Background: mTORC1 is dysregulated in human disease, and there is an interest in the development of mTORC1 inhibitors. Niclosamide inhibits mTORC1 signaling, but its mode of action remains unclear.
Results: Niclosamide extrudes protons from lysosomes, thus lowering cytoplasmic pH and inhibiting mTORC1 signaling.
Conclusion: Cytoplasmic acidification inhibits mTORC1 signaling.
Significance: Our findings may aid the design of niclosamide-based anticancer therapeutic agents.
Keywords: Acidosis, Lysosomes, mTOR Complex (mTORC), Organellar pH Homeostasis, S6 Kinase, Cytoplasmic pH Homeostasis, Niclosamide
Abstract
Mammalian target of rapamycin complex 1 (mTORC1) signaling is frequently dysregulated in cancer. Inhibition of mTORC1 is thus regarded as a promising strategy in the treatment of tumors with elevated mTORC1 activity. We have recently identified niclosamide (a Food and Drug Administration-approved antihelminthic drug) as an inhibitor of mTORC1 signaling. In the present study, we explored possible mechanisms by which niclosamide may inhibit mTORC1 signaling. We tested whether niclosamide interferes with signaling cascades upstream of mTORC1, the catalytic activity of mTOR, or mTORC1 assembly. We found that niclosamide does not impair PI3K/Akt signaling, nor does it inhibit mTORC1 kinase activity. We also found that niclosamide does not interfere with mTORC1 assembly. Previous studies in helminths suggest that niclosamide disrupts pH homeostasis of the parasite. This prompted us to investigate whether niclosamide affects the pH balance of cancer cells. Experiments in both breast cancer cells and cell-free systems demonstrated that niclosamide possesses protonophoric activity in cells and in vitro. In cells, niclosamide dissipated protons (down their concentration gradient) from lysosomes to the cytosol, effectively lowering cytoplasmic pH. Notably, analysis of five niclosamide analogs revealed that the structural features of niclosamide required for protonophoric activity are also essential for mTORC1 inhibition. Furthermore, lowering cytoplasmic pH by means other than niclosamide treatment (e.g. incubation with propionic acid or bicarbonate withdrawal) recapitulated the inhibitory effects of niclosamide on mTORC1 signaling, lending support to a possible role for cytoplasmic pH in the control of mTORC1. Our data illustrate a potential mechanism for chemical inhibition of mTORC1 signaling involving modulation of cytoplasmic pH.
Introduction
Mammalian target of rapamycin (mTOR)5 is a Ser/Thr protein kinase found in two protein complexes known as mTORC1 and mTORC2, each controlling distinct cellular functions. mTORC2 phosphorylates PKB/Akt, serum- and glucocorticoid-regulated kinase 1 (SGK1), and PKCα to regulate cell survival and cytoskeletal organization (1–7), whereas mTORC1 phosphorylates S6K1 and 4E-BP1 to control cell growth and proliferation (8, 9). S6K1 and 4E-BP1 are the best studied mTORC1 targets, but emerging evidence indicates that mTORC1 phosphorylates numerous other proteins (10–12).
mTORC1 is regulated by different signaling cascades, many of which comprise tumor suppressors and proto-oncogenes mutated in human cancer. Tumor suppressors known to function upstream of mTORC1 include PTEN, TSC1/TSC2, LKB1/STK11, NF1, and NF2 (13–19). Mouse embryo fibroblasts lacking TSC1, TSC2, PTEN, LKB1, or NF1 display elevated mTORC1 signaling (14, 16, 19, 20). mTORC1 is also positively regulated by a number of proto-oncogenes, e.g. PI3K, PKB/Akt, and EGF receptor (21, 22). mTOR itself is mutated in a variety of human cancers as reported in the COSMIC (Catalogue Of Somatic Mutations in Cancer) human cancer database (23). Notably, some of these mutations result in mTORC1 hyperactivation and accelerated cell cycling (24). mTORC1 is therefore regarded as an attractive target for cancer therapy, and there is currently a high level of interest in the development of pharmacological inhibitors of mTORC1.
mTORC1 is inhibited allosterically by the natural product rapamycin (sirolimus), which interacts with the FKBP12-rapamycin-binding domain in mTOR in complex with FKBP12 (25, 26). Rapamycin has been shown previously to abolish the binding of raptor to mTOR in phosphate-containing buffers (27, 28). Raptor is thought to function as a scaffolding protein that brings substrates in close proximity to mTOR for efficient phosphorylation (29–31). The ability of rapamycin to dissociate mTOR from raptor may explain the inhibitory effect of rapamycin on S6K1 and 4E-BP1 phosphorylation, but the exact mechanism by which rapamycin perturbs mTORC1 function has not been fully elucidated.
Rapamycin has unfavorable pharmacokinetic properties (32). This realization prompted the development of alternatives to rapamycin with improved pharmacological properties for use in cancer treatment. Such efforts culminated in the development of rapamycin analogs temsirolimus (cell cycle inhibitor-779 (CCI-779)), everolimus (RAD-001), and ridaforolimus (AP23573; also known as MK-8669) with slightly improved bioavailability (33). The first two are approved for human use in the treatment of advanced renal cell carcinoma and are currently in clinical trials for various solid tumors and hematopoietic malignancies (34, 35). AP23573 is currently in clinical trials for advanced soft tissue and bone sarcomas, endometrial cancer, and hematopoietic malignancies (36).
Rapamycin inhibits many, but not all, signaling events mediated by mTORC1. For instance, acute treatment of various cell types with rapamycin readily abrogates the phosphorylation of S6K1 at Thr389 and 4E-BP1 at Ser65 but has little or no effect on the phosphorylation of Thr37/46 in 4E-BP1 (37), whereas mTORC2 activity is insensitive to acute rapamycin treatment. However, mTORC2 is sensitive to chronic rapamycin treatment in certain cell types (38). Recent years have witnessed a heightened interest in the pursuit of alternative mTOR inhibitors that directly target the catalytic activity of mTOR and thus are capable of blocking both rapamycin-sensitive and -insensitive functions of mTOR (39, 40). Five independent studies (41–45) have reported the development of distinct ATP-competitive inhibitors (Torin1, Ku-0063794, WAY-600, WYE-687, WYE-354, AZD8055, and PP242) that potently inhibit all mTOR functions. Other groups (46, 47) have developed ATP-competitive inhibitors (PI-103 and NVP-BEZ235) that target the catalytic activity of both mTOR and PI3K. All the aforementioned compounds differ from rapamycin and rapamycin analogs in that they directly target the catalytic activity of mTOR and thus inhibit both mTORC1 and mTORC2.
We recently identified niclosamide as a structurally distinct mTORC1 signaling inhibitor (48) in the context of a screen for modulators of autophagy, a catabolic cellular process that is inhibited by mTORC1 (49). Niclosamide rapidly and potently induced accumulation of the autophagy marker EGFP-LC3 while simultaneously inhibiting phosphorylation of both 4E-BP1 and S6K1 (48), the most widely used readouts for mTORC1 activation. Niclosamide, however, did not inhibit mTORC2 (48), suggesting that it does not directly inhibit mTOR kinase activity but rather impairs signaling to mTORC1.
Niclosamide is an oral salicylanilide derivative approved since 1960 for human use in the treatment of various tapeworm infections (50). Niclosamide was developed on the basis of activity in animal models of parasitic worm infection without knowledge of its target(s), and its mechanism of action remains poorly defined (51). Niclosamide is believed to owe its antiparasitic activity to its ability to disrupt the pH homeostasis of the parasite (52). In the present study, we examined the mechanism by which niclosamide may inhibit mTORC1 signaling.
Our data show that niclosamide does not directly inhibit the catalytic activity of mTORC1 or other kinases in vitro. Rather, niclosamide induces cytoplasmic acidification by causing an inward flux of protons through the plasma membrane and dissipating proton gradients across the membrane of intracellular organelles, such as lysosomes. Notably, we observed that the chemical features of niclosamide essential for cytoplasmic acidification are also required for inhibition of mTORC1 signaling, suggesting a role for cytoplasmic pH in the control of mTORC1 signaling. Consistent with this idea, modulation of cytoplasmic acidification by means other than niclosamide treatment elicited similar inhibitory effects on mTORC1 signaling.
How does cytoplasmic pH modulate mTORC1 activity? Further experiments will be required to fully characterize the exact signaling events underpinning acidosis-mediated mTORC1 inhibition. Interestingly, however, our data demonstrate that the PI3K/Akt/TSC2/Rheb signaling cascade (a major regulatory axis of mTORC1) is entirely dispensable in this context.
EXPERIMENTAL PROCEDURES
Chemicals
dl-Dithiothreitol (catalog number DTT001.5) was purchased from BioShop Canada, Inc. 5-(N-Ethyl-N-isopropyl amiloride) (EIPA) (catalog number A3085) and 1,1-dimethylbiguanide hydrochloride (metformin) (catalog number D5035) were purchased from Sigma. PD184352 (catalog number P-8499) was bought from LC Laboratories. CGP57380 (catalog number CA60801-314) was purchased from VWR. Torin1 (1-[4-[4-(1-oxopropyl)-1-piperazinyl]-3-(trifluoromethyl)phenyl]-9-(3-quinolinyl)-benzo[h]-1,6-naphthyridin-2(1H)-one) (catalog number 4247) and PP242 (2-[4-amino-1-(1-methylethyl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl]-1H-indol-5-ol) (catalog number 4257) were bought from Tocris Bioscience. Rapamycin (catalog number 553211) was obtained from Calbiochem and niclosamide (catalog number N3510) from Sigma-Aldrich. Niclosamide analog 1 was prepared by carbodiimide-mediated amide coupling between 2-chloro-4-nitroaniline and 3-chlorobenzoic acid. Niclosamide analog 2 was prepared by carbodiimide-mediated amide coupling between 2-chloro-4-nitroaniline and 3-chloro-5-methoxybenzoic acid followed by cleavage of the methyl ether by BBr3. Niclosamide analog 3 was prepared by methylation of niclosamide using the methylating agent dimethyl sulfate and K2CO3. Niclosamide analog 4 was prepared by coupling between 2-acetoxy-5-chlorobenzoic acyl chloride and 2-chloroaniline followed by hydrolysis of the O-acetyl by K2CO3. Niclosamide analog 5 was prepared by methylation of O-acetylniclosamide, which was previously synthesized by reaction of niclosamide and acetic anhydride, using the methylating agent dimethyl sulfate and K2CO3 followed by hydrolysis of the O-acetyl by K2CO3. All analogs were purified by column chromatography using Silicycle Ultra Pure silica gel (230–400 mesh) and characterized by 1H and 13C NMR spectroscopy and high resolution electrospray mass spectrometry.
