution with a BioMate 5 spectrophotometer (Thermo Electron Corp.). Iron concentration was calculated using the following formula: Asample/Astandard × 90 μm = total iron (μm).
RESULTS
Structural Similarities of CTDs of Catalytic Subunits of Pol δ and Pol ζ
We aligned the amino acid sequences and predicted secondary structures of the human Pol δ CTD with the CTDs of Pol α, Pol ϵ, and Pol ζ (Fig. 2). The alignment revealed that the Pol δ CTD (p125C) shares high structural similarity with the Pol ζ CTD (p353C), but not the Pol α (p180C) and Pol ϵ (p261C) CTDs. The CTDs of Pol α and Pol ϵ are significantly larger in size and share similar topology with β-strand-based zinc fingers. In contrast, the predicted secondary structures of the Pol δ and Pol ζ CTDs are all-helical, and three of their metal-coordinating cysteines contribute directly from α-helices.
FIGURE 2.
Amino acid sequence alignments of human Pol δ C-terminal domain with CTDs of Pol α, Pol ϵ, and Pol ζ. The alignment was performed by ClustalW with default parameters and then adjusted manually to align the metal-binding cysteines. Secondary structure predictions were made using Phyre software (45). Conserved metal-binding cysteines, α-helices, and β-strands are highlighted in yellow, red, and cyan, respectively. In p125C and p353C, the first cysteines in MBS1 and MBS2 and the second cysteine in MBS2 are located in the α-helices.
Pol ζ CTD Interacts with Pol δ B-subunit
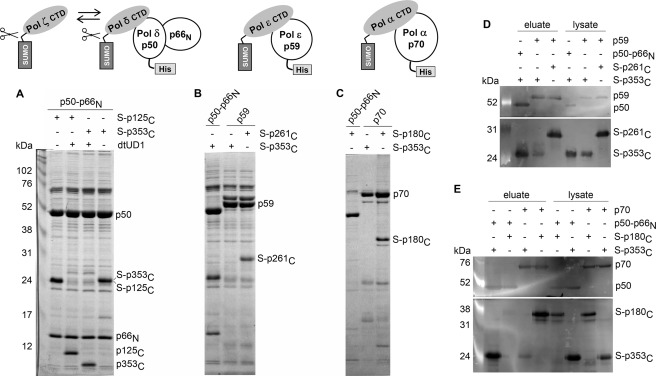
To determine whether the topological similarities of the Pol ζ and Pol δ CTDs result in similar binding properties for the Pol δ B-subunit (p50), we carried out in vitro binding studies. Instead of the p50 subunit alone, we used the p50-p66N complex, which structurally resembles the second subunits of Pol α and Pol ϵ, whose N-terminal parts have significant similarity to the p66N sequence (18). Moreover, the N-terminal domain of the C-subunit stabilizes p50, forming with it a relatively large intersubunit contact area (5398 Å2) (18, 25). To address the problem of low solubility of the CTDs during expression in E. coli, we used N-terminal fusions with a cleavable SUMO tag. Coexpression of the genes encoding the SUMO-tagged CTDs of the Pol δ or Pol ζ catalytic subunits and the Pol δ accessory subunits (p50-p66N; p50 has an N-terminal His6 tag) allowed us to obtain stable ternary complexes by affinity purification on Ni-IDA resin (Fig. 3A). These CTD/B intersubunit interactions are mediated by CTDs only because the SUMO tags were not retained in complexes after their cleavage with the SUMO-specific protease dtUD1. Notably, both CTDs (p125C and p353C) have a similar stoichiometry in these complexes, which is close to 1:1 (Fig. 3A and supplemental Table S1), indicating that they bind p50 with approximately the same affinity. In the next set of experiments, we coexpressed the His6-tagged B-subunits of Pol α and Pol ϵ with their own SUMO-CTDs (SUMO-p180C and SUMO-p261C, respectively) and with SUMO-p353C. All SUMO-CTDs were robustly produced as judged by Western blotting of lysates with anti-SUMO antibodies (Fig. 3, D and E). Ni-IDA pulldown experiments demonstrated that the B-subunits of Pol α and Pol ϵ did not form a complex with SUMO-p353C, but they bound strongly to their own SUMO-CTDs (Fig. 3, B–E). Trace levels of SUMO-p353C detected in partially purified samples of p70 and p59 by Western blotting (Fig. 3, D and E) were due to nonspecific binding with Ni-IDA because SUMO-p353C is aggregation-prone without the proper binding partner. In a reciprocal experiment, coexpression of p50-p66N and the Pol α SUMO-CTD (SUMO-p180C) did not lead to complex formation (Fig. 3, C and E). These results confirm that the interaction between p353C and the B-subunit of Pol δ is highly specific.
