Background: Humans have two enzyme isoforms to produce the universal sulfate donor 3′-phosphoadenosine 5′-phosphosulfate (PAPS).
Results: The main difference between the two PAPS synthases is their stability, which is modulated by nucleotides.
Conclusion: Protein stability is a major contributing factor for PAPS availability.
Significance: Naturally occurring changes in APS concentrations may be sensed by the labile PAPS synthase 2 that might act as a novel biosensor.
Keywords: Biosensors, Heparan Sulfate, Nucleotide, Protein Stability, Sulfotransferase, PAPS Synthase, Phase II Biotransformation, Sulfation
Abstract
Activated sulfate in the form of 3′-phosphoadenosine 5′-phosphosulfate (PAPS) is needed for all sulfation reactions in eukaryotes with implications for the build-up of extracellular matrices, retroviral infection, protein modification, and steroid metabolism. In metazoans, PAPS is produced by bifunctional PAPS synthases (PAPSS). A major question in the field is why two human protein isoforms, PAPSS1 and -S2, are required that cannot complement for each other. We provide evidence that these two proteins differ markedly in their stability as observed by unfolding monitored by intrinsic tryptophan fluorescence as well as circular dichroism spectroscopy. At 37 °C, the half-life for unfolding of PAPSS2 is in the range of minutes, whereas PAPSS1 remains structurally intact. In the presence of their natural ligand, the nucleotide adenosine 5′-phosphosulfate (APS), PAPS synthase proteins are stabilized. Invertebrates only possess one PAPS synthase enzyme that we classified as PAPSS2-type by sequence-based machine learning techniques. To test this prediction, we cloned and expressed the PPS-1 protein from the roundworm Caenorhabditis elegans and also subjected this protein to thermal unfolding. With respect to thermal unfolding and the stabilization by APS, PPS-1 behaved like the unstable human PAPSS2 protein suggesting that the less stable protein is evolutionarily older. Finally, APS binding more than doubled the half-life for unfolding of PAPSS2 at physiological temperatures and effectively prevented its aggregation on a time scale of days. We propose that protein stability is a major contributing factor for PAPS availability that has not as yet been considered. Moreover, naturally occurring changes in APS concentrations may be sensed by changes in the conformation of PAPSS2.
Introduction
All eukaryotic sulfation reactions require the activated sulfate donor 3′-phosphoadenosine 5′-phosphosulfate (PAPS).3 This high-energy sulfate is essential for the build-up of extracellular matrices as well as modification of many extracellular proteins and low-molecular weight substances. The addition of sulfate to biomolecules can be understood as a high-affinity, low-capacity conjugation system (1). This means that the supply of the precursor PAPS becomes rate-limiting, and hence, sulfation processes could be regulated in general by the availability of PAPS (2).
Entrez Gene (3) lists more than 55 current sulfotransferase entries within the human genome; they are located in the cytoplasm (4), the nucleus (5), as well as the Golgi apparatus (6). These diverse enzymes are fed by only two PAPS producing enzymes in vertebrates, the bifunctional PAPS synthases 1 and 2. PAPS synthases consist of an N-terminal adenosine 5′-phosphosulfate (APS) kinase domain and a C-terminal ATP sulfurylase domain connected by a short irregular linker (7). These enzymes have recently come into the focus of biomedical research. Lower animals seem to have only one PAPS synthase gene and this single enzyme is essential at least for the worm Caenorhabditis elegans (8). Recessively inherited defects in the genes for human and mouse PAPS synthase 2 cause bone and cartilage malformation (9, 10) and later articular cartilage lesions of joints (11). PAPS synthases have also been implicated in steroid metabolism (12) and hepatitis B infection (13). Moreover, PAPSS1 has recently been reported to play an as yet undefined role in retroviral infection (14). Overall, mutations in one of the two PAPS synthase genes cause severe disease states.
Why then can one PAPS synthase isoform not complement for the other? Several answers can be derived from existing literature. The different cellular distribution of the two isoforms may perhaps play a role (15), their slightly different enzymatic capabilities might be important (16), or both proteins may be needed to form heterodimers (17). We have addressed these possibilities. First, subcellular localization of both proteins is more intricately regulated than previously anticipated with both protein isoforms having access to both cytoplasm and nucleus (18). Second, whereas a 10-fold higher overall activity for PAPSS2 than PAPSS1 was reported when treating the two-step enzymes as Michaelis-Menten enzymes (16), the difference in the rate-limiting APS kinase reaction is more subtle and may even lead to a more active PAPSS1 (17). Third, the recently discovered heterodimers have not yet revealed any synergistic effects. They simply behave like an additive mixture of the respective homodimers (17). Taken together, localization, specific activity, and the formation of heterodimers do not provide an adequate explanation for why PAPS synthase isoforms cannot substitute for one another.
We have now made the unexpected observation that the two human isoforms of PAPS synthase differ in their protein stability. PAPS synthases are large proteins and strictly dimeric. Hence, they unfold irreversibly via at least one intermediate state. Without addition of ligands, PAPSS2 was found to be less stable under all conditions tested. During synthesis of PAPS several nucleotides encounter these fragile enzymes. Hence, we tested the influence of AMP, ADP, ATP, sulfate, APS, and PAPS on the unfolding behavior of PAPS synthases. Notably, the nucleotide APS stabilized both PAPS synthases and this effect was specific for this intermediate of PAPS synthesis. Moreover, APS effectively prevented aggregation of PAPSS2. Finally, we classified invertebrate PAPS synthases by machine learning as the PAPSS2 type, and tested this prediction biochemically upon cloning and expression of PPS-1, the sole PAPS synthase of C. elegans. Strikingly, it behaved like human PAPSS2 in several aspects. PAPS synthases undergo dramatic changes in their behavior in the presence of physiological concentrations of APS that are expected to change when the cell consumes the sulfate donor PAPS. Hence, PAPS synthases may represent biochemical sensors for the sulfation requirements of a cell.
MATERIALS AND METHODS
Protein Expression
Expression constructs for human PAPS synthases 1 (RefSeq: NM_005443) and 2A (RefSeq: NM_004670) in a modified pET-41 vector with GST-His6 fusion tags and a recognition site for PreScission protease and a deleted ApaI site within the LacI open reading frame have been described previously (17). PPS-1 (RefSeq: NM_069456) was amplified from C. elegans cDNA (kindly provided by Cristina Hartmann-Fatu) using the following primers: 957F, 5′-CAC ACA CGG GCC CCT CAC TCC ACG GGA TGA AAA CAA CG-3′ (ApaI) and 958R, 5′-CCA ACC AAG CTT ATT AGT TTG AGT TTT GTA GTG ATT TGT AGT AGC C-3′ (HindIII). This fragment was then ApaI/HindIII ligated into the modified pET-41 vector described previously (17) and fully sequenced using the internal primers 961F, 5′-GCT CAA GGA GAG TCA AAA TCT TCC-3′ and 962R, 5′-TGT TCC AAA TTG ACG ACA TAC ACG-3′, as well as standard primers. Point mutants of human PAPS synthase 1 and 2A were obtained by site-directed mutagenesis with DpnI digestion of the parental DNA (QuikChange kit, Stratagene, Agilent Technologies, Waldbronn, Germany) using the primers listed in supplemental Table S1. All constructs were verified by sequencing and expressed as described previously (17) including heterologous protein expression in Escherichia coli BL21/Rosetta (DE3) cells, GSH-Sepharose purification, PreScission cleavage, and gel filtration on Superdex 200.
