Background: The role of S100A14 in tumorigenesis and the underlying mechanisms have not been fully understood.
Results: S100A14 affects cell invasiveness by regulating MMP2 transcription in a p53-dependent manner.
Conclusion: S100A14 acts as either an inducer or an inhibitor of cell invasion depending on the p53 status of cells.
Significance: These studies significantly increase our understanding of how S100A14 regulates cell invasiveness.
Keywords: Cancer Biology, Cell Invasion, Chromatin Immunoprecipitation (ChiP), Gene Regulation, Matrix Metalloproteinase (MMP), p53, Transcription Regulation
Abstract
S100 proteins have been implicated in tumorigenesis and metastasis. As a member of S100 proteins, the role of S100A14 in carcinogenesis has not been fully understood. Here, we showed that ectopic overexpression of S100A14 promotes motility and invasiveness of esophageal squamous cell carcinoma cells. We investigated the underlying mechanisms and found that the expression of matrix metalloproteinase (MMP)-2 is obviously increased after S100A14 gene overexpression. Inhibition of MMP2 by a specific MMP2 inhibitor at least partly reversed the invasive phenotype of cells overexpressing S100A14. By serendipity, we found that S100A14 could affect p53 transactivity and stability. Thus, we further investigated whether the effect of MMP2 by S100A14 is dependent on p53. A series of biochemical assays showed that S100A14 requires functional p53 to affect MMP2 transcription, and p53 potently transrepresses the expression of MMP2. Finally, RT-quantitative PCR analysis of human breast cancer specimens showed a significant correlation between S100A14 mRNA expression and MMP2 mRNA expression in cases with wild-type p53 but not in cases with mutant p53. Collectively, our data strongly suggest that S100A14 promotes cell motility and invasiveness by regulating the expression and function of MMP2 in a p53-dependent manner.
Introduction
Tumor metastasis remains a major problem in the management of cancer and is the main cause of death of patients with cancer (1). Metastasis is an extremely complicated process that occurs by a series of steps, including epithelial-mesenchymal transition, invasion, transportation through vessels, mesenchymal-epithelial transition, and outgrowth of secondary tumors (2–4). Degradation of basement membranes and the stromal extracellular matrix are critical steps for tumor invasion and metastasis. Matrix metalloproteinases (MMPs)2 belong to a large group of proteases responsible for degrading the multiple components of the extracellular matrix. They are implicated in tumorigenesis, cancer invasion, and metastasis (5). Among them, the gelatinases (MMP-2 and MMP9) were identified as the key MMPs involved in tumor invasion, metastasis, and angiogenesis (6, 7).
The S100 protein family is one of the largest subfamily of EF-hand calcium-binding proteins that contribute to multiple key cellular and subcellular processes (8). Some S100 proteins have strong associations with some types of cancer. In particular, it is well known that some S100 proteins (S100A4, S100A8, and S100A9) are closely associated with tumor invasion and metastasis (9, 10), which involves regulating expression and activity of matrix metalloproteinases (MMPs) (11, 12). S100A14 is a member of S100 calcium-binding proteins, which is markedly up-regulated in several tumor tissues, including ovarian, breast, and uterine tumor but is down-regulated in some tumors, such as kidney, colon, rectal, and esophageal tumor (13). Recently, our study demonstrated that extracellular S100A14 affects esophageal cancer cell proliferation and apoptosis via binding to receptor for advanced glycation end products (14). In addition, S100A14 has also been reported to regulate oral squamous cell carcinoma cell proliferation and invasion (15, 16). However, the role of S100A14 in tumorigenesis and progression and the underlying molecular mechanisms need to be further investigated.
p53 is a crucial tumor suppressor whose major function is inducing either growth arrest or apoptosis after cellular stress (17). Emerging evidence shows that p53 also contributes to the modulation of cell invasion (18, 19). The levels and activity of p53 are mainly regulated by post-translational modifications, such as phosphorylation, acetylation, and ubiquitination (20). Several S100 proteins, including S100A2, S100A4, and S100B, have been reported to interact with p53 and have different effects on p53 transcriptional activity and biological functions (21–23). Although the interactive regulation between S100 proteins and p53 is of particular interest, the mechanism requires to be further elucidated.
In this study, we investigated the role of S100A14 in cell motility and invasiveness. The promotion of the invasiveness of cells by S100A14 showed a strong correlation with induction of MMP2. Importantly, our findings showed that both the promotion of cell invasiveness and the induction of MMP2 by S100A14 were dependent on p53 status in cell lines. Furthermore, we confirmed the transcriptional repression of MMP2 by p53. Convincingly, the overexpression of S100A14 was significantly correlated with the up-regulation of MMP2 in clinical breast cancer samples with wild-type p53.
EXPERIMENTAL PROCEDURES
Tissue Specimens
Tissue specimens from 50 patients with breast cancer were analyzed. Patients were consecutively recruited at the Chinese Academy of Medical Sciences Cancer Hospital (Beijing, China). At recruitment, informed consent was obtained from each subject. This study was approved by the Institutional Review Board of the Chinese Academy of Medical Sciences Cancer Institute.
Cell Culture
Human colon carcinoma cell lines HCT116/p53+/+ and HCT116/p53−/− were kindly provided by Dr. Bert Vogelstein of Johns Hopkins University. Human esophageal squamous cell carcinoma cell line EC9706 was established in our own laboratory. MCF-7, HCT116/p53+/+, and HCT116/p53−/− cells were maintained in Dulbecco's modified Eagle's medium (DMEM), whereas H1299, HT1080, and EC9706 cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 units/ml streptomycin, and 100 units/ml penicillin.
Plasmids
Full-length cDNA of human S100A14 was cloned into the mammalian expression vectors pcDNA3.1 and pcDEF. The promoter region of MMP2 (−1759 top +25) was cloned into pGL3-basic vector. The resulting construct was verified by direct sequencing. PCB6 and pCB6-p53 plasmids were provided by Dr. Karen Vousden of NCI, Frederick Cancer Research and Development Center. The p53-Luc plasmid was purchased from Stratagene (La Jolla, CA).
Transfection and Generation of Stable Cell Lines
Transfection and establishment of stable cell lines were performed as described previously (24).
Soft Agar Assay for Colony Formation
Soft agar assay was performed as described previously (24).
Immunofluorescence
The experiment was performed as described previously (24).
Invasion and Migration Assays
These procedures were performed as described (25).
siRNA Transfection
Cells were transfected with siRNAs (25 nm) by HiperFect (Qiagen) and ON-TARGETplus pool siRNAs (25 nm) by DharmaFECT 1 (Dharmacon) following the manufacturers' protocol. The sequences for siRNAs were listed in supplemental Table S1.
RNA Isolation and PCR Analysis
RNA purification and real time RT-PCR were performed as described previously (26). Primers used are listed in supplemental Table S1.
