Background: Lysosomes are important in degradation of expanded polyQ proteins, which accumulate in several neurodegenerative diseases.
Results: Two proteases, cathepsins L and Z, account for the lysosome's capacity to digest polyQ sequences.
Conclusion: These cathepsins are critical in cellular clearance of polyQ protein aggregates.
Significance: Cathepsins L and Z are important in defending against the accumulation and toxicity of polyQ proteins.
Keywords: Huntington Disease, Lysosomes, Neurodegeneration, Polyglutamine, Protease, Cathepsins, Polyglutamine Sequences, PolyQ Proteins
Abstract
In neurodegenerative diseases caused by extended polyglutamine (polyQ) sequences in proteins, aggregation-prone polyQ proteins accumulate in intraneuronal inclusions. PolyQ proteins can be degraded by lysosomes or proteasomes. Proteasomes are unable to hydrolyze polyQ repeat sequences, and during breakdown of polyQ proteins, they release polyQ repeat fragments for degradation by other cellular enzymes. This study was undertaken to identify the responsible proteases. Lysosomal extracts (unlike cytosolic enzymes) were found to rapidly hydrolyze polyQ sequences in peptides, proteins, or insoluble aggregates. Using specific inhibitors against lysosomal proteases, enzyme-deficient extracts, and pure cathepsins, we identified cathepsins L and Z as the lysosomal cysteine proteases that digest polyQ proteins and peptides. RNAi for cathepsins L and Z in different cell lines and adult mouse muscles confirmed that they are critical in degrading polyQ proteins (expanded huntingtin exon 1) but not other types of aggregation-prone proteins (e.g. mutant SOD1). Therefore, the activities of these two lysosomal cysteine proteases are important in host defense against toxic accumulation of polyQ proteins.
Introduction
Several neurodegenerative diseases are caused by the expansion of polyglutamine (polyQ)6 sequences in proteins, such as Huntington disease and spinocerebellar ataxia type 3 (1). These aggregation-prone mutant proteins accumulate in cells as intracellular protein inclusions and cause neuronal disease through toxic gain-of-function mechanisms (2). Most likely, the deleterious species are actually soluble oligomers of the polyQ proteins that also are probably intermediates in the formation of the large intraneuronal inclusions (3). Despite extensive studies, the mechanisms responsible for the accumulation of these proteins in cells are still unclear, but a variety of observations suggest decreased intracellular degradation, perhaps as a consequence of their aggregates. Consequently, ways to increase the clearance of polyQ proteins before they form these toxic aggregates could have therapeutic relevance.
The cellular pathways for digesting normal and expanded polyQ sequences in proteins have been the subjects of controversy. Because polyQ sequences are also found normally in a variety of proteins (e.g. TATA-binding proteins and CREB-binding proteins) that turn over without aggregating and causing disease, the cell must contain systems to degrade efficiently polyQ sequences. Presumably, the longer polyQ tracts may cause disease by exceeding the degradative capacity of these cellular systems. The two main proteolytic systems in mammalian cells are the autophagic/lysosomal system and the ubiquitin-proteasome pathway. The expanded polyQ proteins accumulate in aggregates associated with ubiquitin and proteasomes (4, 5), and inhibitors of proteasome cause accumulation of extended polyQ containing proteins (6–9). However, the proteolytic sites within eukaryotic 20 S proteasomes cannot digest polyQ sequences, whereas they can cleave efficiently most other types of peptide bonds (10, 11). Instead, while digesting such proteins, they must release the undigested aggregation-prone polyQ-rich fragments for hydrolysis by other cellular enzymes.
Normally, peptide products generated by the proteasome range in size between 2 and 24 residues, but most are shorter than 8 residues. These peptides are hydrolyzed rapidly (within seconds) to amino acids by cytosolic endopeptidases and aminopeptidases. Previously, we showed that only one cytosolic peptidase, puromycin-sensitive aminopeptidase is capable of degrading polyQ sequences efficiently (12). Subsequent overexpression of puromycin-sensitive aminopeptidase in cultured cells, muscle, and Drosophila was found to reduce the content and toxic effects of not only proteins with expanded polyQ sequences (e.g. huntingtin exon 1 or ataxin 3) but also other inclusion-prone proteins lacking such repeats (e.g. mutated SOD1). Surprisingly, puromycin-sensitive aminopeptidase was found not to simply catalyze the digestion of proteasome-derived fragments but to have the capacity to activate autophagy and lysosomal degradation through still unknown mechanisms (13). Moreover, a variety of studies suggest that the large aggregates of polyQ proteins are not degraded by proteasomes but instead tend to be destroyed by autophagy (13).
Furthermore, activation of autophagy by treatment with the mTOR inhibitors (e.g. rapamycin) or carbamazepine (14, 15) or by overexpression of transcription factor EB (16) has been shown to accelerate the clearance of polyQ proteins and reduce their cell toxicity. Such activators of autophagy have even been proposed as a potential therapy for these neurodegenerative diseases (2). These findings all entail that lysosomal enzyme(s) may also have the capacity to degrade these polyQ tracts (17). Although these findings imply that the lysosomal enzymes (unlike proteasomes) are able to hydrolyze completely polyQ sequences, even ones in aggregates (14–16), this capacity has not been systematically studied, and the identity of the lysosomal proteases that can degrade polyQ sequences has not been previously investigated to our knowledge. Their identification would clarify the metabolism and mode of turnover of polyQ proteins, and if their levels limit the rates of polyQ protein clearance, then such enzymes might even be possible therapeutic targets.
Lysosomes contain a large variety of enzymes, including many nucleases, lipases, and proteases that can catalyze the complete degradation of cell constituents and engulfed viruses or bacteria. Lysosomal proteases (generally referred to as cathepsins) are acid-optimal enzymes and include both exopeptidases (e.g. the aminopeptidase cathepsin H and the carboxypeptidases cathepsins A and Z) and many endopeptidases, including many cysteine proteases (e.g. cathepsins B, C, F, H, K, L, V, S, and Z), some aspartic proteases (cathepsins D and E), the serine proteases (cathepsins A and G), and a few metalloproteases (18). Most cathepsins are present in all mammalian tissues, whereas others show a specific tissue distribution that has proven important pharmacologically. For example, cathepsin K is especially abundant in osteoclasts, and cathepsin K inhibitors are therefore useful in treating osteoporosis (19).
In these studies, we investigated systematically whether lysosomal proteases can rapidly degrade polyQ proteins and then identified the critical cathepsins that catalyze this process. By using a combination of inhibitors as well as specific RNAi constructs and studies of the recombinant proteins, we found that two cysteine proteases, cathepsins L and Z, are the only lysosomal proteases capable of rapidly hydrolyzing polyQ sequences within peptides, multiple model polyQ proteins, and insoluble polyQ aggregates. These studies indicate that both the proteasomal and the lysosomal pathways involving cathepsins L and Z are implicated in the degradation of polyQ proteins. Moreover, these enzymes appear important in vivo in digesting such polyQ aggregates because depletion of either of these enzymes resulted in increased cell toxicity caused by proteins containing a long polyQ tract, but not a short polyQ sequence or by other aggregation-prone proteins lacking a polyQ repeat.
EXPERIMENTAL PROCEDURES
Peptides and Enzymes
The various polyQ peptides were a kind gift by Prof. Ron Wetzel and were disaggregated, stored, and used as described previously (10, 21). The peptide library RYX19NKTL where X is an equimolar mixture of Ala, Asp, Glu, Phe, Gly, His, Ile, Lys, Leu, Met, Pro, Gln, Arg, Ser, Thr, and Val, was a gift from Prof. Frank Momberg. Fluorogenic substrates used for assaying peptidase activities were purchased from Bachem (Basel, Switzerland). The specific peptidase inhibitors leupeptin, bestatin, pefabloc, pepstatin, and E64 were purchased from Sigma, and bortezomib (Velcade, PS-341) was a gift from Millenium. Recombinant human cathepsins L, Z, B, S, and H were purchased from R&D Systems.