Plasmids
The pRK7 empty and pRK7-FLAG-Rheb Q64L vectors used in this study have been described previously by Dunlop et al. (53).
Mammalian Cell Culture
BT-4T4, Hs578T, and TSC2+/+/p53−/− and TSC2−/−/p53−/− mouse embryo fibroblasts (MEFs) were propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS), 100 units/ml penicillin G, and 100 μg/ml streptomycin sulfate at 37 °C in a 5% (v/v) CO2 humidified incubator. TSC2+/+/p53−/− and TSC2−/−/p53−/− MEFs were a kind gift from Dr. David Kwiatkowski (Boston, MA). MDA-MB-468 cells were maintained in Leibovitz's L-15 medium supplemented with 10% (v/v) FBS, 100 units/ml penicillin G, and 100 μg/ml streptomycin sulfate at 37 °C in a CO2-free incubator. MCF-7 cells were maintained in complete Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% (v/v) FBS, 100 units/ml penicillin G, and 100 μg/ml streptomycin sulfate at 37 °C in a 5% (v/v) CO2 humidified incubator. Serum deprivation was carried out in the same medium lacking serum. Bicarbonate/CO2 withdrawal experiments were carried out by incubating MCF-7 cells in bicarbonate-free RPMI 1640 medium (Wisent Inc., catalog number 350-010-CL) supplemented with 10% (v/v) FBS, 100 units/ml penicillin G, and 100 μg/ml streptomycin sulfate and incubating MEFs in bicarbonate-free DMEM (Wisent Inc., catalog number 219−010-XK) supplemented with 10% (v/v) FBS, 100 units/ml penicillin G, and 100 μg/ml streptomycin sulfate at 37 °C in a CO2-free incubator for the periods of time indicated in the figure legends. Experiments involving modulation of the extracellular pH were carried out by incubating MCF-7 cells in bicarbonate-free RPMI 1640 complete medium (described above) adjusted with HCl to the desired pH value at 37 °C in a CO2-free incubator. MEFs were incubated in bicarbonate-free complete DMEM (described above) under the same temperature and CO2-free conditions. In experiments using 50 mm propionic acid, the NaCl concentration of the medium was reduced to 60 mm to maintain isotonicity.
Cell Lysis and Immunoblotting
Cells were harvested by scraping in the following extraction buffer: 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% (v/v) Triton X-100, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate supplemented with fresh 1 mm Na3VO4, 1 mm dithiothreitol, and 1× Complete protease inhibitor mixture (Roche Applied Science, catalog number 11697498001). Lysates were precleared by centrifugation at 21,000 × g for 10 min at 4 °C. A distinct lysis buffer was used for immunopurification of intact mTORC1 complexes (see “In Vitro mTORC1 Kinase Assay” below for details). Phosphorylation and total protein levels were analyzed by resolving samples of lysates on a 10% (w/v) acrylamide gel containing 0.1% methylene bisacrylamide (w/v) followed by Western blotting with the indicated antisera. Anti-phospho-Thr37/46 4E-BP1 (catalog number 2855), anti-phospho-Thr389 p70S6K1 (catalog number 9234), anti-phospho-Ser371 p70S6K1 (catalog number 9208), anti-phospho-Thr421/Ser424 p70S6K1 (catalog number 9204), anti-phospho-Thr172 AMPK (catalog number 2531), anti-phospho-Thr308 PKB/Akt (catalog number 2965), anti-phospho-Ser473 PKB/Akt (catalog number 9721), anti-phospho-Thr202/Tyr204 p44/42 ERK1/ERK2 (catalog number 9106), anti-4E-BP1 (catalog number 9644), anti-AMPKα (catalog number 2532), anti-p44/42 ERK1/ERK2 (catalog number 9107), anti-PKB/Akt (catalog number 4691), and anti-mTOR (catalog number 2983) antibodies were bought from Cell Signaling Technology. Anti-S6K C-18 (catalog number SC-230) was purchased from Santa Cruz Biotechnology, Inc. Anti-raptor (catalog number 09−217) antibodies were bought from Millipore. Anti-phospho-Ser209 eIF4E (catalog number NB100-79938) and anti-eIF4E (catalog number 610270) were obtained from Novus Biologicals and BD Transduction Laboratories, respectively. Phosphorylation and total protein levels were monitored by enhanced chemiluminescence.
Viability/Proliferation 3-(4,5-Dimethylthiazol-2-yl)-2,5-tetrazoliumbromide Assay
MCF-7 cells were seeded in 96-well microtiter plates at 8,000 cells per well and propagated for 18 h. Cells were then treated with rapamycin, niclosamide or analogs. Cell viability/proliferation was measured 48 h later using the 3-(4,5-dimethylthiazol-2-yl)-2,5-tetrazoliumbromide assay (Sigma, catalog number M2128) as described (54).
Escherichia coli Inner Membrane Vesicle pH Measurement
Inverted inner membrane vesicles (IMVs) were prepared from E. coli strain KM9 as described (55). Fluorescence measurements were performed at 25 °C on a Varian Cary Eclipse spectrofluorometer. IMVs (40 μg) were diluted to 150 μl in 50 mm Tris-SO4, pH 7.9, 25 mm K2SO4, 10 mm MgSO4, 0.2 mg/ml BSA in a quartz cuvette. NADH (1 mm) was added to generate an acidic pH inside the IMVs. IMV lumen acidification was monitored using the pH-sensitive dye 9-amino-6-chloro-2-methoxyacridine (4 μm) at excitation and emission wavelengths of 409 (20-nm slit width) and 474 nm (10-nm slit width), respectively. The stability of the pH gradient was challenged with niclosamide, niclosamide analogs, or carbonyl cyanide p-chlorophenylhydrazone at 1 μm.
EGFP-LC3 Autophagy Assay
MCF-7 cells stably expressing EGFP-LC3 were seeded in PerkinElmer Life Sciences View 96-well microtiter plates at 20,000 cells/well. Eighteen hours after seeding, rapamycin, niclosamide, and analogs were added to each well to the final concentrations indicated in the figure legends. Plates were incubated for 4 h at 37 °C. The medium was then removed, and the cells were fixed with 3% (v/v) paraformaldehyde containing 500 ng/ml Hoechst 33342 for 15 min at room temperature. Fixed cells were washed once with PBS containing 1 mm MgCl2 and 0.1 mm CaCl2 and then stored in the same medium at 4 °C until plates were ready for analysis. Plates were read in a CellomicsTM Arrayscan VTI automated fluorescence imager. Cells were photographed using a 20× objective in the Hoechst and GFP (XF-100 filter) channels. The compartment analysis algorithm was used to identify the nuclei, apply a cytoplasmic mask, and quantitate GFP spots in the GFP channel fixed at 250 pixel intensity units. Fluorescence intensity in the GFP channel was gated at 10 average pixel intensity units inside the cytoplasmic mask to select against cells expressing very low levels of EGFP-LC3. The total pixel intensity for punctate EGFP-LC3 was acquired as “circular spot total intensity channel.”
Measurement of Cytoplasmic pH
MCF-7 cells were propagated on glass coverslips coated with 0.01% (v/v) poly-l-lysine in complete medium for 48 h prior to measurement of intracellular pH. Cells were then loaded with the pH-sensitive dye 2′,7′-bis(2-carboxyethyl)-5(6)-carboxyfluorescein (BCECF) acetomethyl ester by incubating the cells for 10 min at room temperature in buffered saline (120 mm NaCl, 5 mm KCl, 1 mm MgCl2, 2 mm CaCl2, 5 mm glucose, 20 mm HEPES, pH 7.4) containing 2 μm BCECF acetomethyl ester. After rinsing with saline, coverslips with cells attached were mounted on a temperature-controlled recording chamber filled with saline, placed on the microscope, and then superfused at 2 ml/min with saline at 34 °C for the remainder of the experiment. At the times indicated, 10 μm niclosamide or identical concentrations of analog 1 or 2 were added to the perfusate, and changes in cytoplasmic pH were monitored over time. In some experiments, cells were treated with high [K+] medium (50 mm KCl, 75 mm NaCl, 1 mm MgCl2, 2 mm CaCl2, 5 mm glucose, 20 mm HEPES, pH 7.4) either prior to or following addition of 10 μm niclosamide. In another experiment, cells were treated with 100 nm rapamycin for 10 min followed by a brief treatment with isotonic NH4Cl-saline (50 mm NH4Cl, 70 mm choline chloride, 5 mm KCl, 1 mm MgCl2, 2 mm CaCl2, 5 mm glucose, 20 mm HEPES, pH 7.4). To measure pHi, BCECF dye was excited alternately at 452 and 488 nm, and the emission intensity at 515 nm was measured using a fluorescence ratio imaging system (Atto Bioscience, Rockville, MD). Background-corrected BCECF emission intensity ratios were converted into pHi values using the high K+/nigericin technique (56, 57).
Lysosomal Visualization by Wide Field Fluorescence Microscopy
MCF-7 cells were seeded at 5 × 105 cells in 6-well plates in 2 ml of complete medium (RPMI 1640 medium supplemented with 10% (v/v) FBS, 100 units/ml penicillin G, and 100 μg/ml streptomycin sulfate). Cells were propagated at 37 °C and 5% (v/v) CO2 for 48 h until they reached ∼70–80% confluence at which point LysoTracker Red (DND-99) (Molecular Probes, Invitrogen, catalog number L7528) was added directly to the medium at a final concentration of 100 nm. Cells were incubated with the dye for 1 h, and the medium was then replaced with fresh complete medium containing either 0.1% (v/v) DMSO, 10 μm niclosamide, 10 μm analogs, or 100 nm rapamycin. Cells were incubated with the drugs for 2 h at 37 °C and 5% (v/v) CO2, and LysoTracker staining was visualized in a Zeiss microscope. Images were acquired using an AxioCam HR camera and analyzed with Axiovert release 4.8 software.