Camptothecin Treatment and Adenovirus Infection
These experiments were performed as described previously (26).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP was performed as described previously (27). Antibody used was anti-p53 (pAb421) from Oncogene Science (Cambridge, MA).
Western Blot Analysis
Western blots were performed as described previously (25). Antibodies used were anti-p53 (Sc-126, Santa Cruz Biotechnology, Santa Cruz, CA), anti-S100A14 (gifts of Dr. Iver Petersen, University Hospital Charité, Berlin, Germany, and Dr. Youyong Lü, Beijing Cancer Hospital and Institute, Beijing, China), anti-MMP2 (MAB13405, Millipore, Billerica, MA), and β-actin antibody (A5316, Sigma).
Luciferase Assay
Luciferase assay was performed as described previously (26).
Statistical Analysis
We statistically evaluated experimental results using two-tailed paired Student's t test, two-independent sample t test, and χ2 test. All tests of significance were set at p < 0.05.
RESULTS
Altered S100A14 Affects Cell Migration and Invasion
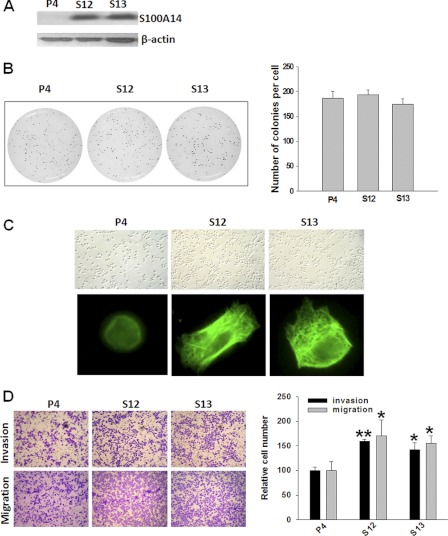
Our previous study has shown that extracellular S100A14 affected esophageal squamous cell carcinoma proliferation and apoptosis by interacting with receptor for advanced glycation end products (14). To characterize the role of intracellular S100A14 in the development of malignant phenotype, we established S100A14-overexpressed stable transfectants in EC9706 cells. Determination of S100A14 immunoreactivity using antibody against S100A14 indicated a high enrichment of S100A14 in stable transfectants (clones S12 and S13) compared with empty vector-transfected cells (clone P4) (Fig. 1A). We analyzed the effect of S100A14 overexpression on cell proliferation of EC9706 cells using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, flow cytometry, and soft agar assay. As shown in Fig. 1B and supplemental Fig. S1, no significant difference on cell proliferation was observed between S100A14-overexpressed cells and empty vector-transfected cells. Because cell shape controls some physiological processes such as cell growth, apoptosis, and motility, and the actin cytoskeleton governs multiple cellular activities (28), we observed cell morphology under microscope images and actin cytoskeleton by phalloidin staining in stable transfectants. As shown in Fig. 1C, S100A14 overexpressed cells appeared more stretched and formed a well organized actin cytoskeleton compared with empty vector-transfected cells (clone P4). Because these processes are closely correlated with cell motility (29), we further investigated the effect of S100A14 overexpression on the cell migration and invasion, and we conducted three-dimensional cell migration and invasion assays using transwell chambers and found that overexpression of S100A14 (S12 and S13) increased motility and invasiveness of EC9706 cells compared with empty vector-transfected cells (P4) (Fig. 1D). Taken together, these results demonstrate that S100A14 plays an important role in cell motility and invasiveness.
FIGURE 1.
Stable overexpression of S100A14 in EC9706 cells promotes cell motility and invasiveness. A, Western blot using S100A14 polyclonal antibody determined S100A14 expression in stable transfectants. P4, empty vector-transfected clone; S12 and S13, S100A14-transfected clones, β-actin served as a loading control. B, soft-agar colony formation assay was performed with EC9706 transfectants (P4, S12, and S13). Left panel shows the representative pictures, and the right panel shows the quantification of colonies. C, cells were cultured for 24 h and then optical images were acquired (top panel). Cells were cultured for 16 h and then were fixed and stained by FITC-labeled phalloidin; fluorescent images were visualized with fluorescent microscope (bottom). D, left panel, representative pictures of invading (top panel) and migrating (bottom panel) cells (P4, S12, and S13). Right panel, relative numbers of migrating and invading cells. Cells were counted in four randomly selected fields. Error bars represent the S.E. of triplicate experiments. *, p < 0.05; **, p < 0.01 two-tailed Student's t test.
S100A14 Modulates Cell Migration and Invasion by Regulating MMP2 Expression
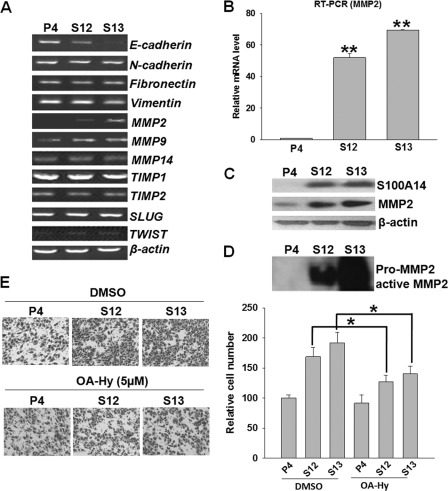
To explore the molecular mechanism of S100A14 promoting cell migration and invasion, we characterized the expression of some metastatic related genes by RT-PCR analysis (Fig. 2A). Interestingly, among the MMPs that were detected, MMP2 expression was dramatically induced in the S100A14-overexpressed cells (S12 and S13). In contrast, expression of MT1-MMP (MMP14), a major activator of MMP2, and TIMP2, an endogenous inhibitor of MMP2, were not significantly altered (30, 31). RT-qPCR and Western blot analysis further confirmed the dramatic increase of MMP2 expression, as well as the secretion and activation of MMP2 in S100A14 stable transfectants (Fig. 2, B–D). To define the role of MMP2 in S100A14-enhanced cell invasion, we pretreated cells with a potent specific MMP2 inhibitor OA-Hy (10 μm, Chemicon, Temecula, CA) and evaluated the effect on cell invasion. We found that pretreatment with OA-Hy significantly attenuated S100A14-enhanced cell invasion (Fig. 2E). Taken together, these data demonstrate that S100A14 controls invasive potential through regulation of MMP2 expression.
FIGURE 2.
Overexpression of S100A14 promoted cell invasiveness by regulating MMP2. A, expression of some metastasis-related genes was examined by RT-PCR in EC9706 transfectants. B, MMP2 mRNA expression levels were further confirmed by RT-qPCR. Western blot analysis showed the expression in cell lysates (C) and the secretion and activation of MMP2 in conditioned medium in EC9706 transfectants (D). E, EC9706 cells (P4, S12, and S13) were pretreated with 10 μm OA-Hy or solvent (DMSO) for 24 h and suspended in 150 μl of serum-free medium containing OA-Hy or solvent. Cells were then seeded in the upper chamber of the Matrigel plate for the invasion assay. Left panel, representative pictures of invading cells. Right panel, relative numbers of invading cells. Cells were counted in four randomly selected fields. Error bars represent the S.E. of triplicate experiments. *, p < 0.05 two-tailed Student's t test.
