Background: Modification of proteins by ubiquitin (Ub) and ubiquitin-like proteins (Ubls) is essential in eukaryotes.
Results: Proteins for tRNA-thiouridine synthesis and other proteins are modified by a bacterial Ubl in Thermus thermophilus.
Conclusion: The existence of a Ub/Ubl homologous conjugation system in Bacteria is demonstrated.
Significance: This suggests an ancient origin of the eukaryotic Ub/Ubl system.
Keywords: Bacteria, RNA Modification, Sulfur, Transfer RNA (tRNA), Ubiquitin
Abstract
Posttranslational modification of proteins with ubiquitin and ubiquitin-like proteins plays important regulatory roles in eukaryotes. Although a homologous conjugation system has recently been reported in Archaea, there is no similar report in Bacteria. This report describes the identification of a ubiquitin-like conjugation system in the bacterium Thermus thermophilus. A series of in vivo analyses revealed that TtuB, a bacterial ubiquitin-like protein that functions as a sulfur carrier in tRNA thiouridine synthesis, was covalently attached to target proteins, most likely via its C-terminal glycine. The involvement of the ubiquitin-activating enzyme-like protein TtuC in conjugate formation and the attachments of TtuB to TtuC and TtuA, which are proteins required for tRNA thiouridine synthesis, were demonstrated. Mass spectrometry analysis revealed that lysine residues (Lys-137/Lys-226/Lys-229) of TtuA were covalently modified by the C-terminal carboxylate of TtuB. Intriguingly, a deletion mutant of a JAMM (JAB1/MPN/Mov34 metalloenzyme) ubiquitin isopeptidase homolog showed aberrant TtuB conjugates of TtuC and TtuA and an ∼50% decrease in thiouridine amounts in tRNA. These results would support the hypothesis that thiouridine synthesis is regulated by TtuB-conjugation.
Introduction
Ubiquitin (Ub)2 and ubiquitin-like proteins (Ubls) are protein modifiers with important roles in proteolysis and the regulation of diverse processes in eukaryotes (1). The breakdown of the Ub/Ubl system is often associated with the development of various diseases. In the first step of conjugation to target proteins, the conserved C-terminal glycine of Ub/Ubl is acyl-adenylated by an activating enzyme (E1) and covalently linked to a cysteine residue of E1 to form a Ub/Ubl-E1 thioester intermediate. The activated Ub/Ubl is next transferred to a conjugating enzyme (E2). Finally, Ub/Ubl is attached to a lysine residue in the target protein by a ligase (E3) (1).
Proteins homologous to eukaryotic Ub/Ubl and E1s exist in almost all members of Bacteria and Archaea (2) (supplemental Fig. S1). Earlier works established that these proteins in bacteria function in the biosynthesis of sulfur compounds such as molybdenum cofactor, thiamine, and tRNA-thiouridine (3). Bacterial Ubls are adenylated by cognate E1 homologs, subsequently bind activated sulfur at their C termini, and finally work as sulfur donors (4–8). These findings imply an evolutionary link between the eukaryotic Ub/Ubl system and the bacterial sulfur transfer reaction (1). Although prokaryotic Ubls have long been believed to function as sulfur carriers in the biosynthesis of sulfur compounds and not as protein modifiers, it was reported recently that the small archaeal modifier proteins (SAMPs) 1 and 2 are conjugated to proteins in an archaeon, Haloferax volcanii (9). SAMP1 seems to function in part as a targeting tag to the proteasome (9). SAMPs are activated by the E1-like UbaA and also function as sulfur carriers in tRNA thiolation and molybdenum cofactor synthesis (10). In eukaryotes, the Ub-related modifier Urm1 is thought to be the most ancient Ubl (11, 12). Together with Uba4 (E1-like enzyme for Urm1), these proteins are closely related to prokaryotic sulfur carriers and E1-like proteins (supplemental Fig. S1). Indeed, Urm1 acts as a sulfur carrier for thiouridine synthesis in tRNA (13–18).
Although bacterial Ubls have not been reported as protein modifiers, actinobacteria can modify proteins by a small protein modifier termed prokaryotic ubiquitin-like protein (Pup), which can target proteins for degradation by proteasomes (19, 20). Pup is an intrinsically disordered protein (21, 22), in contrast to the ordered β-grasp fold of Ub/Ubls. This study focused on investigating whether a bacterial Ubl can function as a protein modifier in the bacterium Thermus thermophilus.
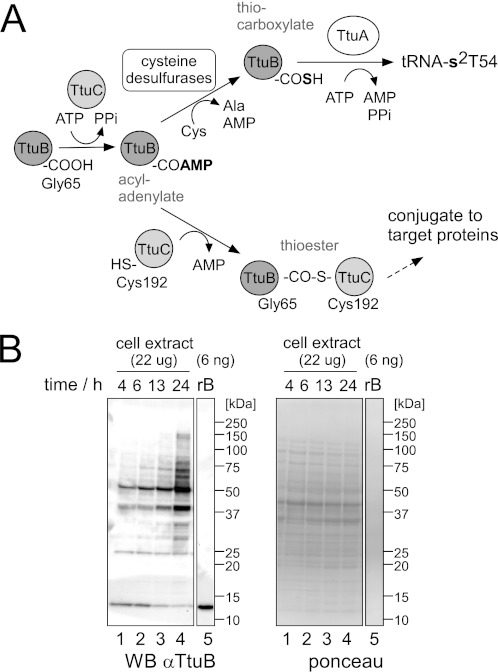
2-Thioribothymidine (s2T), a 2-thiolated derivative of 5-methyluridine (ribothymidine (rT)), is found at position 54 in almost all tRNAs from several thermophiles, such as T. thermophilus and Pyrococcus furiosus (23, 24). The 2-thiolation content of rT54 increases with cultivation temperature (24, 25). Thiolation is assumed to increase the thermal stability of the tRNA and increase its translation efficiency at high temperatures (25, 26). Prior work from our group identified proteins required for tRNA thiolation in T. thermophilus, including cysteine desulfurases, TtuA, TtuB, and TtuC, and the reaction was reconstituted in vitro (Fig. 1A) (27–29). C-terminal Gly (Gly-65) of TtuB is acyl-adenylated by TtuC and is then thiocarboxylated by cysteine desulfurases. The sulfur atom of thiocarboxylated TtuB is transferred to tRNA by TtuA. Homology modeling suggested that TtuB possesses a Ub/β-grasp fold (supplemental Fig. S1B), although the sequence similarity with eukaryotic Ub/Ubls is limited to the area around the C-terminal glycines (supplemental Fig. S1A). TtuC shows significant sequence homology with the adenylation domain of eukaryotic E1s (supplemental Fig. S1C). Interestingly, TtuC has a Cys-192 that corresponds to the “catalytic Cys” of E1, and the TtuB-Gly-65 ∼ TtuC-Cys-192 thioester that was formed in vitro (Fig. 1A) (29) was similar to the Ub/Ubl∼E1 thioester formed in the first step of Ub/Ubl conjugation (1). These findings suggest that there exist Ub/Ubl homologous conjugation systems also in Bacteria.