Enzymatic Assays
After incubation of 5 μm PAPS synthase for 3–12 min at temperatures between 37 and 50 °C, denaturation was stopped by rapid cooling on ice and the remaining kinase or sulfurylase activity was assayed as described previously (17, 19, 20). In short, the kinase activity was measured in a coupled spectrophotometric assay where production of ADP is linked to the consumption of NADH via pyruvate kinase and lactate dehydrogenase with the following conditions according to STRENDA convention: 0.3 mm NADH, 0.8 mm P-enolpyruvate, 25 units of lactate dehydrogenase, 17.5 units of pyruvate kinase (Sigma), 2.5 mm ATP, 1 unit of nuclease P1, 500 nm PAPS synthase, 0.8 mm MgCl2, 20 mm Tris, pH 7.3, 100 mm KCl, 0.1 mm EDTA. Reactions were started by adding 15 μm APS into a total volume of 1 ml at 25 °C.
Sulfurylase activity was measured in reverse direction (toward ATP synthesis) by coupling the formation of ATP to NADPH production via hexokinase and glucose-6-phosphate dehydrogenase (Glc-6-P DH) as the following: 0.3 mm NADP+, 1 mm glucose, 0.5 mm pyrophosphate, 4 units of Glc-6-P DH, 3.5 units of hexokinase (Sigma), 70 nm PAPS synthase, 5 mm MgCl2, 20 mm Tris, pH 7.3, 100 mm KCl, 0.1 mm EDTA. Reactions were started by adding 150 μm APS into a total volume of 1 ml at 20 °C. NADH consumption or NADPH production was monitored for 10 min at 340 nm in a Varian Cary 100 spectrophotometer and the remaining enzyme activity was plotted logarithmically versus incubation time at various temperatures (supplemental Fig. S2) to obtain half-times of inactivation.
Fluorescence Spectroscopy
Unfolding transitions induced by urea, guanidine hydrochloride, or temperature increase were monitored by fluorescence spectroscopy in HEPES buffer (20 mm HEPES, pH 7.3, 150 mm NaCl, 1 mm MgCl2, 0.5 mm EDTA, and 1 mm DTT). For chemical unfolding, an increasing concentration of detergent was added to a constant concentration of 250 nm PAPS synthase and allowed to equilibrate for 5 min. Subsequently, fluorescence emission intensities were recorded at 334 and 376 nm with excitation at 280 nm in a Varian Cary Eclipse fluorimeter (averaging time, 15 s; excitation slit, 10 nm; emission slit, 5 nm). Similarly, thermal unfolding of 500 nm PAPS synthase was followed from 25 to 75 °C with a 0.1 °C data interval and a heating rate of 25 °C/h (average time, 8 s; excitation slit, 10 nm; emission slit, 10 nm).
The difference in fluorescence intensities was normalized according to Equation 1 and then normalized between 0 and 1,
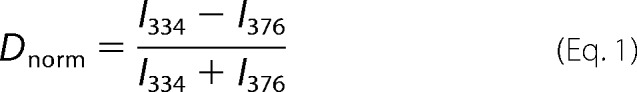 |
where I334 and I376 represent the fluorescence intensities at 334 and 376 nm, respectively.
CD Measurements
Far-UV CD measurements were recorded on a Jasco J-715 spectropolarimeter fitted with a cell holder thermostatted by a PTC 348-WI Peltier unit. CD intensities are presented as the CD absorption coefficient calculated on a mean residue weight basis (ΔϵMRW). All CD measurements were made in 10 mm HEPES, pH 7.3, 50 mm NaCl, 1 mm MgCl2, 0.5 mm EDTA, and 1 mm DTT. CD spectra were typically recorded with 0.2-nm resolution and baseline corrected by subtraction of the appropriate buffer spectrum using fused silica cuvettes with 1-mm path length (Hellma, Jena, Germany). Data processing was achieved with software written in house.4
Thermal unfolding curves were obtained by monitoring the ellipticity at 225 nm using 10-mm path length cuvettes. Thermal unfolding of PAPS synthases was irreversible under our conditions for the large and dimeric PAPS synthase proteins. As expected for an irreversibly unfolding system, midpoints of thermal transitions were dependent on the heating rate for both human PAPS synthases. Hence, identical heating rates of 25 °C/h were applied for all measurements.
Chemical unfolding was induced by addition of 500 nm PAPS synthase to different concentrations of urea in 2-mm cuvettes. Ellipticity at 225 nm was averaged for 60 s (data pitch, 5 s; response time, 4 s; bandwidth, 1 nm) and the titration points were normalized between 0 and 1.
For unfolding kinetics measurements, the ellipticity of 0.5 μm PAPS synthase was monitored at 225 nm with a response time of 8 s using 10-mm cuvettes. Data points were fitted to Equation 2,
where S(t) represents the ellipticity at time t, S0 the ellipticity at the start of the measurement, P the ellipticity of denatured protein, and k the rate constant of unfolding. Half-lives were calculated as t0.5 = ln2/k and extrapolated to 37 °C using an exponential decay fit. Activation energies were determined by a linear regression of 1/K against 1/T, yielding the activation energy in J/mol as slope.
PAPS Synthase Sequences
Orthologous PAPS synthase sequences were downloaded from Ensembl (21). At the time of analysis, this included 45 PAPSS1 and 47 PAPSS2 vertebrate sequences. Moreover, six invertebrate sequences as well as three PAPS synthase protein sequences from zebrafish were obtained, but not included in the training data set. 60% of these sequences contained more than 600 amino acids and were, hence, regarded as full-length sequences. The remaining 37 sequences had an average length of 491 amino acids mostly missing one or two exons. Related sequences from the guide tree feature were then used to manually fill up the missing exons. Sequence information for an alternatively spliced PAPSS2B form could be obtained for 29 PAPSS2 sequences. This additional exon of PAPSS2B comprised 5 amino acids on average, was mostly hydrophobic, but did not display obvious sequence conservation. Whether this insertion plays a functional role analogous to a 5-amino acid insertion in some parvulin proteins (22) remains to be determined. This exon was omitted from the 604 amino acids comprising the PAPS synthase core sequence as was the variable N-terminal 10–20 amino acids. The final set of 101 PAPS synthase core sequences was then used to estimate phylogeny by maximum-likelihood using PhyML 3.0 (23) with 100 bootstraps as previously described (24); including FigTree 1.3.1 for visualization.
Machine Learning
In our study we used a linear support vector machine as well as a random forest (RF) to discriminate PAPSS1 and PAPSS2 sequences. The models were trained using 45 PAPSS1 and 47 PAPSS2 sequences. We used the R packages kernlab and randomForest, respectively (25).
For pre-processing of the amino acid sequences, we used the R package Interpol (26). The sequences were consecutively encoded using all available descriptors in the Interpol package and the resulting importance values of the RF classification were averaged. As importance measurement we used the mean decrease in Gini impurity (27).
Limited Proteolysis
To test for protein stability by an orthologous method, 10 μg of PAPS synthase protein was incubated with varying concentrations of proteinase K (New England Biolabs, Frankfurt am Main, Germany) in 20 μl of buffer (20 mm HEPES, pH 7.3, 150 mm NaCl, 0.5 mm EDTA, 1 mm MgCl2, 1 mm DTT). After incubation for 10 min at 20 °C, proteolysis was stopped by addition of 5 mm phenylmethanesulfonyl fluoride (Roth, Karlsruhe, Germany). Each sample was analyzed by 12.5% SDS-PAGE and Coomassie staining.
Aggregation Monitoring
PAPS synthase aggregation was monitored in a Varian Cary Eclipse fluorimeter as described previously (28). In short, 500 nm PAPS synthase was excited with monochromatic light (280 nm) and the intensity of the scattered light was measured also at 280 nm. Additionally, the protein fluorescence signal was recorded at 340 nm and the aggregation rate was calculated according to Equation 3,
where I280 and I340 represent the intensities of emitted light at 280 and 340 nm, respectively. This ratio is displayed in all aggregation diagrams. Half-times were calculated by monoexponential fitting as described for CD traces.
