Background: Through splicing, squid express a conventional ADAR and a novel form with an extra RNA binding domain.
Results: Chloride inhibits the ability of the conventional, but not the novel, ADAR to bind and edit RNA.
Conclusion: The extra RNA binding domain allows squid, an osmoconformer, to edit in a high salt environment.
Significance: The ability of ADARs to bind RNA is a target for adaptation.
Keywords: Double-stranded RNA, Protein-Nucleic Acid Interaction, RNA-binding Protein, RNA Editing, RNA-Protein Interaction, ADAR, Chloride, Squid
Abstract
A-to-I RNA editing is particularly common in coding regions of squid mRNAs. Previously, we isolated a squid editing enzyme (sqADAR2) that shows a unique structural feature when compared with other ADAR2 family members: an additional double-stranded RNA (dsRNA) binding domain (dsRBD). Alternative splicing includes or excludes this motif, generating a novel or a conventional variant termed sqADAR2a and sqADAR2b, respectively. The extra dsRBD of sqADAR2a increases its editing activity in vitro. We hypothesized that the high activity is due to an increase in the affinity of the enzyme for dsRNA. This may be important because protein-RNA interactions can be influenced by physical factors. We became particularly interested in analyzing the effects of salt on interactions between sqADAR2 and RNA because squid cells have a ∼3-fold higher ionic strength and proportionally more Cl− than vertebrate cells. To date, in vitro biochemical analyses of adenosine deamination have been conducted using vertebrate-like ionic strength buffers containing chloride as the major anion, although the vast majority of cellular anions are known to be organic. We found that squid-like salt conditions severely impair the binding affinity of conventional ADAR2s for dsRNA, leading to a decrease in nonspecific and site-specific editing activity. Inhibition of editing was mostly due to high Cl− levels and not to the high concentrations of K+, Na+, and organic anions like glutamate. Interestingly, the extra dsRBD in sqADAR2a conferred resistance to the high Cl− levels found in squid neurons. It does so by increasing the affinity of sqADAR2 for dsRNA by 30- or 100-fold in vertebrate-like or squid-like conditions, respectively. Site-directed mutagenesis of squid ADAR2a showed that its increased affinity and editing activity are directly attributable to the RNA binding activity of the extra dsRBD.
Introduction
Adenosine deamination is the most common form of RNA editing among eumetazoans, occurring in organisms that differ dramatically in terms of their complexity and the environments in which they inhabit. It is catalyzed by an enzyme family known as adenosine deaminases that act on RNA (ADARs)3 (1), which, by removing a primary amine, convert adenosine to inosine (A-to-I). Biologically, this conversion serves several roles. First, nonspecific deamination in largely double-stranded RNA (dsRNA) is thought to help prevent the invasion of viruses that express dsRNA during their life cycle (2). Second, promiscuous deamination of adenosines in non-coding mRNA regions has been proposed to regulate message stability and gene expression (3–5). Third, because inosine is recognized as guanosine by the molecular machinery (6), site-specific deamination of adenosines can also change codons and protein function (for a review, see Ref. 7). This last role has received the greatest focus by the research community, and mounting evidence points out that it regulates neuronal function in important ways. For example, postsynaptic calcium permeability, G-protein signaling, and repetitive firing are regulated by editing ionotrophic glutamate receptor (GluR-B subunit), serotonin receptor (5-HT2c) and potassium channel (Kv1.1A) transcripts, respectively (8–10). Thus, A-to-I RNA editing expands genetic information and fine tunes protein function, enabling greater complexity in physiological responses.
Although A-to-I editing can change protein function, different organisms use it to different extents. In vertebrates, for example, it is not frequently used to modify protein structure. Numerous bioinformatic screens comparing mammalian transcriptomes and genomes have uncovered only 55 editing sites in coding regions, meaning that less than 0.3% of transcripts appear to be edited (11–15). Recoding events are apparently more abundant in invertebrates. Similar bioinformatic screens in Drosophila have predicted 831 editing sites within the coding regions of ∼500 targets, representing 3–4% of the whole transcriptome (16–19). Cephalopod coding regions seem to undergo exceptional extensive editing. Direct analysis of a handful of mRNAs has revealed each to be heavily edited, including two voltage-dependent K+ channels, a voltage-dependent Na+ channel, a Na+/K+ pump, synaptotagmin I, and two ADARs (20–24).4 More than 100 editing sites have already been found in the open reading frames of less than 10 mRNAs from cephalopods. Furthermore, some of these sites cause substantial effects on protein function. For example, the R87G edit in squid Kv1.1A mRNAs regulates subunit tetramerization (22), and the I877V edit in squid Na+/K+ ATPase mRNAs changes the pump intrinsic voltage dependence, accelerating Na+ release to the extracellular medium (23). At present, neither genome nor transcriptome data are available for cephalopods. However, based on available data, it is expected that bioinformatics will reveal exceptionally high levels of editing.
If editing is indeed high in cephalopods, how are their editing enzymes capable of recognizing a broader set of targets? To approach this question, it is useful to consider a typical ADAR structure. All ADARs have a common domain architecture consisting of a variable number of dsRBDs and a C-terminal deaminase domain. The dsRBDs, which are small (65–70 amino acids), exclusively bind dsRNA and are present in a broad variety of proteins (25, 26). The deaminase domain is large (300–350 amino acids) and catalyzes the deamination reaction. There are two catalytically active ADAR families among vertebrates: ADAR1 and ADAR2. Along with other structural differences, ADAR1 has three dsRBDs, whereas ADAR2 has just two. In fact, every predicted or characterized ADAR2 from cnidarians to humans contains only two dsRBDs except for one. The notable exception is squid ADAR2 (sqADAR2), which contains a third dsRBD (20). Through alternative splicing, this domain can be included or excluded, generating squid ADAR2a and ADAR2b, respectively. We have shown previously that the additional dsRBD of squid ADAR2a increases its ability to edit specific adenosines in vitro that are also edited in vivo. Thus, we concluded that this novel feature explains in part the highly active editing observed in squid (20).
It is reasonable to speculate that the extra dsRBD of sqADAR2a would increase the affinity of the enzyme for dsRNA, leading to higher activity. However, this is only part of the equation. Binding is also affected by physical factors such as ionic strength and composition, temperature, and pH. In this study, we focused on ion concentration and composition for two main reasons. First, salts can influence the electrostatic interactions between positively charged dsRBDs and the negatively charged RNA backbone. Second, as osmoconformers, squid, like all marine invertebrates, have a plasma and cytoplasm that is isotonic with seawater. In contrast, the osmolarity of the plasma and the cytoplasm in osmoregulators like insects and mammals are about 3-fold lower. Despite this range of osmolarities, cells from different organisms maintain a similar overall ionic composition. In each case, the major cation is K+, and relatively large organic molecules constitute the major anions. However, the chloride ion is an exception. In vertebrate cells, it constitutes a small fraction of the total anions (7.5%), whereas in squid axoplasm, it constitutes a significant fraction (20–25%) (27–30). To our knowledge, every previous biochemical study of RNA editing by adenosine deamination has been conducted with buffers that have vertebrate-like ionic strengths and contain chloride as the major anion (9, 31–34). Because one of the splice variants of sqADAR has a novel structure and the other does not, we decided to analyze the effects of ionic concentrations and composition on their binding capacities and deaminase activities. We found that high salt concentrations approximating those found in marine osmoconformers and the Cl− ion severely impair the ability of the canonical isoform to edit. By contrast, the novel isoform maintains high editing activity under the same conditions and binds 30–100-fold more tightly to dsRNA, depending on the conditions.
