Background: Currently, no information is available about the association of the mitochondrial porin pore with the major outer membrane proteins Om14p and Om45p.
Results: Por1p forms complexes with Om14p and Om45p.
Conclusion: Molecular organization of the porin pore and its interaction with the inner membrane is influenced by Om14p and Om45p.
Significance: The newly identified protein complex improves the understanding of mitochondrial transport processes.
Keywords: Membrane Transport, Mitochondria, Protein-Protein Interactions, Transporters, Yeast, Om14p, Om45p, Por1p, TSPO
Abstract
Numerous transport processes occur between the two mitochondrial (mt) membranes due to the diverse functions and metabolic processes of the mt organelle. The metabolite and ion transport through the mt outer membrane (OM) is widely assumed to be mediated by the porin pore, whereas in the mt inner membrane (IM) specific carriers are responsible for transport processes. Here, we provide evidence by means of Blue Native (BN)-PAGE analysis, co-immunoprecipitation, and tandem affinity purification that the two mt OM proteins Om14p and Om45p associate with the porin pore. Porin molecules seem to assemble independently to build the core unit. A subpopulation of these core units interacts with Om14p and Om45p. With preparative tandem affinity purification followed by MS analysis, we could identify interaction partners of this OM complex, which are mainly localized within the mt IM and function as carriers for diverse molecules. We propose a model for the role of the two OM proteins in addressing the porin pore to bind to specific channels in the mt IM to facilitate transport of metabolites.
Introduction
The transport of metabolites and ions through the mt2 OM is widely assumed to occur unspecifically via the porin pore (Por1p in Saccharomyces cerevisiae). In mammalian organisms, this pore is called the voltage-dependent anion channel (VDAC) as it selects for negatively charged molecules like ATP (for review see Refs. 1, 2). Rostovtseva and Colombini (3, 4) provided experimental evidence for the existence of an open and closed state of the porin pore for ATP flux. Although the regulation of the gating mechanism is still under debate, it seems likely that a positively charged mobile domain in the porin wall senses changes of the electric field applied to the membrane. Change of charge moves this voltage sensor to the surface of the membrane and results in a reduced pore diameter and inverted ion selectivity due to an electrostatic barrier (5).
The mt IM is equipped with a large number of specific channels, most of them belonging to the mt carrier family (MCF), that mediate the highly regulated transfer of metabolites and ions. The genome of S. cerevisiae encodes for 34 putative members of MCF. Up to now, the transported molecules for 26 of those are known or are assumed (6, 7).
Transport processes between the OM and IM can occur independently, but contact sites that couple the two membranes and could lead to more efficient transport have been described (for review see Ref. 8). For protein translocation, the coupling of the two translocase complexes of the OM and IM is well known (reviewed in Refs. 8, 9). Exchange of ADP/ATP and creatine/phosphocreatine is facilitated by coupling VDAC and the adenine nucleotide translocator (in mammals, homologous to the yeast Aac1/2/3p isoforms). Creatine kinase and hexokinase are regulating factors of the two membrane-spanning pores (8, 10).
It remains to be elucidated how the porin pore selects and binds to individual transporters residing in the IM. Besides Por1p, there are two additional abundant proteins of the mt OM in S. cerevisiae, Om14p and Om45p (11–13). The function of both proteins is unknown so far, and there are no homologous proteins in other organisms other than budding yeasts (Saccharomycetaceae) (13). Classified protein motifs that could hint at a possible function are lacking. The respective null mutants possess no detectable phenotype (12, 13). However, expression of OM14 and OM45 is tightly co-regulated and increased after diauxic shift (14), suggesting a role in the transition to respiratory growth. Om45p has a molecular mass of 45-kDa and is anchored within the mt OM by an α-helical transmembrane domain at the N terminus (amino acids 5–22). Available data on the orientation of the protein in the membrane are conflicting (11–13, 15, 16). Om14p is a small (14 kDa), cysteine-rich protein with three α-helical transmembrane domains. The N terminus is localized in the cytosol, and the C terminus protrudes in the mt IMS (13). The existence of α-helical transmembrane domains in both OM proteins points toward protein functions procured after the endosymbiotic event, like protein translocation, mt fission, and apoptosis (13).
Here, we report on the physical interaction of the three most abundant mt OM proteins Om14p, Om45p, and porin by means of two-dimensional BN-SDS-PAGE, Co-IP, and TAP. In addition, the associated complex was isolated by preparative TAP, and its components were identified by MS. To elucidate the function of the OM proteins, the impact of OM14 and/or OM45 deletion on the mt proteome was analyzed by two-dimensional difference in gel electrophoresis (DIGE). Our data are summarized in an interaction model suggesting the involvement of Om14p and Om45p in addressing the porin pore to bind to specific channels in the mt IM.
EXPERIMENTAL PROCEDURES
Yeast Strains and Media
S. cerevisiae strains used in this study are listed in Table 1. The double deletion strain Δom14 Δom45 was constructed by substitution of the OM45-ORF with the replacement cassette KlURA3 by homologous recombination within the strain Δom14. Fusion of proteins in the strains BY4741, OM45-cMyc, Δpor1, and Δpor1 OM45-cMyc with either cMyc-, HA-, or TAP-tag was done as described (17–19). Correct integration and expression were confirmed using PCR and Western blot analysis with antibodies directed against the tags, respectively. Yeast complete and minimal media were prepared as described (18, 20).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Ref. |
|---|---|---|
| BY4741/WT | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0 | Euroscarf (accession no. Y00000) |
| OM14-HA | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YBR230c::(YBR230c-3HA-SpHIS5+) | This work |
| OM45-cMyc | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YIL136w::(YIL136w-9cMyc-SpHIS5+) | This work |
| OM14-HA OM45-cMyc | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YBR230c::(YBR230c-3HA-KlURA3), YIL136w::(YIL136w-9cMyc-SpHIS5+) | This work |
| OM14-TAP | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YBR230c::(YBR230c-TAP-KlURA3) | This work |
| Δom14 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YBR230c::kanMX4 | Euroscarf (accession no. Y03370) |
| Δom14 OM45-cMyc | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YBR230c::kanMX4, YIL136w::(YIL136w-9cMyc-SpHIS5+) | This work |
| Δom45 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YIL136w::kanMX4 | Euroscarf (accession no. Y02295) |
| Δom45 OM14-HA | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YIL136w::kanMX4, YBR230c::(YBR230c-3HA-SpHIS5+) | This work |
| Δom14 Δom45 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YBR230c::kanMX4, YIL136w::KlURA3 | This work |
| Δpor1 | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YNL055c::kanMX4 | Euroscarf (accession no. Y07374) |
| Δpor1 OM14-HA | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YNL055c::kanMX4, YBR230c::(YBR230c-3HA-SpHIS5+) | This work |
| Δpor1 OM45-cMyc | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YNL055c::kanMX4, YIL136w::(YIL136w-9cMyc-SpHIS5+) | This work |
| Δpor1 OM14-HA OM45-cMyc | MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0, YNL055c::kanMX4, YBR230c::(YBR230c-3HA-KlURA3), YIL136w:: (YIL136w-9cMyc-SpHIS5+) | This work |
Isolation and Purification of Mitochondria
Yeast cells were grown to the early stationary phase in media containing ethanol as the carbon source. For mechanic isolation of mt, cells were resuspended in lysis buffer (650 mm mannitol, 20 mm Tris-HCl, pH 7.6, 1 mm EDTA, pH 8.0, 1 mm 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF, AppliChem), and 1× proteinase inhibitor mixture (PI-Mix, EDTA-free, Roche Applied Science)) and disrupted by shaking with glass beads (0.25–0.5 mm, Roth) for 5 min. Cell debris and glass beads were pelleted by centrifugation (3,500 × g, 5 min, 4 °C), and the crude mt fraction was obtained by further centrifugation of the supernatant (12,000 × g, 10 min, 4 °C). Enzymatic isolation and purification of mt by two successive sucrose gradient centrifugations were performed as described (21, 22). Protein concentrations were determined with the colorimetric DC protein assay (Bio-Rad) using bovine serum albumin as the standard.
Proteinase K Treatment
25 μg of fresh enzymatically isolated mt proteins were incubated with 2 μg/ml Proteinase K solution (Invitrogen) in buffer (650 mm sorbitol, 10 mm Tris-HCl, pH 7.4) in the presence or absence of 1% Triton X-100 (Roth) for 15 min on ice. Proteinase K activity was stopped by adding AEBSF (1 mm) and PI-Mix (1×).
Two-dimensional Blue Native-SDS-PAGE
BN-PAGE (23, 24) was accomplished as described previously (25). Briefly, 200 μg of highly purified mt proteins were lysed with 4% digitonin (detergent/protein ratio of 4:1, highly pure, Sigma) in lysis buffer (100 mm NaCl, 5 mm 6-aminocaproic acid, 50 mm imidazole, 1 mm AEBSF, and 1× PI-Mix). A high molecular weight gel filtration calibration kit (thyroglobulin, 669 kDa; ferritin, 440 kDa; aldolase 158 kDa; conalbumin, 75 kDa; GE Healthcare) or the NativeMarkTM unstained native protein marker (Invitrogen) were used to determine the apparent molecular weights of the complexes.