FIGURE 3.
Analysis of interaction between B-subunits and CTDs of human DNA polymerases. Samples loaded on and eluted from Ni-IDA were subjected to 12% SDS-PAGE, followed by detection with Coomassie Blue staining (A–C) or by Western blotting (D and E). A. The Pol δ B-subunit binds to the CTDs of Pol δ and Pol ζ. The SUMO (S) tag was cleaved off by dtUD1 prior to binding to the resin. B and D, the Pol ϵ B-subunit binds to the CTD of Pol ϵ and not to the CTD of Pol ζ. C and E, the Pol α B-subunit binds to the CTD of Pol α and not to the CTD of Pol ζ. The Pol δ B-subunit does not bind to the CTD of Pol α. Left lanes in A, D, and E, ECL Plex fluorescent rainbow markers (GE Healthcare). DNA Pol subcomplexes are schematically shown above the A–C. All B-subunits have N-terminal His6 tags.
Human Pol δ and Pol ζ CTDs Contain Iron-Sulfur Clusters
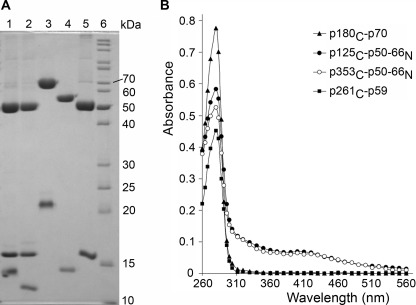
The purified human Pol δ and Pol ζ CTD-p50-p66N complexes exhibit physical properties characteristic of proteins with iron-sulfur clusters: the yellow-brownish color; a broad peak with a maximum at 410 nm in UV-visible spectra (with an A410/A280 ratio of >0.1); and development of a pink color in the presence of the iron-specific indicator Ferrozine, which allows quantification of the iron content as 2.8 and 2.6 per molecule, respectively (Fig. 4 and supplemental Fig. S1) (26, 27). Such stoichiometry is close to what is typically observed for proteins containing [4Fe-4S] clusters, taking into account that one or two iron atoms per molecule were lost during purification (28, 29). Our data are consistent with the recent finding of [4Fe-4S] clusters in yeast Pol δ and Pol ζ (30), in human and yeast primases (29, 31), and in several proteins involved in DNA repair (26–28, 32).
FIGURE 4.
Analysis of purified human DNA Pol subcomplexes. A, purity analysis by electrophoresis on 12% SDS-polyacrylamide with Coomassie Blue staining. Lane 1, p125C-p50-p66N; lane 2, p353C-p50-p66N; lane 3, p180C-p70; lane 4, p261C-p59; lane 5, p50-p66N; lane 6, EZ-Run Rec protein ladder (Fisher Scientific). B, absorbance analysis by UV-visible spectrophotometry.
Absence of Iron-Sulfur Cluster in Human Pol α and Pol ϵ CTD-B Complexes
The samples of similarly purified p180C-p70 and p261C-p59 complexes and p50-p66N alone were colorless, their spectra did not exhibit a peak at 410 nm, and their iron content measured by Ferrozine was close to the background level (Fig. 4 and supplemental Figs. S1 and S2). Consistent with our data, the crystal structure of the yeast Pol α CTD-B complex revealed only zinc ions in both MBSs (13). Recently, an iron-sulfur cluster was detected in partially purified yeast Pol α and Pol ϵ (30). In contrast to our purification scheme, the authors placed a tag on the catalytic subunit, so their partially purified samples contained the polymerase complex itself and an excess of the catalytic subunit. Probably both zinc and iron could be incorporated to CTDs during expression, but only the CTD with the appropriate metal makes a stable complex with the B-subunit. In support of this argument, partially purified human Pol ϵ SUMO-CTD alone has same iron level as Pol ζ SUMO-CTD (supplemental Fig. S3). The absence of iron in pure and stoichiometric Pol α and Pol ϵ CTD-B complexes (Fig. 4) indicates that their CTDs with an inadvertently incorporated iron-sulfur cluster are not able to make a stable interaction with the B-subunit and are removed during purification.