EXPERIMENTAL PROCEDURES
Molecular Biology and Protein Purification
All molecular biology was performed using standard techniques. PCR amplifications used Phusion DNA polymerase (New England Biolabs), and PCR products were gel-purified and directly sequenced. sqADAR2a K89E/K90A/K93A was generated by mutating the conserved KKXXK binding motif of dsRBD1 to EAXXA using the QuikChange mutagenesis kit (Stratagene). Production and purification of recombinant ADARs from Pichia pastoris were carried out as described previously (20).
Editing and Binding Assay Buffers
Editing and binding assays were performed using the following buffers: human-like buffer contained 10 mm NaCl, 140 mm potassium glutamate, 10 mm Tris glutamate, pH 7, and 20% glycerol. The squid-like buffer contained 250 mm potassium glutamate, 50 mm sodium glutamate, 100 mm KCl, 10 mm Tris glutamate, pH 7, and 20% glycerol. Potassium glutamate buffers contained 10–400 mm potassium glutamate, 10 mm Tris glutamate, pH 7, and 20% glycerol, and sodium glutamate buffers contained 10–400 mm sodium glutamate, 10 mm Tris glutamate, pH 7, and 20% glycerol. KCl buffers contained 10–400 mm KCl, 10 mm Tris-Cl, pH 7, and 20% glycerol. In addition, each functional assay contained 100 ng/μl BSA, 50 ng/μl tRNA, 1 mm DTT, 0.5 mm PMSF, 0.7 μg/ml pepstatin A, 0.4 μg/ml leupeptin, and 1 unit/μl RNase inhibitor in a volume of 20 μl.
Nonspecific Editing Assays
Recombinant squid and human ADAR2 purification, radiolabeled dsRNA substrate synthesis, and nonspecific editing assays were performed and analyzed as described previously (20). As before, the dsRNA substrate fragment was derived from the squid Na+ channel GFLN1 (GenBankTM accession number L19979.1; nucleotides 2111–2808 plus 12 nucleotides from both T7 promoter regions, generating a 710-bp fragment RNA duplex). Recombinant proteins (4 pm–17 nm) were incubated with the dsRNA radiolabeled substrate (0.5 pm) in triplicate and incubated from 2 to 16 h at 35 °C under the specified buffer conditions.
Site-specific Editing Assays
Recombinant squid ADAR2 purification, sqKv1.1A cRNA synthesis, and site-specific editing assays were performed and analyzed as described previously (20). Recombinant proteins (2–20 nm) were incubated with cRNA (3.33 pm) for 2 h at 35 °C under the specified buffer conditions. Assays were performed in triplicate.
Filter Binding Assays
dsRNA substrates of different sizes were synthesized in the same manner as for the 710-bp substrate except that different reverse primers were used to amplify the probe template. The 355-bp substrate used primer sqNC25, the 165-bp substrate used primer sqNC26, and the 79-bp substrate used primer sqNC27 (Table 1). RNA binding assays were carried out using recombinant ADAR2s (0.5 pm–128 nm) and four different radiolabeled dsRNA substrates (0.5, 1, 2, and 4 pm 710-, 355-, 165-, and 79-bp fragments, respectively) in triplicate as described previously (35, 36). A Bio-Dot® SF microfiltration apparatus (Bio-Rad) was used, and the nitrocellulose membrane was scanned using a Typhoon 9200 phosphor/fluorescence imager (GE Healthcare). Reactions were incubated from 10 min to 16 h at 35 °C under the specified buffer conditions. Apparent KD values were estimated by fitting the data to a Hill equation. Real KD values were estimated by fitting apparent KD values to Equation 1 (see “Results”) using Origin® software.
TABLE 1.
Reverse primers used for three new smaller dsRNA substrates
| Primer | Sequence (5′ → 3′) |
|---|---|
| sqNC25 | AGTAATACGACTCACTATAGGGAGAGCACCAAACGAAGACAGTAACAAGGC |
| sqNC26 | AGTAATACGACTCACTATAGGGAGACCCATCGCGGAACTTGATTGTCTTC |
| sqNC27 | AGTAATACGACTCACTATAGGGAGAAATACGATGATTCCTAGCACAAGGG |
Electrophoretic Mobility Shift Assays
Electrophoretic mobility shift assays were performed as described previously (37, 38). Recombinant ADAR2s (6 pm–150 nm) were incubated with a 710-bp perfect dsRNA radiolabeled substrate (0.5 pm) for 2 h at 35 °C using the human-like solution (20). Reactions were loaded onto a 6% non-denaturing polyacrylamide gel (19:1 acrylamide:bisacrylamide). After electrophoresis, gels were dried and scanned with the Typhoon 9200 phosphor/fluorescence imager.
RESULTS
Osmoregulator Versus Osmoconformer Ionic Strengths
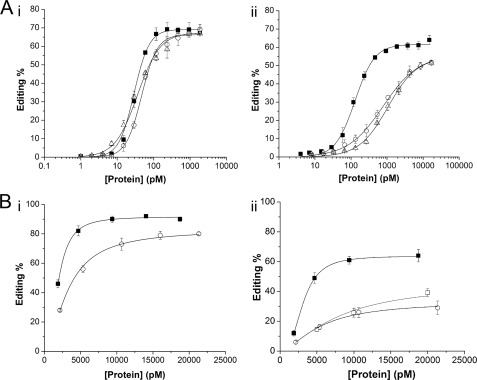
To date, most biochemical analyses of A-to-I RNA editing have been conducted using conditions that approximate vertebrate ionic strength and use Cl− as the major anion. In contrast, the cellular environment of a marine invertebrate has about 3-fold more salt, and all cells strictly limit the concentration of the Cl− ion. Because the cellular environment in squid differs from vertebrates and because squid neurons express two splice variants of sqADAR2 that have different numbers of dsRBDs, we decided to analyze the effects of ionic composition and concentration on the squid isoforms. For this purpose, we patterned two buffers on the major intracellular ions found in vertebrate or squid cells (Table 2; see “Experimental Procedures” for buffer compositions). In each, glutamate, a representative organic anion, replaces Cl− as the major anion. The osmolarity of the squid-like buffer, however, is ∼3 times higher, and it contains substantially more Cl− (27–30). As a first step, we compared the promiscuous editing activity of recombinant sqADAR2a, which contains three dsRBDs, and sqADAR2b and human ADAR2, each of which contains two dsRBDs. A range of concentrations of each enzyme was incubated with a 710-bp perfect RNA duplex that contained 32P-labeled adenosine using both vertebrate and squid buffers. In the vertebrate-like solution, the three enzymes converted a similar amount of adenosines into inosines (65–70% maximum) over a wide range of protein concentrations (Fig. 1A, panel i). The protein concentration required for half-maximal editing was also very similar, ranging from 30 to 45 pm. When assayed under squid-like conditions, however, the editing activities of the conventional ADAR2s were severely impaired, whereas the novel squid variant was able to maintain a high activity (Fig. 1A, panel ii). sqADAR2a was able to convert up to ∼60% of available adenosines to inosines at concentrations greater than ∼1 nm, and its midpoint value was estimated to be ∼7-fold lower than those for the conventional ADAR2s. By switching from vertebrate-like to squid-like conditions, the midpoint values for the conventional ADAR2s increased by >18-fold, whereas those for the novel squid variant rose by only ∼4-fold.
TABLE 2.