RESULTS
PAPS Synthase Isoforms Differ Markedly in Their Stability
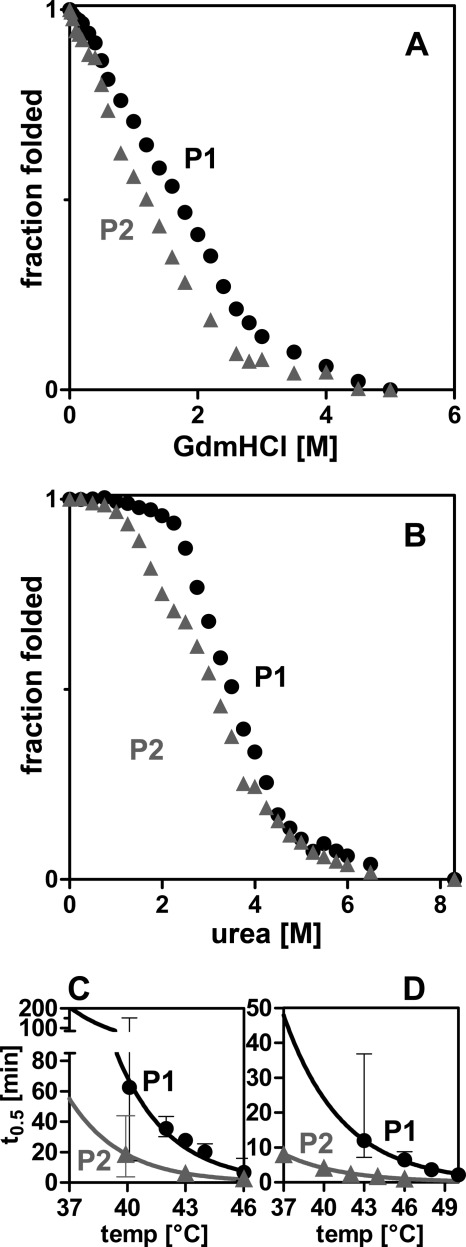
In searching for differences between PAPS synthase isoforms, we analyzed protein stability by chemically induced unfolding of PAPSS1 and -S2 monitored by intrinsic tryptophan fluorescence. First, unfolding was induced by guanidine hydrochloride (GdnHCl) (Fig. 1A). The two proteins differed markedly in their stability against GdnHCl-induced unfolding. 0.5 m less GdnHCl was required to reach the midpoint of unfolding for PAPSS2 than for PAPSS1. Both curves lack a pronounced plateau for the fully folded proteins at low GdnHCl concentrations; this observation can be explained by a distinct salting-in effect of PAPS synthases (supplemental Fig. S1, A and B). On the contrary, urea-induced unfolding resulted in extended plateau phases for both folded proteins, but again the midpoint of unfolding was reached at 0.4 m lower urea concentrations for PAPSS2 than for PAPSS1 (Fig. 1B). Urea-induced unfolding monitored by CD confirmed the higher stability of the PAPSS1 isoform (supplemental Fig. S1C). These findings clearly indicate that PAPSS2 is less stable than PAPSS1 against chemically induced unfolding.
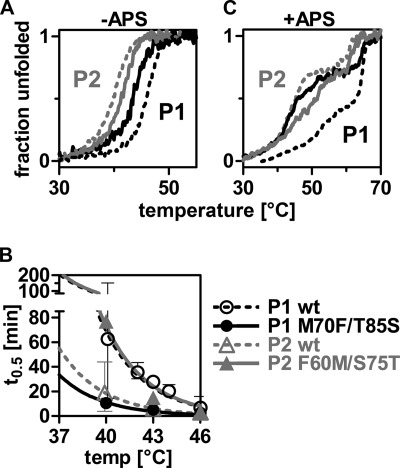
FIGURE 1.
PAPS synthases differ in their protein stability. A and B, chemical unfolding of human PAPS synthases by guanidine hydrochloride (GuaHCl) and urea monitored by intrinsic tryptophan fluorescence. Irreversible unfolding proceeds via at least one intermediate. Midpoints of unfolding were at 1.70 and 1.19 m GdnHCl, as well as 3.50 and 3.11 m urea for PAPS synthases 1 and 2 (P1 and P2), respectively. C and D, loss-of-activity measurements of APS kinase and ATP sulfurylase domains, respectively. Proteins were incubated for increasing times at elevated temperatures. Subsequently, residual activity was measured under standard conditions. Error bars represent 95% confidence intervals.
Structural studies have shown that the kinase part of PAPSS1 behaved differently within the context of the full-length protein and as an isolated protein domain. In one case they were half-loaded with nucleotides in an asymmetric conformation (7); in another they showed nearly perfect C2 symmetry (29). Hence, we wanted to investigate the two domains individually, but within the context of full-length bifunctional PAPS synthases. Therefore, we recorded residual enzymatic activity after thermal inactivation for the APS kinase and ATP sulfurylase, respectively. PAPS synthase proteins were incubated at different temperatures and then assayed under standard conditions using coupled enzymatic assays (supplemental Fig. S2). Half-lives for the irreversible inactivation of the kinase and sulfurylase activities were finally extrapolated to 37 °C (Fig. 1, C and D). Both domains were less stable in PAPSS2 than in PAPSS1. Most notably, half of the sulfurylase activity of PAPSS2 was already gone after 8 min at 37 °C under our conditions. If PAPSS2 is such an unstable protein, it might not be able to function as a proper enzyme. But why then do mutations in the respective genes cause severe diseases? In other words, something needs to stabilize this fragile protein within the cell.
Stabilization of PAPS Synthase by Ligand Binding
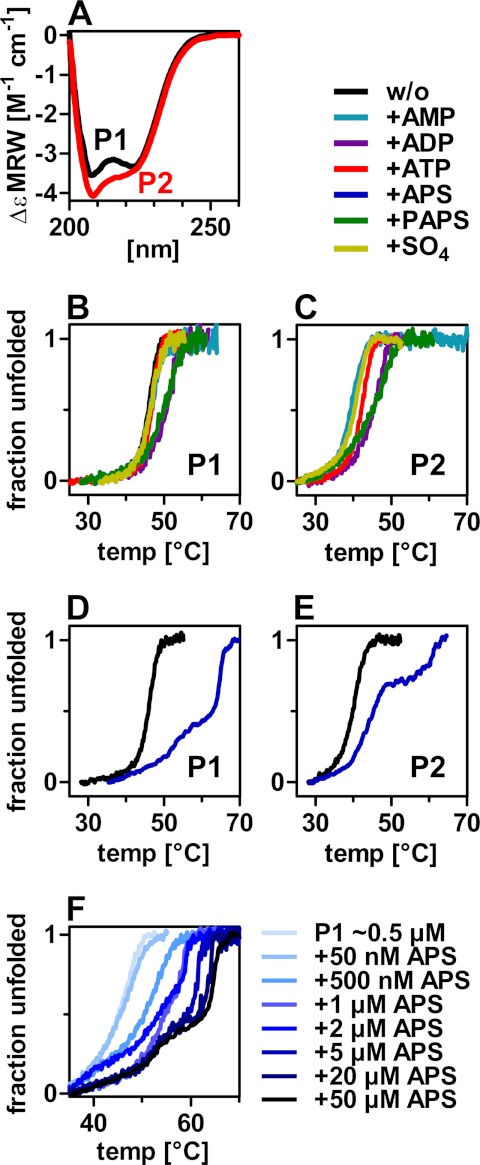
Therefore, we tested the substrates, intermediates, and products of PAPS formation itself for their ability to stabilize the PAPS synthase proteins. PAPS formation is schematically shown in supplemental Fig. S3. A CD spectrum of PAPS synthase 1 indicates a pronounced α-helical secondary structure (Fig. 2A) in agreement with the known crystal structure of PAPSS1 code 1X6V containing 38% α-helix (7). PAPSS2 has a similar CD spectrum, however, with more negative ellipticity at 208 nm indicative of either a slightly different overall secondary structure or local unfolding (Fig. 2A). CD spectra of the two proteins at elevated temperatures are in agreement with the latter (supplemental Fig. S4, A and B). Thermal unfolding of both proteins was monitored using the CD signal at 225 nm in the presence of various ligands (Fig. 2, B and C). Sulfate and AMP had no effect on the unfolding transition of PAPS synthases; ATP had a slight stabilizing effect on PAPSS2 and moderate effects on PAPSS1 and -S2 stability were observed for ADP and PAPS.