Ectopic Overexpression of S100A14 Inhibits Endogenous and Exogenous p53 Protein Expression and Transcriptional Activity of p53
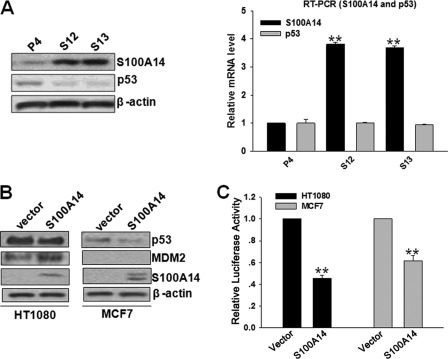
By serendipity, we found that the protein expression of p53 was drastically reduced after S100A14 overexpression in EC9706 cells, but there was no effect on p53 mRNA levels (Fig. 3A). Next, we detected the protein expression of some p53 targets as follows: p21, PCNA, and Bax. Unexpectedly, we did not find any significant effect on the expression of these genes accompanied by down-regulation of p53 upon S100A14 overexpression in EC9706 cells (supplemental Fig. S2). Several possibilities may cause this. (i) S100A14 overexpression might lead to changes of multiple molecules or a number of signaling pathways, and the combinatorial interactions of multiple factors ultimately remain p53 targets such as p21, PCNA, and Bax at stable levels. As a result, the balance of expression of multiple genes regulates the biological behavior and determines the phenotype of cells. (ii) S100A14 may influence p53 promoter selectivity, thereby affecting the transcription of p53-specific target genes (32–34). (iii) The p53 levels, the specific p53 response element (p53RE) sequences, and cell types may be responsible for S100A14-mediated p53 target gene selectivity (33). Because p53 status in EC9706 cells has not been reported yet, we performed p53 coding region sequencing and found two point mutations residing at codons 230 and 1126. To ascertain whether p53 is functional in EC9706 cells, DNA damage experiments were carried out. EC9706 cells were treated with 2 μm camptothecin (CPT) and harvested at 0, 3, 6, 9, 12, and 24 h. Western blot and RT-qPCR analyses showed that p53 target genes (p21, Bax, and MDM2) were promptly induced within 6 h in parallel with the induction of p53 at 6 h, indicating that p53 is functional in EC9706 cells (supplemental Fig. S3). To further confirm the effect of S100A14 on p53 in cell lines with wild-type p53, we determined the effect of S100A14 on p53 in HT1080 and MCF7 cells by transient transfection. The results showed that ectopic overexpression of S100A14 resulted in a reduction of endogenous p53 in HT1080 and MCF-7 cells with WT p53 (Fig. 3B). Moreover, overexpression of S100A14 significantly induced reductions of promoter transactivation of p53-luc (a reporter plasmid expressing firefly luciferase under the control of p53 response element) (Fig. 3C). As p53 is degraded by the ubiquitin-proteasome pathway, we examined whether S100A14 influenced p53 ubiquitination per se. The stability (half-life) of endogenous and exogenous p53 was decreased in cycloheximide-treated S100A14-overexpressed cells compared with control cells (supplemental Fig. S4), indicating that decreased expression occurs through its stabilization. However, the level of ubiquitination of exogenous p53 in H1299 cells did not show any significant changes (data not shown), suggesting that other mechanisms may be responsible for S100A14-decreased p53 levels (35).
FIGURE 3.
S100A14 reduces p53 protein expression and transcriptional activity. A, Western blot and RT-qPCR analyses showed the expression of p53 protein (left panel) and mRNA (right panel) in EC9706 transfectants (P4, S12, and S13). B, MCF-7 and HT1080 cells were transfected with the indicated constructs, and 48 h later, cell lysates were blotted with the indicated antibodies. C, p53-luc construct was cotransfected with the indicated constructs into HT1080 and MCF7 cells, and reporter activity was then determined. Error bars represent the S.E. of triplicate experiments. **, p < 0.01 two-tailed Student's t test.
Altered S100A14 Expression Differently Affects MMP2 Expression and Cell Invasiveness Dependent on p53 Status
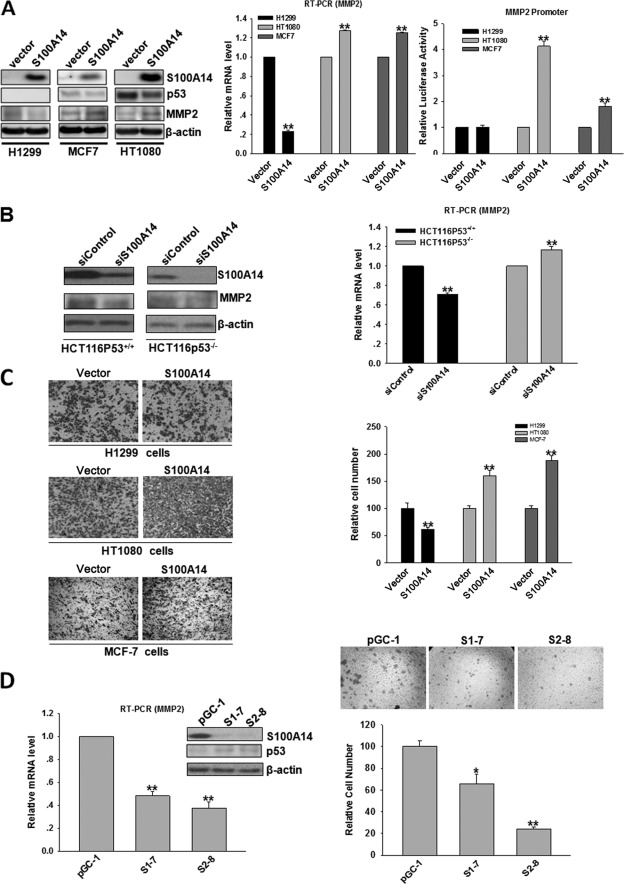
It has been previously shown that p53 positively or negatively regulates the expression of MMP2 (36, 37), and we then suspected that there was a correlation between S100A14-induced MMP2 elevation and S100A14-induced p53 decrease. We subsequently examined whether the regulation of MMP2 by S100A14 is dependent on p53. We selected several cell lines with low levels of S100A14 expression and different p53 status as follows: HT1080 (p53+/+), MCF7 (p53+/+), and H1299 (p53−/−) cells, and we transfected these cells with pcDNA3.1-S100A14 and pcDNA3.1 vectors to establish pooled neomycin (G418)-resistant stable transfectants. The forced expression of S100A14 in HT1080, MCF7, and H1299 cells obviously increased the expression of S100A14, and accordingly, overexpression of S100A14 in HT1080 and MCF7 cells significantly increased the mRNA and protein levels of MMP2, whereas an opposite effect was observed in H1299 cells (Fig. 4A, left and middle panels). To further investigate whether altered S100A14 expression has a direct effect on human MMP2 promoter activity, the MMP2 luciferase reporter construct was transfected into stable transfectants. Remarkably, MMP2 promoter activity was significantly induced in S100A14-overexpressing transfectants in HT1080 and MCF7 cells compared with empty vector-transfected cells. In contrast, the effect was not observed in H1299 cells (Fig. 4A, right panel). These results imply that the induction of MMP2 by S100A14 is dependent on p53. To further verify the above results, small interfering RNAs (siRNAs) targeting S100A14 (ON-TARGETplus SMARTpool) were transfected into HCT116/p53+/+ and HCT116/p53−/− cells. siRNA transfection dramatically reduced cellular S100A14 levels and significantly decreased the mRNA and protein levels of MMP2 in p53 wild-type cells (HCT116/p53+/+), but it slightly increased in p53-null cells (HCT116/p53−/−), indicating that S100A14 affects MMP2 transcription dependent on p53 status (Fig. 4B). Collectively, our data demonstrate that altered S100A14 expression differently affects MMP2 expression dependent on p53 status.
