Background: The typical two-cysteine peroxiredoxin Ahp1 catalyzes the decomposition of alkyl hydroperoxides.
Results: Ahp1 possesses a peroxidatic Cys-62 after resolving Cys-31 and an A-type dimer interface that contributes to the positive cooperativity of hydroperoxide binding.
Conclusion: Ahp1 defines a novel group of thioredoxin-dependent peroxidases.
Significance: Provided is the first structural snapshot of electron transfer from thioredoxin to a thioredoxin-like protein.
Keywords: Crystal Structure, Disulfide, Enzyme Mechanisms, Peroxiredoxin, Protein-Protein Interactions, Thioredoxin, Alkyl Hydroperoxide Reductase, Complex Structure, Positive Cooperativity
Abstract
Peroxiredoxins (Prxs) are thiol-specific antioxidant proteins that protect cells against reactive oxygen species and are involved in cellular signaling pathways. Alkyl hydroperoxide reductase Ahp1 belongs to the Prx5 subfamily and is a two-cysteine (2-Cys) Prx that forms an intermolecular disulfide bond. Enzymatic assays and bioinformatics enabled us to re-assign the peroxidatic cysteine (CP) to Cys-62 and the resolving cysteine (CR) to Cys-31 but not the previously reported Cys-120. Thus Ahp1 represents the first 2-Cys Prx with a peroxidatic cysteine after the resolving cysteine in the primary sequence. We also found the positive cooperativity of the substrate t-butyl hydroperoxide binding to Ahp1 homodimer at a Hill coefficient of ∼2, which enabled Ahp1 to eliminate hydroperoxide at much higher efficiency. To gain the structural insights into the catalytic cycle of Ahp1, we determined the crystal structures of Ahp1 in the oxidized, reduced, and Trx2-complexed forms at 2.40, 2.91, and 2.10 Å resolution, respectively. Structural superposition of the oxidized to the reduced form revealed significant conformational changes at the segments containing CP and CR. An intermolecular CP-CR disulfide bond crossing the A-type dimer interface distinguishes Ahp1 from other typical 2-Cys Prxs. The structure of the Ahp1-Trx2 complex showed for the first time how the electron transfers from thioredoxin to a peroxidase with a thioredoxin-like fold. In addition, site-directed mutagenesis in combination with enzymatic assays suggested that the peroxidase activity of Ahp1 would be altered upon the urmylation (covalently conjugated to ubiquitin-related modifier Urm1) of Lys-32.
Introduction
Peroxiredoxins (Prxs,2 EC 1.11.1.15) are a family of peroxidases that protect cells against reactive oxygen species such as hydrogen peroxide and alkyl peroxides as well as participating in a series of cellular signaling pathways (1–5). Distinct from other peroxidases that require cofactors such as metal ions or prosthetic groups, Prxs reduce peroxides with redox-active cysteines (6, 7). As ubiquitous thiol-specific antioxidant proteins, Prxs have been identified in almost all organisms, are usually expressed at a relatively high level, exist in several isoforms, and possess a high catalytic rate (3–4, 8–10).
Based on sequence homology and structural similarity analyses, Prxs are divided into six subfamilies: AhpC/Prx1, Prx6, Prx5, Tpx, BCP/PrxQ, and AhpE (6, 10, 11). Alternatively, according to their distinct catalytic mechanisms, Prxs are classified into three types: typical 2-Cys, atypical 2-Cys, and 1-Cys Prxs (3, 12). All types of Prxs, however, share a strictly conserved and catalytically essential cysteine called the peroxidatic cysteine (CP), which is usually located at the N-terminal region of Prxs (6, 10). Each cycle of catalysis can be divided into three steps. The first cycle is, peroxidation, where the sulfur atom of the CP residue exerts a nucleophilic attack at the O-O bond of the peroxide substrate to release a corresponding molecule of alcohol or water at the cost of CP being oxidized into the cysteine sulfenic acid form. The second cycle, resolution, is where the second redox-active cysteine called the resolving cysteine (CR), which is usually located at the C-terminal region, attacks the sulfenic acid, resulting in the formation of an inter- or intramolecular disulfide bond for the typical and atypical 2-Cys Prxs, respectively, accompanying the release of a water molecule. However, this step is omitted for 1-Cys Prxs, as they do not have a CR residue. The third cycle, recycling, is where the oxidized Prx is reduced by thiol-containing electron donors such as thioredoxin (Trx), glutathione, or glutathione transferase π (3, 8, 13, 14). The regenerated Prx is then ready for another catalytic cycle.
At a higher level of peroxide, the cysteine sulfenic acid of eukaryotic typical 2-Cys Prxs can further react with a second peroxide molecule to form a cysteine sulfinic acid, which can be turned over by the sulfinic acid reductases, sulfiredoxin or sestrin, in an ATP-dependent manner (15–18). However, once the cysteine sulfinic acid is further oxidized to a cysteine sulfonic acid, the peroxidase activity of the Prx will be completely irreversible (3, 19). This mechanism of reactivation makes Prx robust peroxidases in a broad range of peroxide concentration, whereas the mechanism of inactivation enables catalytically inactive Prxs to function as signal molecules or molecular chaperones at a relatively higher peroxide concentration (4, 7, 9, 19–24).
The alkyl hydroperoxide reductase Ahp1 is one of five Prxs (Tsa1, Tsa2, Prx1, Dot5, and Ahp1) in the yeast Saccharomyces cerevisiae (25). Sequence comparison suggests that Ahp1 is a member of the Prx5 subfamily. A previous report showed that Ahp1 was a 2-Cys Prx with an intermolecular disulfide bond involving the CP (Cys-62) and CR (Cys-120) (26). It possesses a lower catalytic efficiency toward hydrogen peroxide compared with the organic hydroperoxide substrates such as t-butyl hydroperoxide (t-BOOH) and cumene hydroperoxide (26). Moreover, the ahp1 null mutant yeast strain is sensitive to t-BOOH (27). Thus, Ahp1 has long been regarded as an important cytoplasmic thioredoxin-dependent alkyl hydroperoxide reductase involved in oxidative stress (28). However, it has recently been demonstrated that Ahp1 acts as a receptor of alkyl hydroperoxides to activate Cad1, a Yap1-family transcription factor (29). In addition, the residue Lys-32 of Ahp1 can be covalently conjugated to ubiquitin-related modifier Urm1 (30, 31); however, the biological consequence of this modification remains unclear.
Since the first crystal structure of Prx (human Prx6) was determined in 1998 (32), many structures of Prxs have been reported (10). All these structures share a conserved thioredoxin-like fold and three highly conserved residues (Pro, Thr/Ser, and Arg) at the active site. Most of them exist as a dimer or oligomer, with two different interfaces called A- and B-type dimer interfaces (33). Remarkably, some typical 2-Cys Prxs can further assemble into toroid-shaped homodecameric complexes after redox change (7, 34).