Iron-Sulfur Cluster Is Located in MBS2
To map the [4Fe-4S] cluster in Pol ζ, we purified His6-SUMO-CTD variants with changes of all four conserved cysteines in MBS1 or MBS2 to alanines (supplemental Fig. S4A). The changes in MBS2 resulted in a decrease in iron content close to the background level, suggesting that the four cysteines in the MBS2 domain are involved in [4Fe-4S] cluster coordination (supplemental Fig. S4B). Our data are consistent with [4Fe-4S] cluster mapping to MBS2 (CysB) in yeast Pol δ (30).
MBS2 Is Required for TLS in Vivo
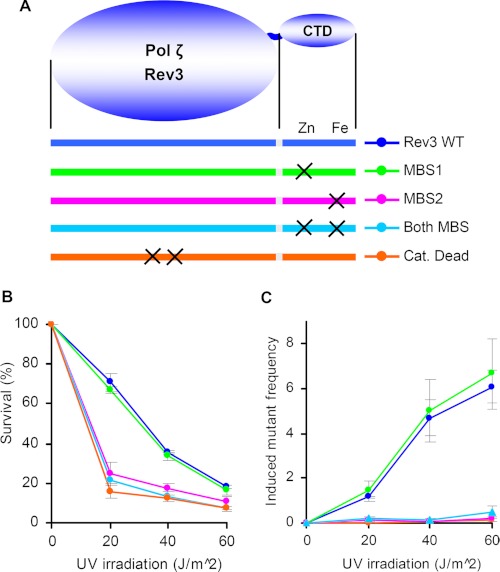
To evaluate the contribution of MBS1 or MBS2 of the Pol ζ CTD to TLS, we used the well established yeast model system. The C-terminal parts of human and yeast Rev3 are conserved (33). The mutation in REV3 leading to the change of four cysteines at MBS1 did not affect UV mutagenesis, whereas the mutations leading to disruption of MBS2 reduced UV mutagenesis to a level observed for catalytically dead Pol ζ (Fig. 5). The observed effects are due to functional defects because none of the aforementioned mutations resulted in a decreased level of Rev3 in yeast extracts (supplemental Fig. S5). The integrity of Pol δ MBS2 is also critical for induced mutagenesis in yeast (16). Our results reveal the critical role of the iron-sulfur cluster-binding domain in Pol ζ function and suggest a similar mechanism of p50 binding by the Pol δ and Pol ζ CTDs.
FIGURE 5.
A, schematic presentation of Rev3 protein and location of amino acid changes in its catalytic domain and CTD. B and C, effect of rev3 mutations on yeast survival and induced mutagenesis, respectively. Catalytically (Cat.) dead Rev3 has a double amino acid change (D1142A/D1144A) at the active site. Induced mutation frequencies (multiplied by 105) in the CAN1 gene were calculated by subtracting the spontaneous frequency from the corresponding frequencies after irradiation.
DISCUSSION
The absence of a B-subunit analog in Pol ζ and the high structural similarity between Pol δ and Pol ζ CTDs prompted us to hypothesize that the Pol δ B-subunit could be a Pol ζ binding partner. Moreover, the crystal structure of the human Pol δ p50-p66N subcomplex (18) revealed several disordered loops in p50, providing a wide flexible surface (supplemental Fig. S6). Mutations disrupting the interaction of the B-subunit with the catalytic subunit were mapped mainly to this disordered surface, indicating that it is a docking site for the Pol δ CTD (34). The flexibility of this surface indicates that p50 could interact with either the Pol δ or Pol ζ CTD. Our biochemical experiments confirmed this idea.
We summarize our main findings and their consequences for TLS as follows: (i) the B-subunit of Pol δ binds equally well to the catalytic subunit of either Pol δ or Pol ζ; (ii) the CTDs of Pol δ and Pol ζ contain a [4Fe-4S] cluster, which is critical for binding to the B-subunit and for UV light-induced mutagenesis; and (iii) both the Pol α and Pol ϵ catalytic subunits lack the [4Fe-4S] cluster, and their B-subunits do not bind the C terminus of Pol ζ. We propose a plausible mechanism for the p125 ↔ Pol ζ switch during TLS (Fig. 6). When Pol δ stalls at a DNA lesion, it induces a signal leading to the dissociation of the Pol δ catalytic subunit. The exact sequence of events leading to this is unknown but involves proliferating cell nuclear antigen (PCNA) ubiquitylation and recruitment of Rev1 (8, 35). The B- and C-subunits that are bound to PCNA remain on the DNA and recruit Pol ζ. The latter, with the assistance of Y-family TLS polymerases, bypasses the DNA lesion and is then replaced again by the catalytic subunit of Pol δ to continue processive replication. According to the crystal structure of the Pol α CTD-B complex (13) and the Pol α-Pol δ CTD alignment results (Fig. 2), the [4Fe-4S] cluster in Pol δ and Pol ζ is likely not buried in the CTD structure and is directly involved in the interaction with p50. Furthermore, the presence of a [4Fe-4S] cluster specifically in the Pol δ and Pol ζ CTDs suggests that it not only plays a structural role but also mediates regulation of the p125 ↔ Pol ζ switch via a change in its oxidation state.