Comparison of ion concentrations inside typical osmoregulator and osmoconformer cells
| [Intracellular] |
||
|---|---|---|
| Osmoregulator (human) | Osmoconformer (squid) | |
| mm | ||
| Cations | ||
| K+ | 140 | 310–360 |
| Na+ | 5–15 | 40–80 |
| Org. base | 60–100 | |
| Anions | ||
| Org. anions | 140 | 350–390 |
| Cl− | 5–15 | 110–170 |
| Total salt | 145–155 | 410–560 |
FIGURE 1.
Effects of vertebrate and squid ionic compositions on promiscuous and site-selective editing activities of ADAR2s. A, promiscuous editing of a 710-bp radiolabeled perfect dsRNA duplex by recombinant ADAR2s was estimated using vertebrate-like (panel i) and squid-like (panel ii) solutions. Overall A-to-I conversion percentages were estimated for a wide range of protein concentrations (1–2000 pm) using a fixed amount of dsRNA (0.5 pm). The midpoint values for sqADAR2a (■), sqADAR2b (○), and hADAR2 (△) were 29 ± 1, 45 ± 2, and 36 ± 1 pm using vertebrate-like solution and 129 ± 5.5, 793 ± 65, and 1044 ± 88 pm using squid-like solution, respectively. B, site-selective editing of adenosine 190 in sqKv1.1 mRNA (GenBank accession number U50543.1) by recombinant sqADAR2s using vertebrate-like (panel i) and squid-like (panel ii) solutions. Editing percentages for nt 190 were quantified for a wide range of protein concentrations (4–17,000 pm). Maximum editing percentages for sqADA2a and sqADAR2b were 92 ± 1 and 80 ± 1, respectively, using vertebrate-like solution. Maximum editing percentages for sqADA2a, sqADAR2b, and sqADAR2a EAA (□) were 64 ± 4, 29 ± 5, and 39 ± 3, respectively, using squid-like solution. Data points were fitted to a Hill equation of the form E = Emax·([ADAR]n/(Kn + [ADAR]n)) where E refers to editing percentage, Emax refers to the maximum editing percentage, and K refers to the midpoint value. n is the Hill coefficient. R2 ≥ 0.98. Error bars, S.D.; n ≥ 3.
Having shown that squid-like conditions affect the ability of ADAR2 to edit nonspecific substrates and that this effect is particularly acute for conventional isoforms, we proceeded to test whether this effect was also observed in site-selective editing of specific substrates. For this purpose, both sqADAR2 isoforms were incubated with full-length squid Kv1.1 mRNA, an mRNA that is naturally edited at 20 sites in the squid nervous system (22).5 Editing was then estimated by RT-PCR followed by direct DNA sequencing. hADAR2 was not included in this assay because it does not naturally edit this substrate. sqADAR2a was able to edit five naturally occurring sites under vertebrate-like conditions (nt 139, 190, 394, 395, and 418). For sqADAR2a under squid-like conditions or sqADAR2b under both conditions, only two sites were edited (nt 139 and 190). Fig. 1B shows the editing frequencies at nt 190, the most highly edited position for both enzymes in both conditions. This position is edited extensively by each enzyme in vertebrate-like conditions; a maximum editing of ∼90% was observed for sqADAR2a, and a maximum editing of ∼80% was observed for sqADAR2b (Fig. 1B, panel i). Squid-like conditions impaired overall site-specific editing activity although far less so for sqADAR2a than for sqADAR2b. For sqADAR2a, maximum editing was reduced from ∼90 to ∼65%, and for sqADAR2b, it was reduced from ∼80 to ∼30% (Fig. 1B, panel ii). The same trends were seen for editing at other sites (supplemental Fig. 1). The additional exon that distinguishes sqADAR2a from sqADAR2b encodes an additional dsRBD but also 16 additional amino acids. It is reasonable to speculate that the enhanced activity of sqADAR2a is due to the added binding capacity of the extra dsRBD; however, there are other possibilities. For example, dsRBDs are known to serve other roles besides RNA binding (for a review, see Ref. 25), or the extra 16 amino acids could serve an unknown function. To exclude these possibilities, we specifically mutated the RNA binding activity of the extra dsRBD. All dsRBDs are known to contain a highly conserved KKXXK motif at their interface with RNA. Mutating the lysines to EAXXA completely abolishes binding without altering protein folding (39, 40). We made the same mutations in the first dsRBD of sqADAR2a (K89E/K90A/K93A (EAA)). In squid-like conditions, sqADAR2a EAA edited nt 190 almost identically to sqADAR2b (Fig. 1B, panel ii), supporting the idea that the enhanced activity of sqADAR2a is due to the binding activity of the extra dsRBD.
High Chloride Concentrations Severely Impair ADAR Activity
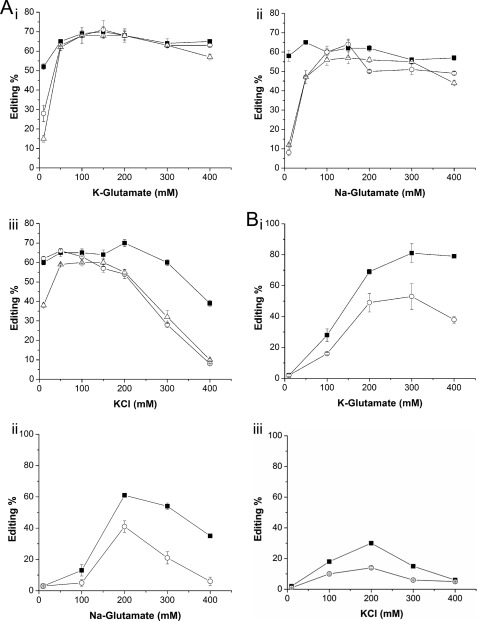
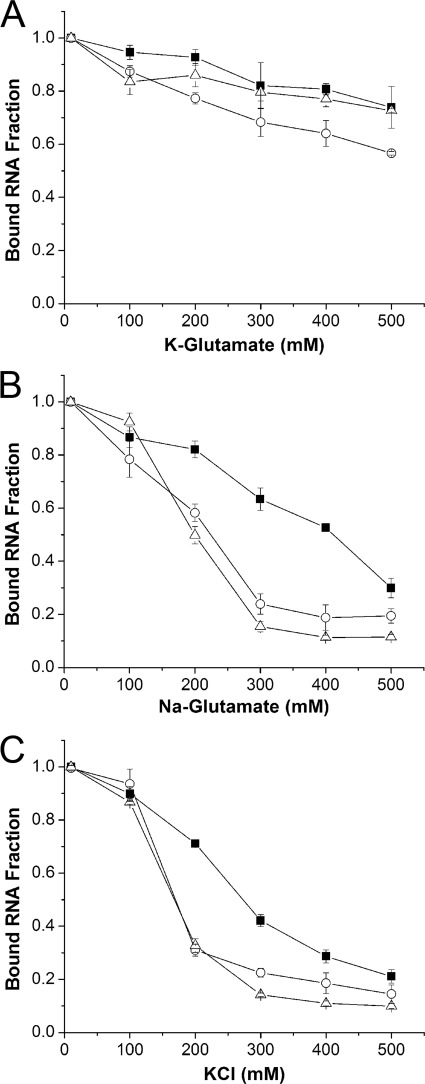
Data thus far clearly show that squid-like conditions inhibit editing. Why? Besides having a 3-fold higher total ionic strength, cells of marine osmoconformers also contain about 6- and 10-fold more Na+ and Cl−, respectively (Table 2). Therefore, we decided to test which of these factors had the greatest effect on editing by using reaction buffers that varied in the identity and concentration of the major cation and anion species. Increasing concentrations of K+ and glutamate had virtually no effect on the promiscuous editing activity of any of the ADAR2s (Fig. 2A, panel i). Similarly, high Na+ and glutamate concentrations had only slightly negative effects on ADAR2 activities, reducing their maximum editing frequencies from ∼60 to ∼50% (Fig. 2A, panel ii). By contrast, high K+ and Cl− concentrations caused a strongly negative effect on A-to-I conversion, particularly at concentrations above ∼150–200 mm (Fig. 2A, panel iii). From this, we conclude that Cl− causes the greatest disruption. Note that the additional dsRBD of sqADAR2a decreased its sensitivity to Cl−. Although the activities of sqADAR2b and human ADAR2 were almost abolished by 400 mm chloride, sqADAR2a was still able to edit up to ∼40% of adenosines. In addition, both squid ADAR2b and human ADAR2 were also highly sensitive to low ionic strengths for all ions tested (<50 mm), whereas sqADAR2a was not. Next, we decided to extend our analysis to site-selective editing at nt 190 of squid Kv1.1. As before, increasing K+ and glutamate ion concentrations from 200 to 400 mm had little effect on any ADAR2 (Fig. 2B, panel i). Unlike in the nonspecific assay, however, 400 mm Na+ impaired sqADAR2a maximal activity by ∼1.5-fold and basically abolished that of sqADAR2b (Fig. 2B, panel ii). Cl− was even more detrimental as only 300 mm was enough to decrease maximal A-to-I conversion rates for sqADAR2a by ∼2-fold and virtually eliminate those of sqADAR2b (Fig. 2B, panel iii). Finally, sqADAR2 activities were highly sensitive to low ionic strengths, but sqADAR2a was more resistant than sqADAR2b. In summary, a higher Cl− concentration explains most of the negative effects that squid-like conditions have on editing. Na+ also plays a role. K+ and glutamate have little effect. Both Cl− and Na+ have more severe effects on specific substrates than on perfect duplexes.
