Background: NDRG1 is an iron-regulated metastasis suppressor which is up-regulated by iron depletion and may be involved in the epithelial-mesenchymal transition (EMT).
Results: NDRG1 is involved in the EMT through the SMAD and Wnt pathways.
Conclusion: Iron chelators could inhibit the TGF-β-induced EMT via NDRG1.
Significance: The results are important for understanding the molecular roles of iron in proliferation and metastasis.
Keywords: Iron, Iron Metabolism, Metals, Protein Metal Ion Interaction, Transport Metals, Protein-Metal Ion Interactions, Iron, Iron Chelator, Iron Metabolism, Iron-regulated Metastasis Suppressor, NDRG1
Abstract
The epithelial-mesenchymal transition (EMT) is a key step for cancer cell migration, invasion, and metastasis. Transforming growth factor-β (TGF-β) regulates the EMT and the metastasis suppressor gene, N-myc downstream-regulated gene-1 (NDRG1), could play a role in regulating the TGF-β pathway. NDRG1 expression is markedly increased after chelator-mediated iron depletion via hypoxia-inducible factor 1α-dependent and independent pathways (Le, N. T. and Richardson, D. R. (2004) Blood 104, 2967–2975). Moreover, novel iron chelators show marked and selective anti-tumor activity and are a potential new class of anti-metabolites. Considering this, the current study investigated the relationship between NDRG1 and the EMT to examine if iron chelators can inhibit the EMT via NDRG1 up-regulation. We demonstrated that TGF-β induces the EMT in HT29 and DU145 cells. Further, the chelators, desferrioxamine (DFO) and di-2-pyridylketone-4,4-dimethyl-3-thiosemicarbazone (Dp44mT), inhibited the TGF-β-induced EMT by maintaining E-cadherin and β-catenin, at the cell membrane. We then established stable clones with NDRG1 overexpression and knock-down in HT29 and DU145 cells. These data showed that NDRG1 overexpression maintained membrane E-cadherin and β-catenin and inhibited TGF-β-stimulated cell migration and invasion. Conversely, NDRG1 knock-down caused morphological changes from an epithelial- to fibroblastic-like phenotype and also increased migration and invasion, demonstrating NDRG1 knockdown induced the EMT and enhanced TGF-β effects. We also investigated the mechanisms involved and showed the TGF-β/SMAD and Wnt pathways were implicated in NDRG1 regulation of E-cadherin and β-catenin expression and translocation. This study demonstrates that chelators inhibit the TGF-β-induced EMT via a process consistent with NDRG1 up-regulation and elucidates the mechanism of their activity.
Introduction
Iron plays a crucial role in proliferation and DNA synthesis and neoplastic cells have an increased requirement for iron as shown by their markedly elevated expression of the transferrin receptor 1 and enhanced uptake of iron (1). Recently, it has been suggested that alteration in the regulation of iron metabolism characterizes the malignant state, with an iron regulatory gene signature predicting outcome in breast cancer (2). However, the precise molecular pathways involved remain unclear and are important to elucidate particularly in terms of the mechanisms involved in metastasis, which is a major problem in cancer treatment.
The epithelial-mesenchymal transition (EMT)3 is a highly conserved process required for embryonic development, tissue remodeling and wound repair (3). In addition, there is increasing evidence that it is a key initial step for cancer cell migration, invasion and metastasis (4). During the process of the EMT, cells lose their epithelial characteristics such as epithelial morphology, cell polarity, cell-cell contact, and gain mesenchymal properties such as fibroblastic morphology, increased migration and invasion, thus causing cancer cell metastasis (5).
Transforming growth factor-β (TGF-β) was first identified as an inducer of the EMT in normal mammary epithelial cells (6) and subsequently recognized as a chief regulator of the EMT in a variety of cell-types and tissues including cancer cells (7). There are reports showing that TGF-β was increased in various cancer cell types and that this plays an important role in the process of metastasis via the induction of the EMT (8, 9).
Iron chelators are a relatively new class of potential anti-metabolites that show marked and selective anti-tumor activity (1, 10), although their molecular targets and mechanisms of action remain to be completely elucidated. We reported a series of novel thiosemicarbazone iron chelators of the dipyridyl thiosemicarbazone class (e.g. di-2-pyridylketone-4,4-dimethyl-3-thiosemicarbazone, Dp44mT; Fig. 1A) as anti-tumor compounds which are effective against belligerent tumors in vivo by the intravenous and oral routes (11, 12).
FIGURE 1.
Line drawing of the chemical structures of: (A) Dp44mT, (B) Dp2mT, and (C) DFO.
This series of compounds were demonstrated to induce cell cycle arrest and apoptosis through a mechanism involving iron depletion which affects a variety of targets including: (i) iron-dependent enzymes such as ribonucleotide reductase (RR), which is critical for DNA synthesis (13); (ii) the metastasis suppressor, N-myc downstream regulated gene 1 (NDRG1) (11); (iii) p53 (14); (iv) cyclin D1 (15); (v) p21CIP1/WAF1 (16) etc. The other major mechanism of thiosemicarbazone activity is mediated through the redox cycling of their iron and copper complexes (17), causing cell death and apoptosis through targeting lysosome integrity (18) and oxidizing glutathione and the sulfhydryl groups of proteins involved in reducing systems (19).
One of the most interesting genes regulated by iron chelators is NDRG1 (11, 20), which is a well-known metastasis suppressor in various cancer cell types (21–26). Significantly, it has also been reported that NDRG1 overexpression is correlated with a lower metastatic rate and increased 5-year survival in clinical studies (21, 23, 27). Hence, NDRG1 is a promising molecular target for cancer therapy that is modulated by novel iron chelators (11, 12, 28). However, the detailed mechanisms for the anti-cancer effects of NDRG1 are not well elucidated and further investigation is required.
Considering the potent anti-metastatic effect of NDRG1 in various cancer types and the role TGF-β plays in cancer metastasis, we examined whether iron chelators could inhibit the cancer cell EMT induced by TGF-β and whether this effect takes place via up-regulation of NDRG1. In this study, we established four stable transfectants with NDRG1 overexpression and knock-down in two cancer cell types, namely colon cancer HT29 and prostate cancer DU145. We then investigated the role and mechanism of NDRG1 in the TGF-β-induced EMT and its related biological functions. Our study shows that cellular iron-depletion inhibits the TGF-β-induced EMT via up-regulation of NDRG1.
EXPERIMENTAL PROCEDURES
Cell Culture and Cell Treatments
Human prostate cancer DU145 cells were grown in RPMI 1640 medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (FBS, Invitrogen). The HT29 human colon cancer cells were grown in McCoy's 5A medium (Invitrogen) supplemented with 10% (v/v) FBS. Cells were obtained from the American Type Culture Collection (ATCC) and used within 2 months of purchase after resuscitation of frozen aliquots. Cell lines were authenticated on the basis of viability, recovery, growth, morphology, and also cytogenetic analysis, antigen expression, DNA profile and iso-enzymology by the provider. Human recombinant TGF-β1 was obtained from R&D Systems and used at a final concentration of 5 ng/ml. The cells were incubated in serum-free medium overnight, and then treated with TGF-β for 48 h and 96 h for DU145 and HT29 cells, respectively, to induce the EMT.
The chelator, Dp44mT (Fig. 1A), and the negative control compound, di-2-pyridylketone 2-methyl-3-thiosemicarbazone (Dp2mT; Fig. 1B), which cannot bind iron, were synthesized and characterized using standard methods (17, 29). Desferrioxamine (DFO; Fig. 1C) was purchased from Novartis. We utilized concentrations of 10 μm for Dp44mT and Dp2mT and 100 μm for DFO in 10% (v/v) FBS supplemented medium. The greater concentration of DFO was implemented due to its limited ability to permeate the cell membrane (30). The chelator Dp44mT was utilized at a lower concentration since this ligand shows far higher membrane permeability and demonstrates marked iron chelation efficacy (29). Both Dp2mT and Dp44mT were freshly dissolved in dimethyl sulfoxide (DMSO) and diluted in culture media (final [DMSO]: ≤ 0.1% (v/v)).
