Background: A universal mechanism for the secretion ATPase superfamily has been proposed.
Results: Magnesium regulates the catalytic cycle of the type IV secretion ATPase TrwD.
Conclusion: Physiological magnesium concentrations maintain the enzyme at slow ATP turnover, preventing a futile ATP hydrolysis cycle.
Significance: The regulatory role of magnesium provides new insights into the catalytic mechanism of the secretion ATPase superfamily.
Keywords: ATPases, Bacterial Conjugation, Enzyme Mechanisms, Magnesium, Secretion
Abstract
TrwD, the VirB11 homologue in conjugative plasmid R388, is a member of the large secretion ATPase superfamily, which includes ATPases from bacterial type II and type IV secretion systems, type IV pilus, and archaeal flagellae assembly. Based on structural studies of the VirB11 homologues in Helicobacter pylori and Brucella suis and the archaeal type II secretion ATPase GspE, a unified mechanism for the secretion ATPase superfamily has been proposed. Here, we have found that the ATP turnover of TrwD is down-regulated by physiological concentrations of magnesium. This regulation is exerted by increasing the affinity for ADP, hence delaying product release. Circular dichroism and limited proteolysis analysis indicate that magnesium induces conformational changes in the protein that promote a more rigid, but less active, form of the enzyme. The results shown here provide new insights into the catalytic mechanism of the secretion ATPase superfamily.
Introduction
Type IV secretion systems (T4SSs)6 are used by Gram-negative bacteria to transport DNA between bacteria and protein effectors into host cells (1). T4SSs are macromolecular assemblies formed at least by 12 different proteins, named after their homologues in Agrobacterium tumefaciens: VirB1–11 and VirD4 (2). Three of these proteins (VirD4, VirB4, and VirB11) are hexameric ATPases that provide the energy for substrate transport and for T4SS biogenesis (3–6). VirB11 proteins are thought to belong to a large family of AAA+ hexameric traffic ATPases, which also includes ATPases from type II secretion systems, type IV pili and archaeal flagellar biogenesis systems (7). The atomic structures of two nonconjugative VirB11 homologues, Helicobacter pylori HP0525 (8, 9) and Brucella suis VirB11 (10), revealed that these proteins form hexameric ring structures in which the N-terminal domain likely interacts with the bacterial membrane, whereas the C-terminal domain, containing a RecA-like domain, is involved in ATP hydrolysis. Although there is not a crystal structure of TrwD, which is the VirB11 homolog in plasmid R388, electron microscopy analysis of this protein revealed that it also forms hexameric ring assemblies (11).
Despite the structural information, little is known about the structure-function relationship and the biological function of the different components of T4SSs. In an effort to gain a further biological understanding of T4SSs, we have characterized the biochemical and genetic properties of the conjugative ATPases of the model plasmid R388 (3, 4, 6, 12, 13). VirB11 is thought to play a chaperone role in the traffic of effectors through T4SSs, using energy from ATP hydrolysis to assemble/disassemble type IV components for biogenesis of the secretion system and/or for substrate translocation across the membrane (14). However, this role, inferred mainly through structural and comparative genomics analysis, is not matched by biochemical evidence. Therefore, the purpose of this work is to provide biochemical evidence that supports the functional placement of VirB11 proteins among the AAA+ traffic ATPase superfamily. To this end, we developed a new purification protocol that renders pure TrwD protein with much higher ATP hydrolase rates than in any previously reported VirB11 analysis (6, 11, 15). Interestingly, we found that the ATP turnover of the enzyme is decreased in the presence of physiological concentrations of Mg2+. Inhibition is not coupled to ATP concentration but to an increase in the affinity for ADP, which in turn results in a delay of product release. Thermal denaturation analysis by circular dichroism spectroscopy showed that magnesium is able to induce conformational changes to the apo-, nucleotide-free form of the enzyme, resulting in a cooperative transition. Limited proteolysis with papain allowed us to discriminate a region within the C terminus of the protein that could promote the closing and aperture of the nucleotide binding pocket. Altogether, these observations could help to gain a further understanding of the catalytic mechanism of this large secretion ATPase superfamily.
EXPERIMENTAL PROCEDURES
Cloning
R388 trwD gen was cloned by PCR using plasmid R388 as template and primers AAAATCATATGTCTACAGTCTCGAAAGC and AAAATGGATCCTAAGCATCTTGGAC (forward and reverse, respectively). The resulting 1,076-bp fragment was digested with NdeI and BamHI and inserted into the same sites of the pET3a expression vector (Novagen, Madison, WI).
Overexpression and Purification
TrwD was expressed into Escherichia coli C41(DE3) strain (16). Cells were isolated and solubilized as reported (4). Lysates were precipitated in a saturated ammonium sulfate solution (60% w/v) and centrifuged at 100,000 × g for 30 min at 4 °C. Pellets were resuspended in buffer A (20 mm Hepes/NaOH, pH 6.8, 0.1 m NaCl, 10% glycerol (w/v), 1 mm DTT, 1 mm PMSF), dialyzed, and applied to a HiTrap Q-Sepharose (5-ml) column (GE Healthcare). The flow-through was dialyzed in buffer A at pH 7.6 and applied to a Resource Q-Sepharose (6-ml) column. Protein was eluted in a linear gradient of NaCl, concentrated, and loaded onto a HiLoad 16/60 Superdex 200 column. Fractions were eluted in 20 mm Hepes/NaOH, pH 7.6, 0.2 mm NaCl, 1 mm PMSF, and 5% glycerol (w/v) and stored at −80 °C.