In Vitro mTORC1 Kinase Assay
mTORC1 kinase assays were performed as described previously (58). Briefly, MCF-7 cells were lysed for 30 min on ice in lysis buffer containing 40 mm HEPES, pH 7.5, 0.3% (w/v) CHAPS, 120 mm NaCl, 1 mm EDTA, 10 mm sodium pyrophosphate, and 10 mm β-glycerophosphate. Lysates were precleared by centrifugation at 10,000 × g for 10 min. mTORC1 complexes were immunoprecipitated from MCF-7 cell lysates using an antibody against raptor (Millipore, catalog number 09-217). Immune complexes were washed twice with lysis buffer (described above) followed by one wash with lysis buffer containing 0.4 m NaCl and two additional washes with kinase buffer (25 mm HEPES, pH 7.4, 50 mm KCl, 10 mm MgCl2). Recombinant 4E-BP1, which was previously cleaved from a fusion with GST, was used as a substrate for mTORC1 at a final concentration of 7.5 ng/μl. Kinase reactions on the immobilized immune complexes were carried out in 20-μl reaction volumes in kinase buffer containing 250 μm ATP. Where indicated, immune complexes were incubated with chemicals in kinase buffer for 10 min prior to commencement of the kinase assay at the indicated concentrations. Inhibitors were included at the same concentrations in the kinase reactions. Reactions were carried out at 30 °C for 20 min and stopped by the addition of 5× SDS-PAGE sample buffer.
Specificity Kinase Panel
Niclosamide was tested against a panel of 95 protein kinases by the National Centre for Protein Kinase Profiling (University of Dundee) using a [33P]ATP filter binding assay as described (59).
RESULTS
Niclosamide Inhibits mTORC1 Signaling Independently of Akt, ERK, and AMPK
We have shown previously that niclosamide (Fig. 1A) inhibits mTORC1 signaling by an unknown mechanism (48). Niclosamide reduces the phosphorylation of 4E-BP1 at Thr37/46, Ser65, and Thr70, and it impairs the assembly of the eIF4F complex and abolishes the phosphorylation of p70S6K1 at Thr389 (48). In the present study, we systematically evaluated several potential mechanisms by which niclosamide may inhibit mTORC1 signaling.
FIGURE 1.
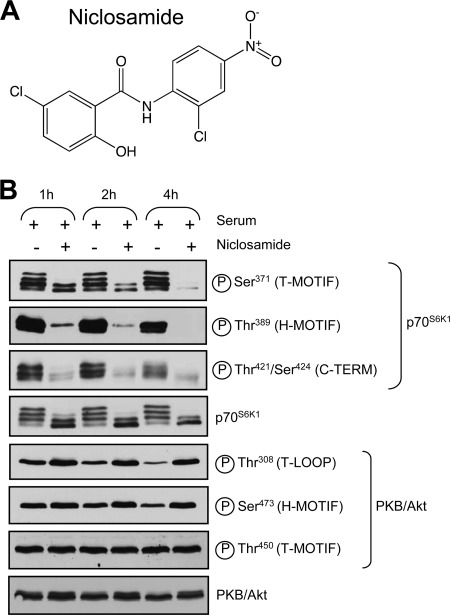
Niclosamide inhibits mTORC1 signaling. A, structure of niclosamide. B, MCF-7 cells were propagated in complete medium for 24 h at which point cells were treated with 10 μm niclosamide for 1, 2, or 4 h in complete medium. Cells lysates were analyzed by immunoblotting using the indicated antibodies. T-MOTIF, turn motif; T-LOOP, theonine loop; H, hydrophobic; TERM, terminus.
First, we examined whether niclosamide interferes with the activation of upstream signaling cascades that are normally involved in mTORC1 activation in response to serum. We began by testing the ability of niclosamide to block PKB/Akt signaling in MCF-7 cells, which were selected for their elevated S6K1 protein levels (supplemental Fig. 1A) (60–62). Exposure of MCF-7 cells to 10 μm niclosamide reduced serum-stimulated phosphorylation of p70S6K1 at several sites, including Thr389 (within the hydrophobic motif), Ser371 (within the turn motif), and Thr421/Ser424 (near the C terminus) (Fig. 1B). Inhibition was profound after 1 h and essentially complete after 4 h of incubation with niclosamide.
By contrast, exposure to niclosamide augmented serum-induced phosphorylation of PKB/Akt at Thr308 (catalyzed by 3-phosphoinositide-dependent protein kinase 1 (PDK1)) and Ser473 (catalyzed by mTORC2) (Fig. 1B) likely due to inactivation of the negative feedback loop linking S6K1 to IRS-1 (63–65). Phosphorylation of Thr450 (dependent on mTORC2) did not change upon serum stimulation or niclosamide treatment (Fig. 1B). The observation that niclosamide inhibited mTORC1 signaling while simultaneously increasing mTORC2 signaling rules out the possibility that the inhibitory effect of niclosamide on mTORC1 signaling resulted from a blockade of the PKB/Akt signaling pathway.
The MEK/ERK signaling pathway has also been previously linked to the activation of mTORC1 in certain cell types (66, 67). To determine whether niclosamide impaired mTORC1 activation through inhibition of MEK/ERK, we monitored MEK activity in niclosamide-treated MCF-7 cells using a phosphospecific antibody against Thr202/Tyr204 on ERK1/ERK2. PD184352 potently blocked ERK phosphorylation, whereas niclosamide did not (supplemental Fig. 1B). The MAPK-interacting kinases Mnk1 and Mnk2 are each activated by both ERK and p38 mitogen-activated protein kinases α and β (68, 69) and phosphorylate Ser209 on eIF4E (70–72), a central component of the translation initiation machinery (73). Inhibition of Mnk1 and Mnk2 activity with the pharmacological agent CGP57380 markedly reduced phosphorylation of eIF4E at Ser209 in MCF-7 cells (supplemental Fig. 1C). By contrast, niclosamide did not affect Mnk catalytic activity when used at concentrations that potently inhibit mTORC1 signaling (supplemental Fig. 1C). These data indicate that niclosamide does not inhibit mTORC1 by interfering with MEK/ERK/Mnk signaling.
AMPK functions as a sensor for energy availability in the cell. Under conditions of energy insufficiency caused by either decreased ATP production or heightened expenditure, AMPK is activated and inhibits mTORC1 to restrict cellular processes that consume high levels of ATP, such as protein synthesis (74). Phosphorylation of AMPK on Thr172 by LKB1 markedly increases the catalytic activity of AMPK (75) and is widely used as a readout of AMPK enzymatic activity. To test whether niclosamide inhibited mTORC1 signaling by mimicking energy stress, MCF-7 cells were incubated with 10 μm niclosamide, 100 nm rapamycin, or 20 mm metformin; the latter is known to increase cytosolic levels of AMP and AMPK activity (76). Metformin increased Thr172 phosphorylation, but rapamycin and niclosamide did not (supplemental Fig. 1D). Therefore, niclosamide does not inhibit mTORC1 signaling by activating AMPK.
Niclosamide Does Not Affect mTOR Binding to Raptor or Catalytic Activity of mTORC1 in Vitro
Because niclosamide does not appear to inhibit major control pathways upstream of mTORC1, we next examined whether it directly interfered with mTORC1 assembly. Rapamycin has been shown previously to disrupt binding of mTOR to raptor in phosphate-containing buffers (27, 28) through a poorly characterized mechanism. To determine whether niclosamide similarly affects mTOR-raptor association, lysates obtained from cells treated with rapamycin or niclosamide were subjected to immunoprecipitation with raptor antibody, and immunoprecipitates were probed for raptor and mTOR by Western blot. Acute rapamycin treatment caused dissociation of mTOR from raptor (supplemental Fig. 2A, upper panel), consistent with earlier studies (27, 28). By contrast, niclosamide did not cause any measurable dissociation of mTOR from raptor (supplemental Fig. 2A, upper panel). Immunoblotting of lysates confirmed that mTORC1 signaling was inhibited in cells treated with either drug as judged by the lack of phosphorylation of S6K1 at Thr389/412 (supplemental Fig. 2A, lower panel). Therefore, niclosamide does not impair mTORC1 signaling by disrupting mTOR-raptor binding.
To determine whether niclosamide directly inhibits the catalytic activity of mTORC1, raptor immunoprecipitates were preincubated with different concentrations of niclosamide. Bacterially expressed recombinant 4E-BP1 was then added to the immunoprecipitates in the presence of ATP. Phosphorylation of 4E-BP1 was monitored by Western blot using a phosphospecific antibody against Thr37/46. Niclosamide had no detectable effect on mTORC1 activity in vitro at any of the concentrations tested, whereas Torin1, PP242, and the rapamycin-FKBP12 complex all efficiently inhibited mTORC1 kinase activity (supplemental Fig. 2B). Immunoblotting of raptor immunoprecipitates confirmed the presence of equal amounts of raptor and mTOR in all reactions (supplemental Fig. 2B). Taken together, these data demonstrate that niclosamide inhibits mTORC1 signaling but does not directly block mTORC1 kinase activity, nor does it disrupt the mTOR-raptor association.
Niclosamide was additionally tested in vitro against a panel of 95 protein kinases, and it did not significantly inhibit any of these kinases at 1 or 10 μm (supplemental Table 1). These concentrations efficiently inhibit mTORC1 signaling in cells, indicating that the mechanism of action of niclosamide is probably not direct kinase inhibition.
Niclosamide Has Protonophoric Activity
Niclosamide is a salicylanilide derivative used in the treatment of parasitic infections. The mechanism of action of niclosamide is not well understood, but its antiparasitic properties are thought to be due to the ability of this drug to transport protons across biological membranes (herein referred to as protonophoric activity), thus disrupting the pH homeostasis of the parasite (52). Next, we examined whether niclosamide shows protonophoric activity using a cell-free system. IMVs prepared from an E. coli strain lacking a functional F0/F1 ATPase can establish a proton gradient via their electron transport chain (55). Indeed, addition of NADH to IMVs caused rapid acidification of the IMV lumen as measured by decreased fluorescence of the pH-sensitive dye 9-amino-6-chloro-2-methoxyacridine (Fig. 2C). The pH gradient was subsequently challenged with niclosamide or with the well characterized protonophore carbonyl cyanide p-chlorophenylhydrazone. At 1 μm, both carbonyl cyanide p-chlorophenylhydrazone and niclosamide rapidly dissipated the pH gradient, demonstrating that niclosamide is an effective protonophore.
FIGURE 2.