Plasmid Construction and Transfection
For NDRG1 overexpression, we used pCMV-tag2-FLAG-NDRG1 (GenHunter) and the empty pCMV-tag2-FLAG vector (Stratagene) as a negative control. Both plasmids contained a G418 resistance marker. The shRNA and negative control plasmids were obtained from Qiagen (Cat. KH02202H) and contained a Hygromycin resistance marker. All cells were transfected using Lipofectamine 2000® (Invitrogen) following the manufacturer's protocol. The cells were selected for more than 4 weeks by incubation with G418 (400 ng/ml for DU145 and 1000 ng/ml for HT29) for overexpression clones, or Hygromycin (500 ng/ml for DU145 and 1000 ng/ml for HT29) for knock-down clones. Stable single clones were selected and NDRG1 expression assessed using Western blot. Two single clones of each stable transfectant were utilized in this study.
Protein Extraction and Western Blotting
Whole cell protein lysates were extracted using lysis buffer with proteinase inhibitor mixture (Cat. 11836170001; Roche Applied Science) and PhosSTOP (Cat. 04906845001; Roche Applied Science) according to standard methods (20). Western blotting was performed via established protocols (20). Primary antibodies were against: ferritin (Cat. ab69090), NDRG1 (Cat. ab37897), vimentin (Cat. ab8978), occludin (Cat. ab31721), ZEB2 (Cat. ab25837), and twist (Cat. ab50581) from Abcam; E-cadherin (Cat. 3195), β-catenin (Cat. 9562), ZO-1 (Cat. 8193), Snail (Cat. 3879), Slug (Cat. 9585), SMAD2 (Cat. 3122), pSMAD2 (serine 465/467) (Cat. 3101), pSMAD3 (Cat. 9520), and SMAD4 (Cat. 9515) were from Cell Signaling Technology and cyclin D1 (Cat. SC8396) was from Santa Cruz Biotechnology. The secondary antibodies used were horseradish peroxidase (HRP)-conjugated anti-goat (Cat. A5420), anti-rabbit (Cat. A6154) and anti-mouse (Cat. A4416) from Sigma-Aldrich; Alexa Fluor® 555 conjugated anti-rabbit (Cat. 4413) and Alexa Fluor® 488 conjugated anti-mouse (Cat. 4408) from Cell Signaling Technology. Primary antibody against β-actin (Cat. A1978) was from Sigma-Aldrich and used as a loading control.
Immunofluorescence
Immunofluorescence was performed as described (31). Briefly, cells seeded on coverslips were fixed with 4% (w/v) paraformaldehyde (Sigma-Aldrich) for 10 min and permeabilized with 0.1% (v/v) Triton X-100 for 5 min at room temperature. The cells were then incubated overnight with primary antibodies at 4 °C, followed by incubation with fluorescent secondary antibody for 1 h at room temperature. After final washes with PBS, the coverslips were mounted using an anti-fade mounting solution containing 4′,6-diamidino-2-phenylindole (DAPI; Cat. P36935, Invitrogen) and images were examined and captured using an Olympus Zeiss AxioObserver Z1 fluorescence microscope (Olympus) with a 63× oil objective. Raw images were analyzed using Olympus Axiovision software (Olympus).
Cell Migration and Invasion Assay
Cell migration was assessed using established wound healing and transwell migration assays (31). Briefly, for the wound healing assay, cellular monolayers at 90–95% confluence were serum-starved for 24 h and scratched using a sterile 20 μl pipette tip. After washing (three times with complete medium) and removing the detached cells, the plates were incubated at 37 °C for 12 h, and the wounds were photographed and analyzed using the Olympus Zeiss AxioObserver Z1 fluorescence microscope. The quantitative migration assay was performed using the CytoSelectTM 24-Well Cell Migration Assay kit from Cell Biolabs, according to the manufacturer's protocol. Images were taken, and the migration abilities were quantified by optical absorbance at 560 nm using a PerkinElmer 1420 multi-label plate reader.
Cell invasion was assessed using the CytoSelectTM 96-well Cell Invasion Assay kit (Cell Biolabs) according to the manufacturer's protocol. Invasion values were reported as mean relative fluorescence units (RFUs) of quadruplicate samples measured at 480 nm/520 nm using the PerkinElmer plate reader.
Statistical Analysis
Data are mean ± S.D. and were statistically analyzed using Student's t test. Results were considered significant when p < 0.05.
RESULTS
TGF-β Induces the EMT in HT29 and DU145 Cells
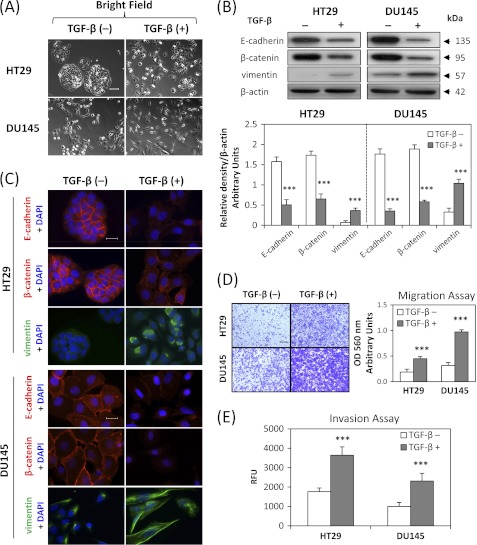
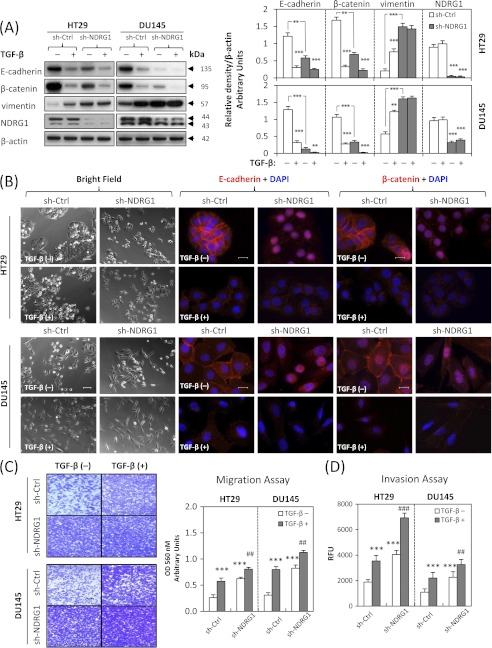
To determine whether TGF-β can induce a mesenchymal phenotype consistent with the EMT in DU145 and HT29 cell-types, we incubated these cells with TGF-β at a physiological dose of 5 ng/ml (32) for 48 h or 96 h, respectively. These different incubation periods were shown in preliminary experiments to demonstrate maximum efficacy at inducing the EMT in each cell type. Treatment with TGF-β resulted in marked morphological changes in the HT29 and DU145 cell types as shown in Fig. 2A. In fact, the cells became more isolated and demonstrated markedly decreased intercellular contacts, with this being particularly notable for HT29 cells (Fig. 2A). Furthermore, for DU145 cells, incubation with TGF-β resulted in more spindle-shaped, fibroblast-like cells. These morphological characteristics were consistent with cells undergoing the EMT after incubation with TGF-β (32, 33).
FIGURE 2.