FIGURE 4.
Effects of altered S100A14 expression on MMP2 expression and the promoter activity of MMP2 are dependent on p53 status. A, H1299, HT1080, and MCF-7 cells were transfected with the indicated constructs, and pooled neomycin (G418)-resistant colonies were established as stable transfectants. The protein expression of S100A14, p53, and MMP2 was determined by Western blot, and β-actin served as a loading control (left panel); the mRNA expression of MMP2 was determined by RT-qPCR (middle panel); and MMP2 transcriptional activity was detected by MMP2-luc reporter assay (right panel). B, siRNAs targeting S100A14 (25 nm) (ON-TARGETplus SMARTpool) or control siRNAs were transfected into HCT116/p53+/+ and HCT116/p53−/− cells, and 72 h later, proteins and mRNA were extracted and subjected to Western blot or RT-qPCR. C, cell invasion assay was performed in S100A14-overexpressed H1299, HT1080, and MCF7 cells. Representative pictures (left panel) and relative numbers of invading cells (right panel) are shown. Cells were counted in four randomly selected fields. Error bars represent the S.E. of triplicate experiments. *, p < 0.05; **, p < 0.01 two-tailed Student's t test. D, stable-silenced S100A14 HCT116 cells were obtained, and proteins and RNA were extracted and subjected to Western blot or RT-qPCR (left panel). pGC-1, control shRNA-transfected clone; S1-7 and S2-8, shRNA targeting S100A14-transfected clones. Cell invasion assay was performed in stable-silenced S100A14 HCT116 cells (right panel).
Because the regulation of MMP2 by S100A14 is dependent on p53 status, we next analyzed whether the effect of S100A14 on cell invasiveness is correlated with its p53 status. In vitro invasion assays showed that the forced expression of S100A14 increased the invasiveness of HT1080 and MCF7 cells but decreased the invasiveness of H1299 cells (Fig. 4C). Meanwhile, we also created stable silencing of S100A14 HCT116 cells using two different shRNA target sequences that have already been verified to highly reduce S100A14 expression by siRNA transfection, and S100A14 expression was effectively suppressed in stable transfectants (S1-7 and S2-8) compared with control cells (pGC-1). In line with S100A14 ablation, p53 expression was significantly increased, and MMP2 expression was dramatically decreased (Fig. 4D, left panel). Accordingly, cell invasiveness was significantly inhibited (Fig. 4D, right panel). Taken together, these data further support that S100A14 affects MMP2 expression and cell invasiveness in a p53-dependent manner.
MMP2 Is Repressed at Transcriptional Level following the Increased Expression of p53
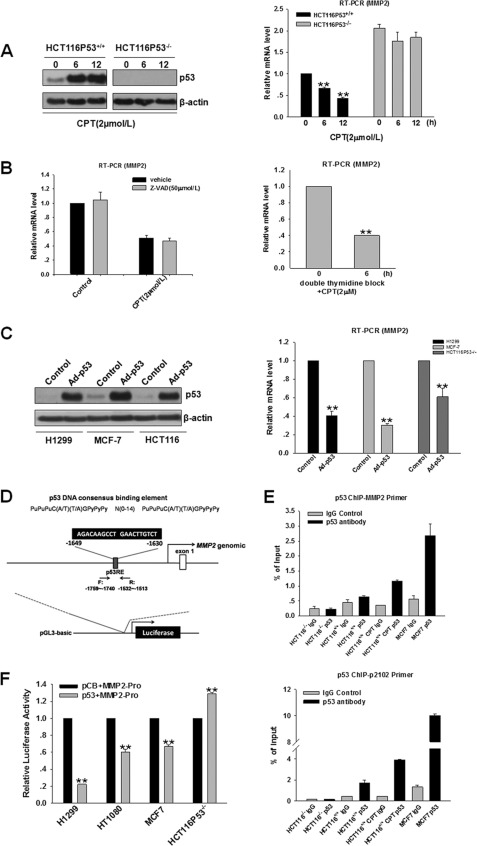
To define the effect of p53 on MMP2 expression, we treated p53 wild-type cells (HCT116/p53+/+) and p53 null cells (HCT116/p53−/−) with 2 μm CPT. After 6 and 12 h, MMP2 mRNA levels were significantly decreased in parallel with increased p53 accumulation in HCT116/p53+/+ cells (Fig. 5A); however, no effect was observed in HCT116/p53−/− cells, confirming that MMP2 down-regulation is dependent on p53. We also compared mRNA expression of MMP2 in HCT116/p53+/+ cells with HCT116/p53−/− cells. As expected, MMP2 mRNA level was significantly greater in HCT116/p53−/− cells versus HCT116/p53+/+ cells. To rule out the possibility that decreased MMP2 was the result of p53-induced apoptosis, HCT116/p53+/+ cells were treated with CPT in combination with benzyloxycarbonyl-VAD-fluoromethyl ketone (50 μm), and RT-qPCR analysis was performed. As shown in Fig. 5B, left panel, benzyloxycarbonyl-VAD-fluoromethyl ketone treatment did not prevent the inhibition of MMP2, indicating that p53 indeed contributes to the regulation of MMP2. To further investigate whether CPT-mediated decrease in MMP2 mRNA expression is dependent on CPT-induced cell cycle arrest, we synchronized HCT116/p53+/+ cells to G1/S phase by using double-thymidine block (38) and examined the effect of CPT on MMP2 mRNA level in synchronized HCT116/p53+/+ cells. Double-thymidine block caused a complete G1/S arrest, which was not further enhanced by CPT treatment (supplemental Fig. S5). Significantly, treatment of HCT116/p53+/+ cells with CPT decreased MMP2 mRNA level in HCT116/p53+/+ cells already arrested in G1/S phase (Fig. 5B, right panel). These results indicate that CPT can directly decrease MMP2 mRNA expression in a cell cycle arrest-independent manner. Collectively, these data strongly suggest that activated endogenous p53 inhibited MMP2 mRNA expression. To further investigate the effect on MMP2 mRNA expression by exogenous p53 overexpression, several cell lines were infected with 40 multiplicities of infection of adenovirus expressing wild-type p53, and the levels of MMP2 mRNA expression were significantly repressed by exogenous p53 at 48 h post-infection (Fig. 5C). We next analyzed p53 response elements (p53REs) in the promoter region of the MMP2 gene (from −3000 to +1 site) by the Jaspar data base (supplemental Table 2) (39) and identified a potential p53RE located at position −1649 to −1630 (Fig. 5D). This p53RE has been previously reported; however, the interaction between p53 and the p53RE-containing DNA region has not yet been