FIGURE 2.
Thermal unfolding of PAPS synthases in the presence of several ligands. A, far CD spectra of human PAPS synthases at 20 °C. Increased negative ellipticity of PAPSS2 (P2) at 208 nm may be attributed to partial unfolding. B–E, thermal unfolding of PAPS synthases monitored by CD spectroscopy at 225 nm in the presence of several ligands. 50 μm AMP, 100 μm ADP, 100 μm ATP, 50 μm APS, 50 μm PAPS, or 2 mm sulfate were added to about 0.5 μm recombinant protein (P1/P2 as indicated). F, thermal unfolding of PAPS synthase 1 (P1) in the presence of increasing APS concentrations.
The most prominent effect on PAPS synthase unfolding, however, had the nucleotide APS, which is the intermediate of overall PAPS formation (Fig. 2, D and E). 50 μm APS caused changes in the overall form of the unfolding transitions as it stabilized an unfolding intermediate by more than 16 °C in both PAPS synthases. From the analysis of several point mutants described below, the stabilized transition was assigned to the unfolding of the sulfurylase domain of bifunctional PAPS synthases.
The distinct shapes of the unfolding transitions of the two PAPS synthases in the presence of APS may arise from the simultaneous versus consecutive unfolding of kinase and sulfurylase domains in PAPSS1 and -S2, respectively. The kinase domain of PAPSS1 was found to be more stable than that of PAPSS2 as seen in the inactivation measurements described above. Assuming this also translates into different thermodynamic stability for the two domains, PAPSS2 kinase unfolding is complete when the APS-stabilized sulfurylase unfolds. In contrast, PAPSS1 kinase unfolding is coincident with unfolding of the APS-stabilized PAPSS1 sulfurylase.
As APS had a significant influence on PAPS synthase stability, thermal unfolding of 0.5 μm PAPSS1 protein was recorded with increasing APS concentrations (Fig. 2F). At equimolar concentrations, APS already had a remarkable effect on the stability of PAPSS1, increasing the midpoint of the unfolding transition by 4 °C. At higher APS concentrations, the shape of the transition changed and an unfolding intermediate becomes visible as described above. Similar changes to the shape of the unfolding transitions were observed for PAPSS2 (supplemental Fig. S4C). The stabilizing effect of APS was additionally confirmed by chemical denaturation (supplemental Fig. S4D). As the affinity for any ligand is a temperature-dependent parameter, it was not possible to derive KD values for APS binding. However, our measurements suggest two effects of APS on the protein: one with high and one with moderate affinity.
Classifying Invertebrate PAPS Synthases
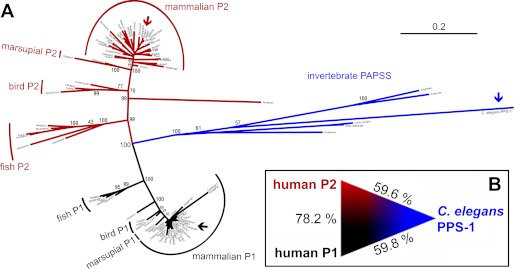
Human PAPS synthase proteins differ significantly in their stability. Is it possible to determine which of the two isoforms is evolutionarily older? We collected all available vertebrate PAPS synthase protein sequences as well as six sequences from invertebrates (see “Materials and Methods” for details) and used these data to compute a maximum-likelihood phylogenetic tree (Fig. 3A). Within this tree, PAPSS1-type, PAPSS2-type, and the invertebrate sequences form three distinct groups separated by a bootstrap value of 100. Moreover, the percentage of amino acid identity (59.8 and 59.6% for human PAPSS1 and -S2, respectively) was found to be nearly identical when directly comparing the two human protein sequences with the sole PAPS synthase PPS-1 of the roundworm C. elegans (Fig. 3B), indicating that this was a nontrivial problem.
FIGURE 3.
Relationship between the human PAPS synthases and their counterpart from C. elegans. A, un-rooted maximum likelihood phylogenetic tree of vertebrate and invertebrate sequences for bifunctional PAPS synthases. The three groups of proteins, PAPSS1-type (black), PAPSS2-type (red), and invertebrate PAPS synthases (blue) are clearly separated by a bootstrap of 100. PAPS synthases from man and worm are marked by arrows. B, amino acid identity between C. elegans PPS-1 and the two human PAPS synthase isoforms indicates that it is not possible to derive further phylogenetic relationships for these proteins.
Still eager to address this question, the collected vertebrate PAPS synthase protein sequences were used to train machine learning models to discriminate between PAPSS1- and PAPSS2-type proteins. A linear support vector machine and a RF were able to distinguish clearly between these two classes. PAPS synthases from the zebrafish Danio rerio were not included in the training set, as this genome contained two annotated PAPSS2 sequences. This is in agreement with one additional round of genome duplication in the line of teleost fish (30). Hence, the three zebrafish sequences could be used as a test case. In agreement with their annotation as PAPSS1, PAPSS2-A, and -S2-B, they were classified by RF as one PAPSS1-type and two PAPSS2-type proteins. Class probability is expressed as P2 = 0 and P1 = 1, and average scores for the three PAPS synthase sequences from zebrafish were 0.94, 0.17, and 0.12 for the sequences annotated as PAPSS1, -S2A, and -S2B, respectively.
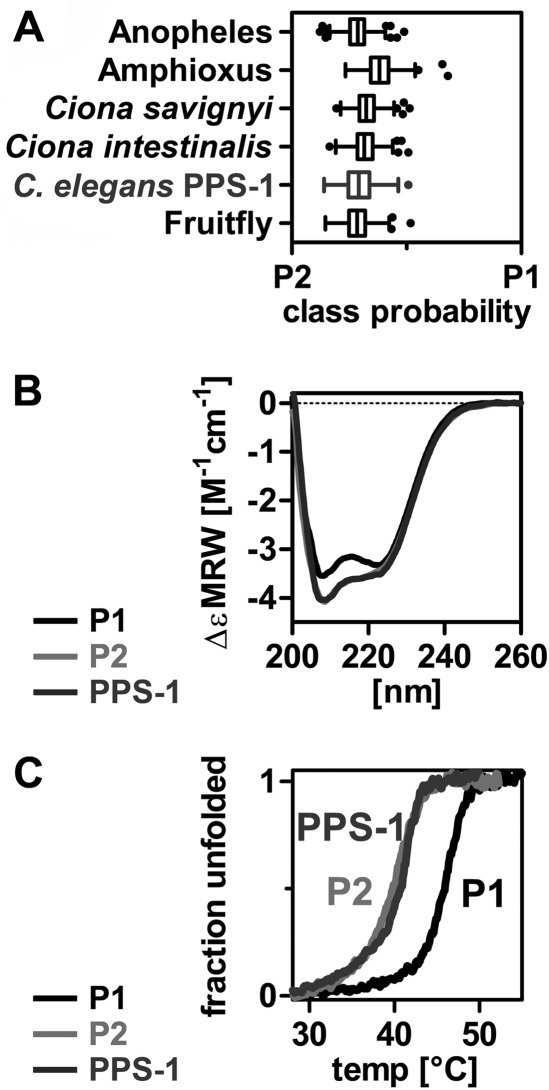
Next, six invertebrate PAPS synthase sequences including the PPS-1 protein from C. elegans were subjected to machine learning-based classification. Notably, they were all classified as PAPSS2-type proteins (Fig. 4A). To test this prediction biochemically, we cloned and expressed the C. elegans PPS-1 protein. Its CD spectrum matches the spectrum of human PAPSS2 including the increased negative ellipticity at 208 nm (Fig. 4B). Strikingly, PPS-1 had a midpoint of thermal unfolding similar to human PAPSS2 (Fig. 4C). Accordingly, its half-life and activation energy for unfolding as well as its stabilization by APS binding resembled the values of human PAPSS2 (supplemental Fig. S5). These data suggest that the PAPS synthase isoform that we found to be less stable is evolutionarily older.