TGF-β induces characteristics of the epithelial-mesenchymal transition (EMT) in HT29 and DU145 cells. HT29 and DU145 cells were treated in the presence or absence of TGF-β (5 ng/ml) for 96 h and 48 h, respectively, to induce the EMT. A, bright field images were taken to show cell morphological changes after TGF-β treatment. Scale bars: 100 μm. B, whole cell lysates were extracted and Western blotting was performed to investigate changes to molecular markers of the EMT. Densitometric analysis is expressed relative to the loading control, β-actin. C, merged images were taken to show immunofluorescence staining of E-cadherin (red), β-catenin (red), and vimentin (green) accompanied by the cell nucleus (blue) stained by DAPI. Scale bars: 20 μm. D, HT29 and DU145 cells were pretreated for 72 h and 24 h, respectively, with TGF-β (5 ng/ml), then seeded at 100,000 cells/well and incubated for 12 h/37 °C in the presence or absence of TGF-β (5 ng/ml). Migratory cells on the bottom of the polycarbonate membrane were stained (purple blue) and quantified at A560 after extraction according to the manufacturer's protocol (Cell Biolabs). Scale bars: 200 μm. E, cells were treated with TGF-β as described in D above, and 50,000 cells/well were seeded in the transwell chamber coated with extra-cellular matrix and incubated for 12 h/37 °C. The invasion values were reported as mean relative fluorescence unit (RFU) of quadruplicate samples. Data are typical of 3–5 experiments, and the histogram values are mean ± S.D. (3–5 experiments). ***, p < 0.001, relative to control cells without TGF-β.
We then investigated the molecular alterations in the expression of well-established EMT markers (4) by Western blotting and immunofluorescence. Consistent with the morphological changes, there was a significant (p < 0.001) 2–4-fold decrease in the expression of the epithelial markers, E-cadherin and β-catenin (7, 32), and a significant (p < 0.001) 3–5-fold increase in the expression of the mesenchymal marker, vimentin (7, 32), after TGF-β treatment of HT29 and DU145 cells (Fig. 2B). This was further confirmed by immunofluorescence staining, which demonstrated a pronounced reduction of membrane E-cadherin and β-catenin levels (red fluorescence) and an induction of mesenchymal vimentin expression (green fluorescence) upon incubation with TGF-β (Fig. 2C). Notably, E-cadherin and β-catenin form the cadherin complex at the cell membrane that is essential for formation of the adherens junction which maintains intercellular integrity (34).
Considering that the EMT is characterized by enhanced cellular motility and invasion (4, 5), we then evaluated the changes to migratory capacity and invasive potential of HT29 and DU145 cells after TGF-β treatment. As shown in Fig. 2, D and E, using the cell migration and invasion assays, TGF-β significantly (p < 0.001) increased migration and invasion of HT29 and DU145 cells, when compared with untreated control cells. Collectively, these observations indicate that TGF-β induces the EMT in HT29 and DU145 cells.
Iron Chelators Attenuate the TGF-β-induced EMT in HT29 and DU145 Cells
We have reported that novel series of iron chelators function as potent anti-tumor agents, among which Dp44mT is one of the most effective (12, 29). As the EMT plays an important role during cancer cell progression and metastasis (4), we examined whether Dp44mT could act against the TGF-β-induced EMT. At the same time, in order to clarify whether this effect was dependent on iron depletion, we used Dp2mT (29). This compound has a similar chemical structure to Dp44mT, but in contrast, cannot bind cellular iron and is thus an appropriate negative control (29) (Fig. 1, A and B). Moreover, as a further control, we also examined the well-characterized iron chelator, DFO (Fig. 1C), which binds iron with high affinity, but does not redox cycle like Dp44mT (1, 17). This latter control was important to determine whether the effects observed were due to iron depletion or redox activity.
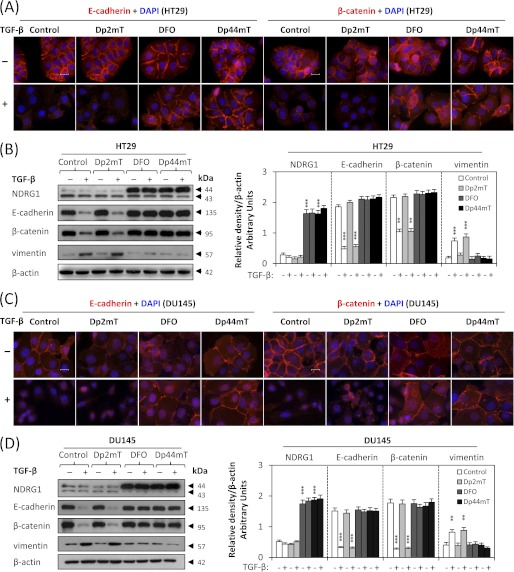
As shown by immunofluorescence and Western blotting studies (Fig. 3, A–D), TGF-β markedly reduced E-cadherin and β-catenin levels in both HT29 (Fig. 3, A and B) and DU145 cells (Fig. 3, C and D), which was in good agreement with the results in Fig. 2, B and C. However, when combined with DFO (100 μm) or Dp44mT (10 μm), the TGF-β-induced loss of E-cadherin and β-catenin was clearly attenuated (Fig. 3, A–D). Compared with Dp44mT and DFO, the negative control compound, Dp2mT, did not have any significant effect on attenuating the TGF-β-induced a decrease of E-cadherin and β-catenin (Fig. 3, A–D). This demonstrates the importance of iron-depletion on the ability of chelators to inhibit the TGF-β-induced EMT. This was further confirmed by pre-treating chelators with iron (as FeCl3) to form iron complexes that cannot bind cellular iron. These complexes prevented the iron depletion-mediated up-regulation of NDRG1, but also inhibited the ability of chelators to attenuate the TGF-β-induced EMT, as shown by the analysis of E-cadherin, β-catenin and vimentin expression (supplemental Fig. S1). Furthermore, the fact that DFO and Dp44mT have similar effects indicates that iron depletion is key to the mechanism involved, rather than the redox activity of Dp44mT (17, 29). Phenotypic analysis demonstrated that HT29 and DU145 cells treated with TGF-β plus Dp2mT showed similar morphology to those treated with TGF-β alone (supplemental Fig. S2). However, DFO or Dp44mT antagonized the effect of TGF-β (supplemental Fig. S2), indicating interrupted TGF-β signaling.
FIGURE 3.
Iron chelators attenuate the TGF-β-induced EMT in HT29 and DU145 cells. Cells were pretreated in the presence or absence of TGF-β (5 ng/ml) for 72 h (HT29) or 24 h (DU145) and followed by co-incubation with either DFO (100 μm), Dp2mT (10 μm) or Dp44mT (10 μm) for another 24 h. A and C, merged immunofluorescence images demonstrate membrane E-cadherin (red) and β-catenin (red) with the cell nucleus (blue) stained by DAPI. Scale bars: 20 μm. B and D, Western analysis of NDRG1 and EMT marker expression in HT29 (B) and DU145 cells (D). Densitometric analysis is expressed relative to the loading control, β-actin. Data are typical of 3–5 experiments, and the histogram values are mean ± S.D. (3–5 experiments). ***, p < 0.001, **, p < 0.01, relative to control cells without TGF-β.
In agreement with the ability of Dp44mT and DFO to attenuate the TGF-β-induced reduction of E-cadherin and β-catenin, these chelators also significantly (p < 0.001) decreased the TGF-β-induced up-regulation of the mesenchymal marker, vimentin, in HT29 (Fig. 3B) and DU145 cells (Fig. 3D). Together, these data demonstrate that chelators can block the TGF-β-induced EMT in HT29 and DU145 cells and that this is dependent on chelation of cellular iron.