Although the catalytic mechanisms of Prxs have been extensively studied, insights into their turnover by thioredoxins remain unclear. Here we systematically investigated the catalytic cycle of Ahp1 using a series of enzymatic assays in combination with three crystal structures: oxidized Ahp1 at 2.40 Å, reduced Ahp1 at 2.91 Å, and the Ahp1-Trx2 complex at 2.10 Å. Our results suggest that Ahp1 represents a novel group of typical 2-Cys Prxs, and the members of this group are distributed in the Prx5 subfamily in organisms such as fungi and bacteria. Enzymatic kinetics assays showed the positive cooperativity of the substrate t-BOOH binding to Ahp1 homodimer. This may enable Ahp1 to eliminate hydroperoxide at a much higher efficiency during oxidative stress. Moreover, the Ahp1-Trx2 complex structure revealed for the first time the electron transfer from the electron donor thioredoxin to a receptor peroxidase, which possesses a thioredoxin-like fold. In addition, our study also gave some hints on the biological consequence of Ahp1 urmylation.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Ahp1 and Mutants
The DNA sequence encoding Ahp1 was amplified by PCR from the genomic DNA of S. cerevisiae S288C (sequences of primers used were shown in supplemental Table 1). The PCR product was cloned into a pET28a-derived vector (Novagen). This construct encoding a hexahistidine (His6) tag at the N terminus of the recombinant protein (supplemental Table 2) was overexpressed in E. coli Rosetta (DE3) strain (Novagen) in 2× YT medium (Oxoid Ltd). The transformed cells were grown at 37 °C to an A600 nm of 0.6–1.0 and induced with 0.2 mm isopropyl-β-d-thiogalactoside for another 4 h at 37 °C. Cells were collected by centrifugation at 4000 × g for 5 min and resuspended in the lysis buffer (20 mm Tris-HCl, pH 7.5, 200 mm NaCl). After three cycles of freezing-thawing followed by sonication, the disrupted cells were centrifuged at 16,000 × g for 30 min. The supernatant containing the target protein was collected and loaded onto a nickel-nitrilotriacetic acid column (Qiagen) equilibrated with the binding buffer (20 mm Tris-HCl, pH 7.5, 200 mm NaCl). The target protein was eluted with the binding buffer containing 300 mm imidazole and further loaded onto a HiLoad 16/60 SuperdexTM 75 column (GE Healthcare) equilibrated with the binding buffer. Fractions containing the target protein were collected and concentrated. The purity of the protein was assessed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the protein sample was stored at −80 °C until use.
Site-directed mutagenesis was conducted using the QuikChange site-directed mutagenesis kit (Stratagene) with the plasmid encoding the wild-type Ahp1 as the template (sequences of primers used are shown in supplemental Table 1). The mutant proteins were prepared according to a similar procedure.
Enzymatic Assays
Peroxidase activity was determined by spectrophotometric assays that monitored the consumption of NADPH at 340 nm, as described previously (35). The S. cerevisiae thioredoxin Trx2 and thioredoxin reductase Trr1 were prepared as previously described (36, 37). The reaction was carried out in a final volume of 200 μl of reaction mix containing 50 mm HEPES-NaOH, pH 7.0, 250 μm NADPH, 1.13 μm Trx2, 0.38 μm Trr1, 0.15 μm Ahp1 (Ahp1C31S, Ahp1C62S, or Ahp1C120S), and 200 μm t-BOOH. The mix without t-BOOH was incubated at 30 °C for 3 min, and then the reaction was triggered by the addition of t-BOOH. The consumption of NADPH was monitored for 3 min with a step of 20 s with a DU800 spectrophotometer (Beckman Coulter, Fullerton, CA). The assays were performed three times, and error bars indicate the standard deviations.
Apparent kinetic parameters of Ahp1 and mutants toward t-BOOH and Trx2 were determined at 0.15 μm enzyme, 10 μm Trx2, 15–300 μm t-BOOH, and 300 μm t-BOOH, 0.2–7.5 μm Trx2. The consumption of NADPH was monitored for 2 min, with a step of 10 s, and the reaction velocity was calculated using the linear section of the plot. All assays were performed three times, and error bars indicated the S.E. The kinetic constants were determined with OriginPro (OriginLab) by fitting to the nonlinear Hill equation,
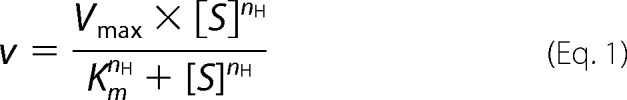 |
where nH is the Hill coefficient.
SDS-PAGE Analysis of Complexes between Ahp1 Mutants and Trx2C34S
The complex of Ahp1-Trx2, linked via an intermolecular disulfide bond, was prepared according to previous procedures (38, 39). Ahp1C31S, Ahp1C62S, and Trx2C34S were purified and reduced by 2 mm dithiothreitol (DTT). After desalting, Trx2C34S was mixed with a 20-fold molar excess of 5,5′-dithiobis-(2-nitrobenzoic acid) at 25 °C to introduce a disulfide bond between the active-site Cys-31 of Trx2 and TNB. After removing the excess 5,5′-dithiobis-(2-nitrobenzoic acid) from Trx2C34S, five samples (Ahp1C31S, Ahp1C62S, Trx2C34S-TNB, Ahp1C31S+Trx2C34S-TNB, and Ahp1C62S+Trx2C34S-TNB) were prepared and incubated at 25 °C for 30 min. Each sample was separated into two parts with or without 5 mm DTT and subjected to SDS-PAGE to detect the intermolecular disulfide bond formation.
Preparation of Ahp1-Trx2 Complex
Trx2 with a His6 tag and Ahp1C62S without a tag were purified in the presence of 2 mm DTT, separately. After removing DTT by desalting, Trx2C34S was mixed with a 20-fold molar excess of 5,5′-dithiobis-(2-nitrobenzoic acid) at 25 °C for 30 min to produce a disulfide bond between the active-site Cys-31 of Trx2 and TNB. Then, Ahp1C62S was incubated with 5-fold molar excess Trx2C34S-TNB to form the Ahp1C62S-Trx2C34S complex linked by an intermolecular disulfide bond. The mixed sample was further purified by nickel-nitrilotriacetic acid column (Qiagen) and HiLoad 16/60 SuperdexTM 75 column to remove the non-cross linked Ahp1C62S and the excess Trx2C34S. The purity of the complex protein was checked by SDS-PAGE.
Crystallization, Data Collection, and Processing
Crystals were grown at 16 °C using hanging drop vapor-diffusion techniques for oxidized and reduced Ahp1 or sitting drop vapor-diffusion techniques for Trx2-complexed Ahp1. In each drop, a 1-μl protein sample (9.5, 7.5, and 7.0 mg/ml for the oxidized, reduced, and Trx2-complexed forms, respectively; 10 mm DDT was added for reduced form) was mixed with 1 μl of reservoir solution. Crystallization conditions were 25% w/v polyethylene glycol monomethyl ether 2000, 0.2 m ammonium sulfate, 0.1 m sodium acetate, pH 4.6, for the oxidized form, 25% w/v polyethylene glycol 3350, 0.2 m ammonium sulfate, 0.1 m HEPES-NaOH, pH 7.5 for the reduced form, and 28% v/v polyethylene glycol 400, 0.2 m calcium chloride, 0.1 m HEPES-NaOH, pH 7.5, for the Trx2-complexed form. The crystals appeared in 2 days and reached the maximum size in 1 week. The crystals of oxidized and reduced Ahp1 were soaked in the cryoprotectant (reservoir solution supplemented with 25% v/v glycerol). All crystals were flash-frozen in the liquid nitrogen before data collection. All diffraction data were collected at 100 K in a liquid nitrogen stream using the beamline 17U with a Quantum Q315r CCD (Area Detector Systems Corporation, USA) at the Shanghai Synchrotron Radiation Facility. All diffraction data were indexed, integrated, and scaled with the program HKL2000 (40).