FIGURE 4.
Machine learning classifies invertebrate PAPS synthases as PAPSS2 types. A, a support vector machine and a random forest were trained to discriminate between PAPSS1- and -S2-type sequences using 45 PAPSS1 and 47 PAPSS2 sequences from vertebrates. The predicted classes for six invertebrate sequences including C. elegans PPS-1 are shown for the random forest output. All these proteins are clearly classified as PAPSS2-type by both machine learning approaches. B, the CD spectrum of C. elegans PPS-1 resembles the one from human PAPSS2 including increased negative ellipticity at 208 nm. C, the thermal unfolding transition of C. elegans PPS-1 is shown. It is very similar to the one of human PAPSS2.
Differences in Amino Acid Content of PAPS Synthase Isoforms
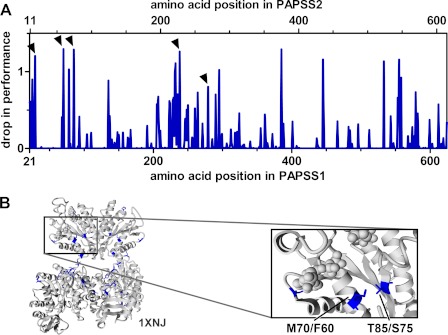
The multiple sequence alignment of the 604 amino acid core sequences from more than 90 PAPS synthases contained 144 invariant positions, 165 positions with some homology, and 295 positions without apparent conservation. Which of these differences account for the above described difference in protein stability? In addition to the support vector machine, we also trained a RF on our dataset. RFs are not only able to classify the data into PAPSS1- and -S2 types correctly, but also give position-specific drop-in performance values (27). These are calculated by iterative sampling of all positions within the multiple sequence alignment monitoring the performance of classification. This allows one to select amino acids important for classification that are difficult to find manually (Fig. 5A). With this approach, 23 positions showed drop-in performance values greater than a cut-off value of 0.62%. Numbering in the two human PAPS synthases differs by 10 amino acids due to the varying length of their N termini. These 23 positions were then mapped onto the crystal structure 1X6V of human PAPSS1 (Fig. 5B).
FIGURE 5.
Sequence positions important for PAPSS1 and S2 discrimination. A, machine learning by random forest yields so called drop-in performance values. They are obtained by leaving out iteratively every single position of the multiple sequence alignment. Average values for all used sequence descriptors are plotted. B, 23 positions with the highest drop-in-performance values are mapped on PDB code 1XNJ. The amino acid pair Met-70/Phe-60 and Thr-85/Ser-75 being close together in space is specially highlighted (inset).
One of the amino acid positions picked up by this approach was Met-70 of PAPSS1, which is close to the ADP/ATP nucleotide binding site of the APS kinase domain. Another highlighted position in spatial proximity to Met-70 was Thr-85. These two residues are replaced by Phe-60 and Ser-75 in human PAPSS2 (Fig. 5B). We swapped these two residues by mutagenesis and investigated their effect on protein stability by CD spectroscopy. The two residues from PAPSS2 destabilized the full-length PAPSS1 protein by 3.0 °C. On the contrary, substituting the residues of PAPSS1 also stabilized PAPSS2 by 1.5 °C (Fig. 6A). Considering the moderate nature of these exchanges, 1.5 °C stabilization is a large effect. Other highlighted residues that we targeted by mutagenesis did not result in comparable changes in protein stability (supplemental Fig. S6). The effect of the Met ⇔ Phe and Thr ⇔ Ser double mutants became even more pronounced when looking at loss-in-activity measurements at different temperatures. There, the two substituted residues resulted in a complete reversal of inactivation behavior for the APS kinases (Fig. 6B).
FIGURE 6.
Characterization of the Met ⇔ Phe/Thr ⇔ Ser double mutants. A, thermal unfolding transitions monitored by CD spectroscopy at 225 nm. B, loss-of-activity measurement of the APS kinase activity of wild-type and mutant proteins. C, thermal unfolding transitions in presence of 50 μm APS. The legend applies to all three graphs.
Looking at thermal unfolding in the presence of APS, mutating M70F and T85S in PAPSS1 resulted in a severely destabilized protein (Fig. 6C). The unfolding transition at the lower temperature nearly resembled that of the PAPSS2 protein. The APS-stabilized second transition, however, overlaid with that of the wild-type PAPSS1 protein. The effect of the appropriate double mutant in PAPSS2 was again observed for the thermal transition at around 50 °C that was slightly stabilized. The APS-stabilized unfolding intermediate at high temperature overlapped with the PAPSS2 wild-type protein (Fig. 6C). Taken together, the kinase mutations modified the first transition, leaving the second unchanged. As the mutations introduced into the kinase domain should have a direct effect only on that domain, we assigned the first transition to the unfolding of the kinase domain. The second APS-stabilized transition should then correspond to unfolding of the sulfurylase domain.
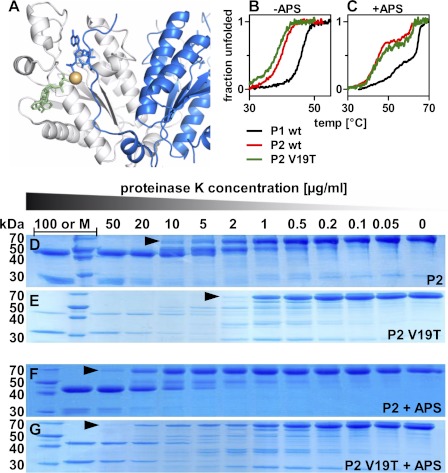
A third important residue in close proximity to the ligand binding site was Thr-29 within the 2OFX structure of the APS kinase domain of PAPSS1 (29). This residue is positioned close to the bound magnesium ion, but lies in an N-terminal part from the other dimeric subunit (Fig. 7A). The swapping mutant V19T in PAPSS2 destabilized the protein by 1.7 °C (Fig. 7B). In the presence of APS, however, this effect is more than rescued (Fig. 7C). The corresponding T29V mutation in PAPSS1 fails to stabilize the protein in the presence of APS (supplemental Fig. 7, A and B).
FIGURE 7.
The V19T mutant of PAPSS2. A, highlighting Thr-29 within the crystal structure PDB code 2OFX of the kinase domain of human PAPSS1. This position of this residue is occupied by valine in PAPSS2. It was also highlighted by random forest analysis. B, unfolding transition for the V19T mutant of PAPSS2. The midpoint of unfolding is lowered from 39.8 to 38.2 °C. C, in presence of APS, however, unfolding is more than rescued. There, the midpoint of the unfolding transition is even shifted to higher temperatures from 45.3 to 47.0 °C indicative of a specific role of threonine at this sequence position. D and E, proteolysis of PAPSS2 by limiting amounts of proteinase K (PK). The amount of PK needed to cleave the full-length proteins differs more than 5-fold for wild-type and V19T mutant proteins. F and G, addition of 50 μm APS stabilized the V19T mutant protein nearly to the wild-type level.
The molecular phenotype of the PAPSS2 V19T mutant was also evident in limited proteolysis studies. V19T was more susceptible toward limited amounts of proteinase K. As seen in Fig. 7, D and E, more than 5-fold proteinase K was needed to cleave wild-type PAPSS2 than for the V19T mutant. Also here, APS binding rescued this mutant, so that the same proteinase K concentration was needed for wild-type and V19T mutant digestion (Fig. 7, F and G). Notably, it was the full-length proteins that were stabilized by APS in these experiments, indicating that this stabilization did not directly mirror the stabilizing effect seen in CD. In this context, APS was not specific: ADP, ATP, and PAPS protected PAPS synthase proteins to a similar extent (data not shown). Nevertheless, a threonine at position 29/19 contributes to protein stability in the presence of APS, a role that cannot be performed by valine. Within the above mentioned crystal structure 2OFX (29), Thr-29 contacts the OH group of C2′ of the ribose moiety of the bound PAPS cofactor via one bridging water molecule. Nothing is yet known about the spatial arrangement of Val-19 in PAPSS2, because this part of the sequence is disordered in the only available PAPSS2 crystal structure of 2AX4.