Co-immunoprecipitation
Co-IP experiments were done as described (18) with the following modifications: 16 μg of cMyc, HA (both Roche Applied Science), or Por1p (Invitrogen) antibodies were used for immobilization. 500 μg of enzymatically isolated mt were lysed in buffer (see two-dimensional BN-PAGE) containing 0.625% n-dodecyl-β-d-maltoside (detergent/protein ratio of 2.5:1, Sigma). Eluate was precipitated by the methanol/chloroform method (26).
Tandem Affinity Purification
TAP method was performed essentially as described (27, 28) with the following specifications: samples of 500 μg (analytical scale) or 4 mg (preparative scale) of mechanically isolated mt were lysed with 1% digitonin (detergent/protein ratio of 4:1) in lysis buffer (100 mm NaCl, 20 mm Tris-HCl, pH 7.4) for 20 min on ice. The extract was cleared by centrifugation for 20 min at 18,000 × g and incubated with an IgG matrix (GE Healthcare) for 2 h. Cleavage by tobacco etch virus (TEV) protease (20 units of AcTEVTM, Invitrogen) was performed in lysis buffer for 2 h at 16 °C. Final concentrations of 7 mm CaCl2, 1 mm imidazole, 1 mm magnesium acetate, and 10 mm β-mercaptoethanol were added to the supernatant before incubation with a calmodulin matrix (GE Healthcare) at 4 °C for 2 h. Elution was done with buffer containing 200 mm potassium acetate, 20 mm Tris-HCl, pH 7.4, 20 mm EGTA, pH 7.9, 1 mm imidazole, 1 mm magnesium acetate, 10 mm β-mercaptoethanol. The eluate was precipitated by the methanol/chloroform method (26). All steps were performed in the presence of 0.1% digitonin, 1 mm AEBSF, and 1× PI-Mix.
SDS-PAGE and Western Blot Analysis
Preparation of SDS-polyacrylamide gels for protein electrophoresis was carried out according to Laemmli (29). Proteins were transferred onto a PVDF membrane (Millipore), probed with primary antibodies, and detected with either horseradish peroxidase-conjugated secondary antibodies and the ECL PlusTM kit (GE Healthcare) or with Cy5-coupled secondary antibodies and the Typhoon Trio scanner (GE Healthcare). Primary antibodies were directed against Aco1p (aconitase; kind gift of R. Lill, Marburg, Germany), Ccp1p (cytochrome c peroxidase; kind gift of W. Neupert, Munich, Germany), cMyc-tag (Roche Applied Science), HA-tag (Roche Applied Science), Cox1p (cytochrome c oxidase subunit I; Invitrogen), Cox2p (cytochrome c oxidase subunit II; Invitrogen), Por1p (porin; Invitrogen), TAP tag (directed against the calmodulin-binding peptide; Open Biosystems), Tom22p (subunit of OM translocase; kind gift of D. Mokranjac, Munich, Germany), and Tom40p (subunit of OM translocase; kind gift of J. Brix, Freiburg, Germany). PageRulerTM Plus Prestained Protein Ladder (Fermentas) or, for the preparative TAP, SpectraTM Multicolor Broad Range Protein Ladder (Fermentas) were used as standards. Quantification of fluorescence signals was done with ImageQuant Version 5.2 software (GE Healthcare).
Two-dimensional Difference in Gel Electrophoresis
Two-dimensional DIGE was essentially performed according to the manual “2D Electrophoresis” from GE Healthcare, and all chemicals were obtained from that company. Briefly, for each sample 50 μg of highly purified mt proteins were lysed in 10 μl of thiourea buffer containing 4% CHAPS for 30 min on ice. Differential labeling of samples was achieved by addition of 1 μl of Cy2-, Cy3-, or Cy5-dye (200 μm) within lysis, stopped by quenching with l-lysine (1 mm final concentration), and incubated for 5 min on ice. Three samples were mixed, and 100 μg of unlabeled mt proteins from each sample were added to adjust the protein content to 450 μg. One volume of 2× sample buffer containing 4% CHAPS, 2 mm DTT, and 4% Pharmalyte, pH 3–10, was added. Two-dimensional isoelectric focusing/SDS-PAGE was performed as described previously (18). Fluorescence signals were detected with a Typhoon Trio scanner (GE Healthcare) and analyzed with Delta2D 4.0 software (Decodon). For visualization of protein spots, gels were stained using a colloidal Coomassie® staining procedure (30).
Mass Spectrometry
Protein bands were excised, washed, in-gel reduced, S-alkylated, and digested with trypsin (Promega) as described (31). MALDI-MS measurements using an Ultraflex MALDI-TOF/TOF mass spectrometer (Bruker Daltonics), spectra processing, and peak list generation were performed as described previously (32). Nano-LC-MS/MS experiments were performed using an UltiMate 3000 nano-HPLC system (Dionex) equipped with a PepMap C18 analytical column (3 μm, 100 Å, 15 cm × 75 μm inner diameter) directly coupled to the nanoelectrospray source (Proxeon) of an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific). Peptides were eluted with an 80-min linear gradient of 5–45% acetonitrile in 0.1% formic acid at 200 nl/min. Mass spectra were acquired in a data-dependent mode with one MS survey scan (resolution of 60,000) in the Orbitrap and MS/MS scans of the eight most intense precursor ions in the LTQ. MS raw files were processed using Proteome Discoverer 1.2.0.208 software. Database searches were performed using an in-house Mascot server version 2.1 (Matrix Sciences Ltd.) for MALDI-TOF/TOF data and Proteome Discoverer applying Sequest algorithm for LTQ Orbitrap XL data. Search criteria were as follows: (i) taxonomy, S. cerevisiae; (ii) enzyme specificity, trypsin; (iii) mass accuracy, 10 ppm for precursor ion mass tolerance (Orbitrap), 50 ppm for peptide mass fingerprinting, and 0.8 Da for fragment ion analysis; (iv) fixed and variable modifications, cysteine carbamidomethylation, methionine oxidation, asparagine, and glutamine deamidation, respectively; (v) maximum of one missed cleavage site; and (vi) databases, NCBI version 20110312 (39953 S. cerevisiae sequences) and SwissProt 2011_02 (6583 S. cerevisiae sequences).
RESULTS
Om14p, Om45p, and Por1p Interact with Each Other
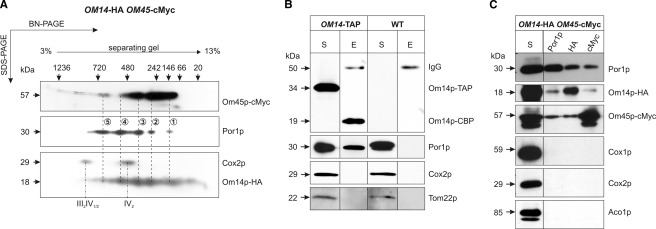
The co-localization and similar steady state levels of Por1p, Om14p, and Om45p in the mt OM (11–13) led us to speculate that these proteins might interact. To test this hypothesis, we compared the molecular organization of the proteins by two-dimensional BN-SDS-PAGE analysis. For that purpose, mt of a strain co-expressing tagged versions of both proteins (OM14-HA OM45-cMyc, see Table 1) were used. Detection of Por1p using specific antibodies revealed five distinct signals with molecular mass of ∼145 (signal 1), 300 (signal 2), 420 (signal 3), 550 (signal 4), and 700 kDa (signal 5) (Fig. 1A). This indicates heterogeneous assembly forms in distinct complexes. Interestingly, Om14p and Om45p show similar separation profiles with focal accumulations in the range of 60–700 kDa (Om45p) and 40–700 kDa (Om14p), respectively. Most strikingly, most of the Por1p spots overlap with the position of signals of either Om14p or Om45p (see dashed lines for assignment in Fig. 1A), suggesting that these proteins are organized at least partially in common complexes. Detection of the expected signals of the respiratory chain supracomplexes III2IV1/2 and the dimer of complex IV (33, 34) using an antibody against Cox2p (a subunit of the mt respiratory complex IV) confirmed both, the sufficient lysis of membrane proteins and the efficient two-dimensional separation.
FIGURE 1.
Om14p, Om45p, and Por1p interact with each other. 200 μg of highly purified mt proteins of the strain OM14-HA OM45-cMyc were solubilized with 4% digitonin and separated by BN-PAGE on a 3–13% gradient gel in the first dimension and by SDS-PAGE on a 12% gel in the second dimension (A). Physical interactions of Om14p, Om45p, and Por1p were analyzed by TAP (B) and Co-IP (C) experiments. For the TAP, 500 μg of mechanically isolated mt of OM14-TAP and the wild type (WT) strain were lysed with 1% digitonin and subjected to the two consecutive TAP-affinity steps. For co-IP, 500 μg of highly purified mt of the strain OM14-HA OM45-cMyc were lysed with 0.625% n-dodecyl-β-d-maltoside. The lysate was incubated with immobilized antibodies against either Por1p, HA, or cMyc. Proteins of the eluate (E) and an aliquot of the respective lysate (S) were separated on a 12% SDS-polyacrylamide gel and transferred onto a PVDF membrane. Immunodetection was done with the indicated antibodies. IV2, dimer of respiratory chain complex IV; III2IV1/2, supracomplexes of respiratory chain.