As a positive control for cellular iron depletion, we examined NDRG1 expression which is increased by chelator-mediated iron deprivation via hypoxia inducible factor-1α (HIF-1α)-dependent and -independent mechanisms (11, 35). As shown in Fig. 3, B and D, examining cells incubated with control medium alone, NDRG1 appeared as 2 bands at ∼43 and ∼44 kDa (36), which may correspond to its different phosphorylation states (36, 37). The upper band is responsive to iron-depletion with chelators (Fig. 3, B and D) and is thought to represent the active form of this molecule (37). Incubation with TGF-β alone did not significantly (p > 0.05) alter NDRG1 expression relative to untreated control cells (Fig. 3, B and D). In addition, Dp2mT did not have any significant effect on NDRG1 expression relative to the control, since it cannot bind cellular iron (29) (Fig. 3, B and D). However, incubation with either DFO or Dp44mT caused a significant (p < 0.001) increase in the ∼44 kDa NDRG1 band relative to the control in the presence or absence of TGF-β, demonstrating that iron chelators induce NDRG1 expression in these cells. Apart from the up-regulation of NDRG1 after iron-depletion, we also demonstrated that under the same incubation conditions, both DFO and Dp44mT down-regulate ferritin, while Dp2mT had no effect (supplemental Fig. S3). Ferritin is well known to be regulated in this manner by iron depletion (1, 13). Indeed, previous studies have clearly demonstrated the ability of DFO and Dp44mT to reduce iron uptake from transferrin by cells and induce cellular iron mobilization, leading to iron depletion (18, 29, 30).
Considering the marked up-regulation of NDRG1 after iron depletion and its functional role as a metastasis suppressor (21–26), we then hypothesized that NDRG1 may be involved in the process of the TGF-β-induced EMT. To examine this, we established NDRG1 overexpression and knock-down models in HT29 and DU145 cells.
Overexpression of NDRG1 Attenuates the TGF-β-induced EMT
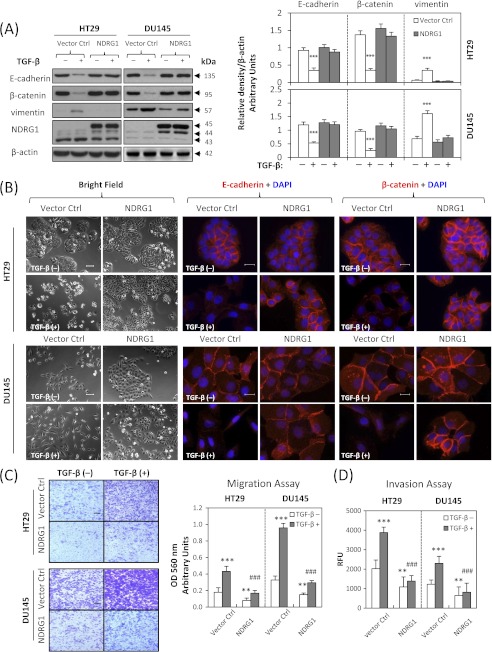
We established stable NDRG1 overexpression transfectants of HT29 and DU145 cells to determine whether NDRG1 plays a role in the EMT. Two stable single clones of each cell-type were selected, namely clones 2 and 3 for HT29 cells and clones 7 and 16 for DU145 cells. For illustrative purposes, the results for clones 2 (HT29) and 7 (DU145) are shown in Fig. 4 and demonstrate the same response as clones 3 (HT29) and 16 (DU145), respectively (data not shown). As shown in Fig. 4A, in NDRG1 overexpressing clones, there was a pronounced and significant (p < 0.001) increase in the expression of FLAG-tagged NDRG1 (at ∼45 kDa) as well as the ∼44 kDa band relative to the empty vector control cells. Notably, there was no marked difference in the morphology of HT29 cells after NDRG1 overexpression (Fig. 4B), probably because their parental cells already exhibited an epithelial phenotype (Fig. 2A). In contrast, it was clear that NDRG1 overexpression in DU145 cells led to different morphology (Fig. 4B). Indeed, there was a transition from isolated fibroblastic cells to “cobblestone”-like epithelial cells that formed colonies upon NDRG1 overexpression (Fig. 4B). After TGF-β treatment, HT29 cells transfected with NDRG1 still exhibited an epithelial morphology where the cells clustered together in groups, while the relative empty vector-transfected cells appeared more isolated. Incubation with TGF-β caused the vector control DU145 cells to become more fibroblastic (Fig. 4B). On the other hand, DU145 cells overexpressing NDRG1 retained their epithelial morphology, but appeared more isolated upon treatment with TGF-β (Fig. 4B). Hence, in both cell-types, NDRG1 overexpression attenuated the ability of TGF-β to induce morphological changes consistent with the EMT (32, 33).
FIGURE 4.
Overexpression of NDRG1 attenuates the TGF-β-induced EMT. NDRG1 overexpressing HT29 cells (clone 2), DU145 cells (clone 7), and their empty vector control cells were treated in the presence or absence of TGF-β (5 ng/ml) for 96 h and 48 h, respectively. A, Western analysis of NDRG1 and EMT marker expression. The FLAG-tagged NDRG1 was strongly expressed in both cell types (∼45 kDa). In NDRG1-overexpressing clones, there was a marked increase in the expression of FLAG-tagged NDRG1 (at ∼45 kDa) as well as the ∼44 kDa band. Densitometric analysis is expressed relative to the loading control, β-actin. B, bright field and immunofluorescence images were taken and E-cadherin (red), β-catenin (red), and nucleus (blue) were stained as described above. Scale bars: 100 μm for bright field and 20 μm for immunofluorescence. C and D, the cell migration and invasion assays were performed as described in Fig. 2, D and E. Scale bars: 200 μm. Data were repeated for 3–5 times, and the values in histograms represent mean ± S.D. (3–5 experiments). *, relative to vector control without TGF-β; #, relative to vector control with TGF-β. **, p < 0.01; ***, p < 0.001; ###, p < 0.001.
We then explored the molecular changes of NDRG1 overexpression against TGF-β stimulation in HT29 and DU145 cells. As shown by immunofluorescence in Fig. 4B, NDRG1 overexpression inhibited TGF-β downstream effects. In fact, membrane expression of the epithelial markers, E-cadherin and β-catenin, was maintained in cells overexpressing NDRG1 in clear contrast to those transfected with the empty vector control after TGF-β treatment (Fig. 4B). We examined the expression of the EMT markers (E-cadherin, β-catenin, and vimentin) using Western blot and demonstrated that NDRG1 overexpression inhibited the changes of EMT markers after incubation with TGF-β (Fig. 4A). In fact, E-cadherin and β-catenin expression was significantly (p < 0.001) decreased after TGF-β treatment in empty vector control cells, while no significant alteration was observed in NDRG1 overexpressing cells. Conversely, expression of the mesenchymal marker, vimentin, was markedly and significantly (p < 0.001) increased after TGF-β treatment of the empty vector control cells. In contrast, vimentin expression was not significantly altered in NDRG1 overexpressing HT29 or DU145 cells (Fig. 4A).
Properties of metastasis were then evaluated by migration and invasion assays. These results revealed that NDRG1 overexpression not only significantly (p < 0.01) decreased migratory and invasive potential of HT29 and DU145 cells in the absence of TGF-β, but also significantly (p < 0.001) reduced the TGF-β-stimulated migration and invasion (Fig. 4, C and D). The wound healing assay further showed a significant (p < 0.01–0.001) inhibitory effect of NDRG1 overexpression on TGF-β-induced migration in HT29 cells (supplemental Fig. S4) and DU145 cells (supplemental Fig. S5). These studies indicate NDRG1 acts to impair migration and invasion and also inhibits TGF-β-induced tumor progression by preventing the EMT.
NDRG1 Knock-down Mimics the TGF-β-induced EMT
Considering the results above and that NDRG1 is down-regulated in many cancers (21, 38, 39), we next used sh-RNA to reduce NDRG1 expression in HT29 and DU145 cells to determine its effects on motility and invasion (Fig. 5). Two stable single clones of each cell-type were selected, namely clones 1 and 6 for HT29 and clones 5 and 12 for DU145. Again, for illustrative purposes, sh-NDRG1 clones 1 (HT29) and 5 (DU145) are shown in Fig. 5, and these demonstrated the same results as clones 6 (HT29) and 12 (DU145), respectively (data not shown).
FIGURE 5.