Protonophoric activity of niclosamide, analogs and their effects on mTORC1, signaling. A, model showing the neutral and anionic forms of niclosamide and the structural features that enable delocalization of the negative charge of the anionic form to maintain its hydrophobicity. B, structures of niclosamide analogs 1–5. C and D, protonophoric activity assay. NADH (1 mm) was added to IMVs at the time indicated by the arrow to generate a pH gradient. One minute later, test chemicals were added at 1 μm. pH changes were monitored using 9-amino-6-chloro-2-methoxyacridine (ACMA) fluorescence. E and F, MCF-7 cells were treated with the indicated concentrations of niclosamide or with 30 nm rapamycin (Rapa) for 4 h in complete medium containing 10% (v/v) FBS. Cell lysates were probed with the indicated antibodies. A.U., absorbance units; CCCP, carbonyl cyanide p-chlorophenylhydrazone.
Niclosamide resembles the well characterized protonophore S13 (77–79). Based on a model for S13 (80), the structural groups that confer protonophoric activity to niclosamide are the weakly acidic OH group with a pKa in the physiological pH range and a number of chemical features that help delocalize the negative charge of the anionic form of the molecule to maintain its hydrophobicity and membrane association, such as the electron-withdrawing NO2 moiety and the NH group that participates in an intramolecular H-bond (Fig. 2A). To test these predictions experimentally, we synthesized and tested five analogs (Fig. 2B). Analog 1 lacking the OH group and hence without a dissociable proton lacked protonophoric activity (Fig. 2C). Analog 2 with the OH group at the meta position to prevent hydrogen bonding to the NH group also lacked protonophoric activity (Fig. 2C). O-Me analog 3 lacking a dissociable proton was also completely inactive as was analog 4 lacking the strongly electron-withdrawing nitro group that makes the dissociable proton less acidic (Fig. 2D). Lastly, analog 5 with an N-methyl substituent incapable of forming the intramolecular H-bond required to maintain the hydrophobicity of the anionic form of niclosamide was also completely devoid of protonophoric activity in vitro (Fig. 2D). These results are in complete agreement with the Terada (78) model for protonophoric activity.
Structural Features of Niclosamide Required for Protonophoric Activity Are Also Needed for Inhibition of mTORC1 Signaling
Having identified the structural features required for protonophoric activity, we next assessed the ability of each analog to reduce mTORC1 signaling in MCF-7 cells (Fig. 2, E and F). None of the analogs showed mTORC1 inhibition at 1 μm at which concentration niclosamide potently inhibits mTORC1 signaling. Analogs 1, 3, and 5 were completely inactive at all concentrations tested. Analog 2 with the dissociable proton at a meta position and analog 4 lacking the nitro group only showed activity at high concentrations. Therefore, the structural requirements for protonophoric activity by niclosamide closely parallel those required for mTORC1 inhibition. Each niclosamide analog was also tested for its ability to modulate autophagy and inhibit cell proliferation. Remarkably, the chemical features of niclosamide required for inhibition of mTORC1 signaling were also essential for modulation of autophagy and inhibition of proliferation (supplemental Fig. 3, A and B) in agreement with the well established roles of mTORC1 in autophagy and cell proliferation (8, 49).
Niclosamide Induces Cytoplasmic Acidification
Protonophores are known to induce acidification of the cytoplasm driven by the negative-inside plasma membrane potential, forcing the inward flux of protons even when cells are maintained in culture medium at physiological pH (81). We therefore examined whether niclosamide can induce cytoplasmic acidification using the pH-sensitive dye BCECF. Switching cells from bicarbonate-buffered growth medium to the HEPES-buffered solution used in the pH measurement protocol resulted in a slow drop in cytoplasmic pH from 7.4 ± 0.09 at the beginning of the experiment to a new resting pH of 7.04 ± 0.03 (Fig. 3A), consistent with the existence of a bicarbonate-dependent acid extrusion mechanism that contributes to maintaining steady state intracellular pH in MCF-7 cells. Addition of 10 μm niclosamide caused a very rapid acidification of the cytosol to a new steady state of 6.61 ± 0.05 (Fig. 3A). The acidification induced by niclosamide persisted for up to 2 h, the longest time recorded (data not shown). By contrast, exposure to 10 μm analog 1 or 2 had no significant effect on cytoplasmic pH (Fig. 3A).
FIGURE 3.
Niclosamide elicits rapid cytoplasmic acidification. MCF-7 cells loaded with BCECF were superfused with HEPES-buffered saline, and cytoplasmic pH was measured over the course of the experiment. Each trace shows the mean intracellular pH values obtained from three independent experiments, each including at least 30 cells. A, MCF-7 cells were superfused with saline containing 0.2% DMSO (vehicle), 10 μm niclosamide, 10 μm analog 1, or 10 μm analog 2 at the time indicated by the arrow. B, MCF-7 cells were superfused with isotonic HEPES-buffered saline solution containing 50 mm KCl for the period of time indicated by the “50 mm K+” bar. Niclosamide (10 μm) was then added for the period of time indicated by the “Drug” bar. In the final 3 min, KCl was washed out with HEPES-buffered saline. C, MCF-7 cells were superfused with saline containing 10 μm niclosamide starting at the time indicated by the arrow and with 50 mm KCl solution for the time indicated by the 50 mm K+ bar followed by washout in HEPES-buffered saline solution. D, MCF-7 cells were superfused with saline solution containing 0.2% (v/v) DMSO, 1 μm FCCP, or 1 μm TFA9 at the time indicated by the arrow. E, MCF-7 cells were propagated in complete medium to near confluence at which point cells were switched to fresh complete medium in the presence of DMSO (vehicle), rapamycin (Rapa) (100 nm), or varying concentrations of TFA9 and FCCP for 2 h. Cells were then lysed, and lysates were analyzed for mTORC1 activation with the indicated antisera. F, MCF-7 cells were superfused with 100 nm rapamycin for the time indicated by the Drug bar. Following drug removal, cells were exposed to saline containing 50 mm NH4Cl for the time period indicated by the “NH4Cl” bar followed by washout in HEPES-buffered saline solution.
The membrane potential can be reduced by incubating cells in isotonic medium containing high [K+]. In the presence of high [K+] (50 mm), the ability of niclosamide to decrease cytoplasmic pH was markedly reduced (Fig. 3B), and it was rapidly reestablished when cells were transferred back to physiological saline conditions containing Na+ (Fig. 3B). Similarly, transfer to high [K+] after the addition of niclosamide partially reversed the cytoplasmic acidification (Fig. 3C), suggesting that sustained cytosolic acidification by niclosamide requires a continuous membrane potential. Depolarization, however, did not completely eliminate the ability of niclosamide to induce cytosolic acidification (Fig. 3C), indicating that release of protons from intracellular stores might also be a contributing factor for niclosamide-dependent cytoplasmic acidification. We explored the contribution of intracellular proton stores to cytosol acification as detailed below.
The observation that the structural features in niclosamide required for cytoplasmic acification (Fig. 3A) paralleled those required for inhibition of mTORC1 signaling (Fig. 2E) raised the possibility that niclosamide inhibited mTORC1 signaling by inducing cytosolic acidification. To test this possibility, we asked whether protonophores that are structurally distinct from niclosamide can also inhibit mTORC1. Incubation of cells with the well characterized protonophores carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) and tyrphostin A9 (TFA9) rapidly lowered the cytoplasmic pH of MCF-7 cells as expected (Fig. 3D). Interestingly, both FCCP and TFA9 efficiently reduced p70S6K1 phosphorylation at low micromolar concentrations (Fig. 3E). The observation that three structurally dissimilar chemicals caused intracellular acidification and inhibited mTORC1 strongly argues that protonophoric activity is the mechanism responsible for these effects rather than structure-specific effects on other unrelated targets.
The data obtained so far are consistent with cytoplasmic acidification by niclosamide inhibiting mTORC1 signaling but also with the possibility that cytoplasmic acidification is a consequence of mTORC1 inhibition. To distinguish between these possibilities, MCF-7 cells were incubated with rapamycin, and the cytoplasmic pH was tracked with BCECF. Rapamycin had no effect on cytoplasmic pH (Fig. 3F). Cytoplasmic pH was sensitive to the weak base ammonia (50 mm NH4Cl), indicating that the cells remained responsive to variations in the extracellular pH. Although ineffective at modulating cellular pH, rapamycin efficiently abolished mTORC1 signaling within 10 min (data not shown). These data indicate that the ability of niclosamide to lower cytoplasmic pH is not a consequence of mTORC1 inhibition. Rather, our data on FCCP and TFA9 indicate that the converse may actually be the case; i.e. protonophore-mediated cytoplasmic acidification leads to mTORC1 inhibition.
Niclosamide Dissipates Lysosomal-Cytoplasmic pH Gradient
Niclosamide and other protonophores are known to act not only at the plasma membrane but also on intracellular membranes to release protons from acidic organelles into the cytosol. In fact, certain protonophores are more efficient at dissipating proton gradients across endomembranes than across the plasma membrane; such is the case of carbonyl cyanide p-chlorophenylhydrazone in yeast (82). Next, we asked whether intracellular proton stores contributed to niclosamide-mediated cytoplasmic acidification.
Lysosomes, the most acidic organelles in the cell with pH values ranging from 4.5 to 4.7 (83), are a potential intracellular source of protons and, given that mTORC1 localizes to the lysosomal surface (84), an attractive one as such. To verify whether lysosomes contribute to niclosamide-dependent cytoplasmic acidification, lysosomal pH of MCF-7 cells was monitored with LysoTracker Red, an acidotropic fluorescent dye with high selectivity for lysosomes (85). Vehicle-treated cells displayed bright fluorescent punctate structures of variable size (Fig. 4A, DMSO panel). Exposure of MCF-7 cells to niclosamide considerably decreased punctate fluorescence, indicating that this drug dissipates proton gradients across lysosomal membranes (Fig. 4A, niclosamide panel). Interestingly, the decrease in punctate staining was accompanied by increased diffuse cytoplasmic fluorescence, consistent with our observation that niclosamide causes acidification of the cytoplasm. As expected, niclosamide analogs 1, 2, 3, and 5 had no discernible effect on punctate LysoTracker fluorescence in agreement with their lack of protonophoric activity (Fig. 4A). Niclosamide analog 4 caused a small but detectable increase in diffuse cytoplasmic LysoTracker fluorescence (Fig. 4A) and partial inhibition of mTORC1 signaling (Figs. 2F and 4B). At a higher concentration of 30 μm, analog 4 showed clearer diffuse cytoplasmic fluorescence and strong inhibition of mTORC1 (supplemental Fig. 4).