FIGURE 1.
TtuB forms protein conjugates in T. thermophilus. A, working model for the dual roles of TtuB. TtuB was shown to function as a sulfur donor in s2T synthesis. The protein-modifier function of TtuB was examined in this study. B, left panel, anti-TtuB immunoblotting of extracts (22 μg) prepared from T. thermophilus cells cultured at 70οC in rich medium for the times indicated. Cells reached full growth at ∼13 h. The growth curve is shown in supplemental Fig. S2. Recombinant TtuB (rB) was loaded in lane 5. The Ponceau-stained membrane shows equal amounts of protein loaded (right panel).
In this paper, the modification of cellular proteins by TtuB in the bacterium T. thermophilus was demonstrated. In vivo analyses were used to examine the mechanism of TtuB conjugation and its functions, and the evolutionary implications for the eukaryotic Ub/Ubl system are discussed.
EXPERIMENTAL PROCEDURES
Strains and Media
The strains used were T. thermophilus HB27 (wild-type), NS2710 (ΔttuA) (28), NS2720 (ΔttuB) (28), NS2730 (ΔttuC) (29), NS2715 (ΔttuB-ttuA, see below), and NS2740 (Δttc1133, see below). These strains were cultured in rich and minimal media (30) at 70 °C unless otherwise stated. For selection, 30 mg/liter kanamycin and 10 mg/liter bleomycin were used.
Antibodies
N-terminal His6-tagged TtuA, TtuB, and TtuC were overexpressed in E. coli Rosetta (DE3), purified by Ni2+ resin, and the His6-tags were removed (29). The proteins were used to immunize rabbits and to purify the antibodies (Operon biotechnologies).
Immunoblotting
Late-log phase cells were used unless otherwise stated. Cells were sonicated in H buffer (50 mm HEPES-KOH (pH 7.6), 10 mm KCl, 10 mm MgCl2, 1 mm dithiothreitol, and 0.3 mm phenylmethylsulfonyl fluoride), and cell debris was removed by centrifugation (20,000 × g for 15 min). Protein concentrations were determined with the Bio-Rad protein assay kit with bovine serum albumin as a standard. Lysate was mixed with equal volume of 2× loading buffer (125 mm Tris-HCl (pH 6.8), 2.1% SDS, 0.7 m β-mercaptoethanol, 0.02% bromphenol blue, and 20% glycerol) and incubated at 95 °C for 5 min. Samples were separated by SDS-PAGE (10–20% gel, Wako) and electroblotted to a polyvinylidene fluoride membrane (iBlot, Invitrogen). Primary antibodies (0.7 μg/ml), horseradish peroxidase-conjugated anti-rabbit IgG antibody (1:2000 dilution, TrueBlot, eBioscience), and ECL plus reagent (GE Healthcare) were used for detection. Ponceau S staining of the membranes was used to confirm equivalent protein loading in each lane.
Immunoprecipitation
Lysates were prepared as described above with T buffer (20 mm Tris-HCl (pH 7.6), 150 mm KCl, and 1 mm EDTA), supplemented with 1% Nonidet P-40 and complete protease inhibitors (Roche). Antibodies were cross-linked on Dynabeads Protein G (Invitrogen) by dimethyl pimelimidate (Pierce). Lysates (500 μg) were incubated with cross-linked antibodies (5 μg anti-TtuA or 10 μg anti-TtuC) at 4 °C for 1 h. After the beads were washed three times, the immunoprecipitate was eluted with 30 μl of buffer (T buffer with 2% SDS for TtuA, T buffer with 2% SDS and 2 m urea for TtuC). Immunoprecipitates were resolved by SDS-PAGE and subjected to Western blotting as described above.
Analysis of the Conjugation Site by Mass Spectrometry
Large-scale immunoprecipitation was performed essentially as described above. Lysate was prepared from NS2715 (ΔttuB-ttuA strain) harboring expression plasmid for TtuA and TtuB (S63R). Lysate (5 mg) was immunoprecipitated with 0.1 mg of cross-linked antibody for TtuA. The eluate was resolved by SDS-PAGE. Proteins were visualized using Coomassie Brilliant Blue R-250, and bands of interest were excised and subjected to in-gel alkylation by iodoacetamide and tryptic digestion as described previously (31). The digests were then analyzed by a linear ion trap mass spectrometer (LTQ Orbitrap XL hybrid mass spectrometer, Thermo Fisher Scientific) and equipped with an electrospray ionization source and a splitless nano HPLC system (DiNa, KYA Technologies) with an ODS capillary column (100 × 0.15 mm, 3 μm particle size, KYA Technologies). The tryptic digests were separated in 0.1% formic acid in water using a linear gradient from 2% to 80% of a solvent consisting of 70% acetonitrile and 0.1% formic acid for 40 min at a flow rate of 300 nl/min. Peptides were identified using the MASCOT data base search engine (Matrix Science) and compared with the T. thermophilus HB27 genome data base (NC_005835 and NC_005838 on NCBI). Allowances in the masses of theoretical tryptic peptides were made for dynamic modifications with oxidized methionine (15.995 Da), carbamidomethylated cysteine (57.022 Da), and two glycines on lysine (114.043 Da).
Complementation Experiments
An E. coli-Thermus shuttle vector, pWUR112/77-1 (32), encoding resistance to bleomycin (shble) was used. The Shine-Dalgarno sequence and the respective genes (ttuB or ttc1133) were cloned into the KpnI/EcoRI sites. Transcription from the PslpA promoter upstream of the shble gene continues through the cloned genes, allowing their expression. For the coexpression of TtuB and TtuA, a HindIII site was introduced just after the EcoRI site of pWUR112/77-1 by site-directed mutagenesis (Stratagene). TtuB and ttuA with the Shine-Dalgarno sequences were cloned into the KpnI/EcoRI sites and EcoRI/HindIII sites, respectively. Mutations were introduced into wild-type constructs by site-directed mutagenesis. Primers used are listed in the supplemental data. The NS2720, NS2740, and NS2715 strains were transformed with the pWUR-derived plasmids.