Preparation of Cytosolic and Lysosomal Extracts
To prepare cytosolic extracts (41), HeLa cells or mammalian muscle or liver tissues were resuspended in ice-cold 50 mm sodium phosphate, pH 7.5, 5 mm MgCl2, 2 mm ATP, 1 mm DTT, and 10% glycerol. Cell extracts were prepared by Dounce homogenization with 10 manual strokes on ice. This homogenate was spun first at a low speed of 10,000 × g for 15 min to remove the nuclear fraction and then at a higher speed of 100,000 × g to remove membranous organelles. The resulting supernatant fractions were aliquoted, snap-frozen in liquid nitrogen, and stored at −80 °C until use. Both the 10,000 × g and 100,000 × g supernatants were tested for cytosolic peptidases at pH 7 to ensure that there was no loss of relevant peptidases by the high speed centrifugation.
To prepare the lysosomal extracts (20), fresh mice livers (WT or cathepsin L−/− mice) were homogenized in 20 mm Tris-HCl, pH 7.5, 0.25 m sucrose, 1 mm EDTA and then centrifuged at 1,500 × g for 10 min to remove the nuclear pellet. Supernatant was then centrifuged at 100,000 × g for 10 min to collect the “microsomal” and organelle pellet, which was resuspended, mixed with Percoll (maintaining 20 mm Tris, 0.25 m sucrose, pH 7.5), and centrifuged for 90 min at 40,000 × g to form a gradient. Lysosomes appeared as a dense band near the bottom of the Percoll gradient and were collected, washed several times to remove Percoll, and then sonicated in 20 mm Tris-HCl, pH 7.5, and cleared by low speed centrifugation. The resulting supernatant fractions were aliquoted, snap-frozen in liquid nitrogen, and stored at −80 °C until use.
Peptide Product Analysis by Fluorescamine Assay
Each of the peptides (15 μm) or proteins (20 μm) was incubated with the respective lysate/enzyme in a 20 μl reaction at 37 °C in 50 mm HEPES-KOH, pH 7.5, unless otherwise specified. N termini of new peptides, generated by enzyme digestion, interact with fluorescamine (22). After the incubation, 5 μl of each sample were mixed and rotated with 25 μl of fluorescamine solution in acetone (0.3 mg/ml) for 3 min and then diluted with 200 μl of 0.2 m sodium borate at pH 9. 200 μl of each sample were transferred to the 96-well plate and measured with the FLUOstar Galaxy plate reader (BMG Labtechnologies, Inc., Durham, NC) at an excitation of 380 nm and emission of 480 nm. An equimolar mixture of peptides of known concentration (EAA-NH2, AEAA-NH2, AAEAAG-NH2, AAVVAAG, and TTQRTRALV) was used as the standard to calculate the amount of new peptide products (i.e. new amino groups) generated.
Product Analysis by MALDI-TOF
Aliquots of the reaction mixtures were directly analyzed by MALDI-TOF MS (Applied Biosytems Voyager 4036 at the Dana Farber Core Facility), using the linear mode of analysis to increase the sensitivity of detection, as described (10).
Cell Culture and Transfections
HeLa, HEK293A, NIH3T3, and N1E-115 cells were cultured in DMEM (Invitrogen) containing 10% FBS, 100 units/ml penicillin G, and 100 μg/ml streptomycin. The Ub-R-GFP, Ub-R-GFP-Q16, and Ub-R-GFP-Q65 constructs were a kind gift of Prof. Dantuma and have been described previously (8). HeLa cells were transiently transfected with Superfect (Invitrogen) and collected for analysis at the indicated times. For inhibitor treatment, HeLa cells were incubated for 16 h in medium containing PS-341 (10 μm), NH4Cl (15 mm), or E64 (100 μm), whereas HEK293A cells (transfected with plasmids for GFP-Htt74Q for 48–72h) were incubated for 24 h with calpastatin peptide (Calbiochem) (1 μm), cathepsin L inhibitor I (Calbiochem) (10 μm), E64 (10 μm), E64d (Sigma) (10 μm), or MDL-28170 (Sigma) (10 μm). Total cell lysates were prepared by sonication in 50 mm HEPES/NaOH buffer, pH 7.5, containing 1 mm DTT and the complete protease inhibitor mixture (Roche Applied Science). Soluble cell lysates were prepared by low speed centrifugation at 10,000 × g, separated by SDS-PAGE (4–12%), and analyzed by Western blotting. The following antibodies were used for Western blots: anti-myoglobin antibody (Sigma), anti-polyglutamine antibody (Chemicon), anti-actin antibody (Sigma), and anti-GFP antibody (Sigma).
NIH3T3 cells were transfected with Lipofectamine (Invitrogen) for 48 h with plasmids encoding Htt74Q-mCherry (generated and described in Ref. 13) or CFP-LC3 (a generous gift from Prof. Lippincott-Schwartz). N1E-115 cells were transfected with ExGen500 (Fermentas) for 48 h with the plasmids for Htt74Q-mCherry and RNAi, either control or against cathepsin L or Z. The plasmids for Htt23Q or 74Q fused to GFP were a kind gift from Dr. Ron Kopito (Stanford University), whereas the plasmid for SOD1G93A-GFP was obtained from Dr. Piera Pasinelli (Farber Institute for Neurosciences, Thomas Jefferson University, Philadelphia, PA). Expression plasmids for microRNA targeting cathepsin L or Z genes were created by annealing self-complementary oligonucleotides and cloning into the pcDNA6.2-GW/EmGFP-miR vector (BLOCK-iT Pol II miR RNAi expression vector, Invitrogen). The oligonucleotides used were as follows: Mmi507307_top_CTPL, 5′-TGCTGAAATCCATCAGGCCTCCGTTAGTTTTGGCCACTGACTGACTAACGGAGCTGATGGATTT-3′; Mmi507307_bot_CTPL,5′-CCTGAAATCCATCAGCTCCGTTAGTCAGTCAGTGGCCAAAACTAACGGAGGCCTGATGGATTTC-3′; Mmi558005_top_CTPZ, 5′-TGCTGTCACGATCCTCATCCAGCCTTGTTTTGGCCACTGACTGACAAGGCTGGGAGGATCGTGA-3′;Mmi558005_bot_CTPZ, 5′-CCTGTCACGATCCTCCCAGCCTTGTCAGTCAGTGGCCAAAACAAGGCTGGATGAGGATCGTGAC-3′. pcDNA6.2-GW/EmGFP-miR-neg control plasmid (Invitrogen), containing an insert that forms a hairpin predicted not to target any known vertebrate gene, was used as a control. For toxicity assays, lactate dehydrogenase release was measured with the CytoTox 96 kit (Promega). Fluorescence images were collected at the Nikon Imaging Center at Harvard Medical School.
Murine Muscle Electroporation
Adult CD1 male mice (25–35 g) were anesthetized by intraperitoneal injection of Avertin (0.2 ml/10 g as 1.2% solution). Using a sterile technique, a 0.5–1.0 cm longitudinal incision was made in the skin overlying the tibialis anterior muscle, and the underlying muscle was exposed by blunt dissection of the surrounding skin. A paddle electrode was then introduced between the muscle and the underlying tibia. Plasmid DNA in sterile saline (40 μl, 25 μg) was injected into the muscle. A second paddle electrode was applied gently over the muscle, and electroporation was performed (12 V, 20-ms duration, 5 pulses, 200-ms intervals), using an ECM830 Electro Square Porator (BTX, Harvard Apparatus). The electrodes were removed, and the skin was closed by suture. Postoperative analgesia was administered every 12 h for the first 24 h with buprenorphine (0.05 mg/kg subcutaneously). Additional analgesia was administered thereafter if there was any evidence of continued animal discomfort. The animals were followed for 1 week, after which time they were euthanized by halothane inhalation followed by cervical dislocation, and the muscles were harvested, quickly frozen in liquid nitrogen-cooled isopentane, and stored at −80 °C. 10 μm muscle cryosections were analyzed for GFP/mCherry fluorescence after fixation with 4% paraformaldehyde and Hoechst staining (10 μg/ml in PBS for 10 min) using a Nikon 80i upright microscope at the Nikon Imaging Center at Harvard Medical School. No gross evidence of necrosis or inflammation as a result of the transfection procedure was noted. Inclusion size and number were analyzed with Metamorph using fixed size and intensity thresholds as described (13).