FIGURE 6.
Schematic presentation of p125 ↔ Pol ζ switch during TLS. A detailed explanation is provided under “Discussion.”
This mechanism explains the genetic data on the role of all three subunits of Pol δ in TLS in yeast, especially the involvement of the Pol δ C-subunit (16, 17, 36–39). The importance of the C-subunit in TLS is probably due to its strong interaction with PCNA and also its role in stabilizing the B-subunit. Although Pol δ can bind PCNA via its three subunits simultaneously, the C-subunit plays a critical role in anchoring Pol δ to the replisome (mutasome), especially when the catalytic subunit dissociates during lesion bypass (40, 41). Consistent with our proposed mechanism, the p12 subunit, which binds simultaneously to the p125 and p50 subunits in human Pol δ, was found to dissociate from the replicative complex upon treatment of cells with DNA-damaging agents (42, 43).
Recently, it was shown that yeast Rev1 interacts with the Pol δ C-subunit (Pol32) and may work as a bridge between Pol δ and Pol ζ (44). In these studies, Pol32 was used without its strong binding partner (Pol31, the B-subunit), which may result in altered protein conformation and properties. The importance of this interaction for TLS was not shown. Moreover, the deletions of Pol32 regions 103–142 (allele −3) and 143–182 (allele −4), which encompass the proposed Rev1-binding domain (residues 100–180), do not affect UV light-induced mutagenesis (40).
It is worth mentioning that all cumulative evidence obtained so far points to Pol δ as a central regulator of TLS. The mutations affecting the components of Pol δ responsible for polymerase switches abolish all induced mutagenesis in both leading and lagging DNA strands. This suggests either that Pol δ is a main replicase for both DNA strands or that TLS events on the leading strand, when replicated by Pol ϵ, should include the switch for Pol δ as an initiating event (4, 41, 44).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM082923 from NIGMS (to T. H. T.). This work was also supported by Eppley Cancer Center seed grants. The work related to yeast genetics experiments was supported by National Institutes of Health Grant CA129925 from NCI, Smoking Disease Research Program Department of Health and Human Services Grant 2011-27, and Russian Federal Program “Innovation Scientific Personnel” State Contract 14.740.11.0916 (to Y. I. P.).

This article contains supplemental “Experimental Procedures,” Figs. S1–S6, Table S1, and additional references.
N. Sharma and P. Shcherbakova, unpublished data.
Primer sequences are available upon request.
- Pol
- polymerase
- CTD
- C-terminal domain
- MBS
- metal-binding site
- TLS
- translesion DNA synthesis
- Ni-IDA
- nickel-iminodiacetic acid
- SUMO
- small ubiquitin-like modifier
- dtUD1
- doubly tagged UD1
- PCNA
- proliferating cell nuclear antigen.