Construction of the ΔttuB-ttuA Strain and the Δttc1133 Strain of T. thermophilus HB27
To construct the mutant strains ΔttuB-ttuA (NS2715) and Δttc1133 (NS2740), the homologous recombination method was used (29). In NS2715, the entire 1.2 kbp of the ttuB and ttuA genes was replaced by a kanamycin resistance gene cassette. To construct the DNA fragment for homologous recombination, the 5′- and 3′-flanking regions (0.5 kbp) of the ttuB-ttuA ORFs were amplified by PCR using the T. thermophilus HB27 genome as a template. The htk cassette (kanamycin resistance gene) was amplified from pUC18-pJHK3 (33). The primers used were ttuBA-5′-F and ttuBA-5′-R, ttuBA-3′-F and ttuBA-3′-R, and htk-F and htk-R. The resulting three fragments were ligated by a second PCR with the ttuBA-5′-F and ttuBA-3′-R primers. The ligated fragment was purified by agarose gel electrophoresis and used for transformation. In NS2740, the core 0.3 kbp of the ttc1133 gene was replaced by the htk cassette, and 0.1 kbp each of the 5′ and 3′ ends of ttc1133 were kept unchanged so that the flanking ORFs would not be damaged. To construct the DNA fragment for homologous recombination, the 5′- and 3′-flanking regions (0.5 kbp) of the ttc1133 ORF were amplified by PCR. The primers used were ttc1133–5′-F and ttc1133–5′-R, and ttc1133–3′-F and ttc1133–3′-R. The resulting two fragments and the htk fragment were ligated by a second PCR with the ttc1133–5′-F and ttc1133–3′-R primers, cloned into pCR-BluntII-TOPO (Invitrogen), and sequenced. Primers used are listed in the supplemental data. T. thermophilus HB27 was transformed with the PCR fragment or the plasmid as described previously (34), and the transformants were selected on rich medium containing 300 mg/l kanamycin. Homologous recombination was confirmed by PCR amplification of the target region, followed by restriction enzyme digestion.
Analysis of tRNA Modification
Hydrolysates of tRNAs were prepared and analyzed by an HPLC system with a photo diode array detector (GL-Sciences) as described previously (29). The amount of s2T was quantified by using pseudouridine as a standard.
RESULTS
TtuB Forms Protein Conjugates in T. thermophilus
To analyze the TtuB-conjugates in T. thermophilus, cellular proteins were resolved by SDS-PAGE and subjected to Western blotting by a TtuB-specific antibody. Recombinant TtuB was detected as a single band of ∼13 kDa (Fig. 1B, lane 5), although its theoretical mass is 7.3 kDa. This band was assumed to be free TtuB, considering that SUMO1 (a Ubl) migrates more slowly than expected (35). A series of bands in a ladder pattern was detected in addition to free TtuB in the cell extract from the wild-type strain cultured at 70 °C (Fig. 1B, lanes 1–4). Because samples were denatured by SDS and reduced by β-mercaptoethanol, these bands were assumed to correspond to covalent TtuB-conjugates. Band intensities generally increased in correlation with cell growth. Notably, the major bands were separated by ∼13-kDa intervals, implying the possible formation of TtuB polymers, although further experimentation is required to confirm this. Under different culture temperatures (55–80 °C) and media (minimal medium), the amounts of TtuB conjugates increased in correlation with growth, whereas the conjugation patterns were essentially unchanged in these conditions (supplemental Fig. S2).
TtuC Is Required for the Formation of TtuB Conjugates
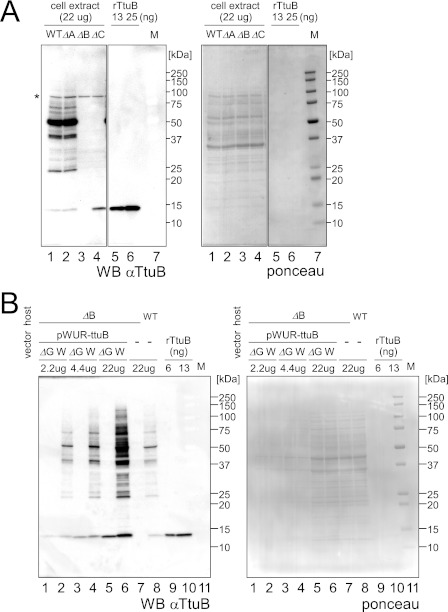
TtuC shares sequence homology with eukaryotic E1s that are required for the first activation of Ub/Ubls (supplemental Fig. S1C), and indeed, TtuC can acyl-adenylate TtuB in vitro (29). Therefore, TtuC may be involved in the formation of TtuB-conjugates in T. thermophilus cells. As expected, only free TtuB was detected in the ttuC deletion strain (ΔttuC) by a TtuB-specific antibody (Fig. 2A, lane 4). By contrast, the TtuB conjugation pattern was almost unchanged in the ΔttuA strain (Fig. 2A, lane 2). The importance of the C terminus of TtuB was examined next. Expression of wild-type TtuB in the ΔttuB cell resulted in a similar pattern of conjugates to that observed in the native strain (Fig. 2B, lanes 2, 4, 6, and 8), but the amount of TtuB-conjugates was ∼10 times greater than in the native strain. When the C-terminal Gly deletion mutant of TtuB (TtuBΔG) was expressed, no band was detected except for free TtuB (Fig. 2B, lanes 1, 3, and 5). These results suggest that TtuB is conjugated to target proteins via its C-terminal Gly and that the E1-like protein TtuC is involved in this process, which is similar to the eukaryotic Ub/Ubl conjugation system.
FIGURE 2.
TtuC is required for the formation of TtuB-conjugates. A, left panel, immunoblotting of cell extracts (22 μg) from wild-type, ΔttuA, ΔttuB, and ΔttuC strains against a TtuB antibody. Recombinant TtuB was loaded in lanes 5 and 6. Molecular weight marker (M) was loaded in lane 7. A nonspecific band is indicated by an asterisk. Right panel, Ponceau-stained membranes indicate equal amounts of protein loaded. B, left panel, immunoblotting of extracts (2.2–22 μg) from the ΔttuB strain transformed with pWUR-ttuB-WT or pWUR-ttuB-ΔG (lanes 1–6) against an anti-TtuB antibody. For comparison, extracts (22 μg) from wild-type and ΔttuB strains were also analyzed (lanes 7 and 8). Recombinant TtuB was loaded in lanes 9 and 10. Right panel, Ponceau-stained membranes show equal amounts of protein loaded. The bands are visible only in lanes 5–8.
Identification of Targets for TtuB Conjugation
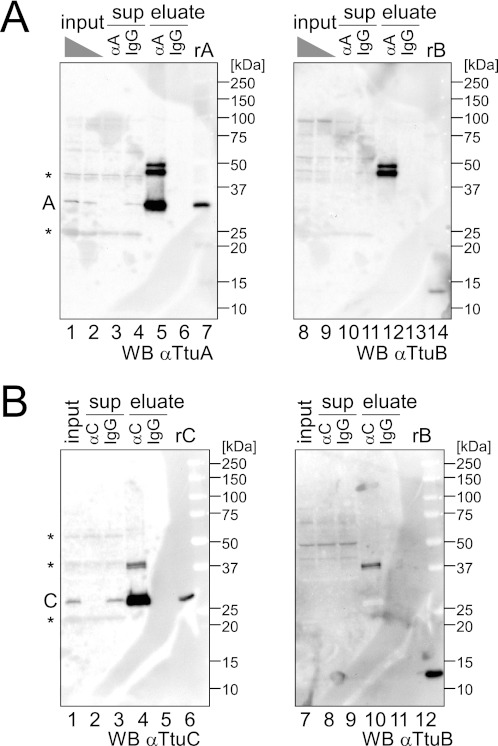
Proteins involved in thiouridine synthesis, such as TtuA and TtuC, may be targets for TtuB conjugation. Using specific antibodies, only free TtuA (∼33 kDa, theoretically 36 kDa; Fig. 3A, lanes 1, 2, and 7) or free TtuC (∼28 kDa, theoretically 30 kDa; Fig. 3B, lanes 1 and 6) could be detected in cell extract from wild-type T. thermophilus. To improve the sensitivity of detection, an immunoprecipitation experiment was performed. Immunoprecipitation of cell extracts using an anti-TtuA antibody resulted in the detection of two bands (∼45 kDa and ∼48 kDa) in addition to free TtuA (∼33 kDa) by the anti-TtuA antibody, and the same bands showed reactivity against an anti-TtuB antibody (Fig. 3A, lanes 5 and 12), confirming that these bands were TtuB-TtuA conjugates. Considering that the TtuB monomer was detected at ∼13 kDa by electrophoresis (Fig. 1B), the ∼45-kDa and ∼48-kDa conjugates may contain one molecule of each protein. Immunoprecipitation using an anti-TtuC antibody resulted in the detection of two species by a TtuC antibody (Fig. 3B, lane 4), which corresponded to free TtuC (∼28 kDa) and an ∼42-kDa band that showed reactivity against a TtuB antibody (Fig. 3B, lane 10), suggesting that this band was a TtuB-TtuC conjugate composed of one molecule of each protein.