Statistical Analysis
A one-way analysis of variance test for multiple comparisons has been applied when distributions of number and size of inclusions were compared. Most data are presented as the mean ± S.D. Statistical comparisons were made using analysis of variance. A p value of <0.05 was considered statistically significant.
RESULTS
Lysosomal Extracts Rapidly Digest PolyQ Repeat Peptides and Proteins
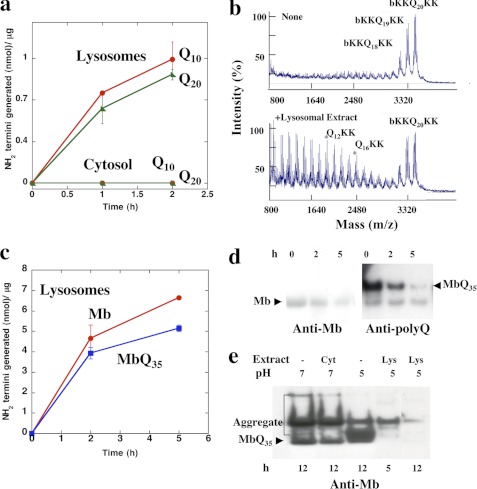
Prior studies have established that cytosolic extracts of mammalian cells lack the ability to efficiently digest polyQ sequence (10). In addition, there is increasing evidence that autophagy and thus lysosomal enzymes are important in the degradation of expanded polyQ-containing proteins (13, 17). To determine if lysosomes have a greater capacity to hydrolyze such sequences than cytosolic enzymes, lysosomes from mouse liver were partially purified by differential centrifugation (20) and sonicated, and their capacity to hydrolyze several polyQ peptides was studied. The model substrates used in these experiments contain 10–30 glutamine residues with flanking lysine, arginine, or aspartate residues to maintain the polyQ-rich sequence solubility (21). The number of cleavages made in these peptides was assayed by measuring the generation of new terminal amino groups with fluorescamine (22). Although there was no digestion of b-KKQ10KK and b-KKQ20KK by lysosomal extracts at pH 7, where lysosomal cathepsins are generally inactive (data not shown), at pH 5, these preparations rapidly digested the polyQ peptides (Fig. 1a). In fact, the rate of digestion of Q20RRGRR by the lysosomal extracts at pH 5 was 100-fold greater than the rate of digestion by the cytosolic extracts at pH 7 (Fig. 1a). The lysosomal extracts digested b-KKQ10KK and the longer b-KKQ20KK at similar rates at pH 5.
FIGURE 1.
Unlike cytosolic extracts, lysosomes rapidly digest polyQ peptides and proteins. a, b-KKQ10KK or b-KKQ20KK (15 μm) was incubated with either 3.2 μg of cytosolic extracts in a 30-μl reaction in 50 mm sodium phosphate, 1 mm DTT, pH 7, or with 3.2 μg of lysosomal extracts in a 30-μl reaction in 50 mm sodium acetate, 1 mm DTT, pH 5, at 37 °C, and the new N termini generated were assayed with fluorescamine. b, b-KKQ20KK (15 μm) was incubated alone or with 3.2 μg of lysosomal extracts in 50 mm sodium acetate, 1 mm DTT, pH 5, at 37 °C for 2 h, and the reaction mixtures were analyzed by MALDI-TOF. b-KKQ20KK alone (same as time (t) = 0 for the reaction) contains contaminating peptides b-KKQ19KK and b-KKQ18KK. c, WT Mb or Mb-Q35 fusion protein (MbQ35) (20 μm) was incubated with 3.2 μg of lysosomal extracts in a 30-μl reaction in 50 mm sodium acetate, 1 mm DTT, pH 5, at 37 °C, and the new N termini generated were assayed with fluorescamine. d, 20 μm WT Mb and Mb-Q35 were incubated with lysosomal extracts, and the reaction aliquots were boiled in 1× SDS sample buffer containing 100 mm DTT, separated by SDS-PAGE, and probed with either a polyclonal antibody against myoglobin or a monoclonal antibody against the polyQ sequence. For Mb-Q35, only the anti-polyQ blot is shown for simplicity. e, Mb-Q35 was preaggregated in 50 mm sodium phosphate, 1 mm DTT, pH 7, for 8 h at 37 °C and then either incubated alone at pH 7 or pH 5 or with 3.2 μg of cytosolic extracts at pH 7 or 3.2 μg of lysosomal extracts in 50 mm sodium acetate, 1 mm DTT, pH 5, at 37 °C. The reaction products were analyzed at the indicated times by SDS-PAGE followed by immunoblotting with anti-myoglobin antibodies. The labeled band corresponds to the monomeric Mb-Q35. Error bars, S.D.
To determine the nature of the digestion products, we used MALDI-TOF mass spectrometry. The lysosomal extracts made multiple cleavages within the polyQ sequence of b-KKQ20KK and generated a variety of fragments, including b-KKQ4, b-KKQ8, Q12KK, and Q16KK (Fig. 1b). These internal cleavages must be due to the actions of one or more lysosomal endopeptidases, although further trimming by amino- or carboxypeptidase(s) is also possible (see below).
To test whether the lysosomal extracts can also digest a polyQ sequence in a full-length soluble protein, we used as a model substrate, myoglobin (Mb), in which a Q35 repeat from ataxin 3 has been inserted (23). With this substrate (Mb-Q35), eukaryotic proteasomes degraded the Mb component completely but released the undigested Q35 sequence (10). (In that study (10) and the present one, we chose to study the proteolytic susceptibility of these polyQ aggregates because they are biochemically much more homogenous than those isolated from polyQ-rich inclusions present in cells.) By contrast, the lysosomal extracts rapidly digested both the WT recombinant Mb and Mb-Q35 (Fig. 1c), whose breakdown was assayed by measuring the generation of new N termini (Fig. 1c) as well as with antibodies against Mb and the polyQ component (Fig. 1, d and e).
Like pathogenic polyQ-expanded proteins, Mb-Q35 tends to form SDS-resistant aggregates upon incubation at 37 °C (10). Because aggregated proteins tend to resist digestion by most proteases, we determined the effect of the lysosomal extracts on these insoluble, polyQ-rich aggregates. After incubation at 37 °C for 8 h, the aggregated Mb-Q35 was incubated either at pH 7 with or without the cytosolic extracts or at pH 5 with or without the lysosomal extracts. The reaction products were analyzed by SDS-PAGE followed by Western blot with anti-Mb antibodies. Although cytosolic extracts at pH 7 were unable to degrade these aggregates, the lysosomal extracts digested them rapidly at pH 5. Thus, unlike proteasomes, the lysosomal extracts can completely digest polyQ sequences in peptides and proteins even when in insoluble aggregates (Fig. 1e).
Lysosomal Cysteine Proteases Degrade PolyQ Peptides
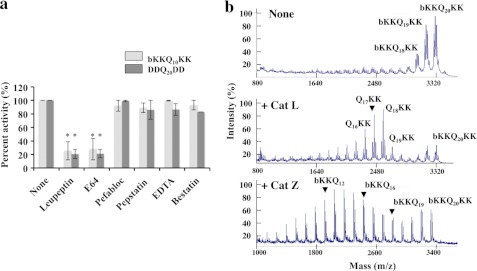
In order to identify the lysosomal proteases responsible for digesting the polyQ repeats, we first tested the effects of different types of protease inhibitors on the hydrolysis of b-KKQ10KK and DDQ20DD by lysosomal extracts. We used leupeptin and E64 to inhibit cysteine proteases, pefabloc to inhibit serine proteases, pepstatin to inhibit aspartyl proteases (e.g. cathepsin D), the chelating agent EDTA to inhibit metalloproteases, or bestatin to inhibit aminopeptidases (24). Lysosomal digestion of b-KKQ10KK and DDQ20DD at pH 5 was completely inhibited by leupeptin and E64 (p < 0.001), whereas there was little or no effect of any other inhibitor (Fig. 2a). Thus, lysosomal cysteine proteases appear to be critical for the digestion of the polyQ repeats.