Further support for an interaction between Om14p and Por1p was obtained by TAP of mt proteins from a strain expressing TAP-tagged Om14p, with wild type cells serving as a negative control. Both mt lysates were subjected to the two specific consecutive steps of the TAP method. Accordingly, samples of the initial lysate and the final eluate were analyzed by Western blot (Fig. 1B). Immunological detection by antibodies recognizing the calmodulin-binding domain of the TAP-tag revealed a signal at the expected mass of 34 kDa in samples of the TAP-tagged strain but not of the wild type, corresponding to the fusion protein Om14p-TAP. The shift of this signal toward lower molecular weight in the final eluate is explained by a successful cleavage of the TAP-tag by the TEV protease, illustrating the efficiency of the applied method. In addition, faint signals at 50 kDa were visible in both eluate fractions due to binding of the secondary antibodies used for detection on co-eluted IgG heavy chains released from the first affinity matrix. Por1p, but none of the hydrophobic mt proteins of the OM (Tom22p) and IM (Cox2p), were co-precipitated and detected in the final eluate of the TAP-tagged strain. This strongly indicates a physical interaction between Por1p and Om14p.
Independent evidence for an interaction between Om14p and the porin pore arises from Co-IP experiments. To this end, proteins from lysates of highly purified mt proteins of a strain co-expressing OM14-HA and OM45-cMyc were incubated with immobilized antibodies directed against either Por1p, HA- or cMyc-, tag (Fig. 1C). For each protein, binding to its specific immobilized antibodies was confirmed by the respective immunological detection. Besides Por1p, the eluate of the Por1p-antibody matrix contained Om14p-HA and Om45p-cMyc. In the corresponding experiments using immobilized HA- or cMyc-antibodies that precipitate Om14p and Om45p, respectively, all three proteins were co-eluted too. This strongly supports the existence of one or more common heteromeric complex(es). Specificity of the Co-IP was proven by the failure to detect the hydrophobic proteins of the mt IM (Cox1p and Cox2p) and the soluble matrix protein Aco1p by the respective antibodies.
Steady State Levels of Om14p and Om45p Depend on Co-expression of Complex Subunits
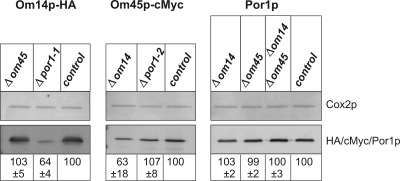
Assembly of proteins in a complex often stabilizes the individual subunits. Thus, loss of a structural complex subunit can disturb the complex assembly and result in reduced steady state levels of unassembled subunits. Hence, we analyzed the steady state levels of the OM proteins in the absence of subunits of the proposed complex. To this end, defined amounts of mt proteins of the respective deletion strains (see Table 1) were separated by SDS-PAGE, transferred onto a PVDF membrane, and subjected to fluorescence quantification based on protein-specific primary and Cy5-coupled secondary antibodies (Fig. 2). Signals were correlated to the intensity of the Cox2p signal, which served as control, and values were normalized to wild type (100%). Steady state concentration of Om14p is significantly reduced to about 60% in the absence of Por1p. In contrast, Om45p had no effect on Om14p abundance (see Fig. 2, left panel). Deletion of OM14 strongly diminished the steady state level of Om45p by about 40%, whereas POR1 knock-out did not (see Fig. 2, middle panel). Strikingly, Por1p concentration is not influenced by either single or double knock-out of OM14 and OM45 (see Fig. 2, right panel). Taken together, these data strongly hint at a role of Por1p as central complex constituent, whereas the other two OM proteins seem to capture accessory positions.
FIGURE 2.
Steady state levels of Om14p, Om45p, and Por1p in deletion strains. 10 μg of mechanically isolated mt proteins from the deletion strains Δom14 OM45-cMyc, Δom45 OM14-HA, Δpor1 OM14-HA (Δpor1-1), Δpor1 OM45-cMyc (Δpor1–2), Δom14 Δom45, and control strain OM14-HA OM45-cMyc were separated on a 15% SDS-polyacrylamide gel and analyzed by Western blot. Specific primary and Cy5-conjugated secondary antibodies were used. Fluorescence images from three independent experiments were quantified using ImageQuant Version 5.2 software. Signals were correlated with the intensity of the Cox2p signal, which served as a control, and values were normalized to control strain (100%).
Deletion of POR1 Affects the Molecular Organization of Om14p and Om45p
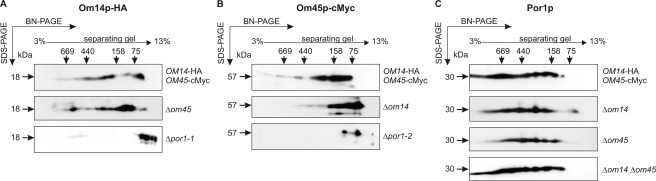
Next we analyzed by two-dimensional BN-SDS-PAGE whether there is also an effect on the molecular organization of the OM proteins in the absence of one or two of the putative binding partners (Fig. 3). In the upper panels of Fig. 3, the two-dimensional BN-SDS-PAGE separation profiles of the three OM proteins in the strain co-expressing the tagged versions of Om14p and Om45p are shown (see also Fig. 1A). These separation profiles are almost unaffected in the absence of Om45p (see Fig. 3, A and C), indicating an accessory role of Om45p rather than being the central subunit. In contrast, deletion of porin led to a marked shift of the signals of both Om14p and Om45p toward a mass below 150 kDa, indicating that high molecular weight complexes (signals 2–5 in Fig. 1A) are either not assembled or degraded (see Fig. 3, A and B, lower panel). Om14p reveals four protein spots with a mass of less than 75 kDa if porin is lacking (Fig. 3A). This hints at the formation of homomultimers of Om14p and/or interaction with Om45p. For Om45p, only two distinct spots of about 60 and 115 kDa were detected in the Δpor1 strain (Fig. 3B). This may be explained by its monomeric and dimeric state or by complex association with Om14p. Interestingly, in the absence of Om14p high molecular weight (>200 kDa) complexes bearing Om45p are only faintly detected (see Fig. 3B, middle panel), although the formation of Por1p complexes is not impaired (see Fig. 3C). Similarly, double deletion of OM14 and OM45 did not change the separation profile of Por1p. The latter results suggest that Om14p is a prerequisite for binding of Om45p onto the complex(es), although the porin complexes seem to assemble independently. A shift of the porin complexes (see signals 1–5 in Fig. 1A) in the absence of either Om14p and/or Om45p is most likely not detected due to the small size of both proteins and their marginal contribution to the molecular weight of the porin complex(es).
FIGURE 3.
Molecular organization of Om14p, Om45p, and Por1p in deletion strains. 200 μg of highly purified mt from the strains Δom45 OM14-HA, Δpor1 OM14-HA (Δpor1-1), Δom14 OM45-cMyc, Δpor1 OM45-cMyc (Δpor1-2), Δom14 Δom45, and OM14-HA OM45-cMyc (control) were lysed with 4% digitonin. High molecular weight complexes were separated by BN-PAGE in a 3–13% gradient gel. Subunit composition of complexes was assessed by a second dimension under denaturing conditions (12% SDS-PAGE). Proteins were transferred onto a PVDF membrane. For detection of Om14p (A), Om45p (B), and Por1p (C), antibodies against HA-, cMyc-tag, and Por1p, respectively, were used. Determination of molecular weight was done with the high molecular weight calibration kit.
Por1p and Om14p Interact Irrespective of Om45p
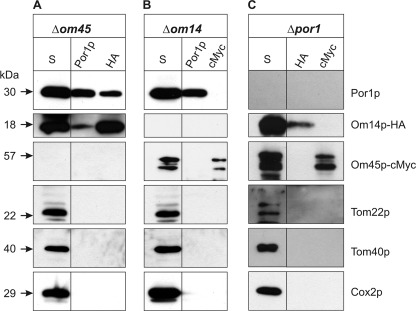
As shown above, deletion of POR1 and OM14 has significant effects on the steady state levels and the molecular organization on the remaining complex subunits. To further confirm their physical interaction, we performed Co-IP experiments using mt lysates from strains devoid of either of the binding partners. To this end, the respective antibodies were immobilized and incubated with mt lysates of the deletion mutants. Successful immobilization of antibodies against the HA-tag (for Om14p), cMyc-tag (for Om45p), Por1p, and subsequent pulldown of the respective target proteins were confirmed by Western blot (Fig. 4). As expected from the result of two-dimensional BN-SDS-PAGE, deletion of OM45 still allows for the co-precipitation of Om14p and Por1p using immobilized antibodies either against Por1p or, in a reciprocal experiment, against the HA-tag (see Fig. 4A). This strongly supports the idea that both proteins interact irrespective of the presence of Om45p. In support of this view, Om45p was not precipitated along with Por1p in the absence of Om14p (see Fig. 4B). Interestingly, after deletion of POR1, neither Om45p nor Om14p could be co-precipitated with antibodies against HA or cMyc, respectively (see Fig. 4C), indicating that Om14p and Om45p do not form a heteromeric subcomplex if Por1p is absent. Specificity of the results is documented by the failure to detect the hydrophobic transport proteins of the mt OM (Tom22p and Tom40p) and IM (Cox2p) by the respective antibodies.