ATP Hydrolysis Assays
ATP hydrolysis activity was measured by using the EnzCheckTM kit (Invitrogen), in a buffer consisting of 50 mm Tris-HCl, pH 8.5, 75 mm potassium acetate, and 10% glycerol (w/v). The reactions were started by the addition of TrwD. ATP and Mg2+ were added at the concentrations indicated in the text, prior to the initiation of the reaction by the addition of TrwD.
Fluorescence Measurements
Nucleotide binding to TrwD was characterized using TNP-ATP, a fluorescent analog of ATP (Molecular Probes, Inc.) (17). Fluorescence emission spectra were recorded at 21 °C using a PerkinElmer Life Sciences MPF-66 spectrofluorometer with the excitation wavelength set at 407 nm and the emission wavelength scanned from 465 to 625 nm. TNP-ATP and TrwD were added at the indicated concentrations in 20 mm Hepes (pH 7.6), 0.2 m NaCl, 5% glycerol (w/v), and 10 μm or 2 mm magnesium acetate.
Circular Dichroism Measurements
Thermal stability experiments were made in the presence or the absence of 2 mm magnesium acetate. Ellipticity was measured at 222 nm on a JASCO (Tokyo, Japan) J-810 spectropolarimeter in the temperature range 20–80 °C at a rate of 60 °C/h, using cells with a 0.1-cm path length. TrwD was added at 5 μm in 50 mm Hepes (pH 8.5), 75 mm potassium acetate, 5% glycerol (w/v) without added magnesium. Magnesium acetate (2 mm) or MgADP (1 mm) was added to the buffer accordingly. Following the acquisition of the measurements at 222 nm, the signal was subtracted from the buffer, and the ellipticity θ (millidegrees) was converted to a residue molar ellipticity [θ] (degree cm2 dmol−1).
Electron Microscopy and Image Analysis
Aliquots (5 μl) of TrwD in the presence of 2 mm magnesium and in its absence were applied to glow-discharged carbon-coated grids and stained for 1 min with 2% uranyl acetate. Images were recorded on Kodak SO163 films at ×60,000 nominal magnification in a JEOL 1200EX-II electron microscope operated at 100 KV. Micrographs were digitized in a Zeiss SCAI scanner with a final sampling rate of 2.33 Å/pixel. Individual particles of TrwD were selected and normalized using the XMIPP image processing software (18). Alignment and classification were performed by maximum likelihood multireference refinement methods.
Papain Proteolysis
Papain digestion was performed at 25 °C in 20 mm Tris (pH 8.5), 75 mm potassium acetate. Papain stocks were dissolved in the same buffer and activated by the addition of 50 mm β-mercaptoethanol (37 °C, 30 min) just before use. TrwD (25 μm) was incubated for 15 min at 25 °C with corresponding concentrations of nucleotide and magnesium according to reaction. Proteolysis was initiated by the addition of papain from the activated stocks at 1:100 papain:TrwD molar ratios. The reaction was stopped after 90 min by the addition of 100 μm E-64 inhibitor (Sigma). Proteolysis products were analyzed by SDS-PAGE (15% polyacrylamide gels) followed by staining with Coomassie Brilliant Blue. Fragments were transferred to an Immobilon-P membrane (Millipore), and N-terminal sequencing determination of proteolytic products was performed by using a Procise 494 automated sequencer (Applied Biosystems).
Molecular Modeling
An atomic model of TrwD was generated by molecular threading using the protein homology and recognition engine Phyre (19) taking the atomic coordinates of B. suis VirB11 (2gza.pdb) (10) as template. Based on the hexameric structure of VirB11, a model of TrwD hexamer was built using the UCSF Chimera package (20).
RESULTS
TrwD ATPase Activity Is Regulated by Magnesium
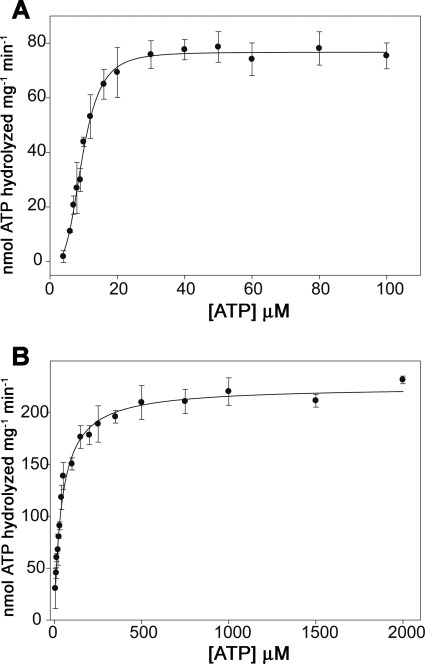
ATPase activity of TrwD was previously reported using a GST-TrwD fusion protein (6). However, because the reported ATP hydrolysis rates were low (4.5 nmol ATP min−1 mg−1), we developed a new purification protocol for wild type TrwD. This new protocol rendered a high yield of pure and homogeneous protein (supplemental Fig. 1). The steady-state kinetic parameters of ATP turnover by TrwD in the presence of 2 mm Mg2+ were 76 nmol min−1 mg−1 (Vmax) and 9.6 μm (Km(app)[ATP]) (Fig. 1A). Surprisingly, at low Mg2+ concentration (10 μm), the ATP turnover was 3-fold higher (216 nmol min−1 mg−1) than that at 2 mm Mg2+ (Fig. 1B), with a significant increase in the Km(app)[ATP] value (45 μm). As expected, the addition of EDTA resulted in a complete inhibition of TrwD ATPase activity.