FIGURE 2.
Lysosomal cysteine proteases are responsible for the digestion of polyQ peptides. a, b-KKQ10KK and DDQ20DD (15 μm) were incubated with 3.2 μg of lysosomal extracts in 50 mm sodium acetate, 1 mm DTT, pH 5, at 37 °C for 1 h, in the presence or absence of inhibitors, and the new N termini generated were assayed. The lysosomal extracts were preincubated with the inhibitors for 20 min at room temperature. The concentrations used were 100 μm for leupeptin, 100 μm for E64, 1 mm for pefabloc, 100 μm for pepstatin, 1 mm for EDTA, and 100 μm for bestatin. *, p < 0.001. b, b-KKQ20KK (15 μm) was incubated with activated pure recombinant human cathepsins B, L, H, S, and Z at amounts varying between 10 and 100 ng under optimal reaction conditions for each cathepsin at 37 °C, and the reaction mixtures were analyzed by MALDI-TOF. Digestion patterns for b-KKQ20KK alone (same as t = 0 of the reaction) and with cathepsin L and cathepsin Z are shown. No digestion of b-KKQ20KK was observed by cathepsins B, S, and H. Cathepsin L acts as an endopeptidase in digesting b-KKQ20KK, whereas cathepsin Z digests b-KKQ20KK as a carboxypeptidase. Error bars, S.D.
The primary cysteine proteases in the lysosomes are well characterized enzymes, especially the endopeptidases cathepsins B, S, and L, the carboxypeptidase cathepsin Z (also known as X or P), and the aminopeptidase cathepsin H, although their substrate specificities remain unclear (25). To identify the cysteine proteases active against the polyQ sequences, we tested the activities of these pure recombinant lysosomal proteases against b-KKQ10KK, b-KKQ20KK, and DDQ20DD. Cathepsins L and Z were found to be the only peptidases capable of digesting these polyQ peptides (Fig. 2b). No degradation was observed with cathepsin B, S, or H when the products were measured with either fluorescamine or MALDI-TOF, although in control experiments, these enzymes were active and hydrolyzed standard fluorogenic peptide substrates, such as benzyloxycarbonyl-GRR-7-amido-4-methylcoumarin (data not shown). Using MALDI-TOF, pure cathepsin L was shown to act as an endopeptidase on b-KKQ20KK, generating Q19KK, Q18KK, Q17KK, and other diverse products. By contrast, pure cathepsin Z acted as a carboxypeptidase on b-KKQ20KK, generating b-KKQ19, b-KKQ18, b-KKQ17, and shorter peptides (Fig. 2b). Thus, it is likely that cathepsin L carries out initial cleavages yielding shorter peptides that are hydrolyzed to amino acids by cathepsin Z.
Lysosomal Extracts from Cathepsin L−/− Mice Degrade PolyQ Peptides and Proteins Less Efficiently than WT
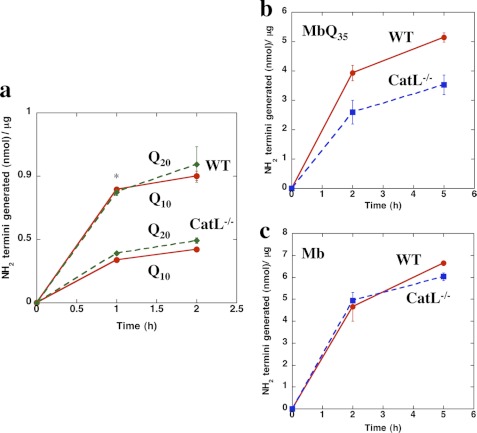
Initially, we attempted to ascertain the individual contributions of cathepsin L or Z to the activity of lysosomal extracts by immunodepletion but could not do so because the available antibodies against cathepsins L and Z were found to cross-react with each other and with other cathepsins. We therefore assessed their specific contributions using lysosomes from available knock-out mice lacking these cathepsins. Cathepsin L-null mice have been described by several groups (26–28) and are characterized by defects in skin epidermal cells and hair growth. Equal amounts of lysosomal extracts from the livers of cathepsin L−/− mice were much less efficient in digesting b-KKQ10KK and b-KKQ20KK than lysosomal preparations from the WT mice (p < 0.01 at 1 h) (Fig. 3a). These findings confirm that cathepsin L is important in the clearance of the polyQ sequences by the lysosomal pathway. Similarly, when tested against the Mb-Q35 fusion protein, the lysosomal extracts from cathepsin L−/− mice were significantly less active than WT extracts (Fig. 3b). By contrast, the lysosomes of cathepsin L−/− and WT mice showed similar activities against the natural protein, Mb, lacking the polyQ repeat (Fig. 3c). These observations confirm that within lysosomes, cathepsin L is a major, probably the only, rate-limiting protease in the digestion of polyQ repeats in proteins, although many other lysosomal proteases can degrade the typical polypeptide sequences in myoglobin. Because cathepsin Z-null mice are not available, we used siRNA approaches to dissect the contribution of this enzyme in the degradation of polyQ-containing proteins (see below).
FIGURE 3.
Lysosomal extracts from Cathepsin L−/− mice cannot efficiently degrade long-polyQ peptides and proteins. a, b-KKQ10KK and b-KKQ20KK (15 μm) were incubated with 3.2 μg of lysosomal extracts prepared from WT mice or cathepsin L−/− mice in 50 mm sodium acetate, 1 mm DTT, pH 5, at 37 °C, and the new N termini generated were assayed. *, p < 0.01. b and c, WT-Mb (c) or Mb-Q35 (b) (20 μm) was incubated with 3.2 μg of lysosomal extracts prepared from WT mice or cathepsin L−/− mice in 50 mm sodium acetate, 1 mm DTT, pH 5, at 37 °C, and the new N termini generated were assayed. Error bars, S.D.
In Vivo Both Proteasomes and Lysosomal Cysteine Proteases Degrade Extended PolyQ Proteins, but Short PolyQ Proteins Are Degraded Only by Proteasomes
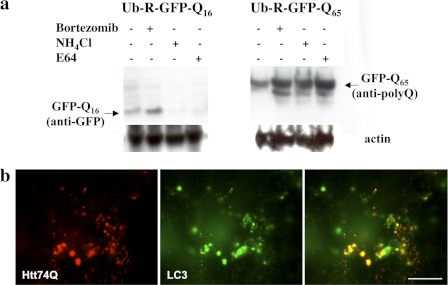
In order to confirm that the lysosomal cysteine proteases are important in vivo, we investigated the degradation of short (Q16) and extended (Q65) polyQ sequences fused to the C terminus of Ub-R-GFP in cultured HeLa cells. In this construct, the ubiquitin moiety is quickly removed, exposing an N-terminal arginine, which is targeted for rapid ubiquitination and proteasomal degradation via the N-end rule pathway (8). Although they have a strong ubiquitination signal targeting them to the proteasomes, we tested whether lysosomes play any role in this rapid degradation of polyQ fused to Ub-R-GFP proteins. Following transient transfections of Ub-R-GFP, Ub-R-GFPQ16 and Ub-R-GFPQ65, we treated the cells with cycloheximide to prevent further protein synthesis and either bortezomib (Velcade, PS-341), a specific inhibitor of proteasomes (29), or NH4Cl, a weak base that inhibits lysosomal proteolysis by increasing the pH within this acidic compartment (30). Soluble extracts of transfected HeLa cells were analyzed by Western blot and probed with antibodies specific to GFP or to polyQ sequences. As expected, full-length Ub-R-GFP (not shown) and Ub-R-GFPQ16 were degraded exclusively by proteasomes in HeLa cells because these proteins accumulated in the presence of bortezomib but not of NH4Cl (Fig. 4a). There was also no measurable accumulation of Q16 peptides upon lysosomal inhibition, presumably because such short polyQ-rich fragments released by the proteasomes are digested in the cytosol by puromycin-sensitive aminopeptidase to amino acids (12).