FIGURE 3.
TtuA and TtuC are the targets of TtuB-conjugation. A, cell extracts from the wild-type strain were subjected to immunoprecipitation with an anti-TtuA antibody (αA) or normal rabbit IgG (IgG). The input (10% (lane 1) or 5% (lane 2)), supernatant (sup) (5%), and eluate were analyzed. Recombinant TtuA (rA) (6 ng) and TtuB (rB) (13 ng) were also analyzed in lane 7. The membrane was probed with an anti-TtuA antibody (left panel) and reprobed with a TtuB antibody (right panel). Nonspecific bands are indicated by asterisks. B, extracts from the wild-type strain were subjected to immunoprecipitation with an anti-TtuC antibody (αC) or normal rabbit IgG (IgG). The input (5%), supernatant (sup) (5%), and eluate were analyzed. Recombinant TtuC (rC) (6 ng) and TtuB (rB) (25 ng) were loaded in lane 6. The membrane was probed with an anti-TtuC antibody (left panel) and reprobed with a TtuB antibody (right panel). Nonspecific bands are indicated by asterisks.
Identification of the Attachment Sites of TtuA-TtuB Conjugates
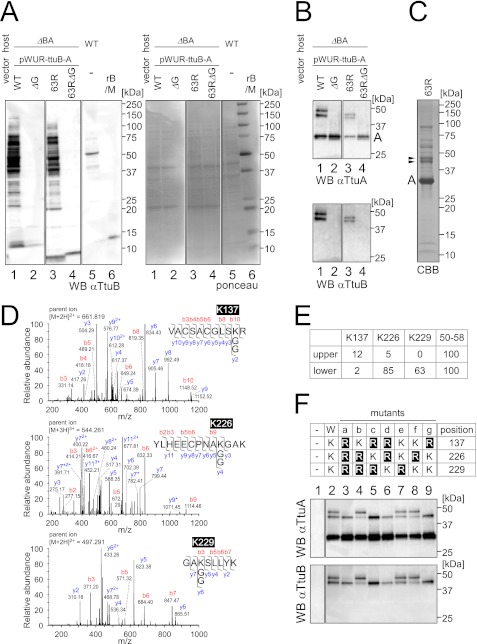
The samples immunoprecipitated from the native strain by anti-TtuA or anti-TtuC antibodies were subjected to MS analysis, but peptides suggesting TtuB-conjugation could not be identified (data not shown). Immunoprecipitation by anti-TtuC antibody using a TtuB-overexpressing strain was performed because overexpression of TtuC was not successful. However, this strategy did not yield sufficient amount of the TtuC-TtuB conjugate for MS analysis (data not shown). To obtain sufficient amounts of TtuA-TtuB conjugates, TtuA and TtuB were simultaneously overexpressed in T. thermophilus ΔttuB-ttuA (NS2715) (Fig. 4A, lanes 1 and 2). In addition, to shorten the TtuB-branched peptide generated after tryptic digestion and improve its detection by MS, arginine was introduced at position 63 (S63R mutant) of TtuB (Fig. 4A, lanes 3 and 4). If an isopeptide bond is formed between the C-terminal carboxylate of TtuB and an amino group of TtuA, TtuB-S63R will leave a “GG footprint” on the target site after trypsin cleavage. This kind of mutation has been used previously for site-specific identification of SUMO-2 targets in human cells (36). The conjugation patterns of wild-type TtuB and the S63R mutant were essentially the same, although the bands with the S63R mutant were moved slightly faster through the gel than bands with wild-type TtuB (Fig. 4A, lanes 1-4). Cell extracts from the strains coexpressing TtuA and TtuB were immunoprecipitated by an anti-TtuA antibody together with detection using anti-TtuA and anti-TtuB antibodies (Fig. 4B). Two TtuB-TtuA conjugates were detected in S63R TtuB cells (Fig. 4B, lane 3) at slightly lower molecular weight positions than wild-type TtuB-TtuA conjugates (lane 1). The formation of TtuB-TtuA conjugates was dependent on the C-terminal glycine of TtuB (Fig. 4B, lanes 2 and 4).
FIGURE 4.
Identification of the attachment sites of TtuA-TtuB conjugates. A, left panel, immunoblotting of extracts (22 μg) from the ΔttuB-ttuA strain transformed with pWUR-ttuB-ttuA against an anti-TtuB antibody. TtuB (wild type (lane 1), C-terminal glycine deletion mutant (lane 2), S63R mutant (lane 3), or S63R with C-terminal deletion mutant (lane 4)) was coexpressed with wild-type TtuA. For comparison, an extract (22 μg) from the wild-type strain was also analyzed (lane 5). Recombinant TtuB (rB) (6 ng) and molecular weight marker (M) were loaded in lane 6. Right panel, the Ponceau-stained membrane shows equal amounts of protein loaded. B, cell extracts (same as in A) were immunoprecipitated with an anti-TtuA antibody, and the eluates were analyzed by Western blot analysis. The membrane was probed with anti-TtuA (upper panel) and reprobed with anti-TtuB (lower panel). C, large-scale immunoprecipitation from ΔttuB-ttuA strain with the expression plasmid for TtuA and TtuB (S63R). The eluate was resolved by SDS-PAGE, and proteins were visualized using Coomassie Brilliant Blue R-250 (CBB). The bands corresponding to TtuA-TtuB conjugates (shown by arrowheads) were excised and subjected to MS analysis. D, lysine 137, lysine 226, and lysine 229 in TtuA were identified as TtuB attachment sites by LC-MS/MS analysis. Three peptides contained a lysine residue modified with two glycines. The collision-induced dissociation spectrum of Lys-137 peptide obtained from the upper band in C is shown in upper panel. For the Lys-226 peptide (center panel) and the Lys-229 peptide (lower panel), the spectra obtained from the lower band are shown. Peptide fragments were identified as b- or y-type ions and deaminated ions (asterisk). E, estimation of the abundance of the modified peptides of TtuA (upper and lower bands in C) by MS analysis. The peptides were roughly quantified by the parent ion mass chromatograms. The values were normalized by using a peptide corresponding to positions 50–58 of TtuA in each band. F, mutational analysis of the identified lysines to arginines in TtuA. The wild type and seven types of mutants (a–g) of TtuA (upper panel) were coexpressed with wild-type TtuB in the T. thermophilus ΔttuB-ttuA strain. The strain with the empty vector was also analyzed (- in lane 1). Cell extracts were immunoprecipitated with an anti-TtuA antibody, and the eluates were analyzed by Western blot analysis. The membrane was probed with anti-TtuA (center panel) and reprobed with anti-TtuB (lower panel).