FIGURE 4.
Niclosamide dissipates protons from lysosomes to cytoplasm. MCF-7 cells were propagated in complete medium for 48 h. LysoTracker dye was then added to the cells at a final concentration of 100 nm. Cells were incubated with LysoTracker for 1 h, and the medium was then replaced with new complete medium containing either DMSO, 10 μm niclosamide, or equimolar amounts of each of the analogs for 2 h. LysoTracker staining was visualized by wide field fluorescence microscopy (A), and the cells were subsequently lysed and analyzed for p70S6K1 phosphorylation (B). Scale bar, 100 μm.
Lysosomal Alkalinization Does Not Affect mTORC1 Signaling
While finalizing this manuscript, an elegant study by Zoncu et al. (86) demonstrated that the vacuolar proton ATPase (v-ATPase), the proton pump responsible for keeping the lumen of lysosomes acidic, plays an important role in mTORC1 activation. Treatment of HEK293T cells with v-ATPase inhibitors, such as concanamycin A or salicylihalamide A, inhibited amino acid-dependent phosphorylation of S6K1 (86). The fact that v-ATPase plays key roles both in lysosomal acidification and in mTORC1 activation raised the intriguing possibility that the inhibitory effect of niclosamide on mTORC1 stemmed from the ability of this drug to raise lysosomal pH. To determine whether lysosomal alkalinization is sufficient to inhibit mTORC1, MCF-7 cells were treated with varying concentrations of bafilomycin A1, a potent v-ATPase inhibitor closely related to concanamycin A (87), and lysosomal pH was monitored with LysoTracker dye. Submicromolar concentrations (0.3 μm) of bafilomycin A1 completely abolished punctate LysoTracker staining (Fig. 5A). However, such concentrations of bafilomycin A1 had only a very mild effect on p70S6K1 phosphorylation (Fig. 5B). This result argues against a role for lysosomal pH in the control of mTORC1 signaling. Higher (10 μm) concentrations of bafilomycin A1 efficiently reduced mTORC1 signaling (Fig. 5B), consistent with the report that similar concentrations of concanamycin A are required to inhibit mTORC1 (86). Interestingly, we observed that cells exposed to high concentrations of bafilomycin A1 showed stronger diffuse cytoplasmic staining when compared with cells treated with submicromolar concentrations of bafilomycin A1 (Fig. 5A). Also, the degree of inhibition of mTORC1 by different concentrations of bafilomycin A1 correlates with cytoplasmic acidification (Fig. 5, cf. A and B).
FIGURE 5.
Lysosomal pH does not regulate mTORC1 signaling. MCF-7 cells were propagated in complete medium for 48 h. LysoTracker dye was then added to the cells at a final concentration of 100 nm. Cells were incubated with LysoTracker for 1 h, and the medium was then replaced with new complete medium containing either DMSO, 10 μm niclosamide, or the indicated concentrations of bafilomycin A1 for 2 h. LysoTracker staining was visualized by wide field fluorescence microscopy (A), and the cells were subsequently lysed and analyzed for p70S6K1 phosphorylation (B). Scale bar, 100 μm.
Cytoplasmic Acidification Causes Inhibition of mTORC1 Signaling
A previous study from our group showed that exposing cells to acidic external pH causes cytoplasmic acidification and inhibits mTORC1 signaling (Ref. 88; see also Fig. 6). However, the relative contributions of external and cytoplasmic acidic pH to mTORC1 inhibition were not investigated. The observation that niclosamide and other protonophores induce cytoplasmic acidification and inhibit mTORC1 signaling in cells maintained at physiological external pH argues that the latter might be sufficient. We next sought to induce cytoplasmic acidification without altering the extracellular pH using propionic acid, a short chain fatty acid that can diffuse through membranes in its neutral form and becomes trapped intracellularly upon dissociation of its proton, thus causing intracellular acidification (89). MCF-7 cells were exposed to cell culture medium containing propionate at pH 7.3 for 1 or 4 h, and phosphorylation of p70S6K1 at Thr389 was monitored. Inhibition of p70S6K1 phosphorylation was observed within 1 h and was sustained for at least 4 h (Fig. 7A), indicating that lowering the cytoplasmic pH is sufficient to impair mTORC1 signaling.
FIGURE 6.
Extracellular acidification inhibits mTORC1 signaling. MCF-7 cells were propagated in complete medium for 48 h at which point cells were transferred to complete medium lacking only sodium bicarbonate (adjusted to the pH values indicated) for 30 min at 37 °C in CO2-free conditions. Lysates were analyzed for mTORC1 activation using the indicated antisera.
FIGURE 7.
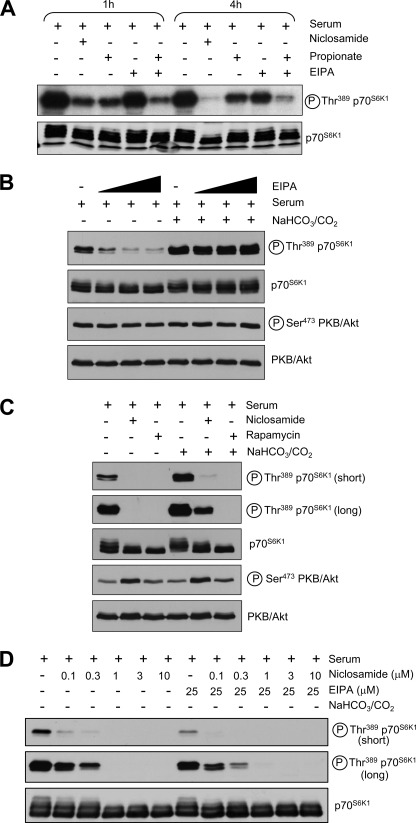
Cytoplasmic acidification inhibits mTORC1 signaling. A, MCF-7 cells were propagated to near confluence in complete medium and then treated with 10 μm niclosamide; 50 mm propionic acid, pH 7.3; and/or 25 μm EIPA for 1 or 4 h in complete medium containing 10% (v/v) FBS (see “Experimental Procedures” for details). Cell lysates were analyzed for mTORC1 activation by immunoblotting with antisera against phospho- and total p70S6K1. B, MCF-7 cells were propagated to near confluence in complete medium (containing sodium bicarbonate) and then transferred to complete medium either containing or lacking sodium bicarbonate in the presence or absence of increasing concentrations of EIPA (25, 50, and 100 μm). Cells were incubated with EIPA for 1 h prior to lysis. Lysates were analyzed for mTORC1 and mTORC2 activation with the indicated antisera. C, cells were incubated with 100 nm rapamycin or 10 μm niclosamide for 1 h in complete medium either containing or lacking sodium bicarbonate and analyzed for mTORC1 and mTORC2 activation. D, MCF-7 cells were cultured for 48 h in complete medium (containing sodium bicarbonate) to near confluence and then treated with varying concentrations of niclosamide in the presence or absence of 25 μm EIPA and/or sodium bicarbonate for 1 h. Lysates were analyzed for mTORC1 activation using the indicated antisera.
NHE1 is an integral plasma membrane protein that plays a major role in maintaining intracellular pH homeostasis by extruding protons that accumulate in the cytoplasm (90). Next, we asked whether inhibition of NHE1 similarly reduced mTORC1 signaling. On its own, the NHE1 inhibitor EIPA did not decrease p70S6K1 phosphorylation after a 1-h incubation. In contrast, EIPA exposure for longer periods of time (4 h) elicited a small inhibitory effect on p70S6K1 phosphorylation (Fig. 7A). The combination of propionate and EIPA potently reduced p70S6K1 phosphorylation at 4 h (Fig. 7A).
Similarly, cytoplasmic acidification induced by switching cells to a bicarbonate-free medium and atmospheric CO2 led to a decrease in p70S6K1 phosphorylation within 30 min (Fig. 7B). Adding EIPA further decreased p70S6K1 phosphorylation in the absence of bicarbonate/CO2 (Fig. 7B). Importantly, mTORC2 activation was not affected by either bicarbonate/CO2 or EIPA exposure, indicating that the inhibitory effects of pH modulation are restricted to mTORC1 signaling (Fig. 7B). Combining niclosamide with bicarbonate/CO2 withdrawal inhibited mTORC1 signaling more strongly than either treatment alone (Fig. 7C), whereas the triple combination of niclosamide, EIPA, and bicarbonate/CO2 withdrawal further inhibited mTORC1 signaling (Fig. 7D). Taken together, these data support a direct role for cytoplasmic pH in the control of mTORC1 signaling.
TSC2 and Rheb Are Not Involved in Regulation of mTORC1 Signaling by Niclosamide or pH
The tuberous sclerosis complex (TSC), comprising the GTPase-activating protein TSC2 and the scaffolding protein TSC1, plays an important role in the control of mTORC1 in response to growth factors and other environmental cues (91, 92). TSC regulates the GTP loading of the small G-protein Rheb (93), which in turn activates mTORC1 through an incompletely understood mechanism. We sought to investigate whether niclosamide reduced mTORC1 signaling via TSC. Serum-starved TSC2 knock-out MEFs displayed high basal mTORC1 activation (Ref. 94; see also Fig. 8A). Niclosamide efficiently blocked both basal and serum-induced mTORC1 activation in both wild type and TSC2 knock-out MEFs (Ref. 48; see also Fig. 8A), demonstrating that niclosamide inhibits mTORC1 signaling independently of TSC. We also verified whether niclosamide inhibited mTORC1 activity by modulating Rheb GTP loading. To address this point, we took advantage of the Q64L mutation on Rheb that renders it constitutively GTP-bound and therefore constitutively active. If niclosamide inhibits mTORC1 signaling by modulating the levels of GTP-bound Rheb, then Rheb Q64L overexpression should block the effect of niclosamide on mTORC1. As expected, Rheb Q64L enhanced p70S6K1 phosphorylation in serum-starved MCF-7 cells (Fig. 8B). Niclosamide retained its ability to block mTORC1 activation in Rheb Q64L-overexpressing cells. Therefore, the drug blocks mTORC1 signaling irrespective of the GTP loading status of Rheb (Fig. 8B). These data are consistent with those obtained with TSC2-null MEFs showing that niclosamide dampened mTORC1 signaling independently of TSC2 (Fig. 8A).
FIGURE 8.