examined. Therefore, we performed a chromatin immunoprecipitation (ChIP) assay to test the functional interaction between p53 and the potential p53RE in the MMP2 regulatory regions. Enriched p53 binding to the MMP2 promoter was observed compared with IgG control in MCF7 and HCT116/p53+/+ cells with WT p53. Moreover, CPT treatment significantly increased the recruitment of p53 to the MMP2 promoter in HCT116/p53+/+ cells. In contrast, the recruitment of p53 to the MMP2 promoter was similar with IgG control in HCT116/p53−/− cells with null p53. The p21 promoter region was used as a positive control for p53 binding (Fig. 5E) (40). These results strongly suggest that p53 regulates MMP2 transcription by binding to the p53RE located in the MMP2 promoter. Moreover, p53 binding was significantly decreased in line with the decrease of p53 protein level in S100A14-overexpressed cells (supplemental Fig. S6). To further explore the role of p53 in regulating MMP2 transcription, we examined whether p53 regulates MMP2 promoter activity. We cotransfected the MMP2 promoter-luciferase construct and p53 expression or control vector into HT1080, MCF7, H1299, and HCT116/p53−/− cells, and p53 expression significantly inhibited the luciferase activity of the MMP2 promoter in HT1080, MCF7, and H1299 cells. Nevertheless, p53 induced slight activation of MMP2 promoter activity in HCT116/p53−/− cells (Fig. 5F). Taken together, these data clearly show that the repression of MMP2 by p53 occurs at the transcriptional level.
FIGURE 5.
p53 trans-represses the expression of MMP2. A, HCT116/p53+/+ and HCT116/p53−/− cells were exposed to CPT, and then protein and RNA were extracted and analyzed by Western blot or RT-qPCR following treatment at the indicated time points. Fold reduction of MMP2 mRNA level was measured by the percentage of HCT116/p53+/+ cells (taken as 1). B, left panel, cells were treated with 50 μm benzyloxycarbonyl-VAD-fluoromethyl ketone for 1 h prior to the addition of CPT, and then RNA was extracted and analyzed by RT-qPCR. Right panel, cells were synchronized to G1/S phase by using double-thymidine block and were treated with 2 μm CPT for 6 h prior to the cell harvest, and the mRNA level of MMP2 was examined. C, H1299, MCF7, and HCT116 cells were infected with Ad-p53 of 40 multiplicities of infection and 48 h later, cells were harvested. Protein and RNA were extracted and analyzed by Western blot or RT-qPCR. D, p53 binds to the promoter of the MMP2 gene. The p53 consensus sequences are shown in capital letters and potential p53 RE are located at −1649 to −1630 of the MMP2 promoter. E, ChIP assay demonstrated that p53 bound to the promoter region of the MMP2 gene. MCF7 and HCT116/p53−/− cells were harvested, and HCT116/p53+/+ cells were collected with or without CPT treatment. ChIP assay was performed with p53 antibody, and anti-mouse IgG antibody was used as a negative control. F, MMP2-luc was cotransfected with the indicated constructs into H1299, HT1080, MCF7, and HCT116/p53−/− cells, and reporter assay was then performed. Error bars represent the S.E. of triplicate experiments. *, p < 0.05; **, p < 0.01 two-tailed Student's t test.
Overexpression of S100A14 Is Correlated with Up-regulation of MMP2 in Clinical Samples with Wild-type p53
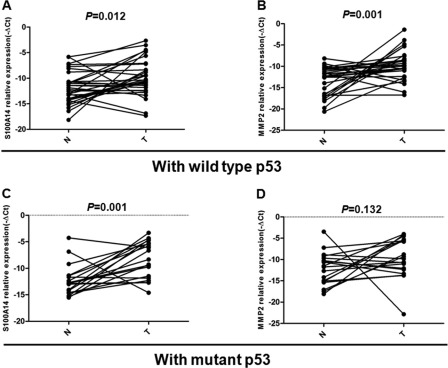
To verify cell studies (MCF-7) in clinical samples, we performed p53 sequence analyses and RT-qPCR in 50 pairs of breast cancer tissues and their adjacent normal tissues. Sequencing analyses revealed that alterations of p53 included point mutation and deletion/insertion, and the mutation rate of p53 gene in breast cancer was 40%. Of 50 cases, 1 case was excluded because p53 mutation was also observed in its adjacent normal tissues. Next, we examined the mRNA expressions of S100A14 and MMP2 simultaneously in 49 cases. The term −ΔCt (Ctβ-actin-CtS100A14 or CtMMP2) was used to describe the expression of S100A14 and MMP2. The results showed that both S100A14 and MMP2 mRNAs were highly expressed in breast cancers (paired t test; p = 0.012 for S100A14; p = 0.001 for MMP2) with wild-type p53 versus their surrounding normal tissues. In contrast, the overexpression of MMP2 was not observed (paired t test; p = 0.132), although S100A14 was also highly expressed (paired t test; p = 0.001) in cases with mutant p53 (Fig. 6). Moreover, we did find a significant correlation between high expression of S100A14 and overexpression of MMP2 in breast cancer samples with wild-type p53 (Table 1, p = 0.035). However, the correlation was not observed in samples carrying mutant p53 (Table 1, p = 0.154). Collectively, these results further supported the regulation of MMP2 by S100A14 is dependent on p53 status in clinical samples.
FIGURE 6.
Expression of S100A14 and MMP2 in clinical samples with different p53 status. RT-qPCR analysis was performed in 49 breast cancer tissues and paired adjacent normal tissues. The term −ΔCt (Ctβ-actin-CtS100A14 or CtMMP2) was used to describe the expression level of S100A14 and MMP2. The expression of S100A14 and MMP2 in breast cancers and their matched surrounding normal tissues with wild-type p53 (A and B) and mutant p53 (C and D) are shown.