FIGURE 1.
Effect of Mg2+ on TrwD ATPase turnover. A and B, ATPase activity of TrwD (1 μm as monomer) was measured at increasing concentrations of ATP in the presence of 2 mm Mg2+ (A) or 10 μm Mg2+ (B). Data were fitted to the Hill equation (Equation 1 in the supplemental material). Error bars in both panels indicate S.D.
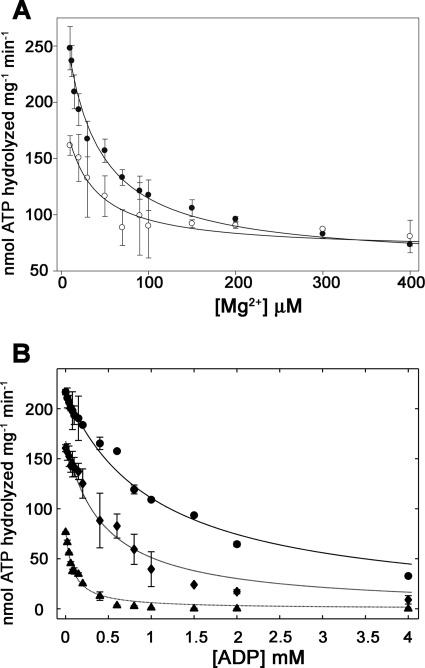
Further analysis of the effect of Mg2+ on TrwD ATP hydrolysis revealed that inhibition by Mg2+ was not affected by ATP concentration (Fig. 2A). The calculated Ki(app)[Mg2+] values were 42 and 35 μm for 1 mm and 100 μm ATP, respectively. Considering the affinity of Mg2+ for ATP (Kd[MgATP] = 20 μm) (21), it would be expected that all Mg2+ was bound to the nucleotide at 1 mm ATP. However, this result indicates that Mg2+ binds the protein with high affinity, which suggests that free Mg2+, rather than MgATP, is responsible of the observed inhibition. Evidence for tight binding of Mg2+ or Mn2+ at or near catalytic sites in the absence of bound nucleotides has been previously reported for other enzymes (22), yet no effect of Mn2+and Zn2+ on TrwD ATPase activity was observed (data not shown).
FIGURE 2.
ATP turnover inhibition by Mg2+ and ADP. A and B, the ATPase activity of TrwD (1.9 μm as a monomer) was measured over a range of Mg2+ (A) and ADP (B) concentrations. A, 1 mm ATP (filled circles) and 100 μm ATP (open circles). B, 1 mm ATP and 10 μm Mg2+ (circles), 100 μm ATP and 10 μm Mg2+ (diamonds), and 100 μm ATP and 2 mm Mg2+ (triangles). Data in A and B were fitted to Equations 2 and 3, respectively (see supplemental material) to determine values for the apparent Ki(app)[Mg2+] in A and the Kd [ADP] and Ki(app) [ADP] in B. Error bars in both panels indicate S.D.
Inhibition of TrwD by ADP Is Modulated by Magnesium
Inhibition of ATP turnover by ADP was studied at low (10 μm) and high (2 mm) Mg2+ concentrations. As expected, under both conditions, ADP exhibited a clear inhibitory effect (Fig. 2B). However, at the same concentration of ATP (100 μm), a 5-fold decrease in the Ki(app)[ADP] value was observed in the presence of 2 mm Mg2+. Moreover, the affinity of TrwD for ADP, Kd(app)[ADP], calculated at different concentrations of ATP and Mg2+, revealed that in the presence of 2 mm Mg2+, the affinity for ADP was increased by ∼20-fold. Therefore, it is likely that Mg2+ inhibition is caused by stabilization of the MgADP inhibited state.