Niclosamide and extracellular pH regulate mTORC1 signaling independently of TSC2 and Rheb. A, TSC2+/+/p53−/− and TSC2−/−/p53−/− MEFs were seeded in complete medium and allowed to propagate for 24 h at which point the cells were then starved of serum for 20 h. Where indicated, cells were incubated with 10 μm niclosamide or 100 nm rapamycin for the last 3 h of serum starvation. After serum starvation/drug treatment, cells were stimulated with 10% (v/v) serum for 30 min and then lysed as described under “Experimental Procedures.” Lysates were analyzed for mTORC1 activation and TSC2 levels. Lysates were also probed for β-actin as a loading control. B, MCF-7 cells were seeded in complete medium and allowed to propagate for 24 h at which point cells were transfected with 2 μg of pRK7-FLAG-Rheb Q64L or the corresponding pRK7 empty vector (E.V.). Cells were incubated for an additional 24 h after transfection to allow for Rheb Q64L expression at which point the cells were then starved of serum for 20 h. Where indicated, cells were incubated with 10 μm niclosamide or 100 nm rapamycin for the last 3 h of serum starvation. Cells were then stimulated with 10% (v/v) serum for 30 min after which they were lysed, and lysates were analyzed for mTORC1 activation. C, TSC2+/+/p53−/− and TSC2−/−/p53−/− MEFs were seeded and propagated in complete medium to near confluence at which point the growing medium was replaced with bicarbonate free-medium (adjusted to different pH values) supplemented with 10% (v/v) serum (cells were incubated with bicarbonate-free medium for 1 h in CO2-free conditions). Cells were then lysed, and lysates were analyzed for mTORC1 activation with the indicated antisera. D, schematic representation of a potential model for niclosamide-mediated cytoplasmic acidification and mTORC1 inhibition.
Because TSC2 is dispensable for niclosamide-mediated mTORC1 inhibition, we wondered whether TSC2 is also dispensable for mTORC1 inhibition by acidosis. As shown in Fig. 8C, acidification of the extracellular medium to a pH value of 6 inhibited mTORC1 signaling of wild type and TSC2-null MEFs in agreement with our data above showing that niclosamide inhibited mTORC1 signaling independently of TSC2.
DISCUSSION
Recent years have seen a strong interest in the development of novel mTORC1 inhibitors (structurally distinct from rapamycin) that target the catalytic activity of mTOR (41–43), several of which will likely turn out to be of therapeutic value (95). An alternative approach to identifying mTORC1 signaling inhibitors with therapeutic potential is to screen for drugs that are already approved for use in humans. This is an attractive strategy because it considerably reduces the time and cost of drug development because pharmacokinetic and safety profiles are already established (96, 97). In the present study, we show that the antiparasitic drug niclosamide strongly inhibited mTORC1 signaling at concentrations as low as 1 μm. Niclosamide is prescribed to eliminate parasites in the gut lumen and was not designed to be absorbed through the intestine. However, after a standard oral dose of 5 mg/kg in rats, a small fraction is absorbed, resulting in a 1.08 μm serum concentration (98), which is within the range that inhibits mTORC1.
Niclosamide shows selective inhibition of mTORC1 over mTORC2, and additional experiments demonstrated that niclosamide did not affect MEK/ERK or LKB1/AMPK signaling. Niclosamide did not inhibit mTORC1 or 95 other protein kinases in vitro, indicating that it must inhibit mTORC1 indirectly, unlike recently developed active site mTOR inhibitors. Examination of the chemical properties of niclosamide showed that it possesses distinctive features of protonophores, including a weakly acidic OH group that can reversibly bind protons in the physiological pH range, a bulky hydrophobic moiety for membrane solubility, and electron-withdrawing moieties that delocalize the negative charge of the anionic form of the protonophore, allowing it to remain associated with membranes (78, 79). These properties enable such chemicals to embed themselves into the plasma membrane and intracellular membranes and carry out rapid cycles of proton binding and release, thereby decreasing proton gradients across membranes. We verified that niclosamide was indeed able to rapidly dissipate proton gradients across membrane vesicles isolated from E. coli.
We observed among niclosamide and five analogs a tight correlation between protonophoric activity and inhibition of mTORC1 signaling, raising the possibility that mTORC1 inhibition is actually due to protonophoric activity. TFA9 and FCCP, which are structurally distinct from niclosamide but are well characterized protonophores, also rapidly and potently inhibited mTORC1, lending strong support to this view.
Protonophores are able to inhibit the coupling between electron transport and oxidative phosphorylation in mitochondria, thus inhibiting ATP synthesis (99). However, inhibition of mTORC1 was not a consequence of ATP depletion because niclosamide did not reduce cellular ATP levels (48) nor did it activate AMPK. Protonophores are also known to transport protons across the plasma membrane (77) and cause net inward proton movement by a mechanism largely driven by the electrochemical proton gradient across the plasma membrane. Niclosamide indeed caused rapid and reversible cytoplasmic acidification by up to 0.5–1 pH unit as did FCCP and TFA9. We attempted to determine whether cytoplasmic acidification was sufficient to inhibit mTORC1 by making use of the weak acid propionic acid, which is widely used to induce cytoplasmic acidification. Propionic acid transiently inhibited mTORC1 signaling, supporting our model that cytoplasmic pH regulates mTORC1. However, a caveat of this experiment is that propionate can also function as a signaling molecule through binding to G-protein-coupled receptors GPR41 and -43 and activation of intracellular signaling cascades that may affect mTORC1 independently of pH (100). Nevertheless, the observation that mTORC1 inhibition by propionic acid was enhanced by co-incubation with the NHE1 inhibitor EIPA, which reduces proton extrusion from cells, indicates that cytoplasmic acidification is likely relevant to the mechanism of action of niclosamide. Moreover, we recently showed that acidification of the extracellular medium, which causes rapid intracellular acidification (88), inhibits mTORC1 signaling. Whether acidic extracellular pH controls mTORC1 directly or as a consequence of cytoplasmic acidification was not examined in that study (88), but the experiments presented here indicate that intracellular acidification is sufficient to inhibit mTORC1 signaling.
Protonophores can dissipate proton gradients across acidic intracellular vesicles (e.g. lysosomes (pH 4.7), late endosomes (pH 5.5), or early endosomes (pH 6.3)). Lysosomes play a pivotal role in the control of mTORC1 signaling: mTORC1 has been shown to localize to lysosomes (84), lysosome localization itself has been shown to coordinate mTORC1 activation (101), and the v-ATPase plays a major role in the activation of mTORC1 by amino acids (86). Thus, we examined whether lysosomal pH also played a role in mTORC1 regulation. Our data show that although the v-ATPase is pivotal for mTORC1 activation lysosomal pH does not regulate mTORC1 signaling in agreement with the findings of Zoncu et al. (86).
In conclusion, the present study shows that cytoplasmic acidification, which can occur both in physiological and pathological conditions (83), inhibits mTORC1 signaling. Our proposed mechanism for niclosamide-mediated mTORC1 inhibition is depicted in Fig. 8D. Although additional work will be required to elucidate how the accumulation of protons within the cytosol silences mTORC1 signaling, so far we have shown that TSC and Rheb are dispensable for mTORC1 regulation by pH. The present study reveals an important new mode of chemical inhibition of mTORC1 signaling involving cytoplasmic acidification.
Supplementary Material
Acknowledgments
We thank Dr. Hilary McLauchlan and members of the Division of Signal Transduction Therapy at the University of Dundee (Scotland, UK) for performing the in vitro protein kinase assay selectivity screen and Dr. Thibault Mayor, Dr. John Church, Dr. Maritza Jaramillo, and Dr. Hilary Anderson for helpful discussions and critical reading of the manuscript.
This work was supported in part by grants from the Canadian Breast Cancer Foundation (to M. R.) and from the Canadian Cancer Society (to N. S.).

This article contains supplemental Figs. 1–4 and Table 1.
- mTOR
- mammalian target of rapamycin
- 4E-BP1
- eIF4E-binding protein 1
- AMPK
- adenosine monophosphate-activated protein kinase
- BCECF
- 2′,7′-bis(2-carboxyethyl)-5(6)-carboxyfluorescein
- EGFP-LC3
- enhanced green fluorescent protein-light chain 3 fusion protein
- FCCP
- carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone
- FKBP12
- FK506-binding protein 12
- MCF-7
- MI Cancer Foundation line 7
- MEF
- mouse embryo fibroblast
- Mnk
- MAPK-interacting kinase or MAPK signal-integrating kinase
- mTORC
- mTOR complex
- PKB/Akt
- protein kinase B/acutely transforming retrovirus AKT8 in rodent T cell lymphoma
- NF
- neurofibromin
- PTEN
- phosphatase and tensin homolog deleted on chromosome 10
- raptor
- regulatory-associated protein of mTOR
- S6K1
- ribosomal protein S6 kinase 1
- TFA9
- tyrphostin A9
- TSC
- tuberous sclerosis complex
- EIPA
- 5-(N-ethyl-N-isopropyl amiloride)
- IMV
- inverted inner membrane vesicle
- v-ATPase
- vacuolar proton ATPase.