Tracing Mutants through Multiple Sequence Alignment of PAPS Synthases
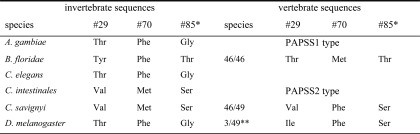
The three residue positions (Thr-29, Met-70, and Thr-85 in PAPSS1) with large effects on protein stability were looked at within the multiple sequence alignment of vertebrate and invertebrate PAPS synthases (Table 1). Strikingly, a Met-70/Thr-85 pair was found in all 46 PAPSS1-type and a Phe-60/Ser-75 pair in all 49 PAPSS2-type vertebrate sequences. Of note, the invertebrate sequences were more heterogeneous at these two positions (Table 1). Val-19 in PAPSS2 is present in 46 sequences, but three species have isoleucine at this position. Thr-29, probably indirectly involved in nucleotide binding, was 100% conserved in all PAPSS1-type sequences. Invertebrate sequences have mostly either Val or Thr at this position (Table 1). C. elegans PPS-1 contains a Thr residue at the N-terminal position, but displays protein stability similar to that of human PAPSS2. In agreement, the midpoint of unfolding of the protein is increased by 9.3 °C in the presence of 50 μm APS. This closely resembles the destabilized V19T mutant of PAPSS2 where APS causes a stabilization by 8.8 °C. Assuming the sulfurylase domain is sufficiently stabilized by endogenous steady-state concentrations of APS, the kinase domain becomes more important for overall protein stability. Here, we have characterized three major sites that contribute to isoform stability by swapping mutagenesis.
TABLE 1.
Tracing the mutants through a multiple sequence alignment of PAPS synthases
* Amino acid numbering is according to human PAPSSI.
** These species are horse (Equus caballus), lesser hedgehog tenrec (Echinops telfairi), and hyrax (Procavia capensis).
Several other positions were also highlighted by the random forest. For all 23 positions mapped onto the crystal structure 1X6V of PAPSS1 in Fig. 5B, we generated sequence logos including all PAPS synthases or subgroups thereof (supplemental Fig. S8). From the logo, one can again appreciate the existence of two PAPS synthase subfamilies. Mostly, invertebrate PAPS sequences follow the sequence motif of the PAPSS2 family, however, with several exceptions. Hence, their classification as PAPSS2-type sequences can be understood at the sequence level. What this means from an evolutionary point of view is discussed further below.
Stabilization of PAPSS2 Also Prevents Protein Aggregation
Until now, the effect of APS was discussed in the context of thermal unfolding. But also at physiological temperatures, the behavior of PAPS synthases was modified by this nucleotide. First, we investigated kinetics of unfolding at constant temperatures by CD spectroscopy (supplemental Fig. 9A). The half-lives of PAPSS2 that were extracted from these traces were clearly shorter than that of PAPSS1 (supplemental Fig. 9B). However, in the presence of 50 μm APS, this value was at least doubled for PAPS synthase 2 (from 12 to more than 27 min at 37 °C). Moreover, residual activity for both the kinase and sulfurylase activity was increased when incubating PAPS synthases with APS at elevated temperatures (data not shown).
Not only is protein stability important within the cell, much more critical is whether the destabilized protein aggregates. We knew from our experiments that PAPS synthases tend to aggregate after prolonged measurement times. Such aggregation needs to be prevented in vivo. Hence, we monitored formation of soluble aggregates by a method described by Travé and co-workers (28) using fluorescence signals at 340 and 280 nm where the latter corresponds to a scattering signal.
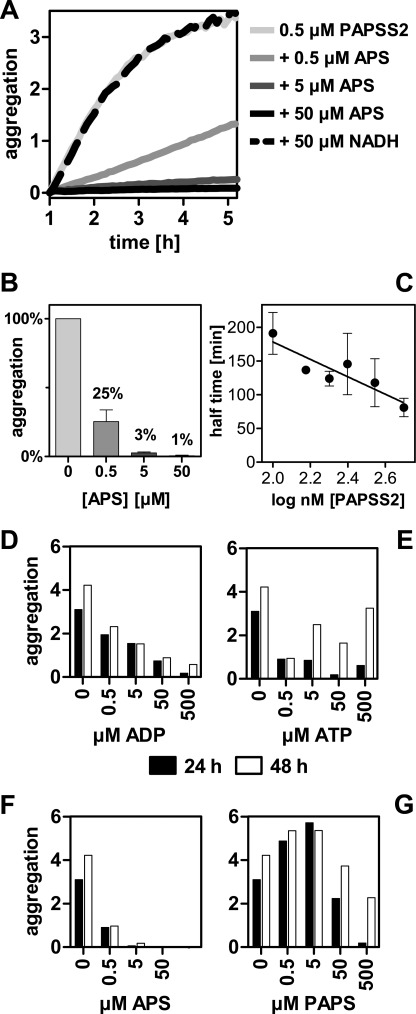
Equimolar concentrations of APS already prevented 70% of aggregation after 24 h at 37 °C (Fig. 8, A and B). Remarkably, this effect was specific for APS. As a negative control, NADH was tested, which had no effect on aggregation at all. The aggregation phenomenon reported here was faster at higher concentrations (Fig. 8C), proving that the effect was due to self-association. The other nucleotides ADP, ATP, and PAPS only had some effect at higher concentrations (Fig. 8, D–G). The aggregates formed during these measurements could not be stained with thioflavin (data not shown) indicating that no amyloid fibers were formed.
FIGURE 8.
APS specifically modifies aggregation behavior of PAPSS2. A, using the side scatter signal at 280 nm normalized by 340 nm fluorescence, aggregation was monitored for PAPSS2 in the presence of varying concentrations of APS at 37 °C. Equimolar concentrations of APS effectively prevented aggregation. The nucleotide cofactor NADH was used as negative control and showed no effect. B, point measurements for the same reactions after 24 h. Equimolar concentrations of APS reduced aggregation to 25%. Error bars represent S.D. of triplicate measurements. C, concentration dependence of the aggregation phenomenon. Aggregation of PAPSS2 was analyzed at concentrations between 100 and 500 nm. Error bars represent S.D. of duplicate measurements. D–G, at low nucleotide concentrations, prevention of aggregation of PAPSS2 was specific for APS. The other nucleotides tested (ADP, ATP, and PAPS) had less effect on preventing protein aggregation at concentrations up to 500 μm.
APS has at least a dual effect on the stability of PAPS synthases. First, it remarkably stabilized an unfolding intermediate and, second, it effectively prevented the proteins' aggregation. Of note, the CD signal during thermal unfolding may have also been changed due to formation of aggregates at higher temperatures. However, chemically induced unfolding (Fig. 1, A and B) needs to have shown protein unfolding as aggregation does not occur at increasing concentrations of chaotropic agents clearly showing that the two isoforms of PAPS synthases differ greatly in their stability.
DISCUSSION
Bifunctional PAPS synthases are responsible for the enzymatic activation of sulfate in all metazoans. These proteins gained attention in biomedical research as mutations in the respective genes caused by a variety of diseases in mouse and man (9, 10, 12–14). The question why two human protein isoforms, PAPSS1 and -S2, exist, but are not able to complement for each other has been around for more than a decade. Recently, we have reported that formation of heterodimers, diverging specific activity, and markedly different cellular localization, all previously assumed to account for different functionality, were not sufficient to explain the above mentioned observation (17, 18).