FIGURE 4.
Short polyQ proteins are degraded by proteasomes, whereas both proteasomes and lysosomal cysteine proteases degrade long polyQ proteins in vivo. a, HeLa cells transiently transfected for 36 h with Ub-R-GFPQ16 and Ub-R-GFPQ65 were treated with specific inhibitors for 12 h (i.e. 10 μm bortezomib, 15 mm NH4Cl, and 100 μm E64). Soluble cell extracts following a 10,000 × g centrifugation were analyzed by immunoblotting with antibodies against GFP or polyQ sequences. Anti-actin blots showed similar intensities for all of the lanes. b, NIH3T3 cells were transfected with Htt74Q-mCherry and CFP-LC3 to stain lysosomes. For simplicity, lysosomes are shown in green, so that areas of co-localization appear in yellow. Scale bar, 20 μm.
By contrast, there was a large accumulation of Ub-R-GFPQ65 upon treatment with either bortezomib or NH4Cl. Thus, both the proteasomal and lysosomal pathways contribute to the degradation of Ub-R-GFPQ65 (Fig. 4a). Despite the strong ubiquitination signal, which targets Ub-R-GFPQ16 exclusively to the proteasomes, the aggregation-prone construct with extended polyQ repeat (i.e. Ub-R-GFPQ65) appears to be hydrolyzed in part by both systems (Fig. 4a). In fact, earlier studies using these fusion proteins had shown that the soluble proteins are degraded efficiently by the proteasomes, but aggregation of these proteins due to extension of the polyQ repeat leads to resistance to proteasomal degradation (8). Ub-R-GFPQ65 should be present both as the monomeric protein and microaggregates, and more likely the lysosomes via autophagic vacuole formation degrade these proteasome-resistant aggregates and microaggregates, whereas the proteasomes eliminate the soluble monomeric proteins. Accordingly, in cultured cells, huntingtin exon 1 with 74Q expansion (Htt74Q-mCherry) was co-localized with the autophagosome marker CFP-LC3 (Fig. 4b).
Moreover, a build-up of Ub-R-GFPQ65 was also observed in the presence of the cysteine protease inhibitor, E64, and the extent of accumulation was similar to that with NH4Cl (Fig. 4a). As expected, there was no accumulation of Ub-R-GFPQ16 upon treatment with the inhibitor E64 (Fig. 4a). These biochemical and morphological results are consistent with our earlier observations that the lysosomal proteases, cathepsins L and Z are the major cellular enzymes functioning in the rapid digestion of extended insoluble polyQ proteins.
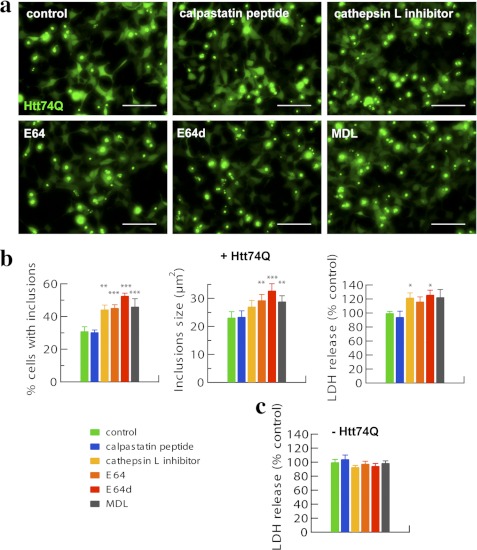
Inhibitors of Lysosomal Cysteine Proteases Enhanced Accumulation of Aggregated, Expanded Huntingtin Exon 1 and Caused Toxicity
Because cells contain other cysteine proteases aside from those in the lysosome that may act on polyQ proteins, such as calpains (31, 32), we used more specific inhibitors to assess their relative contributions. We treated HEK293A cells transfected with Htt74Q-GFP for 48–72 h with inhibitors of calpain I and II (i.e. calpastatin peptide) or cathepsin L inhibitor I and compared their effects with the general inhibitor of cysteine proteases, E64; its more cell-permeable derivative, E64d; or MDL-28170, a leupeptin derivative that blocks calpains and other cysteine proteases. There was a clear increase in cell death when any inhibitor of lysosomal cysteine proteases was combined with transfection of Htt74Q-GFP, as measured by lactate dehydrogenase release (p < 0.05) (Fig. 5b). By contrast, these agents had no such toxic effects in cells not expressing Htt74Q-GFP (Fig. 5c). However, the addition of the selective calpain inhibitors did not induce death in the cells expressing Htt74Q-GFP. Presumably, a failure of the lysosomal enzymes to efficiently degrade the extended polyQ protein has toxic effects and promotes cell death (Fig. 5, a and b).
FIGURE 5.
Cathepsin (cysteine protease) inhibitors but not calpain inhibitors increase the proportion of cells with inclusions, the sizes of inclusions, and the toxicity in HEK293A cells expressing expanded huntingtin exon 1. a, HEK293A cells expressing Htt74Q-GFP for 48–72 h were treated with the indicated inhibitors for 24 h (see “Experimental Procedures”). Representative pictures are shown. Scale bar, 100 μm. b, quantitation of the number of inclusions displayed by HEK293A cells expressing Htt74Q-GFP following different treatments is reported as well as that for sizes of inclusions and related cell toxicity in lactate dehydrogenase (LDH) release assays. Error bars, S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001. n = 6. c, HEK293A cells not expressing Htt74Q-GFP were treated as above, and cell toxicity was measured in lactate dehydrogenase release assays. No evident cell toxicity per se was observed with any of the inhibitors used. n = 6.
To investigate whether this toxicity is due to accumulation of abnormal proteins, we measured the number and size of inclusions containing Htt74Q-GFP. Upon treatments with inhibitors of cysteine proteases, but not inhibitors of calpains, there was a 50–75% increase in the number of cells displaying inclusions (p < 0.001) and a 20–45% increase in their mean areas (p < 0.01) (Fig. 5b). Thus, in those cells that remained viable (i.e. attached to the plate), the inhibition of cysteine proteases, in particular cathepsin L, and the failure to degrade polyQ proteins result in more abundant and larger inclusions containing these proteins. Therefore, the loss of lysosomal degradative enzymes induces a build-up of aggregation-prone expanded huntingtin with toxic consequences (Fig. 5b).
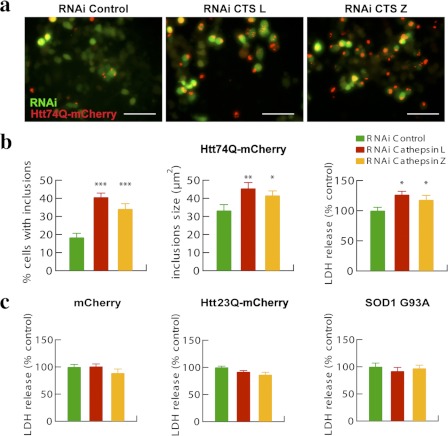
Decreased Cathepsin L or Z Enhances the Aggregation and Toxicity of Expanded Huntingtin Exon 1 in Cultured Cells
To investigate more specifically the roles of cathepsins L and Z in clearing extended polyQ proteins, we transfected murine neuroblastoma N1-E115 cells for 48 h with plasmids for Htt74Q-mCherry and RNAi that down-regulates the expression of cathepsin L or Z or control RNAi constructs. As shown by quantitative PCR, these transfections reduced cathepsin L and Z mRNA by 65–70% (supplemental Fig. S1). We then measured the number of cells showing Htt74Q inclusions and their sizes (Fig. 6, a and b). When cathepsin L was down-regulated for 2 days, the percentage of cells displaying Htt74Q inclusions doubled (p < 0.001). Concomitantly, the mean size of the inclusions in the cathepsin L-deficient cells was about 35% (p < 0.01) greater, and the cell toxicity was about 25% (p < 0.05) higher than in the controls (Fig. 6, a and b). Quite similar increases in the number and sizes of the polyQ inclusions and also in cell death were observed when cathepsin Z was specifically down-regulated with RNAi (Fig. 6, a and b). These pharmacological and genetic approaches nicely confirm that both lysosomal proteases are important in the degradation of polyQ proteins and both help protect cells from polyQ-induced toxicity.
