Background: P450 reductase (POR)-dependent enzymes metabolize numerous endogenous compounds.
Results: Enterocytes of intestinal Por knock-out mice show up-regulation of genes important for immunity and increased geranylgeranyl pyrophosphate levels.
Conclusion: Enterocyte POR-dependent enzymes modulate intestinal expression of major histocompatibility complex class II genes, possibly through intermediates in cholesterol biosynthesis.
Significance: This appears to be the first evidence for a link between POR-dependent enzymes and intestinal immunity.
Keywords: Cholesterol Metabolism, Cytochrome P450, Intestine, Isoprenoid, Major Histocompatibility Complex (MHC), P450 Reductase, Heme
Abstract
NADPH-cytochrome P450 reductase (POR) is essential for the functioning of microsomal cytochrome P450 (P450) monooxygenases and heme oxygenases. The biological roles of the POR-dependent enzymes in the intestine have not been defined, despite the wealth of knowledge on the biochemical properties of the various oxygenases. In this study, cDNA microarray analysis revealed significant changes in gene expression in enterocytes isolated from the small intestine of intestinal epithelium-specific Por knock-out (named IE-Cpr-null) mice compared with that observed in wild-type (WT) littermates. Gene ontology analyses revealed significant changes in terms related to P450s, transporters, cholesterol biosynthesis, and, unexpectedly, antigen presentation/processing. The genomic changes were confirmed at either mRNA or protein level for selected genes, including those of the major histocompatibility complex class II (MHC II). Cholesterol biosynthetic activity was greatly reduced in the enterocytes of the IE-Cpr-null mice, as evidenced by the accumulation of the lanosterol metabolite, 24-dihydrolanosterol. However, no differences in either circulating or enterocyte cholesterol levels were observed between IE-Cpr-null and WT mice. Interestingly, the levels of the cholesterol precursor farnesyl pyrophosphate and its derivative geranylgeranyl pyrophosphate were also increased in the enterocytes of the IE-Cpr-null mice. Furthermore, the expression of STAT1 (signal transducer and activator of transcription 1), a downstream target of geranylgeranyl pyrophosphate signaling, was enhanced. STAT1 is an activator of CIITA, the class II transactivator for MHC II expression; CIITA expression was concomitantly increased in IE-Cpr-null mice. Overall, these findings provide a novel and mechanistic link between POR-dependent enzymes and the expression of MHC II genes in the small intestine.
Introduction
NADPH-cytochrome P450 reductase (CPR2 or POR (P450 oxidoreductase) is a microsomal flavoprotein that serves as an electron donor for multiple enzymes, including microsomal cytochrome P450s (P450 or CYP) (1), heme oxygenases (HOs) (2), and squalene epoxidase (SQLE) (3). POR-dependent enzymes play important roles in the homeostasis of many endogenous compounds, such as bile acids, cholesterol, heme, steroids, and fatty acids. Furthermore, POR-dependent enzymes are essential for fetal development (4). In humans, mutations in the POR gene result in disordered steroidogenesis and the Antley-Bixler syndrome (5).
To circumvent the embryonic lethality of germ line Por knock-out mice, mouse models employing conditional Por gene knock-out have been developed and used for the investigation of the organ-specific functions of POR-dependent enzymes. For example, two liver-specific Por knock-out mouse models have been developed, and, despite the induction of numerous hepatic P450s, both exhibited impaired drug metabolism, decreased serum cholesterol, and enlarged and fatty livers (6, 7). A whole-body Cpr-low mouse was also developed, in which POR levels are reduced by >70% in all tissues examined (8). This latter model was found to have partial embryonic lethality, altered steroid hormone homeostasis, and infertility in females, in addition to impairments in drug metabolism and decreases in serum cholesterol. In both the liver-Cpr-null and the Cpr-low mice, extensive changes in global gene expression were observed in the livers, which revealed the importance of hepatic POR-dependent enzymes in the homeostasis of fatty acids and other lipid metabolites, as well as in the regulation of various metabolic enzymes and transporters (9, 10).
An intestinal epithelium (IE)-specific Por knock-out mouse (named IE-Cpr-null), in which the Por gene is specifically deleted in the enterocytes, was recently obtained (11). The IE-Cpr-null mice do not display any obvious abnormalities in growth, development, or reproduction, and their intestines appear to have normal structure. However, targeted gene expression analysis showed compensatory increases in the expression of several P450 enzymes in the small intestine (SI) (11). Further pharmacological studies revealed deficiencies of the IE-Cpr-null mice in the first-pass metabolism of oral drugs and dietary contaminants (11–13). Given the known ability of various POR-dependent enzymes to metabolize endogenous compounds, the metabolic disturbances detected in the livers of the liver-Cpr-null mice, and the unique chemical environment of the intestinal enterocytes as the portal of entry for lipid molecules entering from either the diet or the bile, we hypothesize that loss of the enterocyte POR expression will impact the homeostasis of endogenous compounds and the expression of genes that have critical biological functions in the SI. Conceivably, the nature of the biochemical consequences of the Por deletion in the intestine may dictate potential functional deficits in the responses to various environmental challenges, including pathogenic infection, in the IE-Cpr-null mice and potentially in people with POR mutations that cause reduced POR expression.