The two bands containing TtuB(S63R)-TtuA conjugates (arrowheads in Fig. 4C) were excised, digested by trypsin, and subjected to MS/MS analysis. In the samples from these two bands, peptide fragments derived from TtuA and TtuB were detected with high-sequence coverage (data not shown), further suggesting that these bands are TtuA-TtuB conjugates. A MASCOT search, which takes into account possible modification of lysine with two glycines (114.04 Da), successfully identified three peptides with a GG footprint on lysines of TtuA (Lys-137, Lys-226, and Lys-229, Fig. 4D). This result also suggests that TtuB is attached to TtuA via its C-terminal carboxylate. The abundance of modified peptides was roughly estimated by the mass chromatograms of the parent ion of the peptides (Fig. 4E). The Lys-137-modified peptide was mainly detected in the upper band, whereas Lys-226 and Lys-229 peptides were predominantly detected in the lower band. The Lys-226-modified peptide detected in the upper band and the Lys-137 peptide detected in the lower band were probably due to cross-contamination of the closely migrating proteins.
Mutational analysis of three lysines identified in TtuA to arginines was performed. Seven types of mutant TtuA (Fig. 4F) were coexpressed with wild-type TtuB in the T. thermophilus ΔttuB-ttuA strain. The TtuA-TtuB conjugates were immunoprecipitated by an anti-TtuA antibody and detected by anti-TtuA and anti-TtuB antibodies. When Lys-137 was mutated to arginine, the upper band of the TtuA-TtuB conjugate disappeared (Fig. 4F, mutants c, d, and g). On the other hand, if Lys-226 and/or Lys-229 were mutated, the intensity of the lower band of the conjugates was reduced (Fig. 4F, mutants b, e, and f). When all three lysines were mutated at the same time, the upper band disappeared, and the lower band was unchanged (Fig. 4F, mutant a). It is possible to hypothesize that a species that is not the Lys-226- or Lys-229-linked conjugate also migrated at the same position to the lower band. Mutating all lysine residues, which is preferentially used for conjugation in normal condition, might result in another residue in the substrate being modified. The MS analysis and the mutational study suggest that Lys-137 in TtuA was modified by TtuB in the upper band and that Lys-226 or Lys-229 in TtuA was modified by TtuB in the lower band.
Aberrant TtuB Conjugation in a JAMM (JAB1/MPN/Mov34 Metalloenzyme) Mutant
Deubiquitinating enzymes responsible for the removal of Ub from target proteins are indispensable for Ub systems in eukaryotes (37, 38). A similar system may function in bacteria, and, if so, the function of Ubl-conjugation can be investigated by disrupting this activity. Among five families of these proteases in eukaryotes, JAMM domain proteins are often found in the Ubl operon, and the domains are sometimes fused with Ubl-related proteins in bacteria (2). In the JAMM motif (EXnHS/THX7SXXD) (supplemental Fig. S3A), the two His and Asp residues coordinate a zinc ion, and the Glu residue is hydrogen-bonded to the aqua ligand in the catalytic center (39).
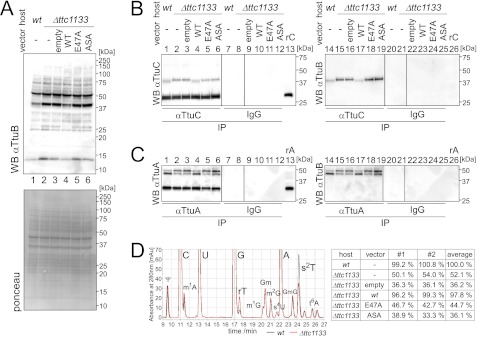
There is one JAMM protein (Ttc1133) in the T. thermophilus genome. Therefore, a ttc1133 deletion mutant (Δttc1133) was constructed, and the TtuB-conjugation in this strain was analyzed. Unexpectedly, the TtuB-conjugation pattern was unchanged in the Δttc1133 strain, except for the ∼42-kDa band, the ∼27-kDa band, and free TtuB (∼13 kDa) (Fig. 5A, lanes 1 and 2). The changes in conjugation pattern were reversed by exogenous expression of wild-type Ttc1133 but not by E47A and ASA (His-101 and His-103 were substituted to Ala) mutants in the catalytic residues of JAMM protease (Fig. 5A, lanes 3–6). On the basis of the proximity of the ∼42-kDa band to the position of the TtuB-TtuC conjugate, an immunoprecipitation experiment with an anti-TtuC antibody was performed. The gel mobility of the TtuB-TtuC conjugate decreased to ∼44 kDa in the Δttc1133 strain (Fig. 5B, lanes 1, 2, 14, and 15). The amount of free TtuC (∼28 kDa) in the Δttc1133 strain was almost unchanged (86 ± 3% of the wild-type value, n = 3). The change in the mobility of TtuB-TtuC was dependent on the activity of Ttc1133 (Fig. 5B, lanes 3–6 and 16–19). An immunoprecipitation experiment with a TtuA-antibody was also performed. The ratio of the amounts of the two TtuB-TtuA conjugates was different in the Δttc1133 strain, which showed a decrease in the ∼45-kDa band in correlation with an increase in the ∼48-kDa band (Fig. 5C, lanes 1, 2, 14, and 15). The amount of free TtuA (∼33 kDa) in the Δttc1133 strain was somewhat decreased (75 ± 7%, n = 3). The changes in the amounts of two TtuB-TtuA conjugates were dependent on the activity of Ttc1133 (Fig. 5C, lanes 3–6 and 16–19).
FIGURE 5.