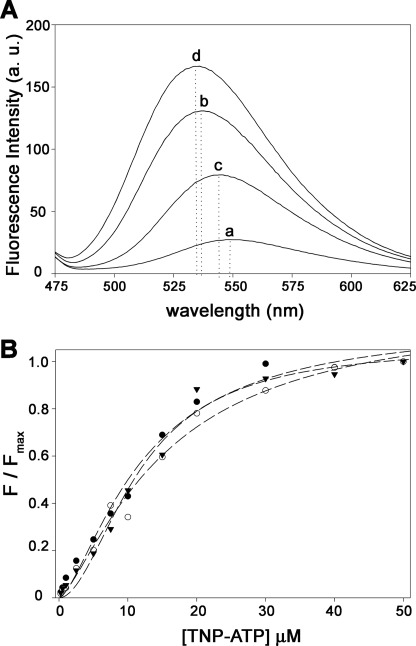
Binding Affinity of TrwD for TNP-ATP Is Not Affected by Magnesium
Nucleotide binding to TrwD was characterized using TNP-ATP (Fig. 3), a fluorescent analog of ATP (17), widely used to characterize binding interactions of ATP with numerous nucleotide-binding proteins (23–25). TNP-ATP is not hydrolyzable by TrwD under the same conditions used for ATP (data not shown). Binding of TNP-ATP to TrwD (5 μm) at different Mg2+ concentrations (Fig. 3B) revealed that the affinity of TNP-ATP for TrwD was not significantly affected, the KdTNP-ATP values being 11 and 9.7 μm in the presence of 2 mm Mg2+ and in the absence of added Mg2+, respectively. Interestingly, the addition of Mg2+ not only resulted in a reduction of fluorescence emission intensity but also in a redshift of the wavelength of maximal emission (Fig. 3A), suggesting that binding of free Mg2+ could induce a conformational change in TrwD.
FIGURE 3.
Effect of Mg2+ on TNP-ATP binding to TrwD. A, fluorescent emission spectra of TNP-ATP (10 μm) in the absence (spectrum a) and in the presence of 5 μm TrwD (spectrum b). Consecutive addition of Mg2+ (2 mm) and EDTA (10 mm) is shown in spectra c and d, respectively. Dotted lines indicate wavelength of maximal emission in each case. a. u., arbitrary units. B, titration curve of TNP-ATP binding to 5 μm TrwD in the absence of Mg2+ (filled triangles) and in the presence of 10 μm (open circles) and 2 mm Mg2+ (filled circles). Titration curves at each magnesium concentrations were normalized to the maximum value of fluorescence emission intensity for each case. (λex = 407 nm).
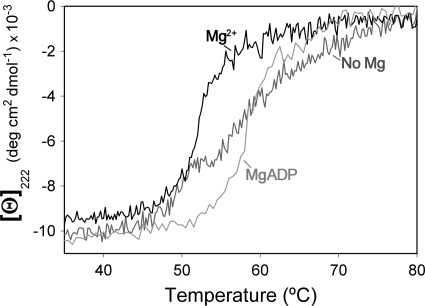
Magnesium Promotes a Conformational Change in TrwD, Reflected in the Thermal Unfolding Profiles Measured by Circular Dichroism
To investigate whether magnesium was exerting its inhibitory effect by inducing a conformational change in TrwD, thermal denaturation curves were obtained by measuring protein ellipticity at 222 nm at increasing temperatures. The thermal unfolding profiles of the protein in the absence or presence of magnesium were significantly different (Fig. 4). In the absence of 2 mm Mg2+, a broad transition between 45 and 75 °C was observed, reflecting denaturation process with low cooperativity. In contrast, binding of magnesium resulted in a highly cooperative denaturation transition centered at 52 °C. In the presence of MgADP, a stabilization of the protein is observed, being the denaturation transition cooperative and centered at 58 °C. These results suggest that both Mg2+ and MgADP induce a conformational change in TrwD that results in a lower ATP turnover.
FIGURE 4.
Thermal denaturation of TrwD followed by CD. Temperature dependence of the molar ellipticity at 222 nm of a sample containing 5 μm TrwD in 50 mm Hepes buffer (pH 8.5) without added magnesium (dark gray) and with 2 mm magnesium acetate (black) or 1 mm MgADP (gray), heated from 20 to 80 °C in a 0.1-cm path length cuvette at a rate of 60 °C/h, deg, degree.
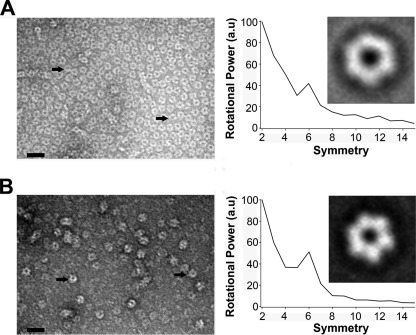
Electron Microscopy
Image analysis of TrwD by electron microscopy revealed that TrwD is able to form hexameric ring structures in the absence of added magnesium (Fig. 5A). A similar proportion of ring structures was also visible in the presence of magnesium (Fig. 5B) and magnesium and nucleotides (data not shown), which indicated that the conformational change induced by magnesium was not affecting the overall oligomeric state of the protein. Interestingly, despite the low resolution, a close inspection of the EM images shows that the inner diameter of the ring is smaller in the presence of magnesium than in its absence, suggesting that magnesium induces a closed conformation of the ring.
FIGURE 5.