FIGURE 2.
Effects of K+, Na+, Cl−, and glutamate ions on promiscuous and site-selective editing activity of ADAR2s. A, promiscuous editing of a 710-bp radiolabeled perfect dsRNA duplex by recombinant ADAR2s was estimated using potassium glutamate (panel i), sodium glutamate (panel ii), or KCl (panel iii) solutions. Overall A-to-I conversion percentages were estimated for a wide range of salt concentrations (10–400 mm). B, site-selective editing of adenosine 190 in sqKv1.1 mRNA by recombinant sqADAR2s was estimated in buffers containing potassium glutamate (panel i), sodium glutamate (panel ii), or KCl (panel iii) as the major ions. Editing percentages were quantified over a wide range of salt concentrations (10–400 mm). Each assay contained 1 nm ADAR2 and 0.5 pm dsRNA. ■, sqADAR2a; ○, sqADAR2b; △, hADAR2. Error bars, S.D.; n ≥ 3.
Reduction in RNA Binding Is Consistent with Reduction in Editing Activity
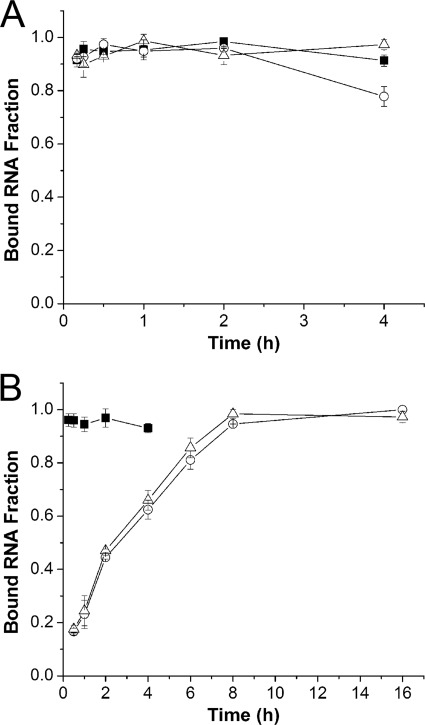
The only difference between sqADAR2a and sqADAR2b is an additional dsRBD. This feature, which confers resistance to squid-like conditions, strongly suggests that high concentrations of Cl− and Na+ disrupt the affinity of ADAR for RNA. Accordingly, we measured ADAR2-RNA binding interactions under vertebrate-like and squid-like conditions using filter binding assays. The RNA was a 710-bp 32P-labeled duplex. Because binding affinities must be measured at equilibrium, we first estimated the amount of time required for the reaction to become saturated under vertebrate-like and squid-like conditions (Fig. 3). Each ADAR2 reached equilibrium after just 15 min in the vertebrate-like solution (Fig. 3A). Although sqADAR2a reached equilibrium after a few minutes in the squid-like solution as well, the conventional ADAR2s required several hours to do so (Fig. 3B).
FIGURE 3.
Effects of vertebrate and squid ionic compositions on binding kinetics of ADAR2s. The fraction of a 710-bp radiolabeled perfect dsRNA duplex bound to different recombinant ADAR2s was tracked over time using vertebrate-like (A) and squid-like (B) solutions. Each assay contained 1 nm ADAR2 and 0.5 pm dsRNA. ■, sqADAR2a; ○, sqADAR2b; △, hADAR2. Error bars, S.D.; n = 3.
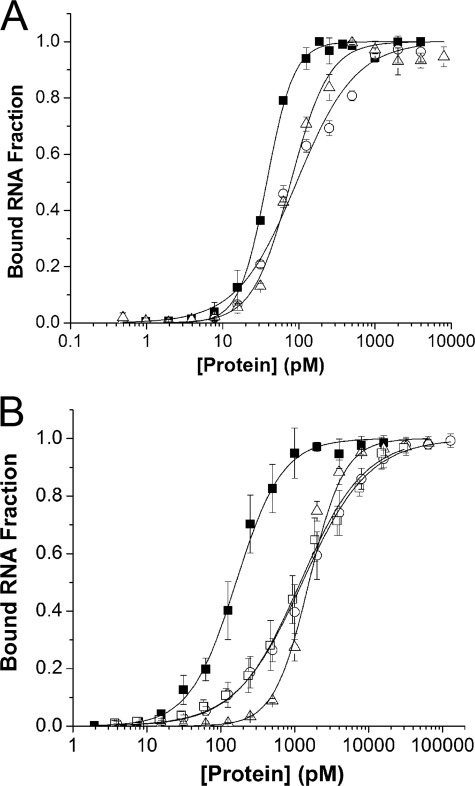
A Hill equation was used to fit ADAR2-RNA binding data and to estimate apparent dissociation constants (Fig. 4). With vertebrate-like conditions, all ADAR2s showed comparable binding. The apparent KD of sqADAR2a was ∼38 pm, which is about 2–3-fold lower than for the other two ADAR2s (Fig. 4A). Squid-like conditions increased the apparent KD for all ADAR2s but to a much greater extent for sqADAR2b and human ADAR2 than for sqADAR2a (Fig. 4B). For example, the apparent KD increased by ∼16-fold for sqADAR2b and human ADAR2 but only by 3.5-fold for sqADAR2a. Significantly, the sqADAR2a EAA mutant showed an apparent KD of 1191 ± 257 pm in squid-like conditions; this value is almost identical to that of sqADAR2b (1266 ± 316 pm). Thus, the high affinity of sqADAR2a can be directly attributable to the binding activity of the extra dsRBD. In summary, the conditions faced by marine osmoconformers severely decrease the affinity of a typical ADAR2, and this is apparent in the drastic change in binding kinetics. The extra dsRBD of sqADAR2a partially offsets this effect.