NDRG1 knock-down mimics the TGF-β-induced EMT. A, NDRG1 knock-down HT29 cells (clone 1), DU145 cells (clone 5), and their respective control cells transfected with scrambled control shRNA were treated with or without TGF-β (5 ng/ml) for 96 h and 48 h, respectively. The efficacy of NDRG1 knock-down and the expression of EMT markers (i.e. E-cadherin, β-catenin, and vimentin) were analyzed by Western blotting. B, bright field microscopy and immunofluorescence showed that NDRG1 knock-down mimics the EMT phenotype in both HT29 and DU145 cells. Scale bars: 100 μm for bright field and 20 μm for immunofluorescence. C and D, cell migration and invasion assays were performed as described in Fig. 2, D and E. Scale bars: 200 μm. Data were repeated 3–5 times, and the values in histograms represent mean ± S.D. (3–5 experiments). *, relative to vector control without TGF-β; #, relative to vector control with TGF-β. **, p < 0.01; ***, p < 0.001; ##, p < 0.01; ###, p < 0.001.
As demonstrated in HT29 cells, NDRG1 expression at ∼44 kDa was almost completely suppressed using NDRG1 sh-RNA, and for DU145 cells, NDRG1 (∼44 kDa) expression was also significantly (p < 0.001) down-regulated three times relative to the scrambled control transfected cells (Fig. 5A). At the same time, in the absence of TGF-β, the cell morphology of HT29 cells with decreased NDRG1 expression changed to more scattered cells (Fig. 5B). Furthermore, immunofluorescence demonstrated that in the absence of TGF-β, after NDRG1 knockdown, both E-cadherin and β-catenin membrane expression was almost abolished, with translocation to the nucleus, being more pronounced in HT29 cells (Fig. 5B). The accumulation of β-catenin in the nucleus is considered to be a tumorigenic effector, as it is thought to act as a transcription factor with oncogenic potential (40, 41). Moreover, the loss of membrane expression and accumulation of nuclear E-cadherin is correlated with higher invasive and metastatic potential (42, 43). Nuclear translocation was not observed in TGF-β-treated cells, indicating that this process occurred independently of the cytokine (Fig. 5B). As E-cadherin and β-catenin are key components of cell adherens junctions (34), their loss from the cell membrane will impair their ability to function in establishing intercellular contacts.
We also examined the EMT markers using Western blotting and revealed that NDRG1 knock-down markedly and significantly (p < 0.001) down-regulated E-cadherin and β-catenin expression 4–5 times in both HT29 and DU145 cells in the absence of TGF-β, while up-regulating vimentin 2–3 times, which was consistent with the EMT (Fig. 5A). Furthermore, these molecular changes for E-cadherin and β-catenin were enhanced by the addition of TGF-β (Fig. 5A). We then investigated metastatic properties via migration and invasion assays. These results demonstrated that NDRG1 knock-down significantly (p < 0.001) increased migration and invasion for HT29 and DU145 cells in the absence of TGF-β and these metastatic potentials were enhanced after incubation with TGF-β (Fig. 5, C and D). Together, these studies indicate that NDRG1 knock-down mimics the EMT in HT29 and DU145 cells and also enhances the downstream effects induced by TGF-β.
Expression of Cell Tight Junction Markers ZO-1 and Occludin Are Not Affected by NDRG1 Expression
Cell morphology and polarity are not only controlled by the adherens junctions, but also by tight junctions (TJs) (44). Hence, we next examined whether cellular TJs were also affected by NDRG1 expression (overexpression and knockdown). In order to investigate this, we examined the expression of well established TJ proteins, including zonula occludin-1 (ZO-1) and occludin (44). However, we saw no significant alterations in the expression of these TJ markers by Western blotting and immunofluorescence in HT29 or DU145 cells in response to alterations in NDRG1 expression (supplemental Fig. S6).
Canonical SMAD/Snail/Slug and Wnt Pathways Are Involved in the NDRG1-mediated Inhibition of the EMT
The above data showed that NDRG1 was involved in the EMT of cancer cells. We then assessed what molecular mechanisms may be involved using the DU145 cell model, as it was more susceptible to TGF-β stimulation and underwent more pronounced morphological changes relative to HT29 cells after NDRG1 overexpression (Fig. 4B).
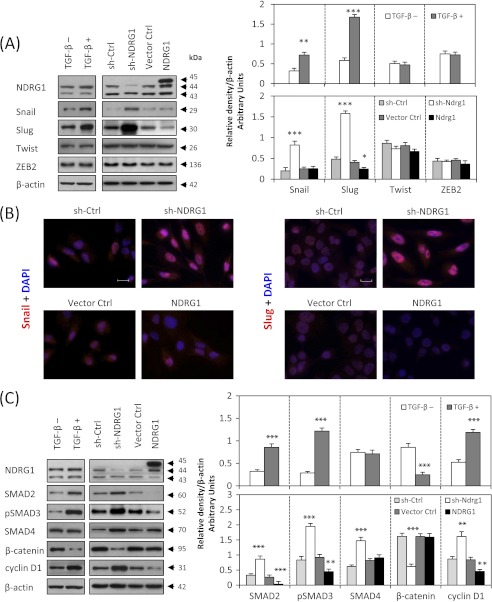
We first examined the expression of nuclear transcriptional repressors responsible for the down-regulation of E-cadherin during the EMT, such as Snail, Slug, Twist and ZEB2 (45). For Twist and ZEB2, there were no significant alterations in expression after NDRG1 knock-down or overexpression (Fig. 6A). In contrast, Snail was significantly (p < 0.001) increased after NDRG1 knock-down, although there was no significant change after NDRG1 overexpression (Fig. 6, A and B). However, NDRG1 knock-down markedly and significantly (p < 0.001) up-regulated the expression of Slug, and NDRG1 overexpression significantly (p < 0.05) reduced Slug expression (Fig. 6, A and B). Hence, the transcriptional E-cadherin repressors, Snail, and Slug, were involved in modulating E-cadherin expression by NDRG1. As a positive control, we also showed that after TGF-β treatment, Snail and Slug expression were significantly (p < 0.01) up-regulated, while there was no significant alteration to Twist or ZEB2 expression (Fig. 6A).
FIGURE 6.
SMAD/Snail/Slug and Wnt pathways are involved in the NDRG1-mediated inhibition of the EMT in DU145 cells. A, Western blotting and B, immunofluorescence staining shows that NDRG1 knock-down increases Snail and Slug expression that could be responsible for E-cadherin repression. C, NDRG1 overexpression decreases the expression of canonical TGF-β/SMAD pathway components (SMAD2 and pSMAD3) and the Wnt-responsive gene, cyclin D1, while NDRG1 knock-down increases their expression. For A and C, cells were also separately incubated in the presence or absence of TGF-β (5 ng/ml) for 48 h as a positive control. Scale bars: 20 μm. Data are typical of 3–5 experiments and the histogram values are mean ± S.D. (3–5 experiments). *, p < 0.05; **, p < 0.01; ***, p < 0.001, relative to control cells without TGF-β.
As shown in this study, NDRG1 overexpression inhibited the TGF-β-induced EMT in DU145 and HT29 cells (Fig. 4) and NDRG1 knock-down mimicked the TGF-β-induced EMT (Fig. 5). We then hypothesized that the TGF-β pathway plays an important role in this process. Hence, we probed for the SMAD proteins that mediate TGF-β signaling (46) to examine whether they were affected by NDRG1. First, we demonstrated that TGF-β significantly (p < 0.001) up-regulated SMAD2 and pSMAD3, although there was no significant difference in SMAD4 expression (Fig. 6C). Then we investigated whether NDRG1 is involved in this pathway. Intriguingly, NDRG1 knock-down in DU145 cells significantly (p < 0.001) increased SMAD2, pSMAD3, and SMAD4 (Fig. 6C), although no pSMAD2 could be detected after exhaustive attempts (data not shown). Furthermore, NDRG1 overexpression significantly (p < 0.01) decreased SMAD2 and pSMAD3 (Fig. 6C), although there was no alteration in SMAD4 expression. Hence, NDRG1 knock-down in DU145 cells activates the TGF-β/SMAD pathway, while NDRG1 overexpression represses it.