Electron microscopy of TrwD oligomers. A and B, protein in 50 mm Hepes buffer (pH 8.5) without added Mg2+ (A) and with 2 mm Mg2+ (B) was stained with uranyl acetate and analyzed by electron microscopy (scale bar, 35 nm). Right panel, rotational power spectra of class averages (106 and 58 particles in A and B, respectively) obtained upon self-organizing neuronal maps classification. a. u., arbitrary units.
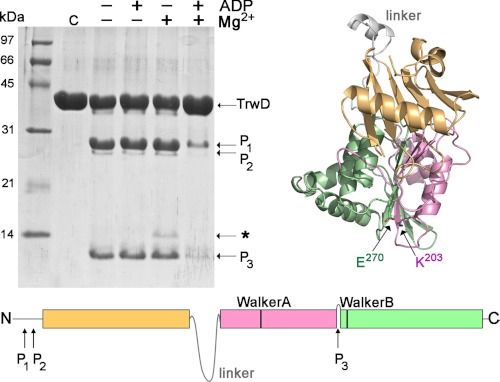
Partial Proteolysis of TrwD
The unspecific protease papain is useful for delimiting structural domains in proteins because it presents a low activity on stable secondary and tertiary structures (26). Therefore, we decided to investigate whether magnesium and/or nucleotides stabilized TrwD conformation by its susceptibility to papain digestion. Fig. 6 shows that in the presence of MgADP, TrwD was protected of papain proteolysis. In its absence, papain digestion resulted in two main bands with molecular masses of roughly 25 and 11 kDa, respectively. These bands were transferred to an Immobilon-P membrane and subjected to N-terminal sequencing determination. The 25-kDa band with an N terminus sequence 12SGNRV16 corresponded to TrwD protein lacking the first 11 amino acid residues (probably by unspecific proteolytic degradation). Running just below this 25-kDa band, another fragment with a N-terminal sequence 18KDQAV22 could be identified. The 11-kDa band at the bottom of the gel has a, N-terminal sequence 257MRQS260, corresponding to the C-terminal end of the protein. This proteolytic cleavage site is located in a loop connecting the Walker A and Walker B domains (Fig. 6 and supplemental Fig. 2). On the hexameric model of TrwD (Fig. 7), this 11-kDa fragment corresponds to a region of the protein facing the interior of the ring. Homologue sequences of this C-terminal region on HP0525 and VirB11 crystal structures show similar disposition within the hexameric rings (Fig. 7). Binding of ADP and magnesium to the protein induces a reorganization of the C-terminal domain (CTD) that impedes papain access to this protein region. This observation is in good agreement with the cooperativity and stabilization of the protein observed by CD. In the absence of nucleotides, and in the presence of magnesium, a weak signal for a band of ∼14 kDa could be observed (marked in Fig. 6 with *). N-terminal sequencing of this band, which was not present in any of the other conditions, resulted in a sequence consistent with a fragment of the protein starting at 210LIEK213, a few residues downstream of the Walker A motif (Lys203). Papain shows very low activity at this site, but the experiment clearly reflects that it is not accessible in the absence of magnesium. This result is in accordance with the CD experiments in which Mg2+ is able to induce conformational changes in the absence of nucleotides.
FIGURE 6.
Papain partial proteolysis of TrwD. Upper left panel, Coomassie Brilliant Blue-stained SDS-PAGE of papain digestion products of TrwD (25 μm) in the absence or presence of nucleotides and Mg2+. Papain:TrwD molar ratio was 1:100, and digestion was performed for 90 min at 25 °C. Bottom panel, papain digestion sites that give rise to P1, P2, and P3 fragments, identified by N-terminal sequencing, are indicated at their corresponding positions. An additional fragment could be appreciated in the condition with only Mg2+ and no nucleotides (marked with *). This peptide, with low signal, started 48 residues upstream of P3. Upper right panel, graphic representation of a TrwD monomer (modeled as explained under “Experimental Procedures”). The P3 cleavage site separates the last 96 residues of the protein, containing the Walker B motif (Glu270) from the rest of the CTD, which contains the Walker A motif (Lys203).
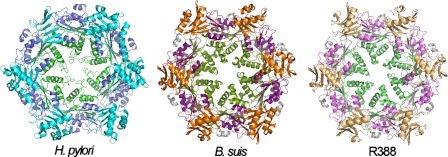
FIGURE 7.
Hexameric structures of VirB11 proteins. The hexameric structures of H. pylori HP0525 (1nlz.pdb), B. suis VirB11 (2gza.pdb), and R388 TrwD (see “Experimental Procedures”) are shown. Each monomer consists of an NTD (cyan, orange, and wheat, respectively) and a CTD (blue, magenta, and pink, respectively), connected by a flexible linker (gray). The region at the C-terminal end identified by papain proteolysis in TrwD (green) and the equivalent positions in B. suis VirB11 (olive) and HP0525 (lime) is facing the interior of the hexameric rings.
DISCUSSION
Based on the crystal structures of the H. pylori and B. suis VirB11 homologues from type IV secretion systems (HP0525 and VirB11, respectively) (8–10) and type II secretion GspE protein of Archaeoglobus fulgidus (afGspE) (27), an universal catalytic mechanism for the secretion ATPase superfamily has been proposed (27). However, biochemical experiments to support this model were still lacking. In this work, we provide new insights into the mode of action of this protein family, using TrwD, a VirB11 homologue of the conjugative plasmid R388, as a working model.