Aberrant TtuB-conjugation in a mutant of a JAMM homolog. A, upper panel, cell extracts (22 μg) from the wild type, Δttc1133, or Δttc1133 complemented with expression vectors were analyzed by anti-TtuB immunoblotting. Empty vector and vectors encoding ttc1133 (wild type, E47A mutant, and ASA mutant) were used. Lower panel, the Ponceau-stained membrane indicates equivalent loading. B, extracts (same as in A) were subjected to immunoprecipitation (IP) with a TtuC antibody or normal rabbit IgG, and eluates were analyzed by Western blotting with a TtuC antibody (left panel) and a TtuB antibody (right panel). Recombinant TtuC (rC) (6 ng) was also loaded (lanes 13 and 26). C, extracts (same as in B) were subjected to immunoprecipitation with a TtuA antibody or normal rabbit IgG, and eluates were analyzed by Western blotting with a TtuA antibody (left panel) and a TtuB antibody (right panel). Recombinant TtuA (rA) (6 ng) was also loaded (lanes 13 and 26). D, analysis of s2T in tRNA by HPLC. Left panel, UV traces (280 nm) for T. thermophilus wild type (black line) and Δttc1133 (red line) are shown. The modified nucleosides were identified by their UV spectra and retention times: Ψ, pseudouridine; m1A, 1-methyladenosine; rT, ribothymidine (precursor for s2T); m1G, 1-methylguanosine; Gm, 2′-O-methylguanosine; m2G, 2-methylguanosine; s4U, 4-thiouridine; GmG, dinucleotide GmpG; s2T, 2-thioribothymidine; t6A, 6-theronylcarbamoyladenosine. Right panel, the amounts of s2T in tRNA from mutants measured in two independent experiments, and the average values are shown.
The alterations in the TtuB-conjugation patterns of TtuA and TtuC in the Δttc1133 strain suggested that the amount of s2T may also be changed. As expected, the s2T content of the tRNA extracted from the Δttc1133 strain was decreased to ∼50% of the wild-type content (Fig. 5D). This result again depended on the catalytic residues of Ttc1133. The Δttc1133 strain showed a similar growth to the wild-type strain between 60–82 °C (supplemental Fig. S3B), suggesting that the residual s2T in tRNA was sufficient for efficient protein synthesis at higher temperatures.
DISCUSSION
Several proteins with homology to the eukaryotic Ub/Ubl system, such as Ub/Ubls, E1s, and JAMM deubiquitinating enzymes, are encoded in prokaryotic genomes (2). In this study, the existence of a Ub/Ubl homologous conjugation system in Bacteria was demonstrated for the first time, implying that the Ub/Ubl conjugation systems may be widespread in other prokaryotes. TtuB, which functions as a sulfur carrier in tRNA thiouridine synthesis in T. thermophilus (29), was covalently attached to target proteins most likely via its C-terminal Gly-65, and the E1-like TtuC enzyme played a role in this process (Figs. 1 and 2). TtuB might be conjugated to the Lys of target proteins through an isopeptide bond as in the case of TtuA (Fig. 4). Although the TtuB-Gly-65 ∼ TtuC-Cys-192 thioester was formed in vitro (29), the importance of this thioester for the formation of TtuB-conjugates in vivo is currently unknown because TtuC complementation experiments were not successful (data not shown).
The modification of proteins by a bacterial Ubl, TtuB, is a more similar phenomenon to ubiquitylation in eukaryotes than pupylation, found in a few species of bacteria, including Actinomycetales (19, 20). TtuB (homology modeling (supplemental Fig. S1B) (29)) and Ub/Ubls in eukaryotes have an ordered β-grasp fold in contrast to the intrinsically disordered Pup protein (21, 22). The mechanism of conjugation of TtuB to target proteins, which utilizes the E1-like TtuC protein, also differs from that of pupylation. In the pupylation reaction, the C-terminal Gln-64 of Pup is deamidated to Glu by the glutamate synthetase-like protein Dop (40–42), and deamidated Pup is conjugated to Lys residues on target proteins by another glutamate synthetase-like protein, PafA (40, 42). The side chain of C-terminal Glu of Pup is coupled to substrates (43).
In an archaeon, Haloferax volcanii (9), SAMPs are activated by the E1-like UbaA and also function as sulfur carriers for molybdenum cofactor and tRNA thiouridine (10). In eukaryotes, the Ub-related modifier Urm1, working with Uba4 (E1-like enzyme for Urm1), acts as a sulfur carrier for thiouridine synthesis in tRNA (13–18). Therefore, bacterial TtuB and archaeal SAMPs are considered to be related to eukaryotic Urm1 and are the most ancient dual-function Ubls. TtuB-conjugation increased after the log phase (Fig. 1B) when the medium presumably becomes more oxidative and fewer nutrients are available. This situation may be somewhat similar to that found in SAMPylation and urmylation, which are induced by nitrogen limitation (9) and by oxidative stress (44), respectively.
TtuB-conjugation did not occur in the ΔttuB and ΔttuC strains but was detected in the ΔttuA strain (Fig. 2A). s2T was not synthesized in these three stains, which were not able to grow at temperatures above 80 °C (29). Therefore, the temperature-sensitive phenotype may be derived from the absence of s2T in tRNA and not from TtuB conjugation.
MS analysis of TtuA-TtuB conjugates identified three lysines in TtuA as the residues modified by C-terminal carboxylate of TtuB (Fig. 4). This is the first identification of the residues modified by Ubl in TtuA/Ncs6 tRNA thiouridylases, which are conserved in the three domains of life. Bacterial TtuA, archaeal TtuA, and eukaryotic Ncs6 constitute the TtuA/Ncs6 family (group II of the TtcA family (45)), which catalyzes the 2-thiolation of specific uridines in tRNA (46). Archaeal HVO_0580 (a TtuA homolog) and eukaryotic ATPBD3 (an Ncs6 homolog) are SAMPylated (9) and urmylated (44), respectively, although the target residues in these proteins have not been identified. Lys-137 in T. thermophilus TtuA is situated just after the central Cys-Xaa-Xaa-Cys motif (130–133), which is strictly conserved in TtuA/Ncs6 proteins. These cysteines are important for the 2-thiolation activity of Ncs6 (47) and a closely related enzyme involved in cytidine 2-thiolation (E. coli TtcA) (45). Thus, Lys-137 may be situated close to the catalytic center of this enzyme family. Lys-137 in TtuA is conserved in related Bacteria and Archaea such as Aquifex, Pyrococcus, Thermococcus, and Metanocaldococcus. On the other hand, this position is occupied by a conserved arginine in eukaryotic Ncs6. Lys-226 and Lys-229 in T. thermophilus TtuA is situated just after Cys-222, which is also strictly conserved in TtuA/Ncs6 proteins. However, Lys-226 and Lys-229 are only conserved in Thermus and a few other species, possibly implying species-specific functions of the conjugation.
Thiouridine biosynthesis proteins were identified as targets of TtuB conjugation (Fig. 3). Intriguingly, in a deletion mutant of the ttc1133 gene, a homolog of the JAMM deubiquitinating enzyme, the conjugation patterns of TtuA and TtuC were altered (Fig. 5, B and C), although the amount of free TtuA and TtuC was little changed (B and C), and the amount of free TtuB was somewhat increased by the mutation (Fig. 5A). The difference between the two conjugates of TtuA was due to differences in the residues that were modified in TtuA (Fig. 4). Ttc1133 might cleave the Lys-226- or the Lys-229-linked conjugate in the lower band more efficiently than the Lys-137-linked conjugate in the upper band. This may be the reason why the conjugation pattern of TtuA was altered. The difference between the two conjugates of TtuC might also be attributed to differences in the residues used for the TtuB conjugation, and Ttc1133 might cleave these conjugates to different extents. Probably because of the changes of the conjugation patterns of TtuA and TtuC, the amount of s2T in tRNA in the ttc1133 mutant was decreased to about half that of the wild-type strain (Fig. 5D). This result may imply that the enzymatic activities of TtuA and TtuC were altered by TtuB conjugation. The biosynthesis of sulfur-containing molecules involves reactive sulfur atoms and, therefore, must avoid nonspecific transfer of sulfur. TtuB conjugation might play a regulatory role to ensure appropriate sulfur transfer in cells. Notably, homologs of TtuA and TtuC are SAMPylated in archaea (9) and urmylated in eukaryotes (44), although the function of these conjugates is unknown. The regulation of thiouridine synthesis might be one of the common functions of these ancient Ubls.