Next, we examined cyclin D1 expression during this process as it is a Wnt-responsive gene that responds downstream of nuclear β-catenin. As shown in Fig. 6C, cyclin D1 expression was significantly (p < 0.001) increased after NDRG1 knock-down and significantly (p < 0.01) decreased upon NDRG1 overexpression. As shown previously, β-catenin was maintained at the cell membrane by NDRG1 overexpression (Fig. 4, A and B), but down-regulated and translocated to the nucleus by sh-NDRG1 (Fig. 5, A and B). TGF-β alone acted similarly to NDRG1 knockdown, leading to significant (p < 0.001) up-regulation of cyclin D1 and significant (p < 0.001) down-regulation of β-catenin (Fig. 6C).
DISCUSSION
We demonstrate that DFO and Dp44mT attenuate the TGF-β-induced EMT in HT29 and DU145 cells. Furthermore, NDRG1 overexpression inhibited the TGF-β-induced EMT and conversely NDRG1 knock-down mimicked the downstream effects of TGF-β. Hence, the effect of chelators on inhibiting the EMT was consistent with their ability to markedly up-regulate NDRG1, a well-known anti-metastatic gene (21–26).
Membrane E-cadherin and β-catenin are important hallmarks of the EMT, as they form the adherens junction complex, which connects adjacent cells (44). We demonstrate that incubation of HT29 and DU145 cells with TGF-β leads to the loss of E-cadherin and β-catenin, which is consistent with the EMT. We further showed that chelators could inhibit the TGF-β-induced loss of E-cadherin and β-catenin at the membrane and that this could be mediated by NDRG1. Indeed, NDRG1 overexpression could maintain membrane localization of E-cadherin and β-catenin, and incubation with iron chelators led to the same result. Conversely, NDRG1 knock-down caused a loss of membrane E-cadherin and β-catenin with some nuclear translocation, particularly in HT29 cells (Fig. 5B).
In agreement with our study, it has been reported that NDRG1 is a Rab4a effector involved in vesicular recycling of E-cadherin and that it plays a role in stabilizing adherens junctions in DU145 cells (47). However, we further revealed that NDRG1 knock-down not only caused decreased E-cadherin expression, but also increased its nuclear translocation and accumulation that is related to higher metastatic and invasive potentials (42, 43). This could explain, in part, the mechanism by which NDRG1 mediates its anti-metastatic effects, as NDRG1 overexpression inhibited migration and invasion, while its knock-down had the opposite effect.
In addition to E-cadherin, the expression of another important adherens junction complex molecule, β-catenin, was also decreased and translocated to the nucleus after NDRG1 knockdown (Fig. 5B). There are generally two pools of β-catenin: one associated with E-cadherin at the cell membrane, while the other is “free” in the cytosol/nucleus (48). The membrane-associated β-catenin is considered to be an epithelial marker, while cytoplasmic/nuclear β-catenin plays a role in transcriptional regulation of the Wnt/β-catenin pathway (49). This investigation shows that knock-down of NDRG1 induced loss of membrane β-catenin and caused its nuclear accumulation. Once in the nucleus, β-catenin combines with the T cell factor/lymphoid enhancer factor 1 (TCF/LEF1) to regulate various Wnt-responsive genes, resulting in tumorigenesis (49). This was confirmed by probing for one of the Wnt-responsive genes, cyclin D1, which is involved in proliferation (50). As shown in our study (Fig. 6C), NDRG1 knock-down increased cyclin D1, while NDRG1 overexpression decreased its level. Hence, this could explain another aspect of how NDRG1 exerts its anti-tumor activity.
We then investigated the mechanisms involved in the NDRG1-mediated inhibition of the EMT to elucidate the role it plays in this process. NDRG1 knock-down increased Snail and Slug expression, but not Twist or ZEB2 levels (Fig. 6, A and B). On the other hand, NDRG1 overexpression decreased only Slug. Both Snail and Slug are members of the super-family of zinc finger transcription factors that bind proximal promoter sequences of E-cadherin, namely E-box elements, thus repressing its transcription (45). Considering this, we sought to elucidate the upstream regulatory mechanisms responsible for the alteration in Snail and Slug expression.
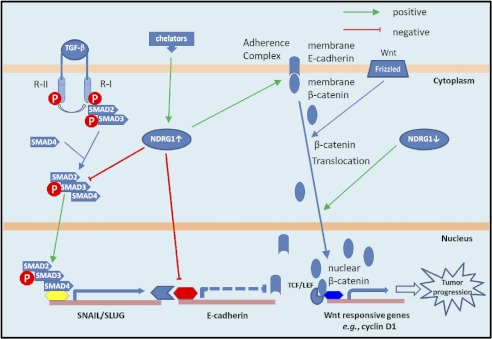
Because NDRG1 overexpression could inhibit the TGF-β-induced effects in HT29 and DU145 cells, we hypothesized the TGF-β/SMAD pathway was involved in the NDRG1-induced alteration in Snail and Slug expression. We showed that TGF-β triggers the EMT primarily via the SMAD pathway, which is activated by the binding of TGF-β to type II TGF-β receptors and subsequent trans-phosphorylation of type I TGF-β receptors on the cell surface (46). After activation, phosphorylated SMAD2/SMAD3 form a complex with the common partner, SMAD4, which translocates to the nucleus to control transcription of target genes like Snail and Slug, in cooperation with other transcriptional factors (46) (Fig. 7). Our investigation showed that NDRG1 knockdown in DU145 cells activated the SMAD pathway via increased SMAD2, pSMAD3, and SMAD4 levels, while NDRG1 overexpression partly inhibits this pathway by reducing SMAD2 and pSMAD3 expression. It is notable that the TGF-β/SMAD pathway acts up-stream of Snail and Slug, promoting their expression (51). In fact, the SMAD complex co-precipitates with Snail and Slug promoters, suggesting a role in their transcription (51). Hence, this leads to a model whereby NDRG1 abrogates SMAD-induced up-regulation of Snail/Slug (Fig. 7). This relieves the repression on E-cadherin expression resulting in enhanced formation of the adherens complex, enabling cell-cell junctions that suppress motility and metastasis (Fig. 7).
FIGURE 7.
Schematic diagram showing the role of NDRG1 in inhibiting the EMT via regulation of the TGF-β/SMAD and Wnt pathways. Iron chelators up-regulate NDRG1 expression and increased NDRG1 inhibits the canonical TGF-β/SMAD pathway through decreasing expression of SMAD2 and pSMAD3. This decreases TGF-β-induced Slug expression which is responsible for E-cadherin repression. Notably, NDRG1 knock-down results in loss of membrane E-cadherin and β-catenin and their translocation into nuclei, thus activating the Wnt pathway and the transcription of cyclin D1 that plays an important role in cell cycle progression and proliferation. Taken together, NDRG1 plays a novel role in regulating the EMT through regulation of the TGF-β and Wnt/β-catenin signaling pathways.
It is notable that NDRG1 overexpression led to decreased expression of the Wnt-responsive gene, cyclin D1, which drives cell cycle progression (50), while NDRG1 knock-down had the opposite effect, leading to cyclin D1 up-regulation (Fig. 6C). These observations are consistent with the finding that NDRG1 enables formation of the E-cadherin/β-catenin adherens complex on the cell membrane. This prevents β-catenin nuclear translocation and its ability to act as a transcription factor to increase cyclin D1 expression that is involved in proliferation (15). In fact, we demonstrate that knockdown of NDRG1 leads to translocation of E-cadherin and β-catenin to the nucleus (Fig. 5B).