FIGURE 6.
Reduced expression of cathepsin L or Z increases the proportion of cells with inclusions, the sizes of inclusions, and toxicity in neuroblastoma N1-E115 cells expressing expanded huntingtin exon 1. a, N1-E115 cells were transfected for 48 h with Htt74Q-mCherry along with GFP-RNAi constructs, either control or against cathepsin L or Z. Representative pictures are shown. Scale bar, 100 μm. b, quantitation of the number of inclusions displayed by N1-E115 cells expressing Htt74Q-mCherry following different treatments is reported as well as that for sizes of inclusions and related cell toxicity in lactate dehydrogenase (LDH) release assays. Error bars, S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001. n = 6. c, N1-E115 cells expressing mCherry, Htt23Q-mCherry, or SOD1G93A were tested for cell toxicity through lactate dehydrogenase release assays. No evident toxicity was observed for any of the proteins expressed. Error bars, S.D. n = 6.
To determine if these enzymes are also critical in defending against other toxic proteins, we also assayed the cell toxicity caused by expression of different model proteins containing an extended (i.e. Htt74Q-mCherry) or normal length polyQ sequence (i.e. Htt23Q-mCherry), mCherry, or a mutated form of superoxide dismutase SOD1G93A that is one of the causes of hereditary amytrophic lateral sclerosis (Fig. 6, b and c). This SOD1 variant was studied because it lacks a polyQ sequence but is a toxic aggregation-prone protein that forms intracellular inclusions and appears to be degraded by similar mechanisms as long polyQ proteins (13). Although decreasing cathepsin L or Z content enhanced the toxicity of the extended form of exon 1-containing 74Qs, it did not increase the cell toxicity associated with overexpression of SOD1G93A as well as of control proteins (Fig. 6c). Thus, cathepsins L and Z are specifically important in protecting cells from the toxic effects of extended polyQ proteins but not all aggregation-prone proteins.
Cathepsins L and Z Appear Important in Clearing Expanded Huntingtin Exon 1 Expressed in Adult Mouse Muscles
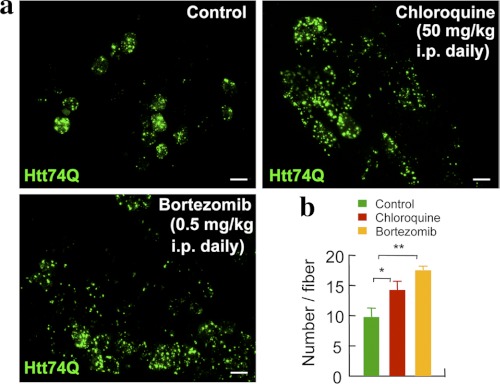
To test whether inhibition of the proteasomal or lysosomal pathway affect the formation of polyQ aggregates in mouse tissues in vivo, we electroporated GFP-tagged huntingtin exon 1 constructs bearing a long (i.e. pathological) 74Q tail into the tibialis anterior muscles of adult mice as described previously (13). This approach has been very useful in studying the role of gene products in muscle growth and atrophy and more recently in the breakdown of aggregation-prone proteins (13). Beginning at the time of electroporation, these mice received daily injections intraperitoneally of either chloroquine to inhibit lysosomal function or bortezomib to inhibit proteasomes in vivo. The doses of bortezomib used (i.e. 0.5 mg/kg body weight, daily) resulted at most in a 50% reduction of the proteasome's chymotrypsin-like activity measured in muscle extracts using Suc-LLVY-7-amido-4-methylcoumarin as the substrate (data not shown). The doses of chloroquine used (i.e. 50 mg/kg daily) have been reported to eliminate malaria infections and not to cause toxicity in mice (33). After 1 week, the inhibition of either pathway caused the number of aggregates per muscle fiber to increase from 10 to about 14 upon chloroquine treatment (p < 0.05) and to about 17 upon bortezomib treatment (p < 0.01) (Fig. 7, a and b). Thus, both lysosomal and proteasomal pathways appear important in vivo for the degradation of proteins containing long polyQ stretches (Fig. 7, a and b), as was also indicated by studies with cultured cells.
FIGURE 7.
In vivo inhibition of the lysosomal or the proteasomal pathway results in increased number of inclusions in mouse muscle fibers expressing expanded huntingtin exon 1. a, mice electroporated with plasmids for Htt74Q-GFP into the tibialis anterior muscle were injected with chloroquine or bortezomib daily for 7 days at the indicated doses. A representative picture for each treatment is shown. Scale bar, 50 μm. b, quantitation of the number of Htt74Q-GFP-positive inclusions per muscle fiber is reported for the different treatments. Error bars, S.D. *, p < 0.05; **, p < 0.01. n = 6.
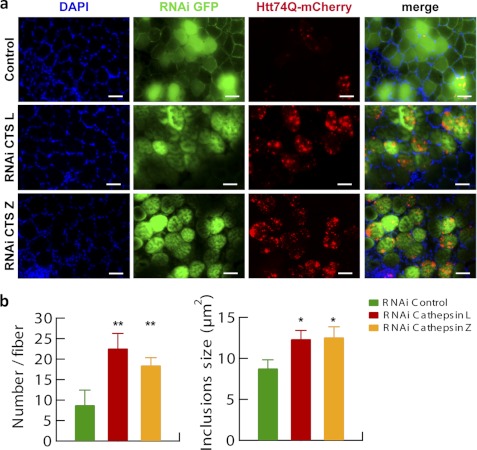
To further confirm the importance of these two lysosomal proteases in a more physiological context than cultured cells, RNAi constructs directed against cathepsin L or Z were electroporated into adult mouse muscles together with Htt74Q-mCherry, and 7 days later, the number of inclusions per muscle fiber and their mean sizes were evaluated. In agreement with the findings in cell culture, down-regulating cathepsin L resulted in a 3-fold greater number of inclusions per muscle fiber (p < 0.01), and these aggregates were about 50% larger than those in fibers expressing a control RNAi (p < 0.05) (Fig. 8, a and b). Similarly, down-regulation of cathepsin Z resulted in a 2-fold larger number of inclusions per fiber (p < 0.01), and these inclusions were about 50% larger than those displayed by control fibers (p < 0.05) (Fig. 8, a and b). Thus, by multiple approaches, in cell-free extracts, cultured cells, and in vivo, we have found that cathepsin L and Z are essential for the efficient clearance of various proteins containing long polyQ stretches because their loss leads to greater protein accumulation of various disease-associated constructs in aggregates and greater toxicity.
FIGURE 8.
Reduced expression of cathepsin L or Z increases the number and size of inclusions in mouse muscle fibers expressing expanded huntingtin exon 1. a, tibialis anterior muscles were electroporated for 7 days with plasmids expressing Htt74Q-mCherry along with GFP-RNAi constructs, either control or against cathepsin L or Z. A representative field of a transverse section of fibers for each condition tested is reported. Nuclei were stained with DAPI. Scale bar, 100 μm. b, quantitation of the number of inclusions per muscle fiber expressing Htt74Q-mCherry under the different treatments is reported as well as that for sizes of inclusions. Larger and more abundant aggregates are observed when cathepsin L or Z is depleted. Error bars, S.D. *, p < 0.05; **, p < 0.01. n = 6.
DISCUSSION
Cathepsins L and Z Are Key Lysosomal Enzymes in Degrading PolyQ Peptides and Proteins
Although a variety of recent cellular studies have indicated that lysosomes are important in the clearance of aggregation-prone polyQ proteins and in protection from their toxic consequences, the precise biochemical basis for lysosomal actions on these unusual proteins has not been clear. The present findings, based initially on cell-free analyses of lysosomal extracts and then on the roles of different cysteine proteases, have indicated a critical role of two lysosomal enzymes, the endopeptidase cathepsin L and the carboxypeptidase cathepsin Z, in the hydrolysis of polyQ repeat sequences, which resist hydrolysis by most proteases and exopeptidases. Although lysosomes contain many (at least 13) acid-optimal hydrolases with overlapping specificities allowing rapid hydrolysis of perhaps all types of proteins (18), cathepsin L and Z seem to be the only two enzymes with clear activity against polyQ sequences in small peptides and proteins. Furthermore, a lack of or a decreased content of either reduces the degradation of such peptides by lysosomal extracts as well as of polyQ proteins in cells.