Structure Solution and Refinement
All structures were determined by molecular replacement with the program MOLREP (41) as a part of the CCP4i program suite (42) using the coordinates of Populus tremula D-type peroxiredoxin (PDB code 1TP9) (43) and S. cerevisiae thioredoxin Trx2 (PDB code 2FA4) (36) as search models. The initial model was refined using the maximum likelihood method implemented in REFMAC5 (44) and rebuilt interactively using the σA-weighted electron density maps with coefficients 2Fo − Fc and Fo − Fc in the program COOT (45). Two TLS groups were introduced in the refinement of Ahp1-Trx2 complex. The final model was validated with the programs MOLPROBITY (46) and PROCHECK (47). The final atomic coordinates and structure factors were deposited in the Protein Data Bank under the accession codes of 4DSQ, 4DSR, and 4DSS. Crystallographic parameters of the three structures are listed in Table 1. All structure figures were prepared with the program PyMOL (48).
TABLE 1.
Crystal parameters, data collection, and structure refinement statistics
| Data processing | Oxidized Ahp1 | Reduced Ahp1 | Ahp1-Trx2 complex |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 0.9793 | 0.9791 | 0.9791 |
| Space group | P21 | H3 | C2221 |
| Unit cell (Å), (°) | a = 40.0, b = 130.6, c = 67.7, α = γ = 90.0, β = 104.3 | a = b = 154.5, c = 91.0, α = β = 90.0, γ = 120.0 | a = 55.7, b = 133.4, c = 78.2, α = β = γ = 90.0 |
| Resolution range (Å)a | 50.00-2.40 (2.44-2.40) | 50.00-2.91 (3.01-2.91) | 50.00-2.10 (2.18-2.10) |
| Unique reflections | 25,277 (1,263) | 16,465 (1642) | 17,691 (1754) |
| Completeness (%) | 97.2 (98.9) | 91.3 (91.5) | 98.5 (100.0) |
| <I/σ(I)> | 4.7 (3.0) | 7.7 (3.0) | 12.8 (4.3) |
| Rmergeb (%) | 17.4 (44.2) | 13.1 (36.9) | 8.0 (33.9) |
| Average redundancy | 4.1 | 4.0 | 5.7 |
| Sigma (I) cutoff | −3.0 | −3.0 | −3.0 |
| Wilson B factors | 39.8 | 70.2 | 32.3 |
| Refinement statistics | |||
| Resolution range (Å) | 46.30-2.40 | 44.20-2.91 | 26.00-2.10 |
| R-factorc/R-freed (%) | 23.61/27.10 | 23.87/27.42 | 21.94/25.89 |
| Number of reflections used in refinement | 23,926 | 15,358 | 16,303 |
| Sigma (F) cutoff | 0 | 0 | 0 |
| Number of protein atoms | 5,328 | 5328 | 2117 |
| Number of water atoms | 184 | 11 | 78 |
| r.m.s.d.e bond lengths (Å) | 0.013 | 0.013 | 0.009 |
| r.m.s.d. bond angles (°) | 1.249 | 1.401 | 1.085 |
| Mean B factors (Å2) | 35.2 | 62.2 | 58.7 |
| Ramachandran plotf (residues, %) | |||
| Most favored (%) | 95.8 | 92.3 | 97.1 |
| Additional allowed (%) | 4.2 | 7.7 | 2.9 |
| Outliers (%) | 0 | 0 | 0 |
| PDB entry | 4DSQ | 4DSR | 4DSS |
a Values in parentheses refer to statistics in the highest bin.
b Rmerge = ΣhklΣi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣIi(hkl), where Ii(hkl) is the intensity of an observation, and 〈I(hkl)〉 is the mean value for its unique reflection; summations are over all reflections.
c R-factor = Σh|Fo(h) − Fc(h)|/ΣhFo(h), where Fo and Fc are the observed and calculated structure-factor amplitudes, respectively.
d R-free was calculated with 5% of the data excluded from the refinement.
e Root mean square deviation from ideal values.
f Categories were defined by Molprobity.
RESULTS
Assignment of CP and CR based on Peroxidase Activity Assays
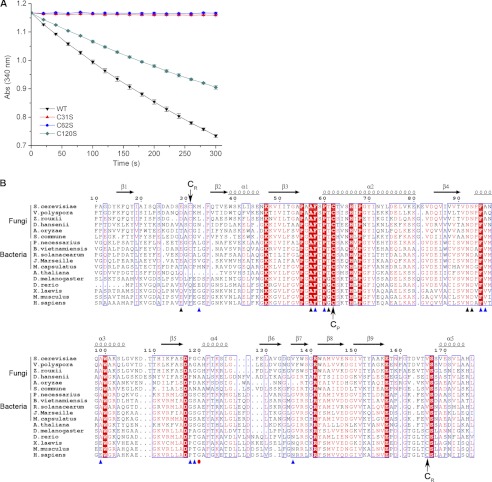
A previous report indicated that Ahp1 is a 2-Cys Prx, and Cys-62 and Cys-120 are the CP and CR, respectively (26). However, our enzymatic assays showed that in the presence of the thioredoxin system (Trx2-Trr1-NADPH), mutants of Ahp1C62S or C31S could not consume t-BOOH, resulting in complete abolition of the peroxidase activity, whereas the C120S mutant could consume t-BOOH as the wild type (Fig. 1A). This clearly indicated that Cys-62 and Cys-31 are catalytic residues. Multiple-sequence alignment of Ahp1 against other Prx5 subfamily proteins also suggested that both Cys-62 and Cys-31, but not Cys-120, are conserved through Prx5 subfamily proteins from several species of fungi and bacteria (Fig. 1B). Moreover, the sequence from Pro-55 to Cys-62 exhibited a conserved active-site motif (PXXXS/TXXC) (8); thus Cys-62 was assigned to CP and, consequently, Cys-31 to CR. It is notable that some homologs of Ahp1 from the higher eukaryotes have a resolving cysteine at the C-terminal region.
FIGURE 1.