FIGURE 4.
Effects of vertebrate and squid ionic compositions on apparent affinities of ADAR2s for dsRNA. Apparent affinities of recombinant ADAR2s for the 710-bp radiolabeled perfect dsRNA duplex were estimated using vertebrate-like (A) and squid-like solutions (B). Fractions of bound RNA were quantified for a wide range of protein concentrations (0.5–8000 pm) using a fixed amount of dsRNA (0.5 pm). The apparent KD values for sqADAR2a, sqADAR2b, and hADAR2 were 38 ± 2, 93 ± 8, and 78 ± 10 pm, respectively, using the vertebrate-like solution. The apparent KD values for sqADA2a, sqADAR2b, hADAR2, and sqADAR2a EAA were 160 ± 31, 1266 ± 316, 1402 ± 104, and 1191 ± 257 pm, respectively, using the squid-like solution. ■, sqADAR2a; ○, sqADAR2b; △, hADAR2. Error bars, S.D.; n ≥ 3. Data were fit using a Hill equation of the form F = 1·([ADAR]n/((KD)n + [ADAR]n)) where F refers to the fraction of bound RNA. R2 ≥ 0.97. Average Hill coefficient values for sqADAR2a, sqADAR2b, hADAR2, and sqADAR2a AEE using vertebrate- and squid-like solutions were 1.8 ± 0.6, 1.1 ± 0.1, 2.1 ± 0.4, and 1.0 ± 0.1, respectively.
As with the editing activity assays, we also analyzed the effects that specific ions exert on RNA binding (Fig. 5). Increasing K+ and glutamate levels caused little effect on ADAR binding to RNA as it was still able to bind a substantial fraction (∼0.7–0.8) of RNA at the highest salt concentration (Fig. 5A). On the other hand, high Na+ and glutamate levels (>300 mm) inhibited the fraction of bound RNA (∼0.2), and although sqADAR2a was less sensitive, just a third of the total bound RNA fraction remained at 500 mm (Fig. 5B). As before, K+ and Cl− caused the most dramatic effect, leading to a sharp decrease of the bound RNA fraction (∼0.3) at just 200 mm for sqADAR2b and hADAR2. For sqADAR2a, a similar decrease was not observed until ∼400 mm KCl (Fig. 5C). Thus, the binding results parallel those observed for the editing activity assays, suggesting that Cl− (and Na+ to a lesser degree) inhibit editing by reducing the affinity of ADAR for RNA.
FIGURE 5.
Effects of K+, Na+, Cl−, and glutamate ions on binding of ADAR2s to dsRNA. Fractional binding of the 710-bp radiolabeled perfect dsRNA duplex to recombinant ADAR2s using potassium glutamate (A), sodium glutamate (B), and KCl (C) solutions. Fractions of bound RNA were quantified for a wide range of salt concentrations (10–500 mm). Each assay contained 1 nm ADAR2 and 0.5 pm dsRNA. ■, sqADAR2a; ○, sqADAR2b; △, hADAR2. Error bars, S.D.; n ≥ 3.
Estimates of True Binding Affinities for Different ADAR2s
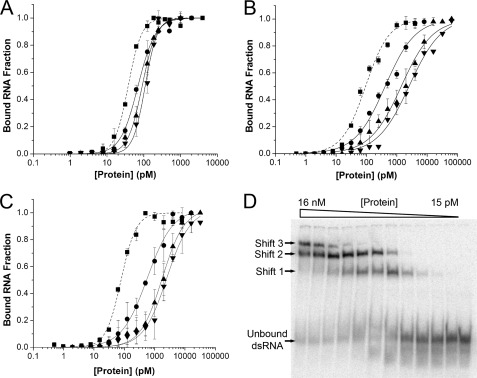
For dsRBD-containing proteins, multiple molecules have been shown to bind to a single long dsRNA molecule (37). In addition, several groups have reported that two protein-RNA complexes are formed when ADAR2 is incubated with a highly double-stranded stem-loop RNA substrate that is 80 nt long (38, 40–43). We expect that numerous ADAR2 molecules are able to bind to the 710-bp perfect duplex used for our studies thus far. Accordingly, our previous estimates of apparent affinities (Fig. 4) were artificially high. By decreasing the size of the dsRNA substrate, the binding stoichiometry should approach unity, and the apparent KD should approach the real KD. Apparent binding constants were estimated for sqADAR2a, sqADAR2b, and hADAR2 (Fig. 6, A–C) using progressively smaller dsRNA substrates (355, 165, and 79 bp) under human-like conditions. As expected, the apparent KD values for each ADAR2 tested increased with smaller substrates. Remarkably, the apparent affinities for sqADAR2b and human ADAR2 decreased by 40–45-fold by moving from the largest to the smallest substrate, whereas that of sqADAR2a decreased by only 3-fold.
FIGURE 6.
Apparent binding affinities of ADAR2s for different sized dsRNAs. Apparent binding affinities of sqADAR2a (A), sqADAR2b (B), and hADAR2 (C) were estimated for perfect dsRNA duplexes of decreasing sizes (355, 165, and 79 bp) using the vertebrate-like solution. Fractions of bound RNA were quantified for a wide range of protein concentrations (1–160,000 pm) using a fixed amount of each dsRNA substrate (1, 2, and 4 pm, respectively). The data points obtained for the 710-bp dsRNA substrate in Fig. 4 were included as a reference (filled squares and dashed lines). ■, sqADAR2a; ●, sqADAR2b; ▴, hADAR2. Error bars, S.D.; n ≥ 3. Data were fit using the same Hill equation as in Fig. 4. R2 ≥ 0.97. Average Hill coefficient values for sqADAR2a, sqADAR2b, and hADAR2 for the four substrates using the vertebrate-like solution were 2.1 ± 0.2, 0.9 ± 0.1, and 1.2 ± 0.4, respectively. D, electrophoretic mobility shift assay for recombinant sqADAR2a and a 79-bp dsRNA substrate using vertebrate-like solution. In similar assays using recombinant sqADAR2b and hADAR2, three shifts were also observed (data not shown).
Finally, we used a simple exponential model to estimate the real affinity constant for the interaction between ADAR2 and dsRNA. As a first step, it was necessary to estimate the footprint of ADAR2 on an RNA molecule. Using the smallest dsRNA substrate (79 nt), we performed electrophoretic mobility shift assays. Three protein-RNA complexes for human and squid ADAR2s were apparent (Fig. 6D). Thus, we concluded that each ADAR2 molecule binds a dsRNA fragment of ∼26 bp. For larger dsRNA substrates, if each ADAR molecule binds independently, then the apparent affinity could be modeled as a binomial probability function. However, others have suggested that ADARs dimerize, leading to possibly cooperative interactions (40, 41, 43–45). In addition, our data show that fits for the apparent affinities of sqADAR2a and hADAR2 have Hill coefficient values of ∼2, suggesting the possibility of cooperative interactions in these cases. For these reasons, we modeled our data using an exponential decay function that makes no assumptions on cooperativity. Equation 1 defines the exponential relationship between apparent (y) and real affinity constants (KD) for a given substrate size in bp (x), which depends on the number of binding sites (x/26) and a generic decay constant (D).