FIGURE 4.
Por1p and Om14p interact irrespective of Om45p. Co-IPs were performed with mt lysates of the deletion strains Δom45 OM14-HA (A), Δom14 OM45-cMyc (B), and Δpor1 OM14-HA OM45-cMyc (C) in combination with the indicated immobilized antibodies against Por1p, HA-, or cMyc-tag, respectively. Proteins of the final eluates and an aliquot of the corresponding supernatant of lysed mt (S) were separated by 12% SDS-PAGE and transferred onto a PVDF membrane prior to immunodetection with specific antibodies against Por1p, Om14p-HA, and Om45p-cMyc. As controls, antibodies against the OM proteins Tom22p and Tom40p and the IM protein Cox2p were used.
OM Complex of Por1p, Om14p, and Om45p Interacts with mt Carrier Proteins of the IM
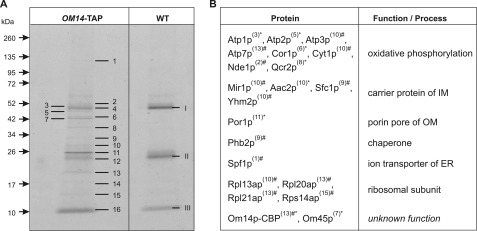
To identify further interacting components, the proposed OM complex of Om14p, Om45p, and Por1p was purified by preparative TAP. Initial studies revealed that Om14p-TAP is not stable during enzymatic mt isolation. This might probably be due to the presence of contaminating enzymes in the zymolyase preparation used, which degrade the TAP-tag of the fusion protein (also described by Puig et al. (28)). Thus, TAP was performed using lysates of 4 mg of mechanically isolated mt of OM14-TAP and wild type. Proteins of the final eluates were separated on an 8–20% gradient SDS-polyacrylamide gel and stained with colloidal Coomassie®. Selected bands of the OM14-TAP lane and the corresponding gel regions of wild type lane were excised and subjected to MALDI-TOF/TOF and nano-LC-MS/MS analysis (Fig. 5A). The three abundant bands (see Fig. 5A, 4/I, 12/II, and 16/III) that appear in both strains, OM14-TAP and wild type, result from contaminations by the IgG matrix used in the first TAP incubation step (heavy and light chain of IgG1) and aprotinin, a trypsin inhibitor that is a component of the used PI-Mix, respectively. The latter PI-Mix mixture was used together with AEBSF during the whole TAP procedure because preliminary studies showed that using these protease inhibitors do not inhibit the TEV cleavage.
FIGURE 5.
Mt interactome of Om14p. The TAP method (A) was used in a preparative scale with 4 mg of mechanically isolated mt of the strains OM14-TAP and wild type (WT) as control. Proteins were separated on an 8–20% gradient SDS-polyacrylamide gel and stained with colloidal Coomassie®. Numbered bands of OM14-TAP lane and the corresponding gel regions of wild type lane (arabic and roman numbers for OM14-TAP and WT, respectively) were subjected to identification by MS (see “Experimental Procedures” and supplemental Table S1). Proteins co-eluted with OM14-TAP (B). Numbers in parentheses refer to the protein bands indicated in A. Information about the proteins was obtained from the database SGD. *, data derived from MALDI-TOF/TOF analysis with a peptide mass fingerprint score of >59 being significant (p < 0.05); #, data arose from nano-LC-MS/MS analysis with a 1% false discovery rate (FDR) and a minimum of two detected unique peptides.
The list of 21 proteins identified after TAP of the OM14-TAP strain is given in Fig. 5B (for MS data see supplemental Table S1). As expected from the Co-IP results and for documentation of the functional TAP procedure, the bait protein Om14p was co-eluted with Om45p and Por1p. Interestingly, among the eluted proteins, 13 are assigned to the mt IM. Four of them (Aac2p, Mir1p, Sfc1p, and Yhm2p) belong to the MCF and function in shuttling of various metabolites across the mt IM (6, 7). Aac2p, also known as Pet9p, is the major isoform of the ADP/ATP carriers in the IM, exchanging matrix ATP and cytoplasmic ADP (35). Mir1p transports inorganic phosphate (Pi) from the IMS into the mt matrix (36). Sfc1p, also known as Acr1p, is responsible for import of cytosolic succinate and export of fumarate (37). The antiporter Yhm2p (Coc1p) transports citrate out of and oxoglutarate into mt (7). A further detected IM protein, Phb2p, is a subunit of the conserved 1.0–1.4-MDa prohibitin complex. It is discussed that it may function as a chaperone, as prohibitin has been shown to interact with nonassembled subunits of the respiratory chain (38, 39). The other eight identified proteins of the IM are engaged in mt oxidative phosphorylation as follows: one of the three NADH dehydrogenases (Nde1p, which is exposed to the IMS (40)), three subunits of respiratory chain complex III (Cor1p, Cyt1p, and Qcr2p), and four subunits of the ATP synthase (Atp1p, Atp2p, Atp3p, and Atp7p). Besides the IM proteins, another ion transporter that is localized in the membrane of the endoplasmatic reticulum, called Spf1p (Cod1p), was co-precipitated with Om14p-TAP. Spf1p is a P-type ATPase that pumps calcium ions (41). Furthermore, four ribosomal proteins (Rpl13ap, Rpl20ap, Rpl21ap, and Rps14ap) were identified in the eluate. Protein band 14 could not be identified as a known yeast protein.
In the wild type control, all these proteins were not detectable. Unfortunately, confirmation of the interaction by using Om45p as bait protein for the TAP procedure proved to be unfeasible, as the TAP-tag interferes with the function of the protein. Strains expressing Om45p-TAP show reduced growth on nonfermentable carbon sources, and the fusion protein is partially mislocalized to the cytoplasmic compartment (data not shown).
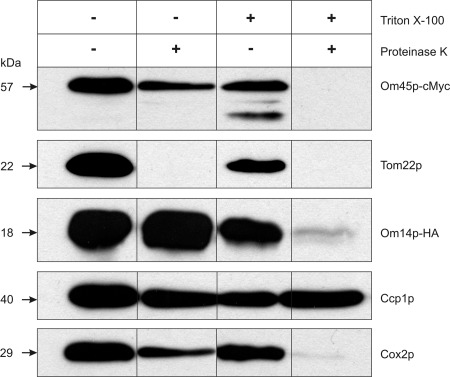
Om45p Is Exposed to the IMS
To reveal the topology of Om45p, we performed Proteinase K treatment of fresh enzymatically isolated mt of a strain bearing Om45p and Om14p C-terminally tagged with cMyc or HA, respectively (Fig. 6). As expected, the protein translocator receptor protein Tom22p of the OM, which is exposed to the cytosol (42, 43), is sensitive to Proteinase K treatment, whereas the soluble IMS protein cytochrome c peroxidase (Ccp1p (22)) and the IM protein Cox2p (44) are resistant. Om45p shows a similar behavior regarding Proteinase K resistance as Ccp1p and Cox2p. The resistance of Om45p is not an inherent property of the protein, because it is degraded in the presence of Triton X-100. These results confirm the data of Riezman et al. (11) according to which Om45p protrudes into the IMS. Additionally, our observation of the Proteinase K resistance of Om14p-HA (Fig. 6) approves the data of Burri et al. (13) that the C-terminal part of Om14p is localized in the IMS.
FIGURE 6.
Topology of Om45p. 25 μg of fresh enzymatically isolated mt proteins of the strain OM14-HA OM45-cMyc were incubated with or without 2 μg/ml Proteinase K in the presence or absence of 1% Triton X-100 for 15 min on ice. Proteinase K activity was stopped by addition of protease inhibitors (AEBSF and PI-Mix). Proteins were separated on a 12% SDS-polyacrylamide gel and analyzed by immunodetection with specific antibodies.