In the present study we describe for the first time PAPS synthases as fragile enzymes. There was a remarkable difference in stability for the two human isoforms, PAPS synthase 1 and 2. PAPSS2 underwent unfolding and aggregation already at physiological temperatures.
The intermediate of overall PAPS synthesis, the nucleotide APS, stabilized PAPS synthase proteins significantly and effectively suppressed the aggregation of PAPS synthase 2. This protein undergoes conformational changes in the presence of 5–50 μm APS at physiological temperatures. These concentrations of APS may readily occur within the cell during PAPS synthesis: Lansdon et al. (19) have approximated a steady state APS concentration of 1.6 μm that can accumulate up to 60 μm under conditions of sulfate excess. These steady state concentrations of APS are exactly in the same range as those significantly stabilizing the PAPSS2 protein. Hence, naturally occurring changes in APS concentrations may be sensed by changes in the conformation of PAPSS2.
Interestingly, Kopriva and colleagues (31) have recently reported increased transcript levels for genes of the glucosinolate biosynthesis in Arabidopsis, when bacterial APS kinase was overexpressed. Glucosinolates are a major class of sulfated secondary metabolites in plants and their biosynthesis requires PAPS as sulfate donor. From their findings they concluded that PAPS or its derivatives may be involved in cellular signaling in plants (31). One such derivative is PAP, the leftover of sulfation reactions (also depicted in supplemental Fig. S3) has recently been implicated in stress signaling in plants (32, 33). PAP also needs to be removed from bacterial (34) and vertebrate (35) cells due to its inhibition of 5′-3′ RNA degradation (36). However, PAP did not have any effect on thermal unfolding behaviour of human PAPS synthases or the enzyme from worm (supplemental Fig. S10).
Nevertheless, we provide here biochemical evidence for a specific effect of APS on PAPS synthase stability. Based on these findings, one could assume a regulatory function that is executed by PAPS synthase 2. The conformational transition of this protein in the presence of physiological levels of APS may be used by eukaryotic cells to sense their sulfation needs that correspond to cellular demands for the active sulfate PAPS. Upon usage of PAPS by any sulfotransferase, APS concentrations are expected to drop and PAPS synthase 2 is then subject to conformational changes. This would still be a paradox situation: when more PAPS is required and the levels of APS decrease, exactly then PAPSS2 unfolds and can no longer function as a proper enzyme. Hence, additional cellular compounds, initiating a signaling cascade that ultimately results in transcriptional up-regulation of genes responsible for sulfate uptake and/or activation may be postulated.
Such regulatory function for PAPSS2 might also explain why two genes for PAPS synthases can be found in nearly all vertebrate genomes investigated. Whole genome duplication events were at the advent of vertebrates. According to Wolfe (37) most of the duplicated gene copies were then lost again. However, one reason for retaining both copies might be a subfunctionalization event (38). Invertebrate PAPS synthases would then share the functions of PAPS production and engagement in some regulation of overall PAPS synthesis that were later parcelled out among the two vertebrate daughter genes.
In line with this assumption, the invertebrate PAPS synthase PPS-1 from worm was classified by machine learning as a PAPSS2-type enzyme and displayed stability most similar to human PAPSS2. Additionally, the highest PAPSS2 expression levels were reported in condensing and proliferating chondrocytes of mouse embryos (39). These cells have a high need for sulfation due to beginning synthesis of the severely sulfated extracellular matrix.
CONCLUSION
Stability of PAPS synthases may be a major contributing factor for the availability of the activated sulfate PAPS. The nucleotide APS, an intermediate of PAPS biosynthesis, exerts a highly specific stabilizing effect on PAPS synthases. This mechanism may be used by metazoans to measure their demand in PAPS, reflecting their sulfation needs. Our study paves the way to a functional understanding of different PAPS synthase isoforms. Finally, the combination of machine learning and biochemical approaches may very well serve to address similar questions for other pairs of enzyme isoforms.
Supplementary Material
Acknowledgments
Alma Rüppel is cordially acknowledged for professional protein purification. Special thanks go to Tina Stratmann for establishing the Thermofluor measurements. Cloning of the C. elegans PPS-1 construct was performed by Raffaela Maltaner. C. elegans cDNA was kindly provided by Cristina Hartmann-Fatu. We are grateful to Christine Kallweit and Gebhard Haberhauer for some of the CD measurements as well as Peter Bayer for his generous support.
This work was supported in part by EMBO Fellowship ASTF 151/2011 (to J. W. M.).

This article contains supplemental Figs. S1–S10 and Tables S1.
S. Martin, unpublished software.
- PAPS
- 3′-phosphoadenosine 5′-phosphosulfate
- APS
- adenosine 5′-phosphosulfate
- PAP
- 3′-phosphoadenosine 5′-phosphate
- PAPSS1/2
- PAPS synthase 1/2
- GdnHCl
- guanidine hydrochloride
- RF
- random forest.
REFERENCES
- 1. Klaassen C. D., Boles J. W. (1997) Sulfation and sulfotransferases 5. The importance of 3′-phosphoadenosine 5′-phosphosulfate (PAPS) in the regulation of sulfation. FASEB J. 11, 404–418 [DOI] [PubMed] [Google Scholar]
- 2. Kauffman F. C. (2004) Sulfonation in pharmacology and toxicology. Drug Metab. Rev. 36, 823–843 [DOI] [PubMed] [Google Scholar]
- 3. Maglott D., Ostell J., Pruitt K. D., Tatusova T. (2011) Entrez Gene, gene-centered information at NCBI. Nucleic Acids Res. 39, D52–D57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Allali-Hassani A., Pan P. W., Dombrovski L., Najmanovich R., Tempel W., Dong A., Loppnau P., Martin F., Thonton J., Edwards A. M., Bochkarev A., Plotnikov A. N., Vedadi M., Arrowsmith C. H. (2007) Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol. 5, e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. He D., Falany C. N. (2006) Characterization of proline-serine-rich carboxyl terminus in human sulfotransferase 2B1b. Immunogenicity, subcellular localization, kinetic properties, and phosphorylation. Drug Metab. Dispos. 34, 1749–1755 [DOI] [PubMed] [Google Scholar]
- 6. Goettsch S., Badea R. A., Mueller J. W., Wotzlaw C., Schoelermann B., Schulz L., Rabiller M., Bayer P., Hartmann-Fatu C. (2006) Human TPST1 transmembrane domain triggers enzyme dimerization and localization to the Golgi compartment. J. Mol. Biol. 361, 436–449 [DOI] [PubMed] [Google Scholar]
- 7. Harjes S., Bayer P., Scheidig A. J. (2005) The crystal structure of human PAPS synthetase 1 reveals asymmetry in substrate binding. J. Mol. Biol. 347, 623–635 [DOI] [PubMed] [Google Scholar]
- 8. Dejima K., Seko A., Yamashita K., Gengyo-Ando K., Mitani S., Izumikawa T., Kitagawa H., Sugahara K., Mizuguchi S., Nomura K. (2006) Essential roles of 3′-phosphoadenosine 5′-phosphosulfate synthase in embryonic and larval development of the nematode Caenorhabditis elegans. J. Biol. Chem. 281, 11431–11440 [DOI] [PubMed] [Google Scholar]
- 9. Faiyaz ul Haque M., King L. M., Krakow D., Cantor R. M., Rusiniak M. E., Swank R. T., Superti-Furga A., Haque S., Abbas H., Ahmad W., Ahmad M., Cohn D. H. (1998) Mutations in orthologous genes in human spondyloepimetaphyseal dysplasia and the brachymorphic mouse. Nat. Genet. 20, 157–162 [DOI] [PubMed] [Google Scholar]