Assignment of the CP and CR of Ahp1. A, shown are peroxidase activity assays. Assays were performed with Ahp1 (black inverted triangles), Ahp1C31S (red triangles), Ahp1C62S (blue circles), and Ahp1C120S (dark cyan diamonds). B, shown is multiple-sequence alignment of Ahp1 against other Prx5 subfamily proteins from different organisms. Both Cys-62 and Cys-31 of Ahp1 are conserved through homologs from several species of fungi and bacteria. In contrast, the homologs from several species of higher eukaryotes lack the corresponding resolving Cys-31 (CR) and have a conserved resolving CR at the C-terminal region. The multi-sequence alignment was performed using programs MultAlin (52) and ESPrint (53). The secondary structural elements of reduced Ahp1 are displayed above the sequences. The residues contributed to dimer interface were indicated with black (hydrogen bonds) and blue (hydrophobic interactions) triangles. Cys-120 was indicated with a red dot. All sequences were downloaded from the NCBI data base (www.ncbi.nlm.nih.gov). The sequences are (NCBI accession numbers codes are in parentheses) S. cerevisiae Ahp1 (NP_013210.1), Vanderwaltozyma polyspora Kpol_1004p59 (XP_001645540.1), Zygosaccharomyces rouxii ZYRO0D06248p (XP_002496705.1), Debaryomyces hansenii DEHA2G17864p (XP_462316.1), Aspergillus oryzae pmp20 (XP_001727651.1), Schizophyllum commune SCHCODRAFT_67750 (XP_003031587.1), Polynucleobacter necessarius redoxin domain-containing protein (YP_001154958.1), Ralstonia solanacearum peroxiredoxin (YP_003744570.1), Methylococcus capsulatus anti-oxidant AhpC-TSA family protein (YP_112582.1), Burkholderia vietnamiensis redoxin domain-containing protein (YP_001118388.1), Janthinobacterium Marseille peroxiredoxin (YP_001354699.1), Arabidopsis thaliana Prx-2D (NP_564763.1), Drosophila melanogaster peroxiredoxin 5 (NP_001027191.1), Danio rerio Prx 5 (NP_001019577.1), Xenopus laevis Prx 5 (NP_001085580.1), Mus musculus Prx 5 (NP_036151.1), and Homo sapiens Prx 5 (NP_036226.1).
Positive Cooperativity of t-BOOH Binding to Ahp1 Homodimer
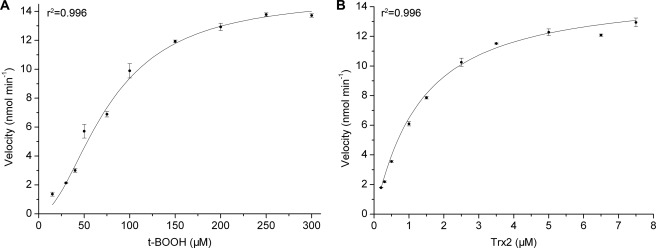
The catalytic cycle begins with recognition of the hydroperoxide substrate and ends with the regeneration of Ahp1 by Trx2. Thus, enzymatic kinetics assays were performed to analyze this bisubstrate reaction. Plots of reaction velocity versus substrate concentration fitted well to the nonlinear Hill equation (r2 = 0.996 for t-BOOH and Trx2) (Fig. 2, A and B). The Hill coefficients (nH) for t-BOOH and Trx2 were 1.9 ± 0.1 and 1.1 ± 0.1, respectively (Table 2, supplemental Fig. S1), suggesting positive cooperativity (nH >1) of t-BOOH and non-cooperativity (nH = 1) of Trx2 binding to Ahp1 homodimer.
FIGURE 2.
The reaction kinetics of Ahp1 toward the substrates. A, t-BOOH. B, Trx2. The plots were fitted by using the Hill equation.
TABLE 2.
Kinetics parameters of Ahp1 and mutants
| Ahp1 |
t-BOOH |
Trx2 |
||||||
|---|---|---|---|---|---|---|---|---|
| Km | kcat | nH | kcat/Km | Km | kcat | nH | kcat/Km | |
| μm | s−1 | ×105m−1s−1 | μm | s−1 | ×106m−1s−1 | |||
| WT | 76.9 ± 5.5 | 8.4 ± 0.3 | 1.9 ± 0.1 | 1.1 | 1.3 ± 0.3 | 8.3 ± 0.6 | 1.1 ± 0.1 | 6.4 |
| K32R | 49.4 ± 3.4 | 5.7 ± 0.1 | 1.5 ± 0.1 | 1.2 | 1.6 ± 0.2 | 6.5 ± 0.3 | 1.0 ± 0.1 | 4.1 |
| K32A | 60.2 ± 9.4 | 5.0 ± 0.5 | 1.6 ± 0.2 | 0.8 | 2.4 ± 0.2 | 5.9 ± 0.2 | 0.9 ± 0.0 | 2.5 |
| K32E | 47.4 ± 3.6 | 3.9 ± 0.2 | 1.5 ± 0.1 | 0.8 | 2.8 ± 0.4 | 5.2 ± 0.3 | 1.0 ± 0.1 | 1.9 |
Overall Structures of Oxidized and Reduced Ahp1
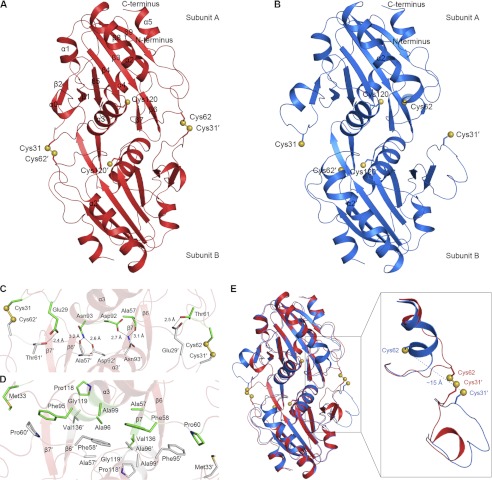
Similar to a previous study (49), gel filtration chromatography indicated that the majority of Ahp1 exists as a dimer in solution (data not shown). We first determined the crystal structure of dimeric Ahp1 in the oxidized form at 2.40 Å. Each asymmetric unit of the crystal contains two Ahp1 dimers, with residues Asp-3–Leu-176 well fitted in the final model of all four chains. Similar to previously reported members of the Prx5 subfamily, each subunit of the dimer adopts a typical Prx-fold consisting of a central twisted seven-stranded β-sheet (β1–β5 and β8–β9), flanked by α-helices α2/3/5 on one side and α1/4 on the other side (Fig. 3A). In addition, Ahp1 contains a short helix α0 inserted between β1 and β2 and a β-hairpin (β6-β7) inserted between α4 and β8.
FIGURE 3.
Structures of the oxidized and reduced Ahp1. A, shown is a schematic of oxidized Ahp1 homodimer structure. The side chains of Cys-31, Cys-62, and Cys-120 are shown as sticks, and sulfur atoms are shown as yellow balls. Residues of subunit B are labeled with a prime. B, shown is a schematic of reduced Ahp1 homodimer structure. The side chains of Cys-31, Cys-62, and Cys-120 are shown as sticks, and sulfur atoms shown as yellow balls. C, shown is a detailed representation of the dimer interface in oxidized Ahp1. Disulfide bonds and hydrogen bonds are indicated in C, and hydrophobic interactions are indicated in D. The involved residues are shown as sticks, and sulfur atoms of cysteines are shown as yellow balls. The two subunits of Ahp1 are shown in green and gray, respectively. E, shown are conformational changes of the oxidized and reduced Ahp1. The oxidized and reduced Ahp1 are colored in red and blue, respectively. The side chains of Cys-31, Cys-62, and Cys-120 are shown as sticks, and sulfur atoms are shown as yellow balls.