Deletion of OM14 and/or OM45 Affects Steady State Levels of mt Proteins That Require Imported Molecules
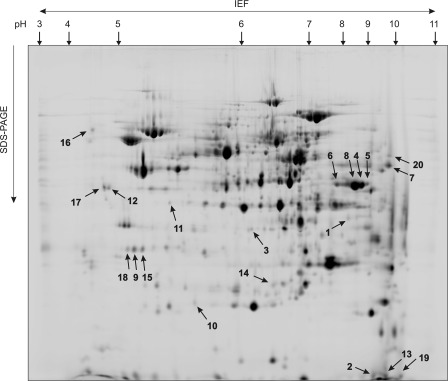
To investigate the impact of Om14p and/or Om45p on other mt proteins, we compared the two-dimensional isoelectric focusing/SDS-PAGE separation profiles of the respective single and double deletion mutants with those of wild type (Fig. 7). For reliable quantification of protein levels, the DIGE approach (see “Experimental Procedure”) was used. Twenty spots with more than 2-fold change in intensity were selected for protein identification by MALDI-TOF/TOF MS (supplemental Table S2). Accordingly, the fluorescence intensity of Om45p is highly reduced in the single (Δom45) and double deletion strains (Δom14 Δom45) (see Fig. 7 and Table 2). Interestingly, Om45p is present in at least four different spots (6, 8, 4, and 5) with pI values of 7.8, 8.6, 8.8, and 9.0, respectively, suggesting post-translational modification(s). Analysis of Om14p was interfered by the co-migration of Pam17p, and hence intensity quantification failed. Furthermore, detection of Om14p in a 12% SDS-polyacrylamide gel was impeded by the low molecular weight of the protein. In line with the one-dimensional analysis of steady state levels in deletion strains (see Fig. 2), the intensity of the protein spots assigned to Om45p are reduced about 2-fold in the deletion strain Δom14 (see Fig. 7 and supplemental Table S2).
FIGURE 7.
Impact of OM14/OM45 deletion on the mt proteome. Two-dimensional DIGE analysis (see “Experimental Procedures”) was performed with differentially labeled mt samples from wild type, Δom14, Δom45, or Δom14 Δom45. The mt proteins were separated by isoelectric focusing (IEF) (pH 3–11, nonlinear) followed by 12% SDS-PAGE. Fluorescence signals of labeled proteins were detected with Typhoon Trio scanner and quantified with Delta2D 4.0 software. Figure shows an image of a wild type sample. Numbered protein spots significantly varied in intensities compared with wild type (threshold, 2-fold) and were subjected to identification by MALDI-TOF/TOF MS.
TABLE 2.
Changes of the mt proteome of the strains Δom14, Δom45, and Δom14 Δom45 in comparison to wild type
Protein spots with significant changes (threshold, 3-fold; −, decreased; +, increased) of their fluorescence intensity in comparison to wild type in the two-dimensional DIGE gels (see Fig. 7) were analyzed by MALDI-TOF/TOF MS (see under “Experimental Procedures” and supplemental Table S2). C, cytoplasm; M, mitochondria; MA, mt matrix; SGD, Saccharomyces Genome Database.
| Spot no. | Protein name | Abundance change | Function/process | Localization | Co-factor/substrate | Carrier for molecule | Ref. |
|---|---|---|---|---|---|---|---|
| Δom14 | |||||||
| 1 | Tdh1p | −4.3 | Glycolysis/glyconeogenesis | C | NAD+/NADH | Ndt1/2p | 6, 45 |
| 2 | Ynl208w-like | −3.2 | Unknown | M | SGD | ||
| 3 | Lsc1p | −3.1 | Citric acid cycle | MA | ADP | Aac1/2/3p | 6, 36, 46 |
| Pi | Mir1p | ||||||
| Δom45 | |||||||
| 4 | Om45p-1 | −24.6 | Unknown | OM | SGD | ||
| 5 | Om45p-4 | −10.1 | Unknown | OM | SGD | ||
| 6 | Om45p-2 | −6.6 | Unknown | OM | SGD | ||
| 8 | Om45p-3 | −6.4 | Unknown | OM | SGD | ||
| 15 | Ccp1p-2 | −5.8 | Oxidative stress response | IMS | Heme | Aac1/2/3p | 48, 54 |
| 7 | Tef2p | −4.8 | Translation elongation factor | C | GDP/GTP | Ggc1p | 6, 47 |
| 9 | Ccp1p-1 | −4.1 | Oxidative stress response | IMS | Heme | Aac1/2/3p | 48, 54 |
| Δom14Δom45 | |||||||
| 1 | Tdh1p | −18.2 | Gluconeogenesis | C | NAD+/NADH | Ndt1/2p | 6, 45 |
| 4 | Om45p-1 | −16.0 | Unknown | OM | SGD | ||
| 5 | Om45p-4 | −10.0 | Unknown | OM | SGD | ||
| 8 | Om45p-2 | −6.8 | Unknown | OM | SGD | ||
| 7 | Tef2p | −5.9 | Translation elongation factor | C | GDP/GTP | Ggc1p | 6, 47 |
| 10 | Pst2p | −4.7 | Stress response | M | FMNa | Unknown | 49, 50, 56, 57 |
| 6 | Om45p-3 | −4.5 | Unknown | OM | SGD | ||
| 11 | Qcr2p | −3.7 | Respiratory chain | IM | Zn2+a | Unknown | 51, 58, 83 |
| 3 | Lsc1p | −3.5 | Citric acid cycle | MA | ADP | Aac1/2/3p | 6, 46 |
| 12 | Lsp1p | −3.4 | Heat stress response | C | Pi | Mir1p | 52 |
| 13 | Om14p/Pam17p | −3.0 | Unknown | OM/IM | SGD | ||
a This is an assumption.
Setting the threshold at 3-fold (Table 2), deletion of OM14 results in decreased levels of Tdh1p (isoenzyme 1 of the glyceraldehyde-3-phosphate dehydrogenase (45)), Ynl208w-like protein (unknown function), and Lsc1p (α subunit of succinyl-CoA ligase, mt enzyme of the citric acid cycle (46)). In the absence of Om45p, steady state concentrations of Tef2p (translational elongation factor EF-1α (47)) and Ccp1p (cytochrome c peroxidase, which degrades reactive oxygen species in mt (48)) are reduced compared to wild type. Consistently, these proteins are also affected in the double deletion strain (see supplemental Table S2). However, the following three additional proteins were identified with decreased abundance if both OM proteins are missing: Pst2p (a protein with similarity to members of a family of flavodoxin-like proteins, which is induced by oxidative stress in a Yap1p-dependent manner (49, 50)); Qcr2p (subunit two of the respiratory chain complex III (51)), and Lsp1p (primary component of eisosomes, which are large immobile patch structures at the cell cortex associated with endocytosis (52)).
Tef2p, Tdh1p, and Lsp1p are cytosolic proteins, suggesting that their association with the OM is maintained by the mt isolation procedure applied avoiding high salt treatment. In line with this, attachment to the mt OM has been shown for Lsp1p (53). Four of the five mt proteins (the other protein is not studied yet) have in common that they have to bind molecules (co-factors or substrates) for their function. These molecules themselves or their precursors have to be transported through the mt membranes (see Table 2). The MCF consists of specific transporter proteins in the IM for these molecules as follows: ADP/ATP are transported through the mt IM by the Aac1/2/3p isoforms, Pi by Mir1p, and the heme precursor protoporphyrin IX by the Aac1/2/3p isoforms as well (6, 54). The carrier protein for flavin was long assumed to be Flx1p (6, 55), but recently it was shown that this carrier only exports FAD out of mt (56). The synthesis of FMN and FAD occurs within mt. Thus, the import of riboflavin, the precursor of FMN, is mandatory. A carrier for riboflavin has not yet been identified (56, 57). The mt import of metal ions, like Zn2+, is also assumed to be carrier-mediated, but a mt transporter protein for zinc has not yet been reported (58). Interestingly, all identified proteins, except for Pst2p, are phosphoproteins according to the UniProt database. Synthesis of ATP, which is necessary for phosphorylation, requires import of Pi and ADP into mt by the IM carriers Mir1p and Aac2p, respectively.
Taken together, our results support the putative involvement of the Om14p-Om45p complex in the selection of specific carriers in the IM to create contact sites between OM and IM. This may lead to an optimization of transport processes through mt membranes.
DISCUSSION
The mt OM proteome of S. cerevisiae is dominated by the presence of three transmembrane proteins Por1p, Om14p, and Om45p (11–13). The molecular organization forms of porin are characterized by five different complexes ranging from 150 to above 700 kDa, reflecting associations of 6–20 or even more porin molecules (59–62). It is of note, however, that the active unit is the monomeric form (63–65). Interestingly, the porin organization pattern as revealed by two-dimensional BN-SDS-PAGE does not change significantly in the absence of Om14p, Om45p, or both. Hence, one can conclude that porin assembly is an independent process, and the porin complex functions as a core unit providing additional binding sites.