REFERENCES
- 1. Pavlov Y. I., Shcherbakova P. V., Rogozin I. B. (2006) Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int. Rev. Cytol. 255, 41–132 [DOI] [PubMed] [Google Scholar]
- 2. Garg P., Burgers P. M. (2005) DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 40, 115–128 [DOI] [PubMed] [Google Scholar]
- 3. Pursell Z. F., Kunkel T. A. (2008) DNA polymerase ϵ: a polymerase of unusual size (and complexity). Prog. Nucleic Acid Res. Mol. Biol. 82, 101–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pavlov Y. I., Shcherbakova P. V. (2010) DNA polymerases at the eukaryotic fork–20 years later. Mutat. Res. 685, 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kunkel T. A., Burgers P. M. (2008) Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 18, 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stone J. E., Kumar D., Binz S. K., Inase A., Iwai S., Chabes A., Burgers P. M., Kunkel T. A. (2011) Lesion bypass by S. cerevisiae Pol ζ alone. DNA Repair 10, 826–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Prakash S., Prakash L. (2002) Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 16, 1872–1883 [DOI] [PubMed] [Google Scholar]
- 8. Lawrence C. W. (2004) Cellular functions of DNA polymerase ζ and Rev1 protein. Adv. Protein Chem. 69, 167–203 [DOI] [PubMed] [Google Scholar]
- 9. Johansson E., Macneill S. A. (2010) The eukaryotic replicative DNA polymerases take shape. Trends Biochem. Sci. 35, 339–347 [DOI] [PubMed] [Google Scholar]
- 10. Hara K., Hashimoto H., Murakumo Y., Kobayashi S., Kogame T., Unzai S., Akashi S., Takeda S., Shimizu T., Sato M. (2010) Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase ζ and REV1. J. Biol. Chem. 285, 12299–12307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tahirov T. H., Makarova K. S., Rogozin I. B., Pavlov Y. I., Koonin E. V. (2009) Evolution of DNA polymerases: an inactivated polymerase-exonuclease module in Pol ϵ and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors. Biol. Direct 4, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sanchez Garcia J., Ciufo L. F., Yang X., Kearsey S. E., MacNeill S. A. (2004) The C-terminal zinc finger of the catalytic subunit of DNA polymerase δ is responsible for direct interaction with the B-subunit. Nucleic Acids Res. 32, 3005–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Klinge S., Núñez-Ramírez R., Llorca O., Pellegrini L. (2009) Three-dimensional architecture of DNA Pol α reveals the functional core of multisubunit replicative polymerases. EMBO J. 28, 1978–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Asturias F. J., Cheung I. K., Sabouri N., Chilkova O., Wepplo D., Johansson E. (2006) Structure of Saccharomyces cerevisiae DNA polymerase ϵ by cryo-electron microscopy. Nat. Struct. Mol. Biol. 13, 35–43 [DOI] [PubMed] [Google Scholar]
- 15. Jain R., Hammel M., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K. (2009) Structural insights into yeast DNA polymerase δ by small angle x-ray scattering. J. Mol. Biol. 394, 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giot L., Chanet R., Simon M., Facca C., Faye G. (1997) Involvement of the yeast DNA polymerase δ in DNA repair in vivo. Genetics 146, 1239–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gerik K. J., Li X., Pautz A., Burgers P. M. (1998) Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 273, 19747–19755 [DOI] [PubMed] [Google Scholar]
- 18. Baranovskiy A. G., Babayeva N. D., Liston V. G., Rogozin I. B., Koonin E. V., Pavlov Y. I., Vassylyev D. G., Tahirov T. H. (2008) X-ray structure of the complex of regulatory subunits of human DNA polymerase δ. Cell Cycle 7, 3026–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pavlov Y. I., Shcherbakova P. V., Kunkel T. A. (2001) In vivo consequences of putative active site mutations in yeast DNA polymerases α, ϵ, δ, and ζ. Genetics 159, 47–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu H., Naismith J. H. (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boeke J. D., LaCroute F., Fink G. R. (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoroorotic acid resistance. Mol. Gen. Genet. 197, 345–346 [DOI] [PubMed] [Google Scholar]
- 22. Orr-Weaver T. L., Szostak J. W. (1983) Yeast recombination: the association between double-strand gap repair and crossing-over. Proc. Natl. Acad. Sci. U.S.A. 80, 4417–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burgers P. M. (1999) Overexpression of multisubunit replication factors in yeast. Methods 18, 349–355 [DOI] [PubMed] [Google Scholar]
- 24. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) in The Proteomics Protocols Handbook (Walker J. M., ed) Humana Press, Totowa, NJ [Google Scholar]