Independent of redox state, Ahp1 forms a homodimer with an A-type dimer interface related by a non-crystallographic 2-fold symmetry axis (Fig. 3, A and B). The dimeric interface of oxidized Ahp1 buries a total surface area of 1760 Å2. Cys-62 (Cys-31) of subunit A and Cys-31′ (Cys-62′) of subunit B (residues of subunit B are labeled with a prime) form two pairs of intermolecular disulfide bonds (Fig. 3, A and C), as shown in the 2Fo − Fc electron density maps (supplemental Fig. S2). Six hydrogen bonds (Glu-29∼Thr-61′, Asn-93∼Ala-57′, Asn-93∼Asp-92′, Asp-92∼Asn-93′, Ala-57∼Asn-93′, and Thr-61∼Glu-29′) contribute to the majority of interface formation (Fig. 3C). The interface is further stabilized by hydrophobic interactions between residues Met-33, Ala-57, Phe-58, Pro-60, Phe-95, Ala-96, Ala-99, Pro-118, Gly-119, and Val-136 from both subunits (Fig. 3D). Multiple-sequence alignment showed that most residues involved in the dimer interface are highly conserved through Prx5 subfamily proteins (Fig. 1B).
In the 2.91 Å structure of reduced Ahp1, the numbers of subunits in an asymmetric unit and of residues traced in the final model are the same as those in the oxidized form. Similar to the previous structures, Cys-62 is folded into the first turn of helix α2 (Fig. 3B). The distance between the sulfur atoms of Cys-62 and Cys-31′ in the homodimer is ∼15 Å. Similar to other members of the Prx5 subfamily, reduced Ahp1 also contains a typical bulge in helix α2. The inserted short helix α0 is solely present in subunit A, and it is obvious that the helix-like segment in subunit B corresponded to helix α0 in subunit A. Reduced Ahp1 also adopts an A-type dimer interface with a buried surface area of 1180 Å2, which is much smaller than the 1760 Å2 interface of the oxidized form.
Conformational Changes upon Oxidation
Superposition of the oxidized Ahp1 on the reduced form yields an overall root mean square deviation (r.m.s.d.) of 0.72 Å over 328 Cα atoms (Fig. 3E), indicating that they share a similar overall structure. In contrast, significant differences were observed within segments juxtaposed to Cys-62 and Cys-31 (Ser-59–His-66 and Gln-23–Lys-32) (Fig. 3E). In the reduced form, the distance between the sulfur atoms of Cys-62 and Cys-31′ in the homodimer is ∼15 Å, which is similar to the distance of other reduced 2-Cys Prxs reported (10, 33). It was proposed that large conformational changes are required for Prx to form the intermolecular disulfide bond between CP and CR (33). Indeed, the two turns of the helix α2 (Ser-59–His-66) of the reduced Ahp1 will be unraveled upon oxidation into loop conformation and shift toward the CR, whereas the segment juxtaposed to CR (Gln-23′–Lys-32′) moves inward, resulting in the approach of CP and CR for the formation of the intermolecular disulfide bonds.
Favored Mixed Disulfide Bond between Ahp1 and Trx2
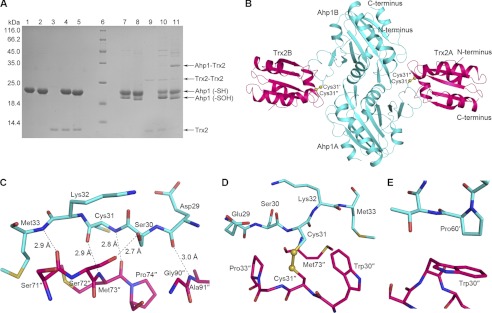
At the stage of Ahp1 regeneration, the intermolecular disulfide bond between Cys-62 and Cys-31′ is subject to attack by Trx2, resulting in the formation of a transient intermolecular disulfide bond between Ahp1 and Trx2-Cys-31. However, which cysteine of Ahp1 (Cys-62 or Cys-31) is favored by Trx2 remains unknown. To ascertain this cysteine, we constructed two mutants, Ahp1C62S and Ahp1C31S, given that the deeply buried Cys-120 could be eliminated (Fig. 3, A and B). SDS-PAGE was employed to determine semiquantitatively the reaction efficiency of Trx2C34S toward these two mutants. The disulfide-linked complex of Trx2C34S with Ahp1C62S is obvious (lane 11, Fig. 4A), whereas that with Ahp1C31S is unclear (lane 10, Fig. 4A). These results suggest that Cys-62 of Ahp1 is favored by Trx2-Cys-31.
FIGURE 4.
Ahp1-Trx2 complex. A, shown is SDS-PAGE analysis of the complexes between Ahp1 mutants and Trx2C34S. The disulfide-linked complex could be reduced by DTT. Lanes 1–5, Ahp1C31S, Ahp1C62S, Trx2C34S, Ahp1C31S+Trx2C34S, and Ahp1C62S+Trx2C34S with 5 mm DTT; lane 6, protein marker; lanes 7–11, samples corresponding to lanes 1–5, respectively, without DTT. B, shown is the overall structure of disulfide-linked Ahp1C62S-Trx2C34S complex. The side chains of Cys-31 are labeled as sticks, and sulfur atoms are labeled as yellow balls. Ahp1 and Trx2 are colored cyan and pink, respectively. Residues of subunit B of Ahp1 are labeled with a prime, and residues of Trx2 are labeled with double prime. C, D, E are shown detailed representations of Ahp1-Trx2 complex interface. Hydrogen bonds (C), a disulfide bond (D), and hydrophobic interactions between subunit A of Ahp1 and subunit A of Trx2 (D). Hydrophobic interactions between subunit B of Ahp1 and subunit A of Trx2 (E). The involved residues are shown as sticks, and sulfur atoms of cysteines shown as yellow balls.
Complex Structure of Ahp1-Trx2
It has been indicated that Ahp1 can be regenerated by thioredoxin via a transient disulfide bond exchange reaction. However, structural insights into the regeneration mechanism remain unclear. Therefore, we purified an Ahp1C62S-Trx2C34S complex linked by an intermolecular disulfide bond between Ahp1-Cys-31 and Trx2-Cys-31 and determined the complex structure at 2.10 Å resolution. Each asymmetric unit contains an Ahp1 (Asp-3–Leu-176) and a Trx2 (Met-1–Val-104) subunit. Symmetric processing enabled us to define a physiologically relevant heterotetramer, which could be regarded as a dimeric Ahp1 attacked by two Trx2 molecules from both sides (Fig. 4B). The 2Fo − Fc electron density maps clearly show the presence of two mixed disulfide bonds (supplemental Fig. S3).