Ttc1133 in T. thermophilus apparently did not work for most of TtuB conjugates (Fig. 5A), implying that there are other proteases for removal of TtuB from the conjugates. Two bacterial JAMM homologs were reported previously to function as peptidase: QbsD in siderophore synthesis in Pseudomonas (48) and Mec+ in cysteine synthesis in Mycobacterium (49). In T. thermophilus, Ttc1133 may also function as peptidase for Ttc1947 and Ttc1835, Ubl-fusion proteins predicted to function in the synthesis of molybdenum cofactor and tungsten cofactor, respectively (29).
In general, Bacteria do not have a counterpart of the eukaryotic proteasome, although it has been found in a few species of Bacteria including Actinomycetales and all Archaea (50). SAMP1 in archaea seems to function in part as a targeting tag to the proteasome (9). In T. thermophilus, the steady-state levels of TtuC remained unchanged regardless of the presence or absence of functional TtuB (supplemental Fig. S4), suggesting that TtuB is not a signal for the degradation of TtuC.
Further detailed characterization of the mechanisms of TtuB conjugate formation and identification of targets by comprehensive methods might lead to a better understanding of the functions of the TtuB conjugation system and its relation with the eukaryotic Ubl system.
Supplementary Material
Acknowledgments
I thank Prof. Tsutomu Suzuki and Ms. Yuriko Sakaguchi (University of Tokyo) for the analysis of TtuB-conjugates by mass spectrometry. I also thank Dr. Stan Brouns (Wageningen University) for providing the plasmid pWUR112/77-1 and Dr. Tetsuro Hirose (AIST) and his lab members, namely Dr. Yasunori Sasaki, and Dr. Kimitsuna Watanabe (Tokyo University of Pharmacy and Life Sciences), for valuable suggestions and discussions.
This study was supported in part by KAKENHI Grant 22770140 of the Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Naito Foundation, and the Kurata memorial Hitachi Science and Technology Foundation.

This article contains supplemental Figs. S1–S4 and experimental procedures.
- Ub
- ubiquitin
- Ubl
- ubiquitin-like protein
- SAMP
- small archaeal modifier protein
- Pup
- prokaryotic ubiquitin-like protein
- s2T
- 2-thioribothymidine
- rT
- ribothymidine
- JAMM
- JAB1/MPN/Mov34 metalloenzyme.
REFERENCES
- 1. Hochstrasser M. (2009) Origin and function of ubiquitin-like proteins. Nature 458, 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Iyer L. M., Burroughs A. M., Aravind L. (2006) The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like β-grasp domains. Genome Biol. 7, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kessler D. (2006) Enzymatic activation of sulfur for incorporation into biomolecules in prokaryotes. FEMS Microbiol. Rev. 30, 825–840 [DOI] [PubMed] [Google Scholar]
- 4. Pitterle D. M., Rajagopalan K. V. (1993) The biosynthesis of molybdopterin in Escherichia coli. Purification and characterization of the converting factor. J. Biol. Chem. 268, 13499–13505 [PubMed] [Google Scholar]
- 5. Taylor S. V., Kelleher N. L., Kinsland C., Chiu H. J., Costello C. A., Backstrom A. D., McLafferty F. W., Begley T. P. (1998) Thiamin biosynthesis in Escherichia coli. Identification of ThiS thiocarboxylate as the immediate sulfur donor in the thiazole formation. J. Biol. Chem. 273, 16555–16560 [DOI] [PubMed] [Google Scholar]
- 6. Lauhon C. T., Kambampati R. (2000) The iscS gene in Escherichia coli is required for the biosynthesis of 4-thiouridine, thiamin, and NAD. J. Biol. Chem. 275, 20096–20103 [DOI] [PubMed] [Google Scholar]
- 7. Leimkühler S., Wuebbens M. M., Rajagopalan K. V. (2001) Characterization of Escherichia coli MoeB and its involvement in the activation of molybdopterin synthase for the biosynthesis of the molybdenum cofactor. J. Biol. Chem. 276, 34695–34701 [DOI] [PubMed] [Google Scholar]
- 8. Zhang W., Urban A., Mihara H., Leimkühler S., Kurihara T., Esaki N. (2010) IscS functions as a primary sulfur-donating enzyme by interacting specifically with MoeB and MoaD in the biosynthesis of molybdopterin in Escherichia coli. J. Biol. Chem. 285, 2302–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Humbard M. A., Miranda H. V., Lim J. M., Krause D. J., Pritz J. R., Zhou G., Chen S., Wells L., Maupin-Furlow J. A. (2010) Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature 463, 54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miranda H. V., Nembhard N., Su D., Hepowit N., Krause D. J., Pritz J. R., Phillips C., Söll D., Maupin-Furlow J. A. (2011) E1- and ubiquitin-like proteins provide a direct link between protein conjugation and sulfur transfer in archaea. Proc. Natl. Acad. Sci. U.S.A. 108, 4417–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Furukawa K., Mizushima N., Noda T., Ohsumi Y. (2000) A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J. Biol. Chem. 275, 7462–7465 [DOI] [PubMed] [Google Scholar]
- 12. Xu J., Zhang J., Wang L., Zhou J., Huang H., Wu J., Zhong Y., Shi Y. (2006) Solution structure of Urm1 and its implications for the origin of protein modifiers. Proc. Natl. Acad. Sci. U.S.A. 103, 11625–11630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmitz J., Chowdhury M. M., Hänzelmann P., Nimtz M., Lee E. Y., Schindelin H., Leimkühler S. (2008) The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry 47, 6479–6489 [DOI] [PubMed] [Google Scholar]
- 14. Nakai Y., Nakai M., Hayashi H. (2008) Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J. Biol. Chem. 283, 27469–27476 [DOI] [PubMed] [Google Scholar]
- 15. Huang B., Lu J., Byström A. S. (2008) A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA 14, 2183–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schlieker C. D., Van der Veen A. G., Damon J. R., Spooner E., Ploegh H. L. (2008) A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc. Natl. Acad. Sci. U.S.A. 105, 18255–18260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leidel S., Pedrioli P. G., Bucher T., Brost R., Costanzo M., Schmidt A., Aebersold R., Boone C., Hofmann K., Peter M. (2009) Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 458, 228–232 [DOI] [PubMed] [Google Scholar]
- 18. Noma A., Sakaguchi Y., Suzuki T. (2009) Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 37, 1335–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pearce M. J., Mintseris J., Ferreyra J., Gygi S. P., Darwin K. H. (2008) Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 322, 1104–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burns K. E., Liu W. T., Boshoff H. I., Dorrestein P. C., Barry C. E., 3rd (2009) Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J. Biol. Chem. 284, 3069–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liao S., Shang Q., Zhang X., Zhang J., Xu C., Tu X. (2009) Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein. Biochem. J. 422, 207–215 [DOI] [PubMed] [Google Scholar]