REFERENCES
- 1. Facchinetti V., Ouyang W., Wei H., Soto N., Lazorchak A., Gould C., Lowry C., Newton A. C., Mao Y., Miao R. Q., Sessa W. C., Qin J., Zhang P., Su B., Jacinto E. (2008) The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 27, 1932–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. García-Martínez J. M., Alessi D. R. (2008) mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem. J. 416, 375–385 [DOI] [PubMed] [Google Scholar]
- 3. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 4. Hresko R. C., Mueckler M. (2005) mTOR·RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J. Biol. Chem. 280, 40406–40416 [DOI] [PubMed] [Google Scholar]
- 5. Ikenoue T., Inoki K., Yang Q., Zhou X., Guan K. L. (2008) Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 27, 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guertin D. A., Stevens D. M., Thoreen C. C., Burds A. A., Kalaany N. Y., Moffat J., Brown M., Fitzgerald K. J., Sabatini D. M. (2006) Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCα, but not S6K1. Dev. Cell 11, 859–871 [DOI] [PubMed] [Google Scholar]
- 7. Yan L., Mieulet V., Lamb R. F. (2008) mTORC2 is the hydrophobic motif kinase for SGK1. Biochem. J. 416, e19–21 [DOI] [PubMed] [Google Scholar]
- 8. Dowling R. J., Topisirovic I., Alain T., Bidinosti M., Fonseca B. D., Petroulakis E., Wang X., Larsson O., Selvaraj A., Liu Y., Kozma S. C., Thomas G., Sonenberg N. (2010) mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328, 1172–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burnett P. E., Barrow R. K., Cohen N. A., Snyder S. H., Sabatini D. M. (1998) RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. U.S.A. 95, 1432–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu Y., Yoon S. O., Poulogiannis G., Yang Q., Ma X. M., Villn J., Kubica N., Hoffman G. R., Cantley L. C., Gygi S. P., Blenis J. (2011) Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332, 1322–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hsu P. P., Kang S. A., Rameseder J., Zhang Y., Ottina K. A., Lim D., Peterson T. R., Choi Y., Gray N. S., Yaffe M. B., Marto J. A., Sabatini D. M. (2011) The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yea S. S., Fruman D. A. (2011) Cell signaling. New mTOR targets Grb attention. Science 332, 1270–1271 [DOI] [PubMed] [Google Scholar]
- 13. Neshat M. S., Mellinghoff I. K., Tran C., Stiles B., Thomas G., Petersen R., Frost P., Gibbons J. J., Wu H., Sawyers C. L. (2001) Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc. Natl. Acad. Sci. U.S.A. 98, 10314–10319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kwiatkowski D. J., Zhang H., Bandura J. L., Heiberger K. M., Glogauer M., el-Hashemite N., Onda H. (2002) A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet. 11, 525–534 [DOI] [PubMed] [Google Scholar]
- 15. Kenerson H. L., Aicher L. D., True L. D., Yeung R. S. (2002) Activated mammalian target of rapamycin pathway in the pathogenesis of tuberous sclerosis complex renal tumors. Cancer Res. 62, 5645–5650 [PubMed] [Google Scholar]
- 16. Onda H., Crino P. B., Zhang H., Murphey R. D., Rastelli L., Gould Rothberg B. E., Kwiatkowski D. J. (2002) Tsc2 null murine neuroepithelial cells are a model for human tuber giant cells, and show activation of an mTOR pathway. Mol. Cell. Neurosci. 21, 561–574 [DOI] [PubMed] [Google Scholar]
- 17. Corradetti M. N., Inoki K., Bardeesy N., DePinho R. A., Guan K. L. (2004) Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 18, 1533–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shaw R. J., Bardeesy N., Manning B. D., Lopez L., Kosmatka M., DePinho R. A., Cantley L. C. (2004) The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6, 91–99 [DOI] [PubMed] [Google Scholar]
- 19. Johannessen C. M., Reczek E. E., James M. F., Brems H., Legius E., Cichowski K. (2005) The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc. Natl. Acad. Sci. U.S.A. 102, 8573–8578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang X., Wullschleger S., Shpiro N., McGuire V. A., Sakamoto K., Woods Y. L., McBurnie W., Fleming S., Alessi D. R. (2008) Important role of the LKB1-AMPK pathway in suppressing tumorigenesis in PTEN-deficient mice. Biochem. J. 412, 211–221 [DOI] [PubMed] [Google Scholar]
- 21. Sekulić A., Hudson C. C., Homme J. L., Yin P., Otterness D. M., Karnitz L. M., Abraham R. T. (2000) A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells. Cancer Res. 60, 3504–3513 [PubMed] [Google Scholar]
- 22. Fan Q. W., Cheng C., Knight Z. A., Haas-Kogan D., Stokoe D., James C. D., McCormick F., Shokat K. M., Weiss W. A. (2009) EGFR signals to mTOR through PKC and independently of Akt in glioma. Sci. Signal. 2, ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Forbes S. A., Bhamra G., Bamford S., Dawson E., Kok C., Clements J., Menzies A., Teague J. W., Futreal P. A., Stratton M. R. (2008) The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr. Protoc. Hum. Genet. Chapter 10, Unit 10.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sato T., Nakashima A., Guo L., Coffman K., Tamanoi F. (2010) Single amino-acid changes that confer constitutive activation of mTOR are discovered in human cancer. Oncogene 29, 2746–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Choi J., Chen J., Schreiber S. L., Clardy J. (1996) Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 273, 239–242 [DOI] [PubMed] [Google Scholar]
- 26. Liang J., Choi J., Clardy J. (1999) Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 55, 736–744 [DOI] [PubMed] [Google Scholar]
- 27. Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 28. Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 29. Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. (2002) Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189 [DOI] [PubMed] [Google Scholar]
- 30. Schalm S. S., Blenis J. (2002) Identification of a conserved motif required for mTOR signaling. Curr. Biol. 12, 632–639 [DOI] [PubMed] [Google Scholar]
- 31. Schalm S. S., Fingar D. C., Sabatini D. M., Blenis J. (2003) TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 13, 797–806 [DOI] [PubMed] [Google Scholar]
- 32. Yatscoff R. W. (1996) Pharmacokinetics of rapamycin. Transplant. Proc. 28, 970–973 [PubMed] [Google Scholar]
- 33. Hartford C. M., Ratain M. J. (2007) Rapamycin: something old, something new, sometimes borrowed and now renewed. Clin. Pharmacol. Ther. 82, 381–388 [DOI] [PubMed] [Google Scholar]
- 34. Gibbons J. J., Abraham R. T., Yu K. (2009) Mammalian target of rapamycin: discovery of rapamycin reveals a signaling pathway important for normal and cancer cell growth. Semin. Oncol. 36, Suppl. 3, S3–S17 [DOI] [PubMed] [Google Scholar]
- 35. Houghton P. J. (2010) Everolimus. Clin. Cancer Res. 16, 1368–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rizzieri D. A., Feldman E., Dipersio J. F., Gabrail N., Stock W., Strair R., Rivera V. M., Albitar M., Bedrosian C. L., Giles F. J. (2008) A phase 2 clinical trial of deforolimus (AP23573, MK-8669), a novel mammalian target of rapamycin inhibitor, in patients with relapsed or refractory hematologic malignancies. Clin. Cancer Res. 14, 2756–2762 [DOI] [PubMed] [Google Scholar]
- 37. Gingras A. C., Raught B., Gygi S. P., Niedzwiecka A., Miron M., Burley S. K., Polakiewicz R. D., Wyslouch-Cieszynska A., Aebersold R., Sonenberg N. (2001) Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15, 2852–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sarbassov D. D., Ali S. M., Sengupta S., Sheen J. H., Hsu P. P., Bagley A. F., Markhard A. L., Sabatini D. M. (2006) Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22, 159–168 [DOI] [PubMed] [Google Scholar]
- 39. Guertin D. A., Sabatini D. M. (2009) The pharmacology of mTOR inhibition. Sci. Signal. 2, pe24. [DOI] [PubMed] [Google Scholar]
- 40. Dowling R. J., Topisirovic I., Fonseca B. D., Sonenberg N. (2010) Dissecting the role of mTOR: lessons from mTOR inhibitors. Biochim. Biophys. Acta 1804, 433–439 [DOI] [PubMed] [Google Scholar]
- 41. Yu K., Toral-Barza L., Shi C., Zhang W. G., Lucas J., Shor B., Kim J., Verheijen J., Curran K., Malwitz D. J., Cole D. C., Ellingboe J., Ayral-Kaloustian S., Mansour T. S., Gibbons J. J., Abraham R. T., Nowak P., Zask A. (2009) Biochemical, cellular, and in vivo activity of novel ATP-competitive and selective inhibitors of the mammalian target of rapamycin. Cancer Res. 69, 6232–6240 [DOI] [PubMed] [Google Scholar]
- 42. García-Martínez J. M., Moran J., Clarke R. G., Gray A., Cosulich S. C., Chresta C. M., Alessi D. R. (2009) Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR). Biochem. J. 421, 29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., Gao Y., Reichling L. J., Sim T., Sabatini D. M., Gray N. S. (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Feldman M. E., Apsel B., Uotila A., Loewith R., Knight Z. A., Ruggero D., Shokat K. M. (2009) Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 7, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chresta C. M., Davies B. R., Hickson I., Harding T., Cosulich S., Critchlow S. E., Vincent J. P., Ellston R., Jones D., Sini P., James D., Howard Z., Dudley P., Hughes G., Smith L., Maguire S., Hummersone M., Malagu K., Menear K., Jenkins R., Jacobsen M., Smith G. C., Guichard S., Pass M. (2010) AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 70, 288–298 [DOI] [PubMed] [Google Scholar]
- 46. Fan Q. W., Knight Z. A., Goldenberg D. D., Yu W., Mostov K. E., Stokoe D., Shokat K. M., Weiss W. A. (2006) A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell 9, 341–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maira S. M., Stauffer F., Brueggen J., Furet P., Schnell C., Fritsch C., Brachmann S., Chène P., De Pover A., Schoemaker K., Fabbro D., Gabriel D., Simonen M., Murphy L., Finan P., Sellers W., García-Echeverría C. (2008) Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol. Cancer Ther. 7, 1851–1863 [DOI] [PubMed] [Google Scholar]
- 48. Balgi A. D., Fonseca B. D., Donohue E., Tsang T. C., Lajoie P., Proud C. G., Nabi I. R., Roberge M. (2009) Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One 4, e7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Díaz-Troya S., Pérez-Pérez M. E., Florencio F. J., Crespo J. L. (2008) The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 4, 851–865 [DOI] [PubMed] [Google Scholar]
- 50. Anand N. (1997) Approaches to Design and Synthesis of Antiparasitic Drugs, p. 240, Elsevier, Amsterdam [Google Scholar]
- 51. Imming P., Sinning C., Meyer A. (2006) Drugs, their targets and the nature and number of drug targets. Nat. Rev. Drug Discov. 5, 821–834 [DOI] [PubMed] [Google Scholar]