The kinetic parameters of ATP hydrolysis at steady-state rates reveal that physiological concentrations of Mg2+ (∼1 mm in bacterial cells (28)) down-regulate the ATP turnover of the enzyme by increasing the affinity for ADP. Inhibition by magnesium has been reported for a large variety of ATPases (25, 29–32). Specially relevant to this work are the reports of ATP hydrolysis inhibition by Mg2+ of SecA (33, 34), a ubiquitous traffic ATPase. Binding of Mg2+ to an allosteric site has been proposed to be a key regulatory step of the catalytic cycle of SecA. Gold et al. (33) reached this conclusion based on the fact that inhibition was not coupled to ATP concentration as the inhibitory effect of Mg2+ remained when ATP was more than 500 times in excess. Likewise, here we show that Mg2+ inhibition of TrwD ATPase activity is not coupled to ATP concentration. Therefore, our data also suggest the existence of a binding site for free Mg2+ different from the nucleotide binding site. However, alternative models that do not require an allosteric binding site could also be compatible with the data. A mechanism in which Mg2+ stabilizes the MgADP form of the enzyme, slowing down turnover, concomitant with a decrease in Km, could also be envisaged.
The effect of Mg2+ and nucleotides on TrwD was further investigated. The affinity of the enzyme for TNP-ATP, a fluorescent analog of ATP, was not affected by the presence of Mg2+ in the buffer. However, the wavelength shift and the variations in fluorescence intensity suggested that Mg2+ could be inducing some kind of conformational change in the enzyme. To confirm this, the effect of Mg2+ and nucleotides on the thermal stability of the enzyme was tested by circular dichroism spectroscopy. We found an increase in the cooperativity of TrwD thermal denaturation profile in the presence of physiological concentrations of Mg2+. These results demonstrate that free Mg2+ is able to induce conformational changes on the protein in the absence of added nucleotides. The addition of nucleotides promotes a stabilization of the enzyme, with an increase of 6 °C in the Tm value. This is compatible with a lower exposure of the C-terminal domain of the protein, which contains the nucleotide binding site, as observed by papain proteolysis.
VirB11 proteins share a common domain organization consisting of two domains, an N-terminal domain (NTD) and an ATPase catalytic CTD, connected by a flexible linker. Comparison of the crystal structures of HP0525 and VirB11 revealed a large domain swap of the NTD pivoting about the linker region over the CTD that does not affect the overall hexameric assembly. Hence, a nucleotide-dependent domain swap has been proposed to be part of the catalytic mechanism of the enzyme (10). Here, we have found a region within the CTD suitable to partial proteolysis in the absence of nucleotides and Mg2+. The obtained fragment corresponds to the last 96 residues of the protein. This region faces the internal channel of the hexameric ring (Fig. 7). Because this band is not observed in the presence of Mg2+ and nucleotides, it is clear that the binding induces a conformational change in the C terminus of the protein that promotes a closed conformation of the enzyme, leaving the proteolysis site inaccessible. This is a novel finding that has not been reflected in any of the published models, providing new insights into the regulatory mechanism of the secretion ATPase superfamily, especially regarding the role of Mg2+ in the catalytic cycle.
In summary, our data show the following. (i) At physiological concentrations of Mg2+, similar to the free Mg2+ found in bacteria (28), the ATP turnover and the Km of the reaction decrease, and (ii) the affinity for ADP increases dramatically; (iii) Mg2+ is able to induce conformational changes in the protein in the absence of nucleotides; (iv) in the presence of Mg2+ and nucleotides, the protein undergoes further conformational changes that make it more stable and less sensitive to papain degradation; and (v) the C-terminal end of the protein, which faces the inner channel in the ring, is a flexible domain that moves upon binding of Mg2+ and nucleotides.
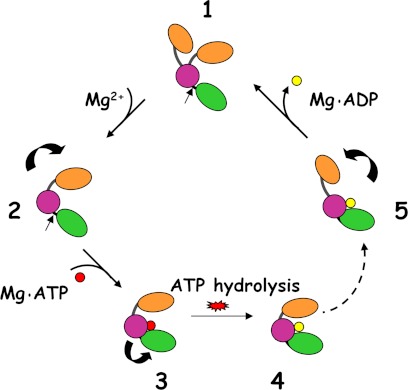
In view of these results and the structural studies of HP0525 (8, 9), VirB11 (10), GspE (27), and other proteins of this family (35, 36), we propose the model shown in Fig. 8. According to this model, the enzyme in the apo state would have high flexibility between the NTD and CTD around its linker (8, 27). At low Mg2+ concentrations, lower than the Kd of Mg2+ for ATP (Kd[MgATP] = 20 μm (21)), the enzyme would bind ATP and Mg2+. Upon ATP hydrolysis and ADP release, the enzyme would return to its initial state. However, at physiological concentrations of Mg2+, the enzyme would be in a closed conformation that would impede ADP release, thus promoting an MgADP-inhibited state of the enzyme. This would imply that under physiological conditions, the enzyme would be inhibited, and only upon a specific signal (for example, substrate binding/release), ADP would be released, thus completing the ATP cycle.