- 25. Baranovskiy A. G., Babayeva N. D., Pavlov Y. I., Tahirov T. H. (2008) Crystallization and preliminary crystallographic analysis of the complex of the second and third regulatory subunits of human Pol δ. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64, 822–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Porello S. L., Cannon M. J., David S. S. (1998) A substrate recognition role for the [4Fe-4S]2+ cluster of the DNA repair glycosylase MutY. Biochemistry 37, 6465–6475 [DOI] [PubMed] [Google Scholar]
- 27. Rudolf J., Makrantoni V., Ingledew W. J., Stark M. J., White M. F. (2006) The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol. Cell 23, 801–808 [DOI] [PubMed] [Google Scholar]
- 28. Hinks J. A., Evans M. C., De Miguel Y., Sartori A. A., Jiricny J., Pearl L. H. (2002) An iron-sulfur cluster in the family 4 uracil-DNA glycosylases. J. Biol. Chem. 277, 16936–16940 [DOI] [PubMed] [Google Scholar]
- 29. Weiner B. E., Huang H., Dattilo B. M., Nilges M. J., Fanning E., Chazin W. J. (2007) An iron-sulfur cluster in the C-terminal domain of the p58 subunit of human DNA primase. J. Biol. Chem. 282, 33444–33451 [DOI] [PubMed] [Google Scholar]
- 30. Netz D. J., Stith C. M., Stumpfig M., Kopf G., Vogel D., Genau H. M., Stodola J. L., Lill R., Burgers P. M., Pierik A. J. (2011) Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 8, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klinge S., Hirst J., Maman J. D., Krude T., Pellegrini L. (2007) An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat. Struct. Mol. Biol. 14, 875–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cunningham R. P., Asahara H., Bank J. F., Scholes C. P., Salerno J. C., Surerus K., Münck E., McCracken J., Peisach J., Emptage M. H. (1989) Endonuclease III is an iron-sulfur protein. Biochemistry 28, 4450–4455 [DOI] [PubMed] [Google Scholar]
- 33. Gan G. N., Wittschieben J. P., Wittschieben B. Ø., Wood R. D. (2008) DNA polymerase ζ n higher eukaryotes. Cell Res. 18, 174–183 [DOI] [PubMed] [Google Scholar]
- 34. Sanchez Garcia J., Baranovskiy A. G., Knatko E. V., Gray F. C., Tahirov T. H., MacNeill S. A. (2009) Functional mapping of the fission yeast DNA polymerase δ B-subunit Cdc1 by site-directed and random pentapeptide insertion mutagenesis. BMC Mol. Biol. 10, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ulrich H. D., Walden H. (2010) Ubiquitin signaling in DNA replication and repair. Nat. Rev. Mol. Cell Biol. 11, 479–489 [DOI] [PubMed] [Google Scholar]
- 36. Hanna M., Ball L. G., Tong A. H., Boone C., Xiao W. (2007) Pol32 is required for Pol ζ-dependent translesion synthesis and prevents double-strand breaks at the replication fork. Mutat. Res. 625, 164–176 [DOI] [PubMed] [Google Scholar]
- 37. Huang M. E., de Calignon A., Nicolas A., Galibert F. (2000) Pol32, a subunit of the Saccharomyces cerevisiae DNA polymerase δ, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr. Genet. 38, 178–187 [DOI] [PubMed] [Google Scholar]
- 38. Sugimoto K., Sakamoto Y., Takahashi O., Matsumoto K. (1995) HYS2, an essential gene required for DNA replication in Saccharomyces cerevisiae. Nucleic Acids Res. 23, 3493–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gibbs P. E., McDonald J., Woodgate R., Lawrence C. W. (2005) The relative roles in vivo of Saccharomyces cerevisiae Pol η, Pol ζ, Rev1 protein, and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct, and T-T cis,syn-cyclobutane dimer. Genetics 169, 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Johansson E., Garg P., Burgers P. M. (2004) The Pol32 subunit of DNA polymerase δ contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 279, 1907–1915 [DOI] [PubMed] [Google Scholar]
- 41. Acharya N., Klassen R., Johnson R. E., Prakash L., Prakash S. (2011) PCNA-binding domains in all three subunits of yeast DNA polymerase δ modulate its function in DNA replication. Proc. Natl. Acad. Sci. U.S.A. 108, 17927–17932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li H., Xie B., Zhou Y., Rahmeh A., Trusa S., Zhang S., Gao Y., Lee E. Y., Lee M. Y. (2006) Functional roles of p12, the fourth subunit of human DNA polymerase δ. J. Biol. Chem. 281, 14748–14755 [DOI] [PubMed] [Google Scholar]
- 43. Zhang S., Zhou Y., Trusa S., Meng X., Lee E. Y., Lee M. Y. (2007) A novel DNA damage response: rapid degradation of the p12 subunit of DNA polymerase δ. J. Biol. Chem. 282, 15330–15340 [DOI] [PubMed] [Google Scholar]
- 44. Acharya N., Johnson R. E., Pagès V., Prakash L., Prakash S. (2009) Yeast Rev1 protein promotes complex formation of DNA polymerase ζ with Pol32 subunit of DNA polymerase δ. Proc. Natl. Acad. Sci. U.S.A. 106, 9631–9636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.