TABLE 1.
The correlation between S100A14 overexpression and MMP2 overexpression in paired breast cancer tissues with different p53 status
These results were analyzed by the Pearson χ2 test. p values with significance are shown as superscripts.
| Characteristics |
S100A14 expression |
Case no. | |
|---|---|---|---|
| Non-overexpressed* | Overexpressed* | ||
| Overall | 49 | ||
| MMP2 expression in cases with wild type p53 | |||
| Non-overexpressed,a, n (%) | 3 (75.0) | 1 (25.0) | 4 |
| Overexpressed,an (%) | 6 (23.1) | 20 (66.9)0.035 | 26 |
| MMP2 expression in cases with mutant p53 | |||
| Non-overexpressed,an (%) | 2 (33.3) | 4 (66.6) | 6 |
| Overexpressed,an (%) | 1 (76.9) | 12 (23.1)0.154 | 13 |
a For S100A14 and MMP2 mRNA expression levels, the matched cancer/normal ratio >1 was taken as overexpressed group, and the ratio ≤1 was taken as the non-overexpressed group.
DISCUSSION
Increasing evidence showed the importance of S100 family in cell migration, invasion, and cancer metastasis. Among them, S100A4 has been identified as a well known metastasis marker (9). In addition, S100A2 has also been reported as a strong metastasis inducer in non-small cell lung cancer (41). S100A8/S100A9 has been implicated in both myeloid cell recruitment and tumor cell invasion in lung cancer (10). Recently, it has been reported that S100A14 low expression in combination with S100A4 high expression was correlated with high colorectal cancer metastatic potential (42). S100A14 has also been identified as an invasion suppressor in oral squamous cell carcinoma cells (13). In this study, we further demonstrated that S100A14 may act as either an inducer or an inhibitor of cell invasion depending on the p53 status of cells.
Importantly, S100 proteins are reported to affect the cell migration and invasion through regulating the expression of MMPs (11, 12, 15). Our study also showed that S100A14 affected cell invasion by regulating the expression of MMP2, a key proteolytic enzyme in the process of cell invasion (7), and MMP2 is an important mediator of S100A14 promotion of EC9706 cell invasion. To our surprise, in line with up-regulation of MMP2, the protein level of p53 is dramatically reduced in S100A14-overexpressed EC9706 cells. Furthermore, we confirmed the effect of S100A14 on p53 expression, DNA binding, and transactivity in HT1080 and MCF7 cells with wild-type p53. Consistent with previous observations that several S100 proteins interact with p53 to perform different effects on p53 activity (21–23), this study showed that S100A14 is a critical negative regulator of p53. In addition, our previous study demonstrated that S100A14 is a target gene of p53 pathway (26). Therefore, a p53-S100A14 negative feedback loop was formed. Although under our experimental conditions, the interaction between S100A14 and p53 was not found, our data demonstrated that p53 regulation by S100A14 is likely mediated by affecting p53 stability. It is well documented that some E3 ubiquitin ligases such as MDM2, COP1, and Pirh2 regulate p53 stability (43), and we also found that MDM2 was slightly induced by overexpression of S100A14 in HT1080 cells. However, we did not detect the change of p53 ubiquitination by co-IP assay (data not shown). Thus, it is possible that S100A14 promotes p53 degradation and limits its activity via the effect on other molecules to regulate p53 stability. The identification of the interactive protein of S100A14 will further improve our standing of the mechanism underlying regulating p53 by S100A14.
Our study further showed that the regulation of MMP2 by S100A14 is dependent on the p53 status of cells. In the presence of p53, S100A14 enhanced MMP2 expression by decreasing p53 protein levels. However, in the absence of p53, S100A14 inhibited MMP2 expression, indicating that other molecules are responsible for mediating inhibition of MMP2. The unknown mechanism needs to be further investigated in future studies. Collectively, our data strongly suggest that S100A14 plays a dual role in regulating the expression of MMP2 and cell invasion in a p53-dependent manner.
Previous studies showed that p53 is involved in cancer progression by specifically regulating cancer invasion (18, 19, 44). Moreover, several reports have demonstrated that p53 trans-represses the expression of distinct MMPs, including MMP1, MMP9, and MMP13 (45–47). However, p53 has a dual effect on regulation of MMP2. Capogrossi and co-workers (36) previously demonstrated that p53 inhibited transcription of MMP2 in cells with mutant p53. In contrast, there is report that p53 plays a dual role in the regulation of MMP2 promoter activity (37). We examined the transcriptional regulation on MMP2 by p53 under our experimental conditions. Luciferase reporter studies showed the introduction of wild-type p53 suppressed MMP2 promoter activity in several cells except for HCT116/p53−/− cells. The disparity of the regulation of MMP2 by p53 may reflect the diverse characteristics of the experimental systems, including cell types. We further investigated the regulation of MMP2 by p53. The results showed that MMP2 mRNA level was significantly decreased in HCT116/p53+/+ but not HCT116/p53−/− cells with CPT treatment, demonstrating that decreased MMP2 expression is dependent on p53 status. In accordance with these results, exogenous p53 overexpression significantly inhibited the MMP2 mRNA levels. Previous studies showed that p53 functioned as a transactivator as well as a “trans-enhancer” to activate MMP2 transcription, and p53RE located at position −1649 to −1630 in the promoter of MMP2 gene is crucial to activate MMP2 transactivity (37). We searched for potential p53-binding sites in the promoter of MMP2 by the Jaspar data base. In agreement with previous reports, we acquired the same p53RE located at the promoter of MMP2. Subsequently, we confirmed that p53 can directly bind to the regulatory regions of MMP2 containing p53RE by ChIP assay. Moreover, p53 binding was significantly decreased in line with the decrease of the p53 protein level in S100A14-overexpressed HT1080 cells (supplemental Fig. S6). Although the mechanism of p53-mediated trans-repression has not been fully elucidated, a number of studies showed that p53 can trans-repress target gene transcription by directly repressing the basal transcriptional machinery, interfering with the functions of coactivators or recruiting corepressors (48). Previous investigators also speculated that p53 may trans-repress MMP2 expression by the element(s) other than p53RE or other transcription factors (37). It will be of great importance to identify and characterize the potential p53 negative regulatory elements or cofactors in the regulatory regions of MMP2 in future studies.
Through its action to p53 and MMP2, S100A14 could function as a metastasis inducer or inhibitor. Moreover, S100A14 overexpression was significantly correlated with MMP2 overexpression in breast cancer tissue samples. In future studies, it will be interesting to examine the correlations among S100A14, p53 status, and MMP2 expression with clinicopathological features, including tumor metastasis, survival, and prognosis in many types of cancers. Moreover, the effect on invasiveness of S100A14 using xenograft mouse models after overexpression of S100A14 will be determined in the future.