FIGURE 8.
Model of the regulatory mechanism by magnesium of VirB11 ATPases. A proposed catalytic mechanism for VirB11 proteins, adapted from Ref. 8 and extended to the secretion ATPase superfamily (27), taking into consideration the effect of Mg2+, is shown. Step 1, the nucleotide-free apo form would be flexible with the NTD (orange) and CTD (magenta/green) in equilibrium between an open and closed conformation. The papain site limiting the C terminus end (green) is indicated by an arrow. Step 2, high Mg2+ concentrations (similar to the free Mg2+ found in bacteria) would induce a conformational change that would be followed by MgATP binding (Step 3), which, in turn, would promote a conformational change in the CTD (the papain site is no longer accessible). After ATP hydrolysis, an inhibited MgADP state (Step 4) would be stabilized, resulting in an ATP turnover decrease. Upon specific signal (for instance, substrate binding or release), the MgADP-inhibited state would be unlocked (Step 5), and hence, the cycle could resume with ADP release, returning to the initial apo form.
Supplementary Material
Acknowledgments
We thank S. Zunzunegui for excellent technical assistance, Dr. Arturo Muga for helpful discussion and assistance with the stability studies, and Dr. J. Martin-Benito for help with the electron microscopy.
This work was supported by the Ministerio de Ciencia e Innovación (MCINN, Spain) Grant BFU2008-00806 (to E. C. and I. A.), Grant BFU2008-00995/BMC from the MCINN, Grant RD06/0008/1012 from the Instituto de Salud Carlos III, and Grant LSHM-CT-2005_019023 from the European VI Framework Program (to F. d. l. C.), and Grant BFU2010-15443 (to F. M.) from the MCINN.

This article contains supplemental text and equations and Figs. 1 and 2.
- T4SS
- type IV secretion system
- TNP-ATP
- 2′(3′)-O-(2,4,6-trinitrophenyl)adenosine 5′-triphosphate
- CTD
- C-terminal domain
- NTD
- N-terminal domain
- CD
- circular dichroism.
REFERENCES
- 1. Cascales E., Christie P. J. (2003) The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1, 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Christie P. J., Atmakuri K., Krishnamoorthy V., Jakubowski S., Cascales E. (2005) Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59, 451–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tato I., Zunzunegui S., de la Cruz F., Cabezon E. (2005) TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA-dependent ATPase. Proc. Natl. Acad. Sci. U.S.A. 102, 8156–8161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arechaga I., Peña A., Zunzunegui S., del Carmen Fernández-Alonso M., Rivas G., de la Cruz F. (2008) ATPase activity and oligomeric state of TrwK, the VirB4 homologue of the plasmid R388 type IV secretion system. J. Bacteriol. 190, 5472–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peña A., Ripoll-Rozada J., Zunzunegui S., Cabezón E., de la Cruz F., Arechaga I. (2011) Autoinhibitory regulation of TrwK, an essential VirB4 ATPase in type IV secretion systems. J. Biol. Chem. 286, 17376–17382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rivas S., Bolland S., Cabezón E., Goñi F. M., de la Cruz F. (1997) TrwD, a protein encoded by the IncW plasmid R388, displays an ATP hydrolase activity essential for bacterial conjugation. J. Biol. Chem. 272, 25583–25590 [DOI] [PubMed] [Google Scholar]
- 7. Planet P. J., Kachlany S. C., DeSalle R., Figurski D. H. (2001) Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. U.S.A. 98, 2503–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Savvides S. N., Yeo H. J., Beck M. R., Blaesing F., Lurz R., Lanka E., Buhrdorf R., Fischer W., Haas R., Waksman G. (2003) VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 22, 1969–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yeo H. J., Savvides S. N., Herr A. B., Lanka E., Waksman G. (2000) Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol. Cell 6, 1461–1472 [DOI] [PubMed] [Google Scholar]
- 10. Hare S., Bayliss R., Baron C., Waksman G. (2006) A large domain swap in the VirB11 ATPase of Brucella suis leaves the hexameric assembly intact. J. Mol. Biol. 360, 56–66 [DOI] [PubMed] [Google Scholar]
- 11. Krause S., Pansegrau W., Lurz R., de la Cruz F., Lanka E. (2000) Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J. Bacteriol. 182, 2761–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Machón C., Rivas S., Albert A., Goñi F. M., de la Cruz F. (2002) TrwD, the hexameric traffic ATPase encoded by plasmid R388, induces membrane destabilization and hemifusion of lipid vesicles. J. Bacteriol. 184, 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tato I., Matilla I., Arechaga I., Zunzunegui S., de la Cruz F., Cabezon E. (2007) The ATPase activity of the DNA transporter TrwB is modulated by protein TrwA: implications for a common assembly mechanism of DNA translocating motors. J. Biol. Chem. 282, 25569–25576 [DOI] [PubMed] [Google Scholar]