In summary, chelators inhibit the TGF-β-induced EMT and this could be mediated by the marked up-regulation of NDRG1. NDRG1 inhibits the TGF-β/SMAD pathway through decreasing SMAD2 and pSMAD3 expression, thus preventing TGF-β-induced Slug expression, which is responsible for repressing E-cadherin (Fig. 7). Hence, then E-cadherin combines with β-catenin on the membrane which forms the adherens complex leading to increased cell-cell contact and decreased invasion. At the same time, due to the localization of β-catenin in the membrane adherens complex, this prevents β-catenin translocation to the nucleus, inhibiting activation of Wnt/β-catenin pathway target genes (e.g. cyclin D1) and halting tumor progression (Fig. 7). This activity explains, in part, the marked anti-tumor activity of chelators, which disrupt TGF-β signal transduction and the EMT.
Supplementary Material

This article contains supplemental Figs. S1–S6.
- EMT
- epithelial-mesenchymal transition
- TGF-β
- transforming growth factor-β
- NDRG1
- N-Myc downstream-regulated gene 1
- DFO
- desferrioxamine
- Dp44mT
- di-2-pyridylketone-4,4-dimethyl-3-thiosemicarbazone
- Dp2mT
- di-2-pyridylketone 2-methyl-3-thiosemicarbazone
- DMSO
- dimethyl sulfoxide
- TJ
- tight junction
- ZO-1
- zonula occludin-1.
REFERENCES
- 1. Kalinowski D. S., Richardson D. R. (2005) The evolution of iron chelators for the treatment of iron overload disease and cancer. Pharmacol. Rev. 57, 547–583 [DOI] [PubMed] [Google Scholar]
- 2. Miller L. D., Coffman L. G., Chou J. W., Black M. A., Bergh J., D'Agostino R., Jr., Torti S. V., Torti F. M. An iron regulatory gene signature predicts outcome in breast cancer. Cancer Res. 71, 6728–6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thiery J. P., Acloque H., Huang R. Y., Nieto M. A. (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 [DOI] [PubMed] [Google Scholar]
- 4. Christiansen J. J., Rajasekaran A. K. (2006) Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 66, 8319–8326 [DOI] [PubMed] [Google Scholar]
- 5. Kalluri R., Weinberg R. A. (2009) The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miettinen P. J., Ebner R., Lopez A. R., Derynck R. (1994) TGF-β induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J. Cell Biol. 127, 2021–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zavadil J., Böttinger E. P. (2005) TGF-β and epithelial-to-mesenchymal transitions. Oncogene 24, 5764–5774 [DOI] [PubMed] [Google Scholar]
- 8. Oft M., Heider K. H., Beug H. (1998) TGFβ signaling is necessary for carcinoma cell invasiveness and metastasis. Curr. Biol. 8, 1243–1252 [DOI] [PubMed] [Google Scholar]
- 9. Pardali K., Moustakas A. (2007) Actions of TGF-β as tumor suppressor and pro-metastatic factor in human cancer. Biochim. Biophys. Acta 1775, 21–62 [DOI] [PubMed] [Google Scholar]
- 10. Rao V. A., Klein S. R., Agama K. K., Toyoda E., Adachi N., Pommier Y., Shacter E. B. (2009) The iron chelator Dp44mT causes DNA damage and selective inhibition of topoisomerase IIα in breast cancer cells. Cancer Res. 69, 948–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Le N. T., Richardson D. R. (2004) Iron chelators with high antiproliferative activity up-regulate the expression of a growth inhibitory and metastasis suppressor gene: a link between iron metabolism and proliferation. Blood 104, 2967–2975 [DOI] [PubMed] [Google Scholar]
- 12. Whitnall M., Howard J., Ponka P., Richardson D. R. (2006) A class of iron chelators with a wide spectrum of potent antitumor activity that overcomes resistance to chemotherapeutics. Proc. Natl. Acad. Sci. U.S.A. 103, 14901–14906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Richardson D. R. (2005) Molecular mechanisms of iron uptake by cells and the use of iron chelators for the treatment of cancer. Curr. Med. Chem. 12, 2711–2729 [DOI] [PubMed] [Google Scholar]
- 14. Liang S. X., Richardson D. R. (2003) The effect of potent iron chelators on the regulation of p53: examination of the expression, localization, and DNA binding activity of p53 and the transactivation of WAF1. Carcinogenesis 24, 1601–1614 [DOI] [PubMed] [Google Scholar]
- 15. Nurtjahja-Tjendraputra E., Fu D., Phang J. M., Richardson D. R. (2007) Iron chelation regulates cyclin D1 expression via the proteasome: a link to iron deficiency-mediated growth suppression. Blood 109, 4045–4054 [DOI] [PubMed] [Google Scholar]
- 16. Fu D., Richardson D. R. (2007) Iron chelation and regulation of the cell cycle: 2 mechanisms of posttranscriptional regulation of the universal cyclin-dependent kinase inhibitor p21CIP1/WAF1 by iron depletion. Blood 110, 752–761 [DOI] [PubMed] [Google Scholar]
- 17. Richardson D. R., Sharpe P. C., Lovejoy D. B., Senaratne D., Kalinowski D. S., Islam M., Bernhardt P. V. (2006) Dipyridyl thiosemicarbazone chelators with potent and selective antitumor activity form iron complexes with redox activity. J. Med. Chem. 49, 6510–6521 [DOI] [PubMed] [Google Scholar]
- 18. Lovejoy D. B., Jansson P. J., Brunk U. T., Wong J., Ponka P., Richardson D. R. (2011) Antitumor activity of metal-chelating compound Dp44mT is mediated by formation of a redox-active copper complex that accumulates in lysosomes. Cancer Res. 71, 5871–5880 [DOI] [PubMed] [Google Scholar]
- 19. Yu Y., Suryo Rahmanto Y., Hawkins C. L., Richardson D. R. (2011) The potent and novel thiosemicarbazone chelators di-2-pyridylketone-4,4-dimethyl-3-thiosemicarbazone and 2-benzoylpyridine-4,4-dimethyl-3-thiosemicarbazone affect crucial thiol systems required for ribonucleotide reductase activity. Mol. Pharmacol. 79, 921–931 [DOI] [PubMed] [Google Scholar]
- 20. Kovacevic Z., Fu D., Richardson D. R. (2008) The iron-regulated metastasis suppressor, Ndrg-1: identification of novel molecular targets. Biochim. Biophys. Acta 1783, 1981–1992 [DOI] [PubMed] [Google Scholar]
- 21. Bandyopadhyay S., Pai S. K., Gross S. C., Hirota S., Hosobe S., Miura K., Saito K., Commes T., Hayashi S., Watabe M., Watabe K. (2003) The Drg-1 gene suppresses tumor metastasis in prostate cancer. Cancer Res. 63, 1731–1736 [PubMed] [Google Scholar]
- 22. Guan R. J., Ford H. L., Fu Y., Li Y., Shaw L. M., Pardee A. B. (2000) Drg-1 as a differentiation-related, putative metastatic suppressor gene in human colon cancer. Cancer Res. 60, 749–755 [PubMed] [Google Scholar]
- 23. Bandyopadhyay S., Pai S. K., Hirota S., Hosobe S., Takano Y., Saito K., Piquemal D., Commes T., Watabe M., Gross S. C., Wang Y., Ran S., Watabe K. (2004) Role of the putative tumor metastasis suppressor gene Drg-1 in breast cancer progression. Oncogene 23, 5675–5681 [DOI] [PubMed] [Google Scholar]
- 24. Maruyama Y., Ono M., Kawahara A., Yokoyama T., Basaki Y., Kage M., Aoyagi S., Kinoshita H., Kuwano M. (2006) Tumor growth suppression in pancreatic cancer by a putative metastasis suppressor gene Cap43/NDRG1/Drg-1 through modulation of angiogenesis. Cancer Res. 66, 6233–6242 [DOI] [PubMed] [Google Scholar]