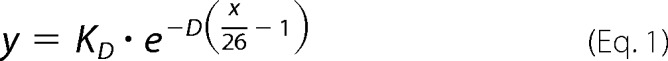 |
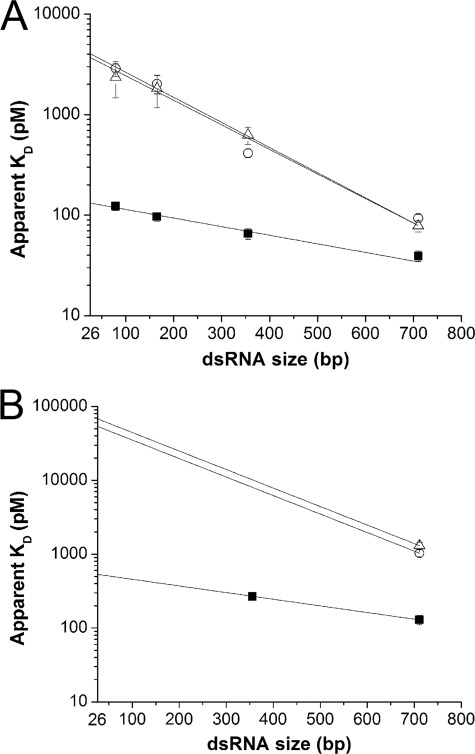
Fig. 7A shows the apparent KD for four different dsRNA substrates fit with this equation using vertebrate-like conditions. Estimates of the real KD suggest that the extra dsRBD increases binding affinity by ∼30-fold from 4065 to 132 pm. Next, we estimated the real KD using squid-like conditions. For sqADAR2b and human ADAR2, we were only able to estimate the apparent KD using the largest substrate (710 bp) because our recombinant protein purification system, which is limited by sqADAR2 self-editing activity as described previously (20), does not produce enough protein. For sqADAR2a, however, because of its higher affinity, we were able to measure the apparent affinity for two substrates (710 and 355 bp). These data showed that the slope factor for sqADAR2a was the same for vertebrate-like and squid-like conditions (∼0.05 pm/bp; Fig. 7B). If we assume the same to be true for sqADAR2b and human ADAR2, then we can estimate their true affinity constants as well based on the data using the 710-bp substrate. Using this assumption, our model estimates that the real KD for squid ADAR2a is ∼100-fold lower than that of sqADAR2b or human ADAR2 under squid-like conditions. Thus, the additional dsRBD of sqADAR2a increases binding affinity to an even larger extent in the conditions encountered within a marine osmoconformer.
FIGURE 7.
Estimates of true binding affinities of ADAR2s for dsRNA. The apparent KD values of recombinant ADAR2s for the 710-, 355-, 165-, and 79-bp radiolabeled perfect dsRNA duplexes using vertebrate-like (A) and squid-like solution (B) are shown. Data were obtained in the same or similar experiments as those shown in Fig. 6 and fit to Equation 1 (see “Results”; r2 ≥ 0.97). The intersection of this fit with the ordinate, which is positioned at 26 bp, is the estimated true KD. ■, sqADAR2a; ○, sqADAR2b; △, hADAR2. Error bars, S.D.; n ≥ 3.
DISCUSSION
Novel Squid ADAR2 Variant Has High Affinity for dsRNA
Previously, we reported that squid ADAR2a, a novel splice variant, has a higher site-specific editing activity than sqADAR2b (20). The fact that sqADAR2a has an additional dsRBD suggests that the increased deaminase activity is due to a higher affinity for dsRNA. In support of this idea, in this study, we estimated the real binding dissociation constant (KD) for both variants on dsRNA so that we could verify whether the extra dsRBD imparts a higher affinity and if so to what extent. Our measurements were complicated by the fact that multiple ADARs can bind to long dsRNA molecules. To overcome this impediment, we measured the apparent KD values of ADARs with progressively smaller dsRNA substrates and extrapolated these results to the presumable minimal substrate. This strategy revealed that the addition of an extra dsRBD to sqADAR2a confers a disproportionally large increase in affinity. For example, if all the dsRBDs were equal, we would predict that by going from two to three dsRBDs the apparent affinity would increase by ∼50%. In contrast, our results showed that the affinity increased by 30–100-fold, depending on the ionic conditions. By mutating the known binding interface of the extra dsRBD (sqADAR2a EAA), we confirmed that its binding activity directly leads to the enhanced binding of sqADAR2a.
Our data suggest that the extra dsRBD of sqADAR2a may have novel features that lead to disproportionately tighter binding. In addition, because the additional dsRBD makes squid ADAR2a less sensitive to high ionic strength, it would seem conceivable that ADARs from other marine osmoconformers may have evolved novel features to compensate for their environment. However, available genome databases show no evidence that the additional dsRBD found in squid is present in other marine invertebrates (sea urchin, limpet, and sea anemone among others). It is interesting to note that vertebrate ADAR1s also contain three dsRBDs. Whether they too exhibit disproportionally high binding is unknown.
Why Does Chloride Impair ADAR Binding?
In this study, we show that increasing Cl− dramatically impaired the affinity of ADAR2 for dsRNA, which by consequence leads to decreased overall activity. In contrast, increasing the organic anion Glu− had virtually no effect on binding or deaminase activity. Similarly, K+, the major cellular cation, had little effect on binding and editing. Finally, although Na+ decreased the binding of ADAR2 to some extent, it had minor effects on the editing activity of the enzymes. Why does Cl−, but not Glu−, K+, and Na+, have such a strong negative effect on editing? Previous studies have shown that besides the overall salt concentration cation valences and the specific composition of anions are key determinants of the stability of protein-nucleic acid interactions (46–48). The interaction surface between nucleic acid-binding proteins like ADARs and their target nucleic acids are typically large. The binding itself is mediated by long range, coulombic interactions between charged side chains in the protein and the phosphate backbone of the RNA and short range, non-coulombic interactions between the hydrocarbon surfaces. Ions tend to affect coulombic interactions generically, depending only on the valence of the ion (49). Non-coulombic interactions, however, are greatly influenced by the specific ion species. Over 100 years ago, Hofmeister (50) classified ions according to how they affect protein solubility and showed that anions have a greater influence than cations. In general, ions can be classified as chaotropes or kosmotropes. The kosmotropes are strongly hydrated, tend to increase the solvent surface tension, and have stabilizing effects on proteins that decreases their solubility. Chaotropes, on the other hand, have the opposite effects (51–53). Thus, ions can affect long range and short range interactions but in different ways.
Our data can be explained by taking both long range and short range ionic effects on binding into account. In general, we saw a gradual decrease in binding and editing as ionic strength increases. This is consistent with a disruption of long range, coulombic interactions. The negative influence of Cl− when compared with Glu−, however, may be considered a Hofmeister effect acting on short range, non-coulombic interactions. Others have shown that Glu− behaves like F−, a well known kosmotrope that is highly excluded from the hydrocarbon surfaces of proteins (52, 54, 55). In contrast, Cl− can be considered a mild chaotrope, having better access to hydrocarbon surfaces. Interestingly, in a cell, both Glu− and Cl− are highly regulated. It is worth noting that although Na+ did not affect the editing activity of ADAR2 it exerted a substantial negative effect on binding. This might be due to the fact that Na+ is slightly more chaotropic than K+ (51). Finally, we should also note that both ion composition and strength may affect the deamination reaction as well as the binding reaction.