Our analysis of cleavage patterns by mass spectrometry indicated a capacity of cathepsin L to cleave within polyQ sequences, which the major endoproteolytic complex in the cytosol, the proteasome, as well as cytosolic peptidases cannot do. Although the six active sites in the 20 S proteasome can cleave nearly all types of bonds in proteins, they must release aggregation-prone polyQ-rich peptides (10, 11), and only one cytosolic peptidase, puromycin-sensitive aminopeptidase, can then digest such sequences (12). However, this cytosolic enzyme is an exopeptidase, whose ability to digest long polyQ sequences is quite limited, as shown here by the very slow cleavage of model polyQ substrates by cytosolic extracts (Fig. 1, a and e). By comparison, the specific activity of lysosomal extracts at pH 5 against both polyQ sequences and insoluble polyQ proteins was much greater (even after the large dilution of the lysosomal enzymes) (Fig. 1).
Most likely, within the lysosome, the major endoprotease, cathepsin L, carries out an initial attack within the polyQ repeat, which generates new C termini, facilitating the actions of the carboxypeptidase cathepsin Z. A recent study has reported that cathepsin Z as well as the cytosolic cysteine aminopeptidase are important for the generation of N-terminal fragments of mutant huntingtin that appear important in HD pathogenesis (34). However, because cathepsin Z is primarily lysosomal, and it functions as a carboxypeptidase, its intracellular role in this process remains to be clarified and confirmed. In our study, down-regulation of cathepsin Z enhanced accumulation and toxicity of expanded huntingtin exon 1, clearly indicating a protective role. However, in their study, cathepsin Z cleavage resulted in increased production of a toxic fragment rather than cell protection (34). Although that study used different cell lines and failed to analyze the effects of other lysosomal hydrolases, our findings in multiple cell extracts, cultured cells, and mature mouse muscles indicate major (and probably synergistic) actions of this endoprotease and endopeptidase in the complete hydrolysis of polyQ sequences within the lysosome or autophagic vacuoles.
Recent studies, especially by Rubinsztein and colleagues (14), have indicated that the clearance of large aggregates of extended polyQ proteins occurs primarily through autophagy and thus lysosomal degradation, which can be accelerated upon treatment with rapamycin (35). Substrate delivery to the lysosome can occur via autophagosomes (14, 36), the double-membranous structures that engulf large parts of the cytoplasm, including organelles and presumably protein aggregates and aggresomes (37, 38), which are formed by the directional movement of ubiquitinated protein aggregates (39) along microtubules. Inhibition of microtubule function impairs both the autophagosome-lysosome fusion as well as aggresome formation and can enhance polyQ-induced toxicity (36, 40). As shown here, only lysosomes have the capacity to rapidly hydrolyze insoluble aggregates as well as soluble aggregates formed by extended polyQ proteins (Figs. 1e and 4a). This capacity appears especially important in light of growing evidence that soluble oligomers of disease-associated toxic proteins are more deleterious than larger aggregates, which may instead even protect cells from protein toxicity (3). The present studies together would suggest that mutations or polymorphisms in cathepsin L or Z, if they do exist, could be a cause of early onset or more aggressive progression of polyQ diseases.
The present study is unusual in its combined use of biochemical, pharmacological, cell biological, and in vivo approaches. Findings initially made on the hydrolysis of model polyQ peptides by lysosomal extracts and pure cloned cathepsins laid the basis for studies on cultured cells that confirmed the importance of cathepsin Z and L in the degradation of polyQ proteins. Selective inhibition of lysosomal cysteine proteases and reducing the levels of either cathepsin L or Z caused greater accumulation of expanded huntingtin exon 1 and concomitantly enhanced its toxic effects, while showing a strong correlation between build-up of the cellular aggregates (i.e. both increased number and size) and cell toxicity (Figs. 5, 6, and 8). Such correlations do not indicate whether the accumulation of inclusions are themselves toxic or if they provide protection for the cell while the non-assayed soluble microaggregates may be the actual cause of cell death.
These findings were also confirmed by our use of a powerful new approach to studying effects of individual proteins (i.e. in vivo electroporation of genes or RNAi into muscle fibers in adult mice to study their intracellular fates or consequences). This approach offers many advantages (e.g. in simplicity and rapidity); it saves major costs of animal handling and avoids possible effects of transgenes or RNAi during development. Electroporation of genes or RNAi has proven very useful in dissecting the mechanisms of muscle atrophy as well as analyzing the importance of puromycin-sensitive aminopeptidase in host defense against misfolded proteins. Moreover, we show for the first time how this approach is useful for establishing effects of small-molecule inhibitors or drugs (e.g. chloroquine and bortezomib) (Fig. 7) on the accumulation of polyQ proteins. This method avoids pharmacological problems due to poor penetration of drug candidates into the brain. Moreover, this approach nicely confirmed in vitro findings precisely on the roles of proteasomes and lysosomal hydrolases, cathepsins L and Z, in protection against polyQ proteins. This agreement between findings in cell-free systems, cultured cells, and electroporated muscles provides further validation of this latter approach, as well as our conclusions about the physiological importance of these two lysosomal proteases.
Supplementary Material
Acknowledgments
We thank the Nikon Imaging Center at Harvard Medical School for help with light microscopy and Mary Dethavong for valuable assistance in the preparation of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AR055255 (to A. L. G.). This work was also supported by grants from the High Q Foundation, the Amyotrophic Lateral Sclerosis Association, and the Packard Center for Amyotrophic Lateral Sclerosis Research at Johns Hopkins.

This article contains supplemental Fig. S1.
- polyQ
- polyglutamine
- CREB
- cAMP-response element-binding protein
- Mb
- myoglobin
- b-
- biotinyl
- Ub
- ubiquitin
- LDH
- lactate dehydrogenase.