- 25. Cangul H. (2004) Hypoxia up-regulates the expression of the NDRG1 gene leading to its overexpression in various human cancers. BMC Genet. 5, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ellen T. P., Ke Q., Zhang P., Costa M. (2008) NDRG1, a growth and cancer-related gene: regulation of gene expression and function in normal and disease states. Carcinogenesis 29, 2–8 [DOI] [PubMed] [Google Scholar]
- 27. Shah M. A., Kemeny N., Hummer A., Drobnjak M., Motwani M., Cordon-Cardo C., Gonen M., Schwartz G. K. (2005) Drg1 expression in 131 colorectal liver metastases: correlation with clinical variables and patient outcomes. Clin. Cancer Res. 11, 3296–3302 [DOI] [PubMed] [Google Scholar]
- 28. Kovacevic Z., Chikhani S., Lovejoy D. B., Richardson D. R. (2011) Novel thiosemicarbazone iron chelators induce up-regulation and phosphorylation of the metastasis suppressor N-Myc downstream-regulated gene 1: a new strategy for the treatment of pancreatic cancer. Mol. Pharmacol. 80, 598–609 [DOI] [PubMed] [Google Scholar]
- 29. Yuan J., Lovejoy D. B., Richardson D. R. (2004) Novel di-2-pyridyl-derived iron chelators with marked and selective antitumor activity: in vitro and in vivo assessment. Blood 104, 1450–1458 [DOI] [PubMed] [Google Scholar]
- 30. Richardson D., Ponka P., Baker E. (1994) The effect of the iron(III) chelator, desferrioxamine, on iron and transferrin uptake by the human malignant melanoma cell. Cancer Res. 54, 685–689 [PubMed] [Google Scholar]
- 31. Bambang I. F., Lu D., Li H., Chiu L. L., Lau Q. C., Koay E., Zhang D. (2009) Cytokeratin 19 regulates endoplasmic reticulum stress and inhibits ERp29 expression via p38 MAPK/XBP-1 signaling in breast cancer cells. Exp. Cell Res. 315, 1964–1974 [DOI] [PubMed] [Google Scholar]
- 32. Pino M. S., Kikuchi H., Zeng M., Herraiz M. T., Sperduti I., Berger D., Park D. Y., Iafrate A. J., Zukerberg L. R., Chung D. C. (2010) Epithelial to mesenchymal transition is impaired in colon cancer cells with microsatellite instability. Gastroenterology 138, 1406–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ellenrieder V., Hendler S. F., Boeck W., Seufferlein T., Menke A., Ruhland C., Adler G., Gress T. M. (2001) Transforming growth factor β1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res. 61, 4222–4228 [PubMed] [Google Scholar]
- 34. Wijnhoven B. P., Dinjens W. N., Pignatelli M. (2000) E-cadherin-catenin cell-cell adhesion complex and human cancer. Br J. Surg. 87, 992–1005 [DOI] [PubMed] [Google Scholar]
- 35. Salnikow K., Davidson T., Zhang Q., Chen L. C., Su W., Costa M. (2003) The involvement of hypoxia-inducible transcription factor-1-dependent pathway in nickel carcinogenesis. Cancer Res. 63, 3524–3530 [PubMed] [Google Scholar]
- 36. Kovacevic Z., Sivagurunathan S., Mangs H., Chikhani S., Zhang D., Richardson D. R. (2011) The metastasis suppressor, N-Myc downstream-regulated gene 1 (NDRG1), up-regulates p21 via p53-independent mechanisms. Carcinogenesis 32, 732–740 [DOI] [PubMed] [Google Scholar]
- 37. Sugiki T., Murakami M., Taketomi Y., Kikuchi-Yanoshita R., Kudo I. (2004) N-Myc down-regulated gene 1 is a phosphorylated protein in mast cells. Biol. Pharm. Bull. 27, 624–627 [DOI] [PubMed] [Google Scholar]
- 38. van Belzen N., Dinjens W. N., Diesveld M. P., Groen N. A., van der Made A. C., Nozawa Y., Vlietstra R., Trapman J., Bosman F. T. (1997) A novel gene which is up-regulated during colon epithelial cell differentiation and down-regulated in colorectal neoplasms. Lab. Invest. 77, 85–92 [PubMed] [Google Scholar]
- 39. Bandyopadhyay S., Pai S. K., Hirota S., Hosobe S., Tsukada T., Miura K., Takano Y., Saito K., Commes T., Piquemal D., Watabe M., Gross S., Wang Y., Huggenvik J., Watabe K. (2004) PTEN up-regulates the tumor metastasis suppressor gene Drg-1 in prostate and breast cancer. Cancer Res. 64, 7655–7660 [DOI] [PubMed] [Google Scholar]
- 40. Behrens J., von Kries J. P., Kühl M., Bruhn L., Wedlich D., Grosschedl R., Birchmeier W. (1996) Functional interaction of β-catenin with the transcription factor LEF-1. Nature 382, 638–642 [DOI] [PubMed] [Google Scholar]
- 41. Suzuki H., Masuda N., Shimura T., Araki K., Kobayashi T., Tsutsumi S., Asao T., Kuwano H. (2008) Nuclear β-catenin expression at the invasive front and in the vessels predicts liver metastasis in colorectal carcinoma. Anticancer Res. 28, 1821–1830 [PubMed] [Google Scholar]
- 42. Elston M. S., Gill A. J., Conaglen J. V., Clarkson A., Cook R. J., Little N. S., Robinson B. G., Clifton-Bligh R. J., McDonald K. L. (2009) Nuclear accumulation of e-cadherin correlates with loss of cytoplasmic membrane staining and invasion in pituitary adenomas. J. Clin. Endocrinol. Metab. 94, 1436–1442 [DOI] [PubMed] [Google Scholar]
- 43. Céspedes M. V., Larriba M. J., Pavón M. A., Alamo P., Casanova I., Parreño M., Feliu A., Sancho F. J., Muñoz A., Mangues R. (2010) Site-dependent E-cadherin cleavage and nuclear translocation in a metastatic colorectal cancer model. Am. J. Pathol. 177, 2067–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hartsock A., Nelson W. J. (2008) Adherens and tight junctions: structure, function, and connections to the actin cytoskeleton. Biochim. Biophys. Acta 1778, 660–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hotz B., Arndt M., Dullat S., Bhargava S., Buhr H. J., Hotz H. G. (2007) Epithelial to mesenchymal transition: expression of the regulators snail, slug, and twist in pancreatic cancer. Clin. Cancer Res. 13, 4769–4776 [DOI] [PubMed] [Google Scholar]
- 46. Heldin C. H., Miyazono K., ten Dijke P. (1997) TGF-β signaling from cell membrane to nucleus through SMAD proteins. Nature 390, 465–471 [DOI] [PubMed] [Google Scholar]
- 47. Kachhap S. K., Faith D., Qian D. Z., Shabbeer S., Galloway N. L., Pili R., Denmeade S. R., DeMarzo A. M., Carducci M. A. (2007) The N-Myc down-regulated Gene1 (NDRG1) Is a Rab4a effector involved in vesicular recycling of E-cadherin. PLoS One 2, e844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu D., Pan W. (2010) GSK3: a multifaceted kinase in Wnt signaling. Trends Biochem. Sci. 35, 161–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reya T., Clevers H. (2005) Wnt signaling in stem cells and cancer. Nature 434, 843–850 [DOI] [PubMed] [Google Scholar]
- 50. Sherr C. J. (1994) G1 phase progression: cycling on cue. Cell 79, 551–555 [DOI] [PubMed] [Google Scholar]
- 51. Brandl M., Seidler B., Haller F., Adamski J., Schmid R. M., Saur D., Schneider G. (2010) IKK(α) controls canonical TGF(β)-SMAD signaling to regulate genes expressing SNAIL and SLUG during EMT in panc1 cells. J. Cell Sci. 123, 4231–4239 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.