However, physical interaction of the mt OM proteins Om14p, Om45p, and Por1p is strongly supported by three independent analyses. Deletion of porin abolishes completely the detection of higher molecular weight assemblies of Om14p and Om45p. This again documents the central role of porin as a core scaffold and suggests an accessory role for Om14p and Om45p. In line with this view are the data on the steady state levels of mt OM proteins, whereas protein levels of porin are almost not affected in strains lacking either of the OM proteins, cells lacking porin contained significantly diminished levels of Om14p. In principle, the proposed porin scaffold(s) could bind both mt OM proteins directly or one by virtue of the other. The latter view is supported by the fact that Om45p does not interact with Por1p if Om14p is absent. Additionally, the deletion of OM14 strongly reduced the steady state level of Om45p but not vice versa. Thus, the presence of Om14p seems to be an essential prerequisite for the stability of Om45p. Most likely, unassembled Om45p molecules are degraded, as it was also shown for the two subunits of the mt prohibitin complex if the gene for either of the subunits was disrupted (66). Interestingly, Om45p and Om14p interact together forming a stable oligomeric structure only in presence of porin. Thereby Om14p mediates the binding to porin, as deletion of OM45 still shows interaction between Om14p and Por1p. Deduced from two-dimensional BN-SDS-PAGE experiments of Δpor1, both mt OM proteins seem to form homo-oligomeric structures. We favor the idea that only a subpopulation of Om14p/Om45p is bound to porin at a certain time and/or a specific physiological state. The remaining mt OM protein molecules may exist as free homomeric structures to provide a pool for porin binding on demand.
To address the possible function of the identified OM complex, we investigated interacting proteins by employing preparative scale TAP. Using Om14p-TAP as bait protein, we were able to purify an associated complex containing, as expected, porin and Om45p but in addition subunits of oxidative phosphorylation complexes, membrane chaperones, and members of the MCF. The identified Om14p interactome includes many proteins of the mt IM indicating a close association between both membranes, possibly facilitating the exchange of metabolites. In line with this assumption are the findings of two-dimensional DIGE experiments aimed at the elucidation of the impact of either OM14 and/or OM45 deletion on the mt proteome. In the deletion mutants protein abundance of at least four mt proteins that require imported co-factors or substrate molecules for their function is diminished. This may hint at an important role for the two OM proteins in assisting or supporting membrane transport through both membranes by mediating attachment of porin scaffolds with the respective carriers in the IM. As the bulk of Om45p is located in the IMS (see Ref. 11 and our data), it might act as a mediator between OM and IM.
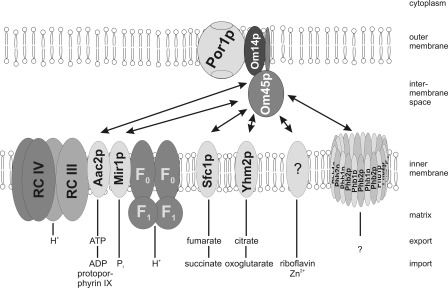
Fig. 8 summarizes our data on interacting proteins. In support of this model, interactions between most of the identified proteins (Por1p, Aac2p, Mir1p, Atp1p, Atp2p, Nde1p, Cor1p, Qcr2p, Cyt1p, and Ggc1p) have already been shown experimentally (67–70). In line with the proposed role in mt complex assembly, Om45p was identified within a mt supramolecular protein complex containing also proteins of the citric acid cycle and the respiratory chain (71).
FIGURE 8.
Model of OM transportome. The mt OM complex of Por1p, Om14p, and Om45p interacts with different carrier proteins of the IM. The transported metabolites are indicated. This model is not to scale. F1/F0, F1/F0 subunit of the mt ATP synthase; RC III/IV, respiratory chain supracomplex III2IV2; Phb1p and Phb2p, prohibitin complex.
The identification of some, but not all, of the respiratory chain complexes might be explained by different affinities toward the precipitated core complex. Additionally, only protein bands detected by Coomassie® were subjected to MS identification, and hence the identified mt OM interactome is currently mostly characterized by proteins of higher abundance.
Association with the giant prohibitin (Phb2p) complex on the one hand and with ribosomal proteins on the other hand may indicate functions in co-translational protein import, assembly, and folding. In line with this, Ohba and Schatz (72) could show that an antiserum against 45-kDa proteins of the mt OM (most likely recognizing Om45p) inhibits mt protein import. However, this finding could not be verified for Om45p by analyzing the protein import of three specific mt proteins (12).
Our data indicate an important function of the OM proteins Om14p and Om45p. However, homologous proteins in higher eukaryotes are lacking, and homology is restricted to budding yeasts (Saccharomycetaceae) (13). As efficient organization of mt import is an essential prerequisite, most likely orthologous proteins may substitute for Om45p and Om14p as was shown for the mt carrier Yhm2p (7). A similar function in mediating membrane-membrane contact can be attributed to the human translocator protein of the mt OM (TSPO) that was identified in particular at the contact sites of the two membranes (73–75). TSPO, like Om14p, is a small protein of 18 kDa (76), hydrophobic (with five transmembrane domains) (74, 77), highly conserved from archaea to metazoans but interestingly is not a component of the S. cerevisiae proteome (78). In analogy, TSPO also interacts with the porin pore (VDAC) and the adenine nucleotide translocator (79, 80) and transports molecules from the cytosol into the mt (76, 81, 82).
In summary, our data support the idea of a highly flexible system of mt to efficiently organize transport processes across two membranes. The coordinated binding of the OM pore system characterized by the different assembly forms of porin toward the IM transporters might be mediated by the activity of the Om14p/Om45p pair. In this scenario, both proteins could serve as a license factor for the predominant positioning of the porin pore in conjunction with the respective mt IM carriers to provide a very efficient system for the exchange of energy metabolites and substrates for respiratory chain. The elevated presence of both proteins during the transition to nonfermentative conditions may help to synchronize the increased demand of metabolites, co-factors, or even proteins for respiration with the availability of pre-disposed channels comprising OM porin pores and IM transporters.
To further validate the functional relevance of the proposed OM complex, the interactions of IM proteins (including carrier proteins) and OM proteins will be systematically analyzed. Moreover, investigation of the transport of small molecules, e.g. of ATP, both in wild type cells and in deletion strains (e.g. Δom14 Δom45) could shed light onto the biological role of Om14p and Om45p.
Supplementary Material
Acknowledgments
We gratefully thank Wolfgang Zachariae (Max Planck Institute of Biochemistry, Munich, Germany) for providing plasmids, and Jan Brix (Albert-Ludwigs-Universität Freiburg, Germany), Roland Lill (Philipps-Universität Marburg, Germany), Dejana Mokranjac (Ludwig-Maximilians-Universität München, Germany), and Walter Neupert (Max Planck Institute of Biochemistry, Munich, Germany) for the kind gifts of antisera. We also greatly appreciate the support with the two-dimensional DIGE method by Uta Gey (Technische Universität Dresden, Germany).

This article contains supplemental Tables S1 and S2.
- mt
- mitochondria(l)
- AEBSF
- 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride
- BN
- Blue Native
- DIGE
- difference in gel electrophoresis
- IM
- inner membrane
- IMS
- inner membrane space
- MCF
- mitochondrial carrier family
- OM
- outer membrane
- PI-Mix
- proteinase inhibitor mixture
- TAP
- tandem affinity purification
- TEV
- tobacco etch virus
- TSPO
- translocator protein of the mt OM
- VDAC
- voltage-dependent anion channel.