Although we used Glu− as a representative organic anion for our studies, cells contain a more diverse array of these molecules, including phosphate, bicarbonate, and aspartate among others. Squid cytoplasm, for example, has a high amount of the isethionate ion (29). Do these anions behave as kosmotropes like Glu−, or do some of them behave as chaotropes like Cl−? Either way, the answer has interesting implications. If they act as kosmotropes, then protein-nucleic acids binding would be scarcely affected by the relatively high ionic strengths encountered within the cells of marine osmoconformers. On the other hand, if the kosmotropic effect is unique for Glu− and a few other ions, then, in the absence of compensatory mechanisms, protein-nucleic acid interactions would be severely affected in marine invertebrates. It is worth remembering that besides its function as an amino acid, Glu− also serves as a neurotransmitter. Perhaps its stabilizing effects on protein-nucleic acid binding is another reason that animals go to great lengths to regulate its concentration.
High Level Editing in Squid
This work supports our previous study suggesting that the extra dsRBD of sqADAR2 allows the enzyme to edit more sites more efficiently despite the marine environment. However, we are still only able to reproduce a subset of the naturally occurring editing sites in vivo. For example, in the mRNA that encodes sqKv1.1A, there are 20 naturally occurring sites. Based on direct sequences of RT-PCR products, only five of these could be recapitulated in vitro using recombinant sqADAR2a. An inability to edit the other sites may be because we have not yet identified all the relevant secondary structures in the mRNA that drive the editing reaction. On the other hand, some of them may only be edited by edited isoforms of sqADAR2 (20) or by different ADAR paralogs. Interestingly, we recently reported an ADAR1 ortholog in squid (56) that may be responsible for expanding the repertoire of editing sites even further. Accordingly, we believe that multiple factors underlie high level editing in squid.
Supplementary Material
Acknowledgments
We thank Dr. Record for useful discussions about the interactions between ions and protein-nucleic acid macromolecules. We also thank Sonia Soto for technical support. Part of this work was conducted in the Molecular Biology Core Facility at the Institute of Neurobiology, which is supported by Research Centers in Minority Institutions Grant G12 RR 03051.
This work was supported, in whole or in part, by National Institutes of Health Grants 2 U54 NS039405-06 and R01 NS064259. This work was also supported by National Science Foundation Grant IBN-0344070.

This article contains supplemental Fig. 1.
J. P. Palavicini, R. A. Correa-Rojas, and J. J. C. Rosenthal, unpublished data.
C. Colina-Prisco, J. P. Palavicini, and J. J. C. Rosenthal, unpublished data.
- ADAR
- adenosine deaminase that acts on RNA
- dsRNA
- double-stranded RNA
- dsRBD
- dsRNA binding domain
- sqADAR2
- squid ADAR2
- hADAR2
- human ADAR2
- nt
- nucleotide(s)
- EAA
- K89E/K90A/K93A
- Glu−
- glutamate
- sqKv1.1
- squid Kv1.1.
REFERENCES
- 1. Bass B. L., Nishikura K., Keller W., Seeburg P. H., Emeson R. B., O'Connell M. A., Samuel C. E., Herbert A. (1997) A standardized nomenclature for adenosine deaminases that act on RNA. RNA 3, 947–949 [PMC free article] [PubMed] [Google Scholar]
- 2. Kumar M., Carmichael G. G. (1997) Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc. Natl. Acad. Sci. U.S.A. 94, 3542–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Athanasiadis A., Rich A., Maas S. (2004) Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2, e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morse D. P., Bass B. L. (1999) Long RNA hairpins that contain inosine are present in Caenorhabditis elegans poly(A)+ RNA. Proc. Natl. Acad. Sci. U.S.A. 96, 6048–6053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morse D. P., Aruscavage P. J., Bass B. L. (2002) RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl. Acad. Sci. U.S.A. 99, 7906–7911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Basilio C., Wahba A. J., Lengyel P., Speyer J. F., Ochoa S. (1962) Synthetic polynucleotides and the amino acid code. V. Proc. Natl. Acad. Sci. U.S.A. 48, 613–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keegan L. P., Gallo A., O'Connell M. A. (2001) The many roles of an RNA editor. Nat. Rev. Genet. 2, 869–878 [DOI] [PubMed] [Google Scholar]
- 8. Bhalla T., Rosenthal J. J., Holmgren M., Reenan R. (2004) Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat. Struct. Mol. Biol. 11, 950–956 [DOI] [PubMed] [Google Scholar]
- 9. Burns C. M., Chu H., Rueter S. M., Hutchinson L. K., Canton H., Sanders-Bush E., Emeson R. B. (1997) Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387, 303–308 [DOI] [PubMed] [Google Scholar]
- 10. Köhler M., Burnashev N., Sakmann B., Seeburg P. H. (1993) Determinants of Ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: diversity by RNA editing. Neuron 10, 491–500 [DOI] [PubMed] [Google Scholar]
- 11. Clutterbuck D. R., Leroy A., O'Connell M. A., Semple C. A. (2005) A bioinformatic screen for novel A-I RNA editing sites reveals recoding editing in BC10. Bioinformatics 21, 2590–2595 [DOI] [PubMed] [Google Scholar]
- 12. Gommans W. M., Tatalias N. E., Sie C. P., Dupuis D., Vendetti N., Smith L., Kaushal R., Maas S. (2008) Screening of human SNP database identifies recoding sites of A-to-I RNA editing. RNA 14, 2074–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Levanon E. Y., Hallegger M., Kinar Y., Shemesh R., Djinovic-Carugo K., Rechavi G., Jantsch M. F., Eisenberg E. (2005) Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 33, 1162–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li J. B., Levanon E. Y., Yoon J. K., Aach J., Xie B., Leproust E., Zhang K., Gao Y., Church G. M. (2009) Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324, 1210–1213 [DOI] [PubMed] [Google Scholar]
- 15. Ohlson J., Ensterö M., Sjöberg B. M., Ohman M. (2005) A method to find tissue-specific novel sites of selective adenosine deamination. Nucleic Acids Res. 33, e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., Yang L., Artieri C. G., van Baren M. J., Boley N., Booth B. W., Brown J. B., Cherbas L., Davis C. A., Dobin A., Li R., Lin W., Malone J. H., Mattiuzzo N. R., Miller D., Sturgill D., Tuch B. B., Zaleski C., Zhang D., Blanchette M., Dudoit S., Eads B., Green R. E., Hammonds A., Jiang L., Kapranov P., Langton L., Perrimon N., Sandler J. E., Wan K. H., Willingham A., Zhang Y., Zou Y., Andrews J., Bickel P. J., Brenner S. E., Brent M. R., Cherbas P., Gingeras T. R., Hoskins R. A., Kaufman T. C., Oliver B., Celniker S. E. (2011) The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoopengardner B., Bhalla T., Staber C., Reenan R. (2003) Nervous system targets of RNA editing identified by comparative genomics. Science 301, 832–836 [DOI] [PubMed] [Google Scholar]
- 18. Stapleton M., Carlson J. W., Celniker S. E. (2006) RNA editing in Drosophila melanogaster: new targets and functional consequences. RNA 12, 1922–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xia S., Yang J., Su Y., Qian J., Ma E., Haddad G. G. (2005) Identification of new targets of Drosophila pre-mRNA adenosine deaminase. Physiol. Genomics 20, 195–202 [DOI] [PubMed] [Google Scholar]
- 20. Palavicini J. P., O'Connell M. A., Rosenthal J. J. (2009) An extra double-stranded RNA binding domain confers high activity to a squid RNA editing enzyme. RNA 15, 1208–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patton D. E., Silva T., Bezanilla F. (1997) RNA editing generates a diverse array of transcripts encoding squid Kv2 K+ channels with altered functional properties. Neuron 19, 711–722 [DOI] [PubMed] [Google Scholar]