- 52. Fairweather I., Boray J. C. (1999) Fasciolicides: efficacy, actions, resistance and its management. Vet. J. 158, 81–112 [DOI] [PubMed] [Google Scholar]
- 53. Dunlop E. A., Dodd K. M., Seymour L. A., Tee A. R. (2009) Mammalian target of rapamycin complex 1-mediated phosphorylation of eukaryotic initiation factor 4E-binding protein 1 requires multiple protein-protein interactions for substrate recognition. Cell. Signal. 21, 1073–1084 [DOI] [PubMed] [Google Scholar]
- 54. Curman D., Cinel B., Williams D. E., Rundle N., Block W. D., Goodarzi A. A., Hutchins J. R., Clarke P. R., Zhou B. B., Lees-Miller S. P., Andersen R. J., Roberge M. (2001) Inhibition of the G2 DNA damage checkpoint and of protein kinases Chk1 and Chk2 by the marine sponge alkaloid debromohymenialdisine. J. Biol. Chem. 276, 17914–17919 [DOI] [PubMed] [Google Scholar]
- 55. Dalal K., Duong F. (2009) The SecY complex forms a channel capable of ionic discrimination. EMBO Rep. 10, 762–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Baxter K. A., Church J. (1996) Characterization of acid extrusion mechanisms in cultured fetal rat hippocampal neurones. J. Physiol. 493, 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smith M. A., Ashford M. L. (1998) Mode switching characterizes the activity of large conductance potassium channels recorded from rat cortical fused nerve terminals. J. Physiol. 513, 733–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sancak Y., Thoreen C. C., Peterson T. R., Lindquist R. A., Kang S. A., Spooner E., Carr S. A., Sabatini D. M. (2007) PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 25, 903–915 [DOI] [PubMed] [Google Scholar]
- 59. Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. (2007) The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bärlund M., Monni O., Kononen J., Cornelison R., Torhorst J., Sauter G., Kallioniemi O. P., Kallioniemi A. (2000) Multiple genes at 17q23 undergo amplification and overexpression in breast cancer. Cancer Res. 60, 5340–5344 [PubMed] [Google Scholar]
- 61. Karni R., de Stanchina E., Lowe S. W., Sinha R., Mu D., Krainer A. R. (2007) The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 14, 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu G. J., Sinclair C. S., Paape J., Ingle J. N., Roche P. C., James C. D., Couch F. J. (2000) 17q23 amplifications in breast cancer involve the PAT1, RAD51C, PS6K, and SIGMA1B genes. Cancer Res. 60, 5371–5375 [PubMed] [Google Scholar]
- 63. Harrington L. S., Findlay G. M., Gray A., Tolkacheva T., Wigfield S., Rebholz H., Barnett J., Leslie N. R., Cheng S., Shepherd P. R., Gout I., Downes C. P., Lamb R. F. (2004) The TSC1–2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J. Cell Biol. 166, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shah O. J., Wang Z., Hunter T. (2004) Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 14, 1650–1656 [DOI] [PubMed] [Google Scholar]
- 65. Um S. H., Frigerio F., Watanabe M., Picard F., Joaquin M., Sticker M., Fumagalli S., Allegrini P. R., Kozma S. C., Auwerx J., Thomas G. (2004) Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431, 200–205 [DOI] [PubMed] [Google Scholar]
- 66. Ma L., Chen Z., Erdjument-Bromage H., Tempst P., Pandolfi P. P. (2005) Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell 121, 179–193 [DOI] [PubMed] [Google Scholar]
- 67. Fonseca B. D., Alain T., Finestone L. K., Huang B. P., Rolfe M., Jiang T., Yao Z., Hernandez G., Bennett C. F., Proud C. G. (2011) Pharmacological and genetic evaluation of proposed roles of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase (MEK), extracellular signal-regulated kinase (ERK), and p90(RSK) in the control of mTORC1 protein signaling by phorbol esters. J. Biol. Chem. 286, 27111–27122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fukunaga R., Hunter T. (1997) MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 16, 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Waskiewicz A. J., Flynn A., Proud C. G., Cooper J. A. (1997) Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 16, 1909–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Waskiewicz A. J., Johnson J. C., Penn B., Mahalingam M., Kimball S. R., Cooper J. A. (1999) Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol. Cell. Biol. 19, 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pyronnet S., Imataka H., Gingras A. C., Fukunaga R., Hunter T., Sonenberg N. (1999) Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 18, 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ueda T., Watanabe-Fukunaga R., Fukuyama H., Nagata S., Fukunaga R. (2004) Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol. Cell. Biol. 24, 6539–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sonenberg N. (2008) eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochem. Cell Biol. 86, 178–183 [DOI] [PubMed] [Google Scholar]
- 74. Steinberg G. R. (2009) Role of the AMP-activated protein kinase in regulating fatty acid metabolism during exercise. Appl. Physiol. Nutr. Metab. 34, 315–322 [DOI] [PubMed] [Google Scholar]
- 75. Hardie D. G. (2004) The AMP-activated protein kinase pathway—new players upstream and downstream. J. Cell Sci. 117, 5479–5487 [DOI] [PubMed] [Google Scholar]
- 76. Zhang L., He H., Balschi J. A. (2007) Metformin and phenformin activate AMP-activated protein kinase in the heart by increasing cytosolic AMP concentration. Am. J. Physiol. Heart Circ. Physiol. 293, H457–H466 [DOI] [PubMed] [Google Scholar]
- 77. McLaughlin S. G., Dilger J. P. (1980) Transport of protons across membranes by weak acids. Physiol. Rev. 60, 825–863 [DOI] [PubMed] [Google Scholar]
- 78. Terada H. (1990) Uncouplers of oxidative phosphorylation. Environ. Health Perspect. 87, 213–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang G. J., Richardson S. R., Thayer S. A. (1995) Intracellular acidification is not a prerequisite for glutamate-triggered death of cultured hippocampal neurons. Neurosci. Lett. 186, 139–144 [DOI] [PubMed] [Google Scholar]
- 80. Shih T. M., Smith R. D., Toro L., Goldin A. L. (1998) High-level expression and detection of ion channels in Xenopus oocytes. Methods Enzymol. 293, 529–556 [DOI] [PubMed] [Google Scholar]
- 81. Grinstein S., Cohen S. (1987) Cytoplasmic [Ca2+] and intracellular pH in lymphocytes. Role of membrane potential and volume-activated Na+/H+ exchange. J. Gen. Physiol. 89, 185–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Beauvoit B., Rigoulet M., Raffard G., Canioni P., Guérin B. (1991) Differential sensitivity of the cellular compartments of Saccharomyces cerevisiae to protonophoric uncoupler under fermentative and respiratory energy supply. Biochemistry 30, 11212–11220 [DOI] [PubMed] [Google Scholar]
- 83. Casey J. R., Grinstein S., Orlowski J. (2010) Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 11, 50–61 [DOI] [PubMed] [Google Scholar]
- 84. Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., Sabatini D. M. (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Han J., Burgess K. (2010) Fluorescent indicators for intracellular pH. Chem. Rev. 110, 2709–2728 [DOI] [PubMed] [Google Scholar]
- 86. Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., Sabatini D. M. (2011) mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science 334, 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Dröse S., Altendorf K. (1997) Bafilomycins and concanamycins as inhibitors of V-ATPases and P-ATPases. J. Exp. Biol. 200, 1–8 [DOI] [PubMed] [Google Scholar]
- 88. Balgi A. D., Diering G. H., Donohue E., Lam K. K., Fonseca B. D., Zimmerman C., Numata M., Roberge M. (2011) Regulation of mTORC1 signaling by pH. PLoS One 6, e21549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sandoval A., Triviños F., Sanhueza A., Carretta D., Hidalgo M. A., Hancke J. L., Burgos R. A. (2007) Propionate induces pHi changes through calcium flux, ERK1/2, p38, and PKC in bovine neutrophils. Vet. Immunol. Immunopathol. 115, 286–298 [DOI] [PubMed] [Google Scholar]
- 90. Brett C. L., Donowitz M., Rao R. (2005) Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. Cell Physiol. 288, C223–C239 [DOI] [PubMed] [Google Scholar]
- 91. Li Y., Corradetti M. N., Inoki K., Guan K. L. (2004) TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem. Sci. 29, 32–38 [DOI] [PubMed] [Google Scholar]
- 92. Pan D., Dong J., Zhang Y., Gao X. (2004) Tuberous sclerosis complex: from Drosophila to human disease. Trends Cell Biol. 14, 78–85 [DOI] [PubMed] [Google Scholar]
- 93. Becker M. L., Remsen E. E., Pan D., Wooley K. L. (2004) Peptide-derivatized shell-cross-linked nanoparticles. 1. Synthesis and characterization. Bioconjug. Chem. 15, 699–709 [DOI] [PubMed] [Google Scholar]
- 94. Zhang H., Cicchetti G., Onda H., Koon H. B., Asrican K., Bajraszewski N., Vazquez F., Carpenter C. L., Kwiatkowski D. J. (2003) Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J. Clin. Investig. 112, 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhang Y. J., Duan Y., Zheng X. F. (2011) Targeting the mTOR kinase domain: the second generation of mTOR inhibitors. Drug Discov. Today 16, 325–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Chong C. R., Sullivan D. J., Jr. (2007) New uses for old drugs. Nature 448, 645–646 [DOI] [PubMed] [Google Scholar]
- 97. O'Connor K. A., Roth B. L. (2005) Finding new tricks for old drugs: an efficient route for public-sector drug discovery. Nat. Rev. Drug Discov. 4, 1005–1014 [DOI] [PubMed] [Google Scholar]
- 98. Chang Y.-W., Yeh T.-K., Lin K.-T., Chen W.-C., Yao H.-T., Lan S.-J., Wu Y.-S., Hsieh H.-P., Chen C.-M., Chen C.-T. (2006) Pharmacokinetics of anti-SARS-CoV agent niclosamide and its analogs in rats. J. Food Drug Anal. 14, 329–333 [Google Scholar]
- 99. Curnock A. P., Thomson T. A., Westwood R., Kuo E. A., Williamson R. A., Yea C. M., Ruuthb E. (2001) Inhibition of stimulated Jurkat cell adenosine 3′,5′-cyclic monophosphate synthesis by the immunomodulatory compound HR325. Biochem. Pharmacol. 61, 227–235 [DOI] [PubMed] [Google Scholar]
- 100. Brown A. J., Goldsworthy S. M., Barnes A. A., Eilert M. M., Tcheang L., Daniels D., Muir A. I., Wigglesworth M. J., Kinghorn I., Fraser N. J., Pike N. B., Strum J. C., Steplewski K. M., Murdock P. R., Holder J. C., Marshall F. H., Szekeres P. G., Wilson S., Ignar D. M., Foord S. M., Wise A., Dowell S. J. (2003) The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 278, 11312–11319 [DOI] [PubMed] [Google Scholar]
- 101. Korolchuk V. I., Saiki S., Lichtenberg M., Siddiqi F. H., Roberts E. A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F. M., O'Kane C. J., Deretic V., Rubinsztein D. C. (2011) Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13, 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.