REFERENCES
- 1. Colombini M. (2004) VDAC. The channel at the interface between mitochondria and the cytosol. Mol. Cell. Biochem. 256, 107–115 [DOI] [PubMed] [Google Scholar]
- 2. Rostovtseva T. K., Bezrukov S. M. (2008) VDAC regulation. Role of cytosolic proteins and mitochondrial lipids. J. Bioenerg. Biomembr. 40, 163–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rostovtseva T., Colombini M. (1996) ATP flux is controlled by a voltage-gated channel from the mitochondrial outer membrane. J. Biol. Chem. 271, 28006–28008 [DOI] [PubMed] [Google Scholar]
- 4. Rostovtseva T., Colombini M. (1997) VDAC channels mediate and gate the flow of ATP. Implications for the regulation of mitochondrial function. Biophys. J. 72, 1954–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Song J., Midson C., Blachly-Dyson E., Forte M., Colombini M. (1998) The sensor regions of VDAC are translocated from within the membrane to the surface during the gating processes. Biophys. J. 74, 2926–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Palmieri F., Agrimi G., Blanco E., Castegna A., Di Noia M. A., Iacobazzi V., Lasorsa F. M., Marobbio C. M., Palmieri L., Scarcia P., Todisco S., Vozza A., Walker J. (2006) Identification of mitochondrial carriers in Saccharomyces cerevisiae by transport assay of reconstituted recombinant proteins. Biochim. Biophys. Acta 1757, 1249–1262 [DOI] [PubMed] [Google Scholar]
- 7. Castegna A., Scarcia P., Agrimi G., Palmieri L., Rottensteiner H., Spera I., Germinario L., Palmieri F. (2010) Identification and functional characterization of a novel mitochondrial carrier for citrate and oxoglutarate in Saccharomyces cerevisiae. J. Biol. Chem. 285, 17359–17370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reichert A. S., Neupert W. (2002) Contact sites between the outer and inner membrane of mitochondrial role in protein transport. Biochim. Biophys. Acta 1592, 41–49 [DOI] [PubMed] [Google Scholar]
- 9. Endo T., Yamamoto H., Esaki M. (2003) Functional cooperation and separation of translocators in protein import into mitochondria, the double membrane-bounded organelles. J. Cell Sci. 116, 3259–3267 [DOI] [PubMed] [Google Scholar]
- 10. Vyssokikh M., Brdiczka D. (2004) VDAC and peripheral channeling complexes in health and disease. Mol. Cell. Biochem. 256, 117–126 [DOI] [PubMed] [Google Scholar]
- 11. Riezman H., Hay R., Gasser S., Daum G., Schneider G., Witte C., Schatz G. (1983) The outer membrane of yeast mitochondria. Isolation of outside-out sealed vesicles. EMBO J. 2, 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yaffe M. P., Jensen R. E., Guido E. C. (1989) The major 45-kDa protein of the yeast mitochondrial outer membrane is not essential for cell growth or mitochondrial function. J. Biol. Chem. 264, 21091–21096 [PubMed] [Google Scholar]
- 13. Burri L., Vascotto K., Gentle I. E., Chan N. C., Beilharz T., Stapleton D. I., Ramage L., Lithgow T. (2006) Integral membrane proteins in the mitochondrial outer membrane of Saccharomyces cerevisiae. FEBS J. 273, 1507–1515 [DOI] [PubMed] [Google Scholar]
- 14. Ohlmeier S., Kastaniotis A. J., Hiltunen J. K., Bergmann U. (2004) The yeast mitochondrial proteome, a study of fermentative and respiratory growth. J. Biol. Chem. 279, 3956–3979 [DOI] [PubMed] [Google Scholar]
- 15. Rapaport D. (2003) Finding the right organelle. Targeting signals in mitochondrial outer-membrane proteins. EMBO Rep. 4, 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Waizenegger T., Stan T., Neupert W., Rapaport D. (2003) Signal-anchor domains of proteins of the outer membrane of mitochondria: structural and functional characteristics. J. Biol. Chem. 278, 42064–42071 [DOI] [PubMed] [Google Scholar]
- 17. Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. (1999) Epitope tagging of yeast genes using a PCR-based strategy. More tags and improved practical routines. Yeast 15, 963–972 [DOI] [PubMed] [Google Scholar]
- 18. Gey U., Czupalla C., Hoflack B., Rödel G., Krause-Buchholz U. (2008) Yeast pyruvate dehydrogenase complex is regulated by a concerted activity of two kinases and two phosphatases. J. Biol. Chem. 283, 9759–9767 [DOI] [PubMed] [Google Scholar]
- 19. Tauche A., Krause-Buchholz U., Rödel G. (2008) Ubiquinone biosynthesis in Saccharomyces cerevisiae. The molecular organization of O-methylase Coq3p depends on Abc1p/Coq8p. FEMS Yeast Res. 8, 1263–1275 [DOI] [PubMed] [Google Scholar]
- 20. Kaiser C., Michaelis S., Mitchell A. (1994) Methods in Yeast Genetics: A Laboratory Course Manual, pp. 207–210, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 21. Meisinger C., Sommer T., Pfanner N. (2000) Purification of Saccharomyces cerevisiae mitochondria devoid of microsomal and cytosolic contaminations. Anal. Biochem. 287, 339–342 [DOI] [PubMed] [Google Scholar]
- 22. Daum G., Böhni P. C., Schatz G. (1982) Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 257, 13028–13033 [PubMed] [Google Scholar]
- 23. Schägger H. (2001) Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol. 65, 231–244 [DOI] [PubMed] [Google Scholar]
- 24. Schägger H., von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 25. Krause-Buchholz U., Schöbel K., Lauffer S., Rödel G. (2005) Saccharomyces cerevisiae translational activator Cbs1p is associated with translationally active mitochondrial ribosomes. Biol. Chem. 386, 407–415 [DOI] [PubMed] [Google Scholar]
- 26. Wessel D., Flügge U. I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 27. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 28. Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001) The tandem affinity purification (TAP) method. A general procedure of protein complex purification. Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 29. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 30. Neuhoff V., Arold N., Taube D., Ehrhardt W. (1988) Improved staining of proteins in polyacrylamide gels, including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9, 255–262 [DOI] [PubMed] [Google Scholar]
- 31. Czupalla C., Nürnberg B., Krause E. (2003) Analysis of class I phosphoinositide 3-kinase autophosphorylation sites by mass spectrometry. Rapid Commun. Mass Spectrom. 17, 690–696 [DOI] [PubMed] [Google Scholar]
- 32. Czupalla C., Mansukoski H., Riedl T., Thiel D., Krause E., Hoflack B. (2006) Proteomic analysis of lysosomal acid hydrolases secreted by osteoclasts. Implications for lytic enzyme transport and bone metabolism. Mol. Cell. Proteomics 5, 134–143 [DOI] [PubMed] [Google Scholar]
- 33. Schägger H., Cramer W. A., von Jagow G. (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217, 220–230 [DOI] [PubMed] [Google Scholar]
- 34. Schägger H., Pfeiffer K. (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19, 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith C. P., Thorsness P. E. (2008) The molecular basis for relative physiological functionality of the ADP/ATP carrier isoforms in Saccharomyces cerevisiae. Genetics 179, 1285–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zara V., Dietmeier K., Palmisano A., Vozza A., Rassow J., Palmieri F., Pfanner N. (1996) Yeast mitochondria lacking the phosphate carrier/p32 are blocked in phosphate transport but can import preproteins after regeneration of a membrane potential. Mol. Cell. Biol. 16, 6524–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Palmieri L., Lasorsa F. M., De Palma A., Palmieri F., Runswick M. J., Walker J. E. (1997) Identification of the yeast ACR1 gene product as a succinate-fumarate transporter essential for growth on ethanol or acetate. FEBS Lett. 417, 114–118 [DOI] [PubMed] [Google Scholar]
- 38. Tatsuta T., Model K., Langer T. (2005) Formation of membrane-bound ring complexes by prohibitins in mitochondria. Mol. Biol. Cell 16, 248–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nijtmans L. G., de Jong L., Artal Sanz M., Coates P. J., Berden J. A., Back J. W., Muijsers A. O., van der Spek H., Grivell L. A. (2000) Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 19, 2444–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Luttik M. A., Overkamp K. M., Kötter P., de Vries S., van Dijken J. P., Pronk J. T. (1998) The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J. Biol. Chem. 273, 24529–24534 [DOI] [PubMed] [Google Scholar]
- 41. Cronin S. R., Rao R., Hampton R. Y. (2002) Cod1p/Spf1p is a P-type ATPase involved in ER function and Ca2+ homeostasis. J. Cell Biol. 157, 1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Becker T., Vögtle F. N., Stojanovski D., Meisinger C. (2008) Sorting and assembly of mitochondrial outer membrane proteins. Biochim. Biophys. Acta 1777, 557–563 [DOI] [PubMed] [Google Scholar]
- 43. Ryan M. T., Wagner R., Pfanner N. (2000) The transport machinery for the import of preproteins across the outer mitochondrial membrane. Int. J. Biochem. Cell Biol. 32, 13–21 [DOI] [PubMed] [Google Scholar]
- 44. Cooper C. E., Nicholls P., Freedman J. A. (1991) Cytochrome c oxidase. Structure, function, and membrane topology of the polypeptide subunits. Biochem. Cell Biol. 69, 586–607 [DOI] [PubMed] [Google Scholar]
- 45. McAlister L., Holland M. J. (1985) Differential expression of the three yeast glyceraldehyde-3-phosphate dehydrogenase genes. J. Biol. Chem. 260, 15019–15027 [PubMed] [Google Scholar]