- 22. Chen X., Solomon W. C., Kang Y., Cerda-Maira F., Darwin K. H., Walters K. J. (2009) Prokaryotic ubiquitin-like protein pup is intrinsically disordered. J. Mol. Biol. 392, 208–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Watanabe K., Oshima T., Saneyoshi M., Nishimura S. (1974) Replacement of ribothymidine by 5-methyl-2-thiouridine in sequence GT psi C in tRNA of an extreme thermophile. FEBS Lett. 43, 59–63 [DOI] [PubMed] [Google Scholar]
- 24. Kowalak J. A., Dalluge J. J., McCloskey J. A., Stetter K. O. (1994) The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry 33, 7869–7876 [DOI] [PubMed] [Google Scholar]
- 25. Watanabe K., Shinma M., Oshima T., Nishimura S. (1976) Heat-induced stability of tRNA from an extreme thermophile, Thermus thermophilus. Biochem. Biophys. Res. Commun. 72, 1137–1144 [DOI] [PubMed] [Google Scholar]
- 26. Yokoyama S., Watanabe K., Miyazawa T. (1987) Dynamic structures and functions of transfer ribonucleic acids from extreme thermophiles. Adv. Biophys. 23, 115–147 [DOI] [PubMed] [Google Scholar]
- 27. Shigi N., Suzuki T., Terada T., Shirouzu M., Yokoyama S., Watanabe K. (2006) Temperature-dependent biosynthesis of 2-thioribothymidine of Thermus thermophilus tRNA. J. Biol. Chem. 281, 2104–2113 [DOI] [PubMed] [Google Scholar]
- 28. Shigi N., Sakaguchi Y., Suzuki T., Watanabe K. (2006) Identification of two tRNA thiolation genes required for cell growth at extremely high temperatures. J. Biol. Chem. 281, 14296–14306 [DOI] [PubMed] [Google Scholar]
- 29. Shigi N., Sakaguchi Y., Asai S., Suzuki T., Watanabe K. (2008) Common thiolation mechanism in the biosynthesis of tRNA thiouridine and sulphur-containing cofactors. EMBO J. 27, 3267–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takada T., Akanuma S., Kotsuka T., Tamakoshi M., Yamagishi A., Oshima T. (1993) Recombination-deficient mutants of an extreme thermophile, Thermus thermophilus. Appl. Environ. Microbiol. 59, 2737–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 32. Brouns S. J., Wu H., Akerboom J., Turnbull A. P., de Vos W. M., van der Oost J. (2005) Engineering a selectable marker for hyperthermophiles. J. Biol. Chem. 280, 11422–11431 [DOI] [PubMed] [Google Scholar]
- 33. Hoseki J., Yano T., Koyama Y., Kuramitsu S., Kagamiyama H. (1999) Directed evolution of thermostable kanamycin-resistance gene: a convenient selection marker for Thermus thermophilus. J. Biochem. 126, 951–956 [DOI] [PubMed] [Google Scholar]
- 34. Koyama Y., Hoshino T., Tomizuka N., Furukawa K. (1986) Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 166, 338–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mahajan R., Delphin C., Guan T., Gerace L., Melchior F. (1997) A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88, 97–107 [DOI] [PubMed] [Google Scholar]
- 36. Matic I., Schimmel J., Hendriks I. A., van Santen M. A., van de Rijke F., van Dam H., Gnad F., Mann M., Vertegaal A. C. (2010) Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell 39, 641–652 [DOI] [PubMed] [Google Scholar]
- 37. Komander D., Clague M. J., Urbé S. (2009) Breaking the chains. Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 38. Wilkinson K. D. (2009) DUBs at a glance. J. Cell Sci. 122, 2325–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ambroggio X. I., Rees D. C., Deshaies R. J. (2004) JAMM. A metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol. 2, E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Striebel F., Imkamp F., Sutter M., Steiner M., Mamedov A., Weber-Ban E. (2009) Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat. Struct. Mol. Biol. 16, 647–651 [DOI] [PubMed] [Google Scholar]
- 41. Imkamp F., Rosenberger T., Striebel F., Keller P. M., Amstutz B., Sander P., Weber-Ban E. (2010) Deletion of dop in Mycobacterium smegmatis abolishes pupylation of protein substrates in vivo. Mol. Microbiol. 75, 744–754 [DOI] [PubMed] [Google Scholar]
- 42. Cerda-Maira F. A., Pearce M. J., Fuortes M., Bishai W. R., Hubbard S. R., Darwin K. H. (2010) Molecular analysis of the prokaryotic ubiquitin-like protein (Pup) conjugation pathway in Mycobacterium tuberculosis. Mol. Microbiol. 77, 1123–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sutter M., Damberger F. F., Imkamp F., Allain F. H., Weber-Ban E. (2010) Prokaryotic ubiquitin-like protein (Pup) is coupled to substrates via the side chain of its C-terminal glutamate. J. Am. Chem. Soc. 132, 5610–5612 [DOI] [PubMed] [Google Scholar]
- 44. Van der Veen A. G., Schorpp K., Schlieker C., Buti L., Damon J. R., Spooner E., Ploegh H. L., Jentsch S. (2011) Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc. Natl. Acad. Sci. U.S.A. 108, 1763–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jäger G., Leipuviene R., Pollard M. G., Qian Q., Björk G. R. (2004) The conserved Cys-X1-X2-Cys motif present in the TtcA protein is required for the thiolation of cytidine in position 32 of tRNA from Salmonella enterica serovar Typhimurium. J. Bacteriol. 186, 750–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Noma A., Shigi N., Suzuki T. (2009) in DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution (Grosjean H. ed) pp 395–408, Landes Bioscience, Austin, TX [Google Scholar]
- 47. Dewez M., Bauer F., Dieu M., Raes M., Vandenhaute J., Hermand D. (2008) The conserved Wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc. Natl. Acad. Sci. U.S.A. 105, 5459–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Godert A. M., Jin M., McLafferty F. W., Begley T. P. (2007) Biosynthesis of the thioquinolobactin siderophore. An interesting variation on sulfur transfer. J. Bacteriol. 189, 2941–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Burns K. E., Baumgart S., Dorrestein P. C., Zhai H., McLafferty F. W., Begley T. P. (2005) Reconstitution of a new cysteine biosynthetic pathway in Mycobacterium tuberculosis. J. Am. Chem. Soc. 127, 11602–11603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Striebel F., Kress W., Weber-Ban E. (2009) Controlled destruction. AAA+ ATPases in protein degradation from bacteria to eukaryotes. Curr. Opin. Struct. Biol. 19, 209–217 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.