In summary, we demonstrated that S100A14 controls cell invasion at least in part through the regulation of MMP2, and the regulation of MMP2 by S100A14 is dependent on p53. We suggest that S100A14 might be a potential biomarker as well as a new therapeutic target in cancer treatment, which could be realized through development of specific S100A14 inhibitors or use of a gene therapy approach.
Supplementary Material
Acknowledgments
We thank Dr. Iver Petersen and Dr. Youyong Lü for the generous gifts of S100A14 antibodies; Dr. Bert Vogelstein and Dr. Karen Vousden for the generous gifts of HCT116/p53+/+ and HCT116/p53−/− cells and pCB6 and pCB6-p53 plasmids, respectively.
This work was supported by National Natural Science Foundation of China Grants 81000954 and 81130043, National Basic Research Program of China Grant 2011CB504205, and Ministry of Education of China Grant 20101106120012.
- MMP
- matrix metalloproteinase
- qPCR
- quantitative PCR
- p53 RE
- p53 response element
- CPT
- camptothecin.
REFERENCES
- 1. Steeg P. S. (2006) Tumor metastasis. Mechanistic insights and clinical challenges. Nat. Med. 12, 895–904 [DOI] [PubMed] [Google Scholar]
- 2. Gupta G. P., Massagué J. (2006) Cancer metastasis. Building a framework. Cell 127, 679–695 [DOI] [PubMed] [Google Scholar]
- 3. Thiery J. P., Acloque H., Huang R. Y., Nieto M. A. (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 [DOI] [PubMed] [Google Scholar]
- 4. Thiery J. P. (2002) Epithelial-mesenchymal transitions in tumor progression. Nat. Rev Cancer 2, 442–454 [DOI] [PubMed] [Google Scholar]
- 5. Roy R., Yang J., Moses M. A. (2009) Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J. Clin. Oncol. 27, 5287–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sato H., Takino T., Okada Y., Cao J., Shinagawa A., Yamamoto E., Seiki M. (1994) A matrix metalloproteinase expressed on the surface of invasive tumor cells. Nature 370, 61–65 [DOI] [PubMed] [Google Scholar]
- 7. Vu T. H., Shipley J. M., Bergers G., Berger J. E., Helms J. A., Hanahan D., Shapiro S. D., Senior R. M., Werb Z. (1998) MMP-9/gelatinase B is a key regulator of growth plate angiogenesis and apoptosis of hypertrophic chondrocytes. Cell 93, 411–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heizmann C. W., Fritz G., Schafer B. W. (2002) S100 proteins. Structure, functions, and pathology. Front. Biosci. 7, d1356–d1368 [DOI] [PubMed] [Google Scholar]
- 9. Boye K., Maelandsmo G. M. (2010) S100A4 and metastasis. A small actor playing many roles. Am. J. Pathol. 176, 528–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hiratsuka S., Watanabe A., Aburatani H., Maru Y. (2006) Tumor-mediated up-regulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat. Cell Biol. 8, 1369–1375 [DOI] [PubMed] [Google Scholar]
- 11. Saleem M., Kweon M. H., Johnson J. J., Adhami V. M., Elcheva I., Khan N., Bin Hafeez B., Bhat K. M., Sarfaraz S., Reagan-Shaw S., Spiegelman V. S., Setaluri V., Mukhtar H. (2006) S100A4 accelerates tumorigenesis and invasion of human prostate cancer through the transcriptional regulation of matrix metalloproteinase 9. Proc. Natl. Acad. Sci. U.S.A. 103, 14825–14830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moon A., Yong H. Y., Song J. I., Cukovic D., Salagrama S., Kaplan D., Putt D., Kim H., Dombkowski A., Kim H. R. (2008) Global gene expression profiling unveils S100A8/A9 as candidate markers in H-ras-mediated human breast epithelial cell invasion. Mol. Cancer Res. 6, 1544–1553 [DOI] [PubMed] [Google Scholar]
- 13. Pietas A., Schlüns K., Marenholz I., Schäfer B. W., Heizmann C. W., Petersen I. (2002) Molecular cloning and characterization of the human S100A14 gene encoding a novel member of the S100 family. Genomics 79, 513–522 [DOI] [PubMed] [Google Scholar]
- 14. Jin Q., Chen H., Luo A., Ding F., Liu Z. (2011) S100A14 stimulates cell proliferation and induces cell apoptosis at different concentrations via receptor for advanced glycation end products (RAGE). PLoS One 6, e19375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sapkota D., Bruland O., Costea D. E., Haugen H., Vasstrand E. N., Ibrahim S. O. (2011) S100A14 regulates the invasive potential of oral squamous cell carcinoma-derived cell lines in vitro by modulating expression of matrix metalloproteinases MMP1 and MMP9. Eur. J. Cancer 47, 600–610 [DOI] [PubMed] [Google Scholar]
- 16. Sapkota D., Costea D. E., Blø M., Bruland O., Lorens J. B., Vasstrand E. N., Ibrahim S. O. (2012) S100A14 inhibits proliferation of oral carcinoma-derived cells through G1-arrest. Oral. Oncol. 48, 219–225 [DOI] [PubMed] [Google Scholar]
- 17. Riley T., Sontag E., Chen P., Levine A. (2008) Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 [DOI] [PubMed] [Google Scholar]
- 18. Sankpal N. V., Willman M. W., Fleming T. P., Mayfield J. D., Gillanders W. E. (2009) Transcriptional repression of epithelial cell adhesion molecule contributes to p53 control of breast cancer invasion. Cancer Res. 69, 753–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hwang C. I., Matoso A., Corney D. C., Flesken-Nikitin A., Körner S., Wang W., Boccaccio C., Thorgeirsson S. S., Comoglio P. M., Hermeking H., Nikitin A. Y. (2011) Wild-type p53 controls cell motility and invasion by dual regulation of MET expression. Proc. Natl. Acad. Sci. U.S.A. 108, 14240–14245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kruse J. P., Gu W. (2009) Modes of p53 regulation. Cell 137, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin J., Yang Q., Yan Z., Markowitz J., Wilder P. T., Carrier F., Weber D. J. (2004) Inhibiting S100B restores p53 levels in primary malignant melanoma cancer cells. J. Biol. Chem. 279, 34071–34077 [DOI] [PubMed] [Google Scholar]
- 22. Mueller A., Schäfer B. W., Ferrari S., Weibel M., Makek M., Höchli M., Heizmann C. W. (2005) The calcium-binding protein S100A2 interacts with p53 and modulates its transcriptional activity. J. Biol. Chem. 280, 29186–29193 [DOI] [PubMed] [Google Scholar]