- 46. Przybyla-Zawislak B., Dennis R. A., Zakharkin S. O., McCammon M. T. (1998) Genes of succinyl-CoA ligase from Saccharomyces cerevisiae. Eur. J. Biochem. 258, 736–743 [DOI] [PubMed] [Google Scholar]
- 47. Carr-Schmid A., Durko N., Cavallius J., Merrick W. C., Kinzy T. G. (1999) Mutations in a GTP-binding motif of eukaryotic elongation factor 1A reduce both translational fidelity and the requirement for nucleotide exchange. J. Biol. Chem. 274, 30297–30302 [DOI] [PubMed] [Google Scholar]
- 48. Charizanis C., Juhnke H., Krems B., Entian K. D. (1999) The mitochondrial cytochrome c peroxidase Ccp1 of Saccharomyces cerevisiae is involved in conveying an oxidative stress signal to the transcription factor Pos9 (Skn7). Mol. Gen. Genet. 262, 437–447 [DOI] [PubMed] [Google Scholar]
- 49. Cardona F., Orozco H., Friant S., Aranda A., del Olmo M. (2011) The Saccharomyces cerevisiae flavodoxin-like proteins Ycp4 and Rfs1 play a role in stress response and in the regulation of genes related to metabolism. Arch. Microbiol. 193, 515–525 [DOI] [PubMed] [Google Scholar]
- 50. Lee J., Godon C., Lagniel G., Spector D., Garin J., Labarre J., Toledano M. B. (1999) Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274, 16040–16046 [DOI] [PubMed] [Google Scholar]
- 51. Braun H. P., Schmitz U. K. (1995) Are the “core” proteins of the mitochondrial bc1 complex evolutionary relics of a processing protease? Trends Biochem. Sci. 20, 171–175 [DOI] [PubMed] [Google Scholar]
- 52. Zhang X., Lester R. L., Dickson R. C. (2004) Pil1p and Lsp1p negatively regulate the 3-phosphoinositide-dependent protein kinase-like kinase Pkh1p and downstream signaling pathways Pkc1p and Ypk1p. J. Biol. Chem. 279, 22030–22038 [DOI] [PubMed] [Google Scholar]
- 53. Zahedi R. P., Sickmann A., Boehm A. M., Winkler C., Zufall N., Schönfisch B., Guiard B., Pfanner N., Meisinger C. (2006) Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell 17, 1436–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Azuma M., Kabe Y., Kuramori C., Kondo M., Yamaguchi Y., Handa H. (2008) Adenine nucleotide translocator transports heme precursors into mitochondria. PLoS ONE 3, e3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tzagoloff A., Jang J., Glerum D. M., Wu M. (1996) FLX1 codes for a carrier protein involved in maintaining a proper balance of flavin nucleotides in yeast mitochondria. J. Biol. Chem. 271, 7392–7397 [DOI] [PubMed] [Google Scholar]
- 56. Bafunno V., Giancaspero T. A., Brizio C., Bufano D., Passarella S., Boles E., Barile M. (2004) Riboflavin uptake and FAD synthesis in Saccharomyces cerevisiae mitochondria: involvement of the Flx1p carrier in FAD export. J. Biol. Chem. 279, 95–102 [DOI] [PubMed] [Google Scholar]
- 57. Pallotta M. L., Brizio C., Fratianni A., De Virgilio C., Barile M., Passarella S. (1998) Saccharomyces cerevisiae mitochondria can synthesize FMN and FAD from externally added riboflavin and export them to the extramitochondrial phase. FEBS Lett. 428, 245–249 [DOI] [PubMed] [Google Scholar]
- 58. Eide D. J. (2006) Zinc transporters and the cellular trafficking of zinc. Biochim. Biophys. Acta 1763, 711–722 [DOI] [PubMed] [Google Scholar]
- 59. Mannella C. A. (1984) Phospholipase-induced crystallization of channels in mitochondrial outer membranes. Science 224, 165–166 [DOI] [PubMed] [Google Scholar]
- 60. Gonçalves R. P., Buzhynskyy N., Prima V., Sturgis J. N., Scheuring S. (2007) Supramolecular assembly of VDAC in native mitochondrial outer membranes. J. Mol. Biol. 369, 413–418 [DOI] [PubMed] [Google Scholar]
- 61. Zalk R., Israelson A., Garty E. S., Azoulay-Zohar H., Shoshan-Barmatz V. (2005) Oligomeric states of the voltage-dependent anion channel and cytochrome c release from mitochondria. Biochem. J. 386, 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hoogenboom B. W., Suda K., Engel A., Fotiadis D. (2007) The supramolecular assemblies of voltage-dependent anion channels in the native membrane. J. Mol. Biol. 370, 246–255 [DOI] [PubMed] [Google Scholar]
- 63. Peng S., Blachly-Dyson E., Colombini M., Forte M. (1992) Determination of the number of polypeptide subunits in a functional VDAC channel from Saccharomyces cerevisiae. J. Bioenerg. Biomembr. 24, 27–31 [DOI] [PubMed] [Google Scholar]
- 64. Rostovtseva T. K., Liu T. T., Colombini M., Parsegian V. A., Bezrukov S. M. (2000) Positive cooperativity without domains or subunits in a monomeric membrane channel. Proc. Natl. Acad. Sci. U.S.A. 97, 7819–7822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. De Pinto V., Reina S., Guarino F., Messina A. (2008) Structure of the voltage-dependent anion channel. State of the art. J. Bioenerg. Biomembr. 40, 139–147 [DOI] [PubMed] [Google Scholar]
- 66. Berger K. H., Yaffe M. P. (1998) Prohibitin family members interact genetically with mitochondrial inheritance components in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 4043–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Brandina I., Graham J., Lemaitre-Guillier C., Entelis N., Krasheninnikov I., Sweetlove L., Tarassov I., Martin R. P. (2006) Enolase takes part in a macromolecular complex associated to mitochondria in yeast. Biochim. Biophys. Acta 1757, 1217–1228 [DOI] [PubMed] [Google Scholar]
- 68. Claypool S. M. (2009) Cardiolipin, a critical determinant of mitochondrial carrier protein assembly and function. Biochim. Biophys. Acta 1788, 2059–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Claypool S. M., Oktay Y., Boontheung P., Loo J. A., Koehler C. M. (2008) Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J. Cell Biol. 182, 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Claypool S. M., Boontheung P., McCaffery J. M., Loo J. A., Koehler C. M. (2008) The cardiolipin transacylase, tafazzin, associates with two distinct respiratory components providing insight into Barth syndrome. Mol. Biol. Cell 19, 5143–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Grandier-Vazeille X., Bathany K., Chaignepain S., Camougrand N., Manon S., Schmitter J. M. (2001) Yeast mitochondrial dehydrogenases are associated in a supramolecular complex. Biochemistry 40, 9758–9769 [DOI] [PubMed] [Google Scholar]
- 72. Ohba M., Schatz G. (1987) Protein import into yeast mitochondria is inhibited by antibodies raised against 45-kDa proteins of the outer membrane. EMBO J. 6, 2109–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Culty M., Li H., Boujrad N., Amri H., Vidic B., Bernassau J. M., Reversat J. L., Papadopoulos V. (1999) In vitro studies on the role of the peripheral type benzodiazepine receptor in steroidogenesis. J. Steroid Biochem. Mol. Biol. 69, 123–130 [DOI] [PubMed] [Google Scholar]
- 74. Gavish M., Bachman I., Shoukrun R., Katz Y., Veenman L., Weisinger G., Weizman A. (1999) Enigma of the peripheral benzodiazepine receptor. Pharmacol. Rev. 51, 629–650 [PubMed] [Google Scholar]
- 75. Lacapère J. J., Papadopoulos V. (2003) Peripheral type benzodiazepine receptor. Structure and function of a cholesterol-binding protein in steroid and bile acid biosynthesis. Steroids 68, 569–585 [DOI] [PubMed] [Google Scholar]
- 76. Papadopoulos V., Baraldi M., Guilarte T. R., Knudsen T. B., Lacapère J. J., Lindemann P., Norenberg M. D., Nutt D., Weizman A., Zhang M. R., Gavish M. (2006) Translocator protein (18 kDa). New nomenclature for the peripheral type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol. Sci. 27, 402–409 [DOI] [PubMed] [Google Scholar]
- 77. Joseph-Liauzun E., Delmas P., Shire D., Ferrara P. (1998) Topological analysis of the peripheral benzodiazepine receptor in yeast mitochondrial membranes supports a five-transmembrane structure. J. Biol. Chem. 273, 2146–2152 [DOI] [PubMed] [Google Scholar]
- 78. Vanhee C., Guillon S., Masquelier D., Degand H., Deleu M., Morsomme P., Batoko H. (2011) A TSPO-related protein localizes to the early secretory pathway in Arabidopsis but is targeted to mitochondria when expressed in yeast. J. Exp. Bot. 62, 497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kinnally K. W., Zorov D. B., Antonenko Y. N., Snyder S. H., McEnery M. W., Tedeschi H. (1993) Mitochondrial benzodiazepine receptor linked to inner membrane ion channels by nanomolar actions of ligands. Proc. Natl. Acad. Sci. U.S.A. 90, 1374–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. McEnery M. W., Snowman A. M., Trifiletti R. R., Snyder S. H. (1992) Isolation of the mitochondrial benzodiazepine receptor. Association with the voltage-dependent anion channel and the adenine nucleotide carrier. Proc. Natl. Acad. Sci. U.S.A. 89, 3170–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Delavoie F., Li H., Hardwick M., Robert J. C., Giatzakis C., Péranzi G., Yao Z. X., Maccario J., Lacapère J. J., Papadopoulos V. (2003) In vivo and in vitro peripheral type benzodiazepine receptor polymerization. Functional significance in drug ligand and cholesterol binding. Biochemistry 42, 4506–4519 [DOI] [PubMed] [Google Scholar]
- 82. Ostuni M. A., Péranzi G., Ducroc R. A., Fasseu M., Vidic B., Dumont J., Papadopoulos V., Lacapere J. J. (2009) Distribution, pharmacological characterization, and function of the 18-kDa translocator protein in rat small intestine. Biol. Cell 101, 573–586 [DOI] [PubMed] [Google Scholar]
- 83. Taylor A. B., Smith B. S., Kitada S., Kojima K., Miyaura H., Otwinowski Z., Ito A., Deisenhofer J. (2001) Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure 9, 615–625 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.