The interface between the Ahp1 dimer and a Trx2 monomer buries a total surface area of 1200 Å2 (840 Å2 between Ahp1A and Trx2B; 360 Å2 between Ahp1B and Trx2B). This is an interface area typical for redox complexes because of their short-lived interactions (50). One side chain (Ser-30-Oγ∼Ser-72″-Oγ) (residues of Trx2 are labeled with double prime) and four main-chain hydrogen bonds (Asp-29–O∼Ala-91″-N, Cys-31-N∼Met-73″-O, Cys-31-O∼Met-73″-N, and Met-33-N∼Ser-71″-O) are involved in interactions between the loop (Asp-29–Met-33) of Ahp1 and the loops (Ser-71″–Pro-74″ and Gly-90″–Ala-91″) of Trx2 (Fig. 4C). Notably, the four main-chain hydrogen bonds are conserved among previously reported Trx2-complexed structures (39). The hydrophobic interactions are mainly contributed by solvent-accessible hydrophobic patches of Trx2 (Trp-30″–Pro-33″ around the redox center and residue Met-73″) and two residues of Ahp1 (Met-33 and Pro-60′) (Fig. 4, D and E).
Structural Insights into Regeneration of Ahp1
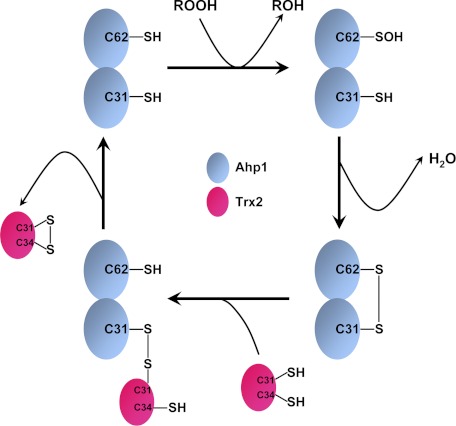
During regeneration, the oxidized Ahp1 should form a transient and unstable complex with reduced Trx2. To obtain a picture of this process, we froze the two proteins by introducing an intermolecular disulfide bond between Ahp1-Cys-31 and Trx2-Cys-31 and determined the complex structure at 2.10 Å resolution (Fig. 4B). To date this is the first Trx-complexed structure of a peroxidase or a Trx-like protein. The complex structure shows the electron transfer from the electron donor Trx to the receptor Prx. Structural comparison demonstrated that Ahp1 in the complex could be well superimposed with the reduced Ahp1 with an overall r.m.s.d. of 0.55 Å over 348 Cα atoms (data not shown). Thus, this complex could mimic the transient state of a reduced Ahp1 just before leaving Trx2. After the attack by Trx2-Cys-34 on the mixed disulfide bond between Ahp1-Cys-31 and Trx2-Cys-31, a regenerated Ahp1 will depart from an oxidized Trx2 for another catalytic cycle (Fig. 5).
FIGURE 5.
A schematic diagram of Ahp1 catalytic cycle.
Lys-32 Is Important for Regeneration of Ahp1 Catalyzed by Trx2
A previous study showed that Ahp1 was the sole identified protein substrate subject to modification by the ubiquitin-related modifier, Urm1, in S. cerevisiae (30). However, the function of urmylated Ahp1 remains unknown. Recent findings indicated that residue Lys-32 of Ahp1 can be covalently conjugated to Urm1 (31). Our results demonstrated that residue Cys-31 is the CR, which plays a crucial role in the peroxidase activity of Ahp1. Furthermore, residue Lys-32 is involved in the interface between Ahp1 and Trx2 (Fig. 4, D and E). Thus, will urmylation of Lys-32 affect the catalysis and the regeneration process of Ahp1? Due to difficulties in preparing the urmylated Ahp1 in vitro, we constructed K32R, K32A, and K32E mutants to detect whether Lys-32 is essential to the peroxidase activity. The reaction kinetics parameters of the mutants toward substrates t-BOOH and Trx2 were determined (supplemental Fig. S1) and compared with those of the wild type. As shown in Table 2, all three mutants had a catalytic efficiency (kcat/Km) toward t-BOOH almost equivalent to that of the wild type, whereas the regeneration efficiency of Ahp1 driven by Trx2 was significantly decreased upon the mutation of Lys-32 to Ala or Glu. These results suggest that Lys-32 might play a significant role in the regeneration process.
DISCUSSION
Ahp1 Represents a Novel Group of Typical 2-Cys Prxs
Ahp1 has three cysteines (Cys-31/62/120). A previous study reported that Ahp1 is classified into 2-Cys Prxs with a CP residue of Cys-62 and a CR residue of Cys-120 (26). However, our enzymatic assays indicated unambiguously that Cys-62 is the CP and Cys-31 is the CR (Fig. 1A). Multiple-sequence alignment also showed that both Cys-62 and Cys-31 are conserved through Prx5 subfamily proteins from several species of fungi and bacteria (Fig. 1B). Notably, the homologs of Ahp1 from the higher eukaryotes lack the corresponding resolving Cys-31 but have a conserved CR at the C-terminal region, as defined in human Prx5 (12).
Furthermore, we observed the formation of an intermolecular disulfide bond between Cys-62 and Cys-31′ in the oxidized Ahp1 structure (Fig. 3A). All Prxs identified to date have a CP residue located in the N-terminal region (6, 10), whereas Ahp1 is the first Prx characterized to have a CR located at the N-terminal region preceding the CP.
Our results also indicated that, from a mechanistic point of view, Ahp1 should be classified into the typical 2-Cys Prxs. However, distinct from the representative members of typical 2-Cys Prxs, Ahp1 possesses an A-type dimer interface that usually exists in atypical 2-Cys Prxs and 1-Cys Prxs (33) and cannot assemble into a toroid-shaped homodecameric complex. Based on sequence homology and structural information, Ahp1 was classified as a member of the Prx5 subfamily. Indeed, the structure of Ahp1 is similar to the other proteins in the Prx5 subfamily (10, 33). Thus, Ahp1 is structurally similar to members of atypical 2-Cys Prxs and 1-Cys Prxs, whereas it is mechanistically similar to members of typical 2-Cys Prxs. Moreover, the oxidized-form structure (Fig. 3A) suggests that Ahp1 is the first identified Prx that possesses an intermolecular disulfide bond crossing an A-type dimer interface. Thus, Ahp1 represents a novel group of typical 2-Cys Prxs. The members of this group are distributed in the Prx5 subfamily and are found in some fungi and bacteria (Fig. 1B).
New Evidence for Evolutionary Advantage of Ahp1 Dimerization
Intriguingly, the kinetic profile of Ahp1 did not follow the Michaelis-Menten plot of velocity versus t-BOOH concentration. Further analyses revealed that it fitted well to the Hill equation (Fig. 2A) with a Hill coefficient of about 2 (Table 2), indicating the positive cooperativity of t-BOOH binding to the dimeric Ahp1. This positive cooperativity enables Ahp1 to eliminate hydroperoxide at a much higher efficiency during oxidative stress and provides new evidence for the evolutionary advantage of Ahp1 dimerization. We were unable to obtain the structure of Ahp1 in complex with the hydroperoxide substrate despite extensive trials. To elucidate the structural basis of the positive cooperativity, we docked t-BOOH to the reduced Ahp1 using the HADDOCK program (51). The t-butyl moiety of t-BOOH at the active-site pocket of subunit A is stabilized by residues Gly-161 and Phe-95′ (from subunit B) via hydrophobic interactions (supplemental Fig. S4). The interaction between the t-butyl moiety and the side chain of Phe-95 ′ may transfer the positive cooperativity signal, crossing the interface, to the active-site pocket of subunit B, resulting in increased binding affinity toward the second t-BOOH molecule.