- 14. Yeo H. J., Waksman G. (2004) Unveiling molecular scaffolds of the type IV secretion system. J. Bacteriol. 186, 1919–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rangrez A. Y., Abajy M. Y., Keller W., Shouche Y., Grohmann E. (2010) Biochemical characterization of three putative ATPases from a new type IV secretion system of Aeromonas veronii plasmid pAC3249A. BMC Biochem. 11, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miroux B., Walker J. E. (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 [DOI] [PubMed] [Google Scholar]
- 17. Hiratsuka T., Uchida K. (1973) Preparation and properties of 2′(or 3′)-O-(2,4,6-trinitrophenyl) adenosine 5′-triphosphate, an analog of adenosine triphosphate. Biochim. Biophys. Acta 320, 635–647 [DOI] [PubMed] [Google Scholar]
- 18. Scheres S. H., Núñez-Ramírez R., Sorzano C. O., Carazo J. M., Marabini R. (2008) Image processing for electron microscopy single-particle analysis using XMIPP. Nat. Protoc. 3, 977–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 20. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 21. Pecoraro V. L., Hermes J. D., Cleland W. W. (1984) Stability constants of Mg2+ and Cd2+ complexes of adenine nucleotides and thionucleotides and rate constants for formation and dissociation of MgATP and MgADP. Biochemistry 23, 5262–5271 [DOI] [PubMed] [Google Scholar]
- 22. Buy C., Girault G., Zimmermann J. L. (1996) Metal binding sites of H+-ATPase from chloroplast and Bacillus PS3 studied by EPR and pulsed EPR spectroscopy of bound manganese(II). Biochemistry 35, 9880–9891 [DOI] [PubMed] [Google Scholar]
- 23. Weber J., Senior A. E. (1996) Binding and hydrolysis of TNP-ATP by Escherichia coli F1-ATPase. J. Biol. Chem. 271, 3474–3477 [DOI] [PubMed] [Google Scholar]
- 24. Stewart R. C., VanBruggen R., Ellefson D. D., Wolfe A. J. (1998) TNP-ATP and TNP-ADP as probes of the nucleotide binding site of CheA, the histidine protein kinase in the chemotaxis signal transduction pathway of Escherichia coli. Biochemistry 37, 12269–12279 [DOI] [PubMed] [Google Scholar]
- 25. Jezewska M. J., Lucius A. L., Bujalowski W. (2005) Binding of six nucleotide cofactors to the hexameric helicase RepA protein of plasmid RSF1010. 1. Direct evidence of cooperative interactions between the nucleotide-binding sites of a hexameric helicase. Biochemistry 44, 3865–3876 [DOI] [PubMed] [Google Scholar]
- 26. Karzai A. W., McMacken R. (1996) A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coli DnaK protein. J. Biol. Chem. 271, 11236–11246 [DOI] [PubMed] [Google Scholar]
- 27. Yamagata A., Tainer J. A. (2007) Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. EMBO J. 26, 878–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Froschauer E. M., Kolisek M., Dieterich F., Schweigel M., Schweyen R. J. (2004) Fluorescence measurements of free [Mg2+] by use of mag-fura 2 in Salmonella enterica. FEMS Microbiol Lett. 237, 49–55 [DOI] [PubMed] [Google Scholar]
- 29. Molnár M., Vas M. (1993) Mg2+ affects the binding of ADP but not ATP to 3-phosphoglycerate kinase: correlation between equilibrium dialysis binding and enzyme kinetic data. Biochem. J. 293, 595–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Berger G., Girault G., Galmiche J. M., Pezennec S. (1994) The role of Mg2+ in the hydrolytic activity of the isolated chloroplast ATPase: study by high-performance liquid chromatography. J. Bioenerg. Biomembr. 26, 335–346 [DOI] [PubMed] [Google Scholar]
- 31. Berger G., Girault G. (2001) Comparison of different cations (Mn2+, Mg2+, Ca2+) on the hydrolytic activity of chloroplast ATPase. J. Bioenerg. Biomembr. 33, 93–98 [DOI] [PubMed] [Google Scholar]
- 32. Schobert B. (1998) Do ATP4− and Mg2+ bind stepwise to the F1-ATPase of Halobacterium saccharovorum? Eur. J. Biochem. 254, 363–370 [DOI] [PubMed] [Google Scholar]
- 33. Gold V. A., Robson A., Clarke A. R., Collinson I. (2007) Allosteric regulation of SecA: magnesium-mediated control of conformation and activity. J. Biol. Chem. 282, 17424–17432 [DOI] [PubMed] [Google Scholar]
- 34. Robson A., Gold V. A., Hodson S., Clarke A. R., Collinson I. (2009) Energy transduction in protein transport and the ATP hydrolytic cycle of SecA. Proc. Natl. Acad. Sci. U.S.A. 106, 5111–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Robien M. A., Krumm B. E., Sandkvist M., Hol W. G. (2003) Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae. J. Mol. Biol. 333, 657–674 [DOI] [PubMed] [Google Scholar]
- 36. Satyshur K. A., Worzalla G. A., Meyer L. S., Heiniger E. K., Aukema K. G., Misic A. M., Forest K. T. (2007) Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure 15, 363–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.