- 22. Rosenthal J. J., Bezanilla F. (2002) Extensive editing of mRNAs for the squid delayed rectifier K+ channel regulates subunit tetramerization. Neuron 34, 743–757 [DOI] [PubMed] [Google Scholar]
- 23. Colina C., Palavicini J. P., Srikumar D., Holmgren M., Rosenthal J. J. (2010) Regulation of Na+/K+ ATPase transport velocity by RNA editing. PLoS Biol. 8, e1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang Y., Lv J., Gui B., Yin H., Wu X., Zhang Y., Jin Y. (2008) A-to-I RNA editing alters less-conserved residues of highly conserved coding regions: implications for dual functions in evolution. RNA 14, 1516–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Doyle M., Jantsch M. F. (2002) New and old roles of the double-stranded RNA-binding domain. J. Struct. Biol. 140, 147–153 [DOI] [PubMed] [Google Scholar]
- 26. St Johnston D., Brown N. H., Gall J. G., Jantsch M. (1992) A conserved double-stranded RNA-binding domain. Proc. Natl. Acad. Sci. U.S.A. 89, 10979–10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. (2002) Molecular Biology of the Cell, 4th Ed., pp. 651–652, Garland Science, New York [Google Scholar]
- 28. Keynes R. D. (1963) Chloride in the squid giant axon. J. Physiol. 169, 690–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koechlin B. A. (1955) On the chemical composition of the axoplasm of squid giant nerve fibers with particular reference to its ion pattern. J. Biophys. Biochem. Cytol. 1, 511–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Villegas J., Villegas L., Villegas R. (1965) Sodium, potassium, and chloride concentrations in the Schwann cell and axon of the squid nerve fiber. J. Gen. Physiol. 49, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bass B. L., Weintraub H. (1988) An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55, 1089–1098 [DOI] [PubMed] [Google Scholar]
- 32. Melcher T., Maas S., Higuchi M., Keller W., Seeburg P. H. (1995) Editing of α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR-B pre-mRNA in vitro reveals site-selective adenosine to inosine conversion. J. Biol. Chem. 270, 8566–8570 [DOI] [PubMed] [Google Scholar]
- 33. O'Connell M. A., Keller W. (1994) Purification and properties of double-stranded RNA-specific adenosine deaminase from calf thymus. Proc. Natl. Acad. Sci. U.S.A. 91, 10596–10600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wagner R. W., Smith J. E., Cooperman B. S., Nishikura K. (1989) A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl. Acad. Sci. U.S.A. 86, 2647–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim U., Wang Y., Sanford T., Zeng Y., Nishikura K. (1994) Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl. Acad. Sci. U.S.A. 91, 11457–11461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lai F., Drakas R., Nishikura K. (1995) Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J. Biol. Chem. 270, 17098–17105 [DOI] [PubMed] [Google Scholar]
- 37. Bass B. L., Hurst S. R., Singer J. D. (1994) Binding properties of newly identified Xenopus proteins containing dsRNA-binding motifs. Curr. Biol. 4, 301–314 [DOI] [PubMed] [Google Scholar]
- 38. Ohman M., Källman A. M., Bass B. L. (2000) In vitro analysis of the binding of ADAR2 to the pre-mRNA encoding the GluR-B R/G site. RNA 6, 687–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ramos A., Grünert S., Adams J., Micklem D. R., Proctor M. R., Freund S., Bycroft M., St Johnston D., Varani G. (2000) RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 19, 997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Valente L., Nishikura K. (2007) RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J. Biol. Chem. 282, 16054–16061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cho D. S., Yang W., Lee J. T., Shiekhattar R., Murray J. M., Nishikura K. (2003) Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J. Biol. Chem. 278, 17093–17102 [DOI] [PubMed] [Google Scholar]
- 42. Jaikaran D. C., Collins C. H., MacMillan A. M. (2002) Adenosine to inosine editing by ADAR2 requires formation of a ternary complex on the GluR-B R/G site. J. Biol. Chem. 277, 37624–37629 [DOI] [PubMed] [Google Scholar]
- 43. Gallo A., Keegan L. P., Ring G. M., O'Connell M. A. (2003) An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. EMBO J. 22, 3421–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chilibeck K. A., Wu T., Liang C., Schellenberg M. J., Gesner E. M., Lynch J. M., MacMillan A. M. (2006) FRET analysis of in vivo dimerization by RNA-editing enzymes. J. Biol. Chem. 281, 16530–16535 [DOI] [PubMed] [Google Scholar]
- 45. Poulsen H., Jorgensen R., Heding A., Nielsen F. C., Bonven B., Egebjerg J. (2006) Dimerization of ADAR2 is mediated by the double-stranded RNA binding domain. RNA 12, 1350–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ha J. H., Capp M. W., Hohenwalter M. D., Baskerville M., Record M. T., Jr. (1992) Thermodynamic stoichiometries of participation of water, cations and anions in specific and non-specific binding of lac repressor to DNA. Possible thermodynamic origins of the “glutamate effect” on protein-DNA interactions. J. Mol. Biol. 228, 252–264 [DOI] [PubMed] [Google Scholar]
- 47. Kowalczykowski S. C., Lonberg N., Newport J. W., von Hippel P. H. (1981) Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. I. Characterization of the binding interactions. J. Mol. Biol. 145, 75–104 [DOI] [PubMed] [Google Scholar]
- 48. Overman L. B., Bujalowski W., Lohman T. M. (1988) Equilibrium binding of Escherichia coli single-strand binding protein to single-stranded nucleic acids in the (SSB)65 binding mode. Cation and anion effects and polynucleotide specificity. Biochemistry 27, 456–471 [DOI] [PubMed] [Google Scholar]
- 49. Pegram L. M., Record M. T., Jr. (2008) Thermodynamic origin of Hofmeister ion effects. J. Phys. Chem. B 112, 9428–9436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hofmeister F. (1888) Zur Lehre von der Wirkung der Salze. Zweite Mittheilung. Arch. Exp. Pathol. Pharmakol. 24, 247–260 [Google Scholar]
- 51. Baldwin R. L. (1996) How Hofmeister ion interactions affect protein stability. Biophys. J. 71, 2056–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vander Meulen K. A., Saecker R. M., Record M. T., Jr. (2008) Formation of a wrapped DNA-protein interface: experimental characterization and analysis of the large contributions of ions and water to the thermodynamics of binding IHF to H′ DNA. J. Mol. Biol. 377, 9–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang Y., Cremer P. S. (2006) Interactions between macromolecules and ions: the Hofmeister series. Curr. Opin. Chem. Biol. 10, 658–663 [DOI] [PubMed] [Google Scholar]
- 54. Kontur W. S., Capp M. W., Gries T. J., Saecker R. M., Record M. T., Jr. (2010) Probing DNA binding, DNA opening, and assembly of a downstream clamp/jaw in Escherichia coli RNA polymerase-λP(R) promoter complexes using salt and the physiological anion glutamate. Biochemistry 49, 4361–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saum S. H., Müller V. (2008) Regulation of osmoadaptation in the moderate halophile Halobacillus halophilus: chloride, glutamate and switching osmolyte strategies. Saline Systems 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Keegan L. P., McGurk L., Palavicini J. P., Brindle J., Paro S., Li X., Rosenthal J. J., O'Connell M. A. (2011) Functional conservation in human and Drosophila of metazoan ADAR2 involved in RNA editing: loss of ADAR1 in insects. Nucleic Acids Res. 39, 7249–7262 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.