REFERENCES
- 1. Zoghbi H. Y., Orr H. T. (2000) Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 23, 217–247 [DOI] [PubMed] [Google Scholar]
- 2. Rubinsztein D. C. (2006) The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443, 780–786 [DOI] [PubMed] [Google Scholar]
- 3. Kitamura A., Kubota H. (2010) Amyloid oligomers. Dynamics and toxicity in the cytosol and nucleus. FEBS J. 277, 1369–1379 [DOI] [PubMed] [Google Scholar]
- 4. Davies S. W., Turmaine M., Cozens B. A., DiFiglia M., Sharp A. H., Ross C. A., Scherzinger E., Wanker E. E., Mangiarini L., Bates G. P. (1997) Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90, 537–548 [DOI] [PubMed] [Google Scholar]
- 5. Waelter S., Boeddrich A., Lurz R., Scherzinger E., Lueder G., Lehrach H., Wanker E. E. (2001) Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell 12, 1393–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bence N. F., Sampat R. M., Kopito R. R. (2001) Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292, 1552–1555 [DOI] [PubMed] [Google Scholar]
- 7. Jana N. R., Zemskov E. A., Wang G., Nukina N. (2001) Altered proteasomal function due to the expression of polyglutamine-expanded truncated N-terminal huntingtin induces apoptosis by caspase activation through mitochondrial cytochrome c release. Hum. Mol. Genet. 10, 1049–1059 [DOI] [PubMed] [Google Scholar]
- 8. Verhoef L. G., Lindsten K., Masucci M. G., Dantuma N. P. (2002) Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Hum. Mol. Genet. 11, 2689–2700 [DOI] [PubMed] [Google Scholar]
- 9. Chai Y., Koppenhafer S. L., Shoesmith S. J., Perez M. K., Paulson H. L. (1999) Evidence for proteasome involvement in polyglutamine disease. Localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum. Mol. Genet. 8, 673–682 [DOI] [PubMed] [Google Scholar]
- 10. Venkatraman P., Wetzel R., Tanaka M., Nukina N., Goldberg A. L. (2004) Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol. Cell 14, 95–104 [DOI] [PubMed] [Google Scholar]
- 11. Holmberg C. I., Staniszewski K. E., Mensah K. N., Matouschek A., Morimoto R. I. (2004) Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J. 23, 4307–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bhutani N., Venkatraman P., Goldberg A. L. (2007) Puromycin-sensitive aminopeptidase is the major peptidase responsible for digesting polyglutamine sequences released by proteasomes during protein degradation. EMBO J. 26, 1385–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Menzies F. M., Hourez R., Imarisio S., Raspe M., Sadiq O., Chandraratna D., O'Kane C., Rock K. L., Reits E., Goldberg A. L., Rubinsztein D. C. (2010) Puromycin-sensitive aminopeptidase protects against aggregation-prone proteins via autophagy. Hum. Mol. Genet. 19, 4573–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ravikumar B., Vacher C., Berger Z., Davies J. E., Luo S., Oroz L. G., Scaravilli F., Easton D. F., Duden R., O'Kane C. J., Rubinsztein D. C. (2004) Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 36, 585–595 [DOI] [PubMed] [Google Scholar]
- 15. Zhang L., Yu J., Pan H., Hu P., Hao Y., Cai W., Zhu H., Yu A. D., Xie X., Ma D., Yuan J. (2007) Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc. Natl. Acad. Sci. U.S.A. 104, 19023–19028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sardiello M., Palmieri M., di Ronza A., Medina D. L., Valenza M., Gennarino V. A., Di Malta C., Donaudy F., Embrione V., Polishchuk R. S., Banfi S., Parenti G., Cattaneo E., Ballabio A. (2009) A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 [DOI] [PubMed] [Google Scholar]
- 17. Williams A., Jahreiss L., Sarkar S., Saiki S., Menzies F. M., Ravikumar B., Rubinsztein D. C. (2006) Aggregate-prone proteins are cleared from the cytosol by autophagy. Therapeutic implications. Curr. Top. Dev. Biol. 76, 89–101 [DOI] [PubMed] [Google Scholar]
- 18. Brix K. (2005) Lysosomal proteases. Revival of the Sleeping Beauty. in Lysosomes (Saftig P., ed) pp. 50–59, Springer, New York [Google Scholar]
- 19. Vasiljeva O., Reinheckel T., Peters C., Turk D., Turk V., Turk B. (2007) Emerging roles of cysteine cathepsins in disease and their potential as drug targets. Curr. Pharm. Des. 13, 387–403 [DOI] [PubMed] [Google Scholar]
- 20. Jonas A. J. (1986) Cystine transport in purified rat liver lysosomes. Biochem. J. 236, 671–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen S., Wetzel R. (2001) Solubilization and disaggregation of polyglutamine peptides. Protein Sci. 10, 887–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Udenfriend S., Stein S., Böhlen P., Dairman W., Leimgruber W., Weigele M. (1972) Fluorescamine. A reagent for assay of amino acids, peptides, proteins, and primary amines in the picomole range. Science 178, 871–872 [DOI] [PubMed] [Google Scholar]
- 23. Tanaka M., Morishima I., Akagi T., Hashikawa T., Nukina N. (2001) Intra- and intermolecular β-pleated sheet formation in glutamine-repeat inserted myoglobin as a model for polyglutamine diseases. J. Biol. Chem. 276, 45470–45475 [DOI] [PubMed] [Google Scholar]
- 24. Umezawa H. (1982) Low molecular weight enzyme inhibitors of microbial origin. Annu. Rev. Microbiol. 36, 75–99 [DOI] [PubMed] [Google Scholar]
- 25. Turk V., Turk B., Turk D. (2001) Lysosomal cysteine proteases: facts and opportunities. EMBO J. 20, 4629–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roth W., Deussing J., Botchkarev V. A., Pauly-Evers M., Saftig P., Hafner A., Schmidt P., Schmahl W., Scherer J., Anton-Lamprecht I., Von Figura K., Paus R., Peters C. (2000) Cathepsin L deficiency as molecular defect of furless. Hyperproliferation of keratinocytes and pertubation of hair follicle cycling. FASEB J. 14, 2075–2086 [DOI] [PubMed] [Google Scholar]
- 27. Benavides F., Starost M. F., Flores M., Gimenez-Conti I. B., Guénet J. L., Conti C. J. (2002) Impaired hair follicle morphogenesis and cycling with abnormal epidermal differentiation in nackt mice, a cathepsin L-deficient mutation. Am. J. Pathol. 161, 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nishimura F., Naruishi H., Naruishi K., Yamada T., Sasaki J., Peters C., Uchiyama Y., Murayama Y. (2002) Cathepsin-L, a key molecule in the pathogenesis of drug-induced and I-cell disease-mediated gingival overgrowth. A study with cathepsin-L-deficient mice. Am. J. Pathol. 161, 2047–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kisselev A. F., Goldberg A. L. (2001) Proteasome inhibitors. From research tools to drug candidates. Chem. Biol. 8, 739–758 [DOI] [PubMed] [Google Scholar]
- 30. Cuervo A. M., Stefanis L., Fredenburg R., Lansbury P. T., Sulzer D. (2004) Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy. Science 305, 1292–1295 [DOI] [PubMed] [Google Scholar]
- 31. Haacke A., Hartl F. U., Breuer P. (2007) Calpain inhibition is sufficient to suppress aggregation of polyglutamine-expanded ataxin-3. J. Biol. Chem. 282, 18851–18856 [DOI] [PubMed] [Google Scholar]
- 32. Gafni J., Hermel E., Young J. E., Wellington C. L., Hayden M. R., Ellerby L. M. (2004) Inhibition of calpain cleavage of huntingtin reduces toxicity. Accumulation of calpain/caspase fragments in the nucleus. J. Biol. Chem. 279, 20211–20220 [DOI] [PubMed] [Google Scholar]
- 33. Amaravadi R. K., Yu D., Lum J. J., Bui T., Christophorou M. A., Evan G. I., Thomas-Tikhonenko A., Thompson C. B. (2007) Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J. Clin. Invest. 117, 326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ratovitski T., Chighladze E., Waldron E., Hirschhorn R. R., Ross C. A. (2011) Cysteine proteases bleomycin hydrolase and cathepsin Z mediate N-terminal proteolysis and toxicity of mutant huntingtin. J. Biol. Chem. 286, 12578–12589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edinger A. L., Linardic C. M., Chiang G. G., Thompson C. B., Abraham R. T. (2003) Differential effects of rapamycin on mammalian target of rapamycin signaling functions in mammalian cells. Cancer Res. 63, 8451–8460 [PubMed] [Google Scholar]
- 36. Webb J. L., Ravikumar B., Rubinsztein D. C. (2004) Microtubule disruption inhibits autophagosome-lysosome fusion. Implications for studying the roles of aggresomes in polyglutamine diseases. Int. J. Biochem. Cell Biol. 36, 2541–2550 [DOI] [PubMed] [Google Scholar]
- 37. Johnston J. A., Ward C. L., Kopito R. R. (1998) Aggresomes. A cellular response to misfolded proteins. J. Cell Biol. 143, 1883–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taylor J. P., Tanaka F., Robitschek J., Sandoval C. M., Taye A., Markovic-Plese S., Fischbeck K H. (2003) Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum. Mol. Genet. 12, 749–757 [DOI] [PubMed] [Google Scholar]
- 39. García-Mata R., Bebök Z., Sorscher E. J., Sztul E. S. (1999) Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 146, 1239–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ravikumar B., Acevedo-Arozena A., Imarisio S., Berger Z., Vacher C., O'Kane C. J., Brown S. D., Rubinsztein D. C. (2005) Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat. Genet. 37, 771–776 [DOI] [PubMed] [Google Scholar]
- 41. Saric T., Graef C. I., Goldberg A. L. (2004) Pathway for degradation of peptides generated by proteasomes. A key role for thimet oligopeptidase and other metallopeptidases. J. Biol. Chem. 279, 46723–46732 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.