- 10. Kurima K., Warman M. L., Krishnan S., Domowicz M., Krueger R. C., Jr., Deyrup A., Schwartz N. B. (1998) A member of a family of sulfate-activating enzymes causes murine brachymorphism. Proc. Natl. Acad. Sci. U.S.A. 95, 8681–8685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ford-Hutchinson A. F., Ali Z., Seerattan R. A., Cooper D. M., Hallgrímsson B., Salo P. T., Jirik F. R. (2005) Degenerative knee joint disease in mice lacking 3′-phosphoadenosine 5′-phosphosulfate synthetase 2 (PAPSS2) activity. A putative model of human PAPSS2 deficiency-associated arthrosis. Osteoarthritis. Cartilage 13, 418–425 [DOI] [PubMed] [Google Scholar]
- 12. Noordam C., Dhir V., McNelis J. C., Schlereth F., Hanley N. A., Krone N., Smeitink J. A., Smeets R., Sweep F. C., Claahsen-van der Grinten H. L., Arlt W. (2009) Inactivating PAPSS2 mutations in a patient with premature pubarche. N. Engl. J. Med. 360, 2310–2318 [DOI] [PubMed] [Google Scholar]
- 13. Shih W. L., Yu M. W., Chen P. J., Wu T. W., Lin C. L., Liu C. J., Lin S. M., Tai D. I., Lee S. D., Liaw Y. F. (2009) Evidence for association with hepatocellular carcinoma at the PAPSS1 locus on chromosome 4q25 in a family-based study. Eur. J. Hum. Genet. 17, 1250–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bruce J. W., Ahlquist P., Young J. A. (2008) The host cell sulfonation pathway contributes to retroviral infection at a step coincident with provirus establishment. PLoS Pathog. 4, e1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Besset S., Vincourt J. B., Amalric F., Girard J. P. (2000) Nuclear localization of PAPS synthetase 1. A sulfate activation pathway in the nucleus of eukaryotic cells. FASEB J. 14, 345–354 [DOI] [PubMed] [Google Scholar]
- 16. Fuda H., Shimizu C., Lee Y. C., Akita H., Strott C. A. (2002) Characterization and expression of human bifunctional 3′-phosphoadenosine 5′-phosphosulfate synthase isoforms. Biochem. J. 365, 497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grum D., van den Boom J., Neumann D., Matena A., Link N. M., Mueller J. W. (2010) A heterodimer of human 3′-phosphoadenosine 5′-phosphosulfate (PAPS) synthases is a new sulfate activating complex. Biochem. Biophys. Res. Commun. 395, 420–425 [DOI] [PubMed] [Google Scholar]
- 18. Schröder E., Gebel L., Eremeev A. A., Morgner J., Grum D., Knauer S. K., Bayer P., Mueller J. W. (2012) Human PAPS synthase isoforms are dynamically regulated enzymes with access to nucleus and cytoplasm. PLoS One 7, e29559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lansdon E. B., Fisher A. J., Segel I. H. (2004) Human 3′-phosphoadenosine 5′-phosphosulfate synthetase (isoform 1, brain). Kinetic properties of the adenosine triphosphate sulfurylase and adenosine 5′-phosphosulfate kinase domains. Biochemistry 43, 4356–4365 [DOI] [PubMed] [Google Scholar]
- 20. Sekulic N., Konrad M., Lavie A. (2007) Structural mechanism for substrate inhibition of the adenosine 5′-phosphosulfate kinase domain of human 3′-phosphoadenosine 5′-phosphosulfate synthetase 1 and its ramifications for enzyme regulation. J. Biol. Chem. 282, 22112–22121 [DOI] [PubMed] [Google Scholar]
- 21. Flicek P., Amode M. R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S., Fitzgerald S., Gil L., Gordon L., Hendrix M., Hourlier T., Johnson N., Kähäri A. K., Keefe D., Keenan S., Kinsella R., Komorowska M., Koscielny G., Kulesha E., Larsson P., Longden I., McLaren W., Muffato M., Overduin B., Pignatelli M., Pritchard B., Riat H. S., Ritchie G. R., Ruffier M., Schuster M., Sobral D., Tang Y. A., Taylor K., Trevanion S., Vandrovcova J., White S., Wilson M., Wilder S. P., Aken B. L., Birney E., Cunningham F., Dunham I., Durbin R., Fernández-Suarez X. M., Harrow J., Herrero J., Hubbard T. J., Parker A., Proctor G., Spudich G., Vogel J., Yates A., Zadissa A., Searle S. M. (2012) Ensembl 2012. Nucleic Acids Res. 40, D84–D90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mueller J. W., Bayer P. (2008) Small family with key contacts. par14 and par17 parvulin proteins, relatives of pin1, now emerge in biomedical research. Perspect. Med. Chem. 2, 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies. Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 [DOI] [PubMed] [Google Scholar]
- 24. Lederer C., Heider D., van den Boom J., Hoffmann D., Mueller J. W., Bayer P. (2011) Single-domain parvulins constitute a specific marker for recently proposed deep-branching archaeal subgroups. Evol. Bioinform. Online. 7, 135–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. R Development Core Team (2010) R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 26. Heider D., Hoffmann D. (2011) Interpol, an R package for preprocessing of protein sequences. BioData Min. 4, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Breiman L. (2001) in Machine Learning, pp. 5–32, Springer, New York [Google Scholar]
- 28. Nominé Y., Ristriani T., Laurent C., Lefèvre J. F., Weiss E., Travé G. (2001) A strategy for optimizing the monodispersity of fusion proteins. Application to purification of recombinant HPV E6 oncoprotein. Protein Eng. 14, 297–305 [DOI] [PubMed] [Google Scholar]
- 29. Sekulic N., Dietrich K., Paarmann I., Ort S., Konrad M., Lavie A. (2007) Elucidation of the active conformation of the APS-kinase domain of human PAPS synthetase 1. J. Mol. Biol. 367, 488–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meyer A., Van de Peer Y. (2005) From 2R to 3R, evidence for a fish-specific genome duplication (FSGD). Bioessays 27, 937–945 [DOI] [PubMed] [Google Scholar]
- 31. Mugford S. G., Lee B. R., Koprivova A., Matthewman C., Kopriva S. (2011) Control of sulfur partitioning between primary and secondary metabolism. Plant J. 65, 96–105 [DOI] [PubMed] [Google Scholar]
- 32. Chen H., Zhang B., Hicks L. M., Xiong L. (2011) A nucleotide metabolite controls stress-responsive gene expression and plant development. PLoS One 6, e26661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Estavillo G. M., Crisp P. A., Pornsiriwong W., Wirtz M., Collinge D., Carrie C., Giraud E., Whelan J., David P., Javot H., Brearley C., Hell R., Marin E., Pogson B. J. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23, 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mechold U., Ogryzko V., Ngo S., Danchin A. (2006) Oligoribonuclease is a common downstream target of lithium-induced pAp accumulation in Escherichia coli and human cells. Nucleic Acids Res. 34, 2364–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Frederick J. P., Tafari A. T., Wu S. M., Megosh L. C., Chiou S. T., Irving R. P., York J. D. (2008) A role for a lithium-inhibited Golgi nucleotidase in skeletal development and sulfation. Proc. Natl. Acad. Sci. U.S.A. 105, 11605–11612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Todeschini A. L., Condon C., Bénard L. (2006) Sodium-induced GCN4 expression controls the accumulation of the 5′ to 3′ RNA degradation inhibitor, 3′-phosphoadenosine 5′-phosphate. J. Biol. Chem. 281, 3276–3282 [DOI] [PubMed] [Google Scholar]
- 37. Wolfe K. H. (2001) Yesterday's polyploids and the mystery of diploidization. Nat. Rev. Genet. 2, 333–341 [DOI] [PubMed] [Google Scholar]
- 38. Conant G. C., Wolfe K. H. (2008) Turning a hobby into a job. How duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950 [DOI] [PubMed] [Google Scholar]
- 39. Stelzer C., Brimmer A., Hermanns P., Zabel B., Dietz U. H. (2007) Expression profile of PAPSS2 (3′-phosphoadenosine 5′-phosphosulfate synthase 2) during cartilage formation and skeletal development in the mouse embryo. Dev. Dyn. 236, 1313–1318 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.