In contrast, this positive cooperativity is not observed for another Ahp1 substrate, Trx2 (Table 2). The regeneration efficiency of Ahp1 driven by Trx2 was 6.4 × 106 m−1 s−1, which is about 60-fold higher than the catalytic efficiency of Ahp1 toward t-BOOH (1.1 × 105 m−1 s−1). Therefore, this higher catalytic regeneration efficiency might ensure that the oxidized Ahp1 would be efficiently reduced in the absence of cooperativity. It also implies that the catalytic efficiency of Ahp1 toward t-BOOH is a rate-limiting step during the catalytic cycle. Whether the other members of this novel subfamily found in fungi and bacteria have a similar positive cooperativity remains to be further investigated.
Some Hints for Function of Urmylation of Ahp1
We found that Lys-32, which is subject to urmylation, is located a distance from the active-site Cys-62 and, thus, may not be involved in the decomposition of t-BOOH. Therefore, Lys-32 mutagenesis does not obviously alter the catalytic efficiency of Ahp1 toward t-BOOH (Table 2). However, mutation of Lys-32 could lower the regeneration efficiency of Ahp1 catalyzed by Trx2 after a decrease in electropositivity (wild-type Ahp1 is about 3-fold higher than K32E mutant). Urm1 is an acidic protein with a theoretical isoelectric point of 4.29, and the K32E mutant is more or less similar to the electrostatic state of the urmylated Ahp1. Lys-32 is located at the loop of Asp-29-Met-33, which is involved in the interface between Ahp1 and Trx2. The decreased apparent Km value of K32E mutant suggested a lower affinity toward Trx2 compared with wild-type Ahp1. On the other hand, covalent conjugation of Urm1 to the Lys-32 of Ahp1 might introduce steric hindrance that will affect Trx2 binding and result in a decreased or even abolished turnover rate. Therefore, we suggest that urmylation of Lys-32 would decrease or abolish the peroxidase activity of Ahp1. Further studies are necessary to elucidate this hypothesis.
Supplementary Material
Acknowledgments
We thank the assistance of the staff at Shanghai Synchrotron Radiation Facility. We also thank Yong-Liang Jiang, Pei-Xin Cui, and Yue Fu for help in data collection and molecular cloning.
This work was supported by the 973 project from the Ministry of Science and Technology of China (Project 2012CB911002) and Chinese National Natural Science Foundation Grant 30870490.

This article contains supplemental Tables S1 and S2 and Figs. S1–S4.
The atomic coordinates and structure factors (codes 4DSQ, 4DSR, and 4DSS) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Prx
- peroxiredoxin
- Ahp
- Alkyl hydroperoxide reductase
- CP
- peroxidatic cysteine
- CR
- resolving cysteine
- Trx
- thioredoxin
- Trr
- thioredoxin reductase
- Urm
- ubiquitin-related modifier
- t-BOOH
- t-butyl hydroperoxide
- nH
- Hill coefficient
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Chae H. Z., Chung S. J., Rhee S. G. (1994) Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem. 269, 27670–27678 [PubMed] [Google Scholar]
- 2. Jacobson F. S., Morgan R. W., Christman M. F., Ames B. N. (1989) An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage. Purification and properties. J. Biol. Chem. 264, 1488–1496 [PubMed] [Google Scholar]
- 3. Wood Z. A., Schröder E., Robin Harris J., Poole L. B. (2003) Structure, mechanism and regulation of peroxiredoxins. Trends. Biochem. Sci. 28, 32–40 [DOI] [PubMed] [Google Scholar]
- 4. Winterbourn C. C. (2008) Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 4, 278–286 [DOI] [PubMed] [Google Scholar]
- 5. Veal E. A., Day A. M., Morgan B. A. (2007) Hydrogen peroxide sensing and signaling. Mol. Cell 26, 1–14 [DOI] [PubMed] [Google Scholar]
- 6. Knoops B., Loumaye E., Van Der Eecken V. (2007) Evolution of the peroxiredoxins. Subcell. Biochem. 44, 27–40 [DOI] [PubMed] [Google Scholar]
- 7. Barranco-Medina S., Lázaro J. J., Dietz K. J. (2009) The oligomeric conformation of peroxiredoxins links redox state to function. FEBS Lett. 583, 1809–1816 [DOI] [PubMed] [Google Scholar]
- 8. Poole L. B. (2007) The catalytic mechanism of peroxiredoxins. Subcell. Biochem. 44, 61–81 [DOI] [PubMed] [Google Scholar]
- 9. Hall A., Karplus P. A., Poole L. B. (2009) Typical 2-Cys peroxiredoxins-structures, mechanisms, and functions. FEBS J. 276, 2469–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hall A., Nelson K., Poole L. B., Karplus P. A. (2011) Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signal. 15, 795–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Soito L., Williamson C., Knutson S. T., Fetrow J. S., Poole L. B., Nelson K. J. (2011) PREX. PeroxiRedoxin classification indEX, a database of subfamily assignments across the diverse peroxiredoxin family. Nucleic Acids Res. 39, D332–D337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seo M. S., Kang S. W., Kim K., Baines I. C., Lee T. H., Rhee S. G. (2000) Identification of a new type of mammalian peroxiredoxin that forms an intramolecular disulfide as a reaction intermediate. J. Biol. Chem. 275, 20346–20354 [DOI] [PubMed] [Google Scholar]
- 13. Manevich Y., Feinstein S. I., Fisher A. B. (2004) Activation of the antioxidant enzyme 1-CYS peroxiredoxin requires glutathionylation mediated by heterodimerization with pi GST. Proc. Natl. Acad. Sci. U.S.A. 101, 3780–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ralat L. A., Manevich Y., Fisher A. B., Colman R. F. (2006) Direct evidence for the formation of a complex between 1-cysteine peroxiredoxin and glutathione S-transferase π with activity changes in both enzymes. Biochemistry 45, 360–372 [DOI] [PubMed] [Google Scholar]
- 15. Biteau B., Labarre J., Toledano M. B. (2003) ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulfiredoxin. Nature 425, 980–984 [DOI] [PubMed] [Google Scholar]
- 16. Budanov A. V., Sablina A. A., Feinstein E., Koonin E. V., Chumakov P. M. (2004) Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science 304, 596–600 [DOI] [PubMed] [Google Scholar]
- 17. Jönsson T. J., Lowther W. T. (2007) The peroxiredoxin repair proteins. Subcell. Biochem. 44, 115–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang T. S., Jeong W., Woo H. A., Lee S. M., Park S., Rhee S. G. (2004) Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J. Biol. Chem. 279, 50994–51001 [DOI] [PubMed] [Google Scholar]