- 23. Grigorian M., Andresen S., Tulchinsky E., Kriajevska M., Carlberg C., Kruse C., Cohn M., Ambartsumian N., Christensen A., Selivanova G., Lukanidin E. (2001) Tumor suppressor p53 protein is a new target for the metastasis-associated Mts1/S100A4 protein. Functional consequences of their interaction. J. Biol. Chem. 276, 22699–22708 [DOI] [PubMed] [Google Scholar]
- 24. Zhang C., Zhu C., Chen H., Li L., Guo L., Jiang W., Lu S. H. (2010) Kif18A is involved in human breast carcinogenesis. Carcinogenesis 31, 1676–1684 [DOI] [PubMed] [Google Scholar]
- 25. Tian Y., Luo A., Cai Y., Su Q., Ding F., Chen H., Liu Z. (2010) MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. J. Biol. Chem. 285, 7986–7994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen H., Yu D., Luo A., Tan W., Zhang C., Zhao D., Yang M., Liu J., Lin D., Liu Z. (2009) Functional role of S100A14 genetic variants and their association with esophageal squamous cell carcinoma. Cancer Res. 69, 3451–3457 [DOI] [PubMed] [Google Scholar]
- 27. Wang Q., Li W., Liu X. S., Carroll J. S., Jänne O. A., Keeton E. K., Chinnaiyan A. M., Pienta K. J., Brown M. (2007) A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol. Cell 27, 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang S., Ingber D. E. (2000) Shape-dependent control of cell growth, differentiation, and apoptosis. Switching between attractors in cell regulatory networks. Exp. Cell Res. 261, 91–103 [DOI] [PubMed] [Google Scholar]
- 29. Pollard T. D. (2003) The cytoskeleton, cellular motility, and the reductionist agenda. Nature 422, 741–745 [DOI] [PubMed] [Google Scholar]
- 30. Itoh Y., Takamura A., Ito N., Maru Y., Sato H., Suenaga N., Aoki T., Seiki M. (2001) Homophilic complex formation of MT1-MMP facilitates pro-MMP-2 activation on the cell surface and promotes tumor cell invasion. EMBO J. 20, 4782–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Itoh Y., Ito A., Iwata K., Tanzawa K., Mori Y., Nagase H. (1998) Plasma membrane-bound tissue inhibitor of metalloproteinases (TIMP)-2 specifically inhibits matrix metalloproteinase 2 (gelatinase A) activated on the cell surface. J. Biol. Chem. 273, 24360–24367 [DOI] [PubMed] [Google Scholar]
- 32. Das S., Boswell S. A., Aaronson S. A., Lee S. W. (2008) p53 promoter selection. Choosing between life and death. Cell Cycle 7, 154–157 [DOI] [PubMed] [Google Scholar]
- 33. Menendez D., Inga A., Resnick M. A. (2009) The expanding universe of p53 targets. Nat. Rev. Cancer 9, 724–737 [DOI] [PubMed] [Google Scholar]
- 34. Vousden K. H., Prives C. (2009) Blinded by the light. The growing complexity of p53. Cell 137, 413–431 [DOI] [PubMed] [Google Scholar]
- 35. Lavin M. F., Gueven N. (2006) The complexity of p53 stabilization and activation. Cell Death Differ. 13, 941–950 [DOI] [PubMed] [Google Scholar]
- 36. Toschi E., Rota R., Antonini A., Melillo G., Capogrossi M. C. (2000) Wild-type p53 gene transfer inhibits invasion and reduces matrix metalloproteinase-2 levels in p53-mutated human melanoma cells. J. Invest. Dermatol. 114, 1188–1194 [DOI] [PubMed] [Google Scholar]
- 37. Bian J., Sun Y. (1997) Transcriptional activation by p53 of the human type IV collagenase (gelatinase A or matrix metalloproteinase 2) promoter. Mol. Cell. Biol. 17, 6330–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fang G., Yu H., Kirschner M. W. (1998) Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell 2, 163–171 [DOI] [PubMed] [Google Scholar]
- 39. Vlieghe D., Sandelin A., De Bleser P. J., Vleminckx K., Wasserman W. W., van Roy F., Lenhard B. (2006) A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 34, D95–D97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu G., Xia T., Chen X. (2003) The activation domains, the proline-rich domain, and the C-terminal basic domain in p53 are necessary for acetylation of histones on the proximal p21 promoter and interaction with p300/CREB-binding protein. J. Biol. Chem. 278, 17557–17565 [DOI] [PubMed] [Google Scholar]
- 41. Bulk E., Sargin B., Krug U., Hascher A., Jun Y., Knop M., Kerkhoff C., Gerke V., Liersch R., Mesters R. M., Hotfilder M., Marra A., Koschmieder S., Dugas M., Berdel W. E., Serve H., Müller-Tidow C. (2009) S100A2 induces metastasis in non-small cell lung cancer. Clin. Cancer Res. 15, 22–29 [DOI] [PubMed] [Google Scholar]
- 42. Wang H. Y., Zhang J. Y., Cui J. T., Tan X. H., Li W. M., Gu J., Lu Y. Y. (2010) Expression status of S100A14 and S100A4 correlates with metastatic potential and clinical outcome in colorectal cancer after surgery. Oncol. Rep. 23, 45–52 [PubMed] [Google Scholar]
- 43. Brooks C. L., Gu W. (2006) p53 ubiquitination. Mdm2 and beyond. Mol. Cell 21, 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mehta S. A., Christopherson K. W., Bhat-Nakshatri P., Goulet R. J., Jr., Broxmeyer H. E., Kopelovich L., Nakshatri H. (2007) Negative regulation of chemokine receptor CXCR4 by tumor suppressor p53 in breast cancer cells. Implications of p53 mutation or isoform expression on breast cancer cell invasion. Oncogene 26, 3329–3337 [DOI] [PubMed] [Google Scholar]
- 45. Sun Y., Sun Y., Wenger L., Rutter J. L., Brinckerhoff C. E., Cheung H. S. (1999) p53 down-regulates human matrix metalloproteinase-1 (Collagenase-1) gene expression. J. Biol. Chem. 274, 11535–11540 [DOI] [PubMed] [Google Scholar]
- 46. Liu J., Zhan M., Hannay J. A., Das P., Bolshakov S. V., Kotilingam D., Yu D., Lazar A. F., Pollock R. E., Lev D. (2006) Wild-type p53 inhibits nuclear factor-κB-induced matrix metalloproteinase-9 promoter activation. Implications for soft tissue sarcoma growth and metastasis. Mol. Cancer Res. 4, 803–810 [DOI] [PubMed] [Google Scholar]
- 47. Sun Y., Cheung J. M., Martel-Pelletier J., Pelletier J. P., Wenger L., Altman R. D., Howell D. S., Cheung H. S. (2000) Wild-type and mutant p53 differentially regulate the gene expression of human collagenase-3 (hMMP-13). J. Biol. Chem. 275, 11327–11332 [DOI] [PubMed] [Google Scholar]
- 48. Ho J., Benchimol S. (2003) Transcriptional repression mediated by the p53 tumor suppressor. Cell Death Differ. 10, 404–408 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.