- 19. Lim J. C., Choi H. I., Park Y. S., Nam H. W., Woo H. A., Kwon K. S., Kim Y. S., Rhee S. G., Kim K., Chae H. Z. (2008) Irreversible oxidation of the active-site cysteine of peroxiredoxin to cysteine sulfonic acid for enhanced molecular chaperone activity. J. Biol. Chem. 283, 28873–28880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Phalen T. J., Weirather K., Deming P. B., Anathy V., Howe A. K., van der Vliet A., Jönsson T. J., Poole L. B., Heintz N. H. (2006) Oxidation state governs structural transitions in peroxiredoxin II that correlate with cell cycle arrest and recovery. J. Cell Biol. 175, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klomsiri C., Karplus P. A., Poole L. B. (2011) Cysteine-based redox switches in enzymes. Antioxid. Redox Signal. 14, 1065–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jang H. H., Lee K. O., Chi Y. H., Jung B. G., Park S. K., Park J. H., Lee J. R., Lee S. S., Moon J. C., Yun J. W., Choi Y. O., Kim W. Y., Kang J. S., Cheong G. W., Yun D. J., Rhee S. G., Cho M. J., Lee S. Y. (2004) Two enzymes in one. Two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 117, 625–635 [DOI] [PubMed] [Google Scholar]
- 23. Chuang M. H., Wu M. S., Lo W. L., Lin J. T., Wong C. H., Chiou S. H. (2006) The antioxidant protein alkylhydroperoxide reductase of Helicobacter pylori switches from a peroxide reductase to a molecular chaperone function. Proc. Natl. Acad. Sci. U.S.A. 103, 2552–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. D'Autréaux B., Toledano M. B. (2007) ROS as signaling molecules. Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8, 813–824 [DOI] [PubMed] [Google Scholar]
- 25. Wong C. M., Siu K. L., Jin D. Y. (2004) Peroxiredoxin-null yeast cells are hypersensitive to oxidative stress and are genomically unstable. J. Biol. Chem. 279, 23207–23213 [DOI] [PubMed] [Google Scholar]
- 26. Jeong J. S., Kwon S. J., Kang S. W., Rhee S. G., Kim K. (1999) Purification and characterization of a second type thioredoxin peroxidase (type II TPx) from Saccharomyces cerevisiae. Biochemistry 38, 776–783 [DOI] [PubMed] [Google Scholar]
- 27. Lee J., Spector D., Godon C., Labarre J., Toledano M. B. (1999) A new antioxidant with alkyl hydroperoxide defense properties in yeast. J. Biol. Chem. 274, 4537–4544 [DOI] [PubMed] [Google Scholar]
- 28. Park S. G., Cha M. K., Jeong W., Kim I. H. (2000) Distinct physiological functions of thiol peroxidase isoenzymes in Saccharomyces cerevisiae. J. Biol. Chem. 275, 5723–5732 [DOI] [PubMed] [Google Scholar]
- 29. Iwai K., Naganuma A., Kuge S. (2010) Peroxiredoxin Ahp1 acts as a receptor for alkylhydroperoxides to induce disulfide bond formation in the Cad1 transcription factor. J. Biol. Chem. 285, 10597–10604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goehring A. S., Rivers D. M., Sprague G. F., Jr. (2003) Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryot. Cell 2, 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Van der Veen A. G., Schorpp K., Schlieker C., Buti L., Damon J. R., Spooner E., Ploegh H. L., Jentsch S. (2011) Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc. Natl. Acad. Sci. U.S.A. 108, 1763–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Choi H. J., Kang S. W., Yang C. H., Rhee S. G., Ryu S. E. (1998) Crystal structure of a novel human peroxidase enzyme at 2.0 Å resolution. Nat. Struct. Biol. 5, 400–406 [DOI] [PubMed] [Google Scholar]
- 33. Karplus P. A., Hall A. (2007) Structural survey of the peroxiredoxins. Subcell. Biochem. 44, 41–60 [DOI] [PubMed] [Google Scholar]
- 34. Parsonage D., Youngblood D. S., Sarma G. N., Wood Z. A., Karplus P. A., Poole L. B. (2005) Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry 44, 10583–10592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kang S. W., Baines I. C., Rhee S. G. (1998) Characterization of a mammalian peroxiredoxin that contains one conserved cysteine. J. Biol. Chem. 273, 6303–6311 [DOI] [PubMed] [Google Scholar]
- 36. Bao R., Chen Y., Tang Y. J., Janin J., Zhou C. Z. (2007) Crystal structure of the yeast cytoplasmic thioredoxin Trx2. Proteins 66, 246–249 [DOI] [PubMed] [Google Scholar]
- 37. Zhang Z., Bao R., Zhang Y., Yu J., Zhou C. Z., Chen Y. (2009) Crystal structure of Saccharomyces cerevisiae cytoplasmic thioredoxin reductase Trr1 reveals the structural basis for species-specific recognition of thioredoxin. Biochim. Biophys. Acta 1794, 124–128 [DOI] [PubMed] [Google Scholar]
- 38. Wang P. F., Veine D. M., Ahn S. H., Williams C. H., Jr. (1996) A stable mixed disulfide between thioredoxin reductase and its substrate, thioredoxin. Preparation and characterization. Biochemistry 35, 4812–4819 [DOI] [PubMed] [Google Scholar]
- 39. Ma X. X., Guo P. C., Shi W. W., Luo M., Tan X. F., Chen Y., Zhou C. Z. (2011) Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1. J. Biol. Chem. 286, 13430–13437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 41. Vagin A., Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 42. Collaborative Computational Project, Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 43. Echalier A., Trivelli X., Corbier C., Rouhier N., Walker O., Tsan P., Jacquot J. P., Aubry A., Krimm I., Lancelin J. M. (2005) Crystal structure and solution NMR dynamics of a D (type II) peroxiredoxin glutaredoxin and thioredoxin-dependent. A new insight into the peroxiredoxin oligomerism. Biochemistry 44, 1755–1767 [DOI] [PubMed] [Google Scholar]
- 44. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 45. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 46. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) Procheck. A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 48. DeLano W. L. (2002) The PyMOL Molecular Graphics System, Version 0.99rc2, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 49. Trivelli X., Krimm I., Ebel C., Verdoucq L., Prouzet-Mauléon V., Chartier Y., Tsan P., Lauquin G., Meyer Y., Lancelin J. M. (2003) Characterization of the yeast peroxiredoxin Ahp1 in its reduced active and overoxidized inactive forms using NMR. Biochemistry 42, 14139–14149 [DOI] [PubMed] [Google Scholar]
- 50. Janin J., Bahadur R. P., Chakrabarti P. (2008) Protein-protein interaction and quaternary structure. Q. Rev. Biophys. 41, 133–180 [DOI] [PubMed] [Google Scholar]
- 51. de Vries S. J., van Dijk M., Bonvin A. M. (2010) The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 5, 883–897 [DOI] [PubMed] [Google Scholar]
- 52. Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gouet P., Robert X., Courcelle E. (2003) ESPript/ENDscript. Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31, 3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.