In the present study, we performed comparative analyses of global gene expression in enterocytes from wild-type (WT) and IE-Cpr-null mice using the Affymetrix Mouse Expression Set 430A 2.0 GeneChip arrays. Groups of genes exhibiting differing expression levels between the IE-Cpr-null and WT mice were identified. Subsequent pathway analysis, conducted using Gene Map Annotator and Pathway Profiler (available from the GenMAPP Web site), led to the identification of “antigen presentation/processing” among the gene ontology (GO) terms that contained the most significantly changed gene expression. A detailed analysis of gene expression changes related to antigen processing and presentation led us to propose a mechanistic scheme to explain the unexpected increase in genes related to this pathway in the intestine. Additional studies of the levels of relevant endogenous metabolites (including cholesterol, 24-dihydrolanosterol (24-DHL), farnesyl pyrophosphate (FPP), geranylgeranyl pyrophosphate (GGPP), heme, and bilirubin) and signaling proteins (including STAT1 and CIITA (class II major histocompatibility complex transactivator)) provided evidence in support of a novel link between POR-dependent enzymes and expression of the major histocompatibility complex class II (MHC II) genes, which are important for intestinal immunity.
EXPERIMENTAL PROCEDURES
Animals
Adult (2.5–4.0-month-old) male IE-Cpr-null mice and WT littermates were used in all experiments. Protocols for breeding and genotyping were as reported previously (11). Animals were maintained at 22 °C with a 12-h on, 12-h off light cycle and were allowed free access to water and a standard laboratory diet. Animal use protocols were approved by the Institutional Animal Care and Use Committee of the Wadsworth Center.
RNA Preparation and Microarray Hybridization
Tissues from IE-Cpr-null and WT mice (8 for each group) were collected for RNA preparation. SI was processed immediately after dissection for enterocyte isolation and RNA preparation, essentially as described (14), except that, following removal of lumenal content, the SI was placed in PBS, pH 7.2, containing 1.5 mm EDTA, 3 units/ml heparin, and 0.5 mm dithiothreitol for 10 min, in order to loosen the mucosa before the epithelial cells were collected by scraping. The harvested cells were placed directly into TRIzol (Invitrogen). Total RNA was prepared from enterocytes of individual mice, using TRIzol; the samples were further purified using RNeasy minicolumns (Qiagen, Valencia, CA). The integrity and purity of the RNA preparations were determined using a RNA 6000 Nano Assay kit on a model 2100 Bioanalyzer (Agilent, Santa Clara, CA). The average of RNA integrity number (RIN) values was 8.2 ± 0.5.
Mouse Expression Set 430A 2.0 GeneChip arrays (Affymetrix, Santa Clara, CA) were used for microarray analyses. Each array contained 22,690 probe sets, representing ∼14,870 distinct genes. Each probe set consisted of 11 pairs of 25-mer oligonucleotides. Four RNA samples, each prepared by pooling equal amounts of enterocyte RNA from two mice of the same genotype, were analyzed for each group (WT or IE-Cpr-null). Synthesis of biotinylated antisense RNA, array hybridization, staining, washing, and collection of expression data were performed within the Microarray Core Facility of the Wadsworth Center, as described previously (9).
Microarray Gene Expression Data Analysis
The experimental data sets were normalized using the Guanine Cytosine Robust Multichip Analysis (for fold change analysis) or Microarray Analysis Suite 5.0 (for absent/present calls) programs of the Genespring 10.0 software package (Agilent). The data for WT mice were used as the base line. Probe sets with raw intensity below the 20th percentile in all eight samples were eliminated, leaving 18,443 probe sets for further analysis. Analysis for significance was performed using the unpaired t test in Genespring 10.0. The ratios of averaged values for each group were used to calculate fold change between two groups. Genes with significantly changed expression were tabulated, along with gene symbol, gene name, transcript identification number, and fold change values, and the data were further examined for reproducibility among multiple probe sets for a given gene, where available. Two programs, MAPPFinder (15) and GenMAPP 2.0 (16), were used to group genes having significantly changed expression according to the GO hierarchy at the level of biological processes, cellular components, and molecular functions as described previously (9).
Determination of Cholesterol Levels in Plasma and Enterocytes
The levels of total cholesterol in plasma and enterocytes were determined using a cholesterol assay kit (including esterase for hydrolysis; Cayman, Ann Arbor, MI) according to the manufacturer's instructions. For enterocytes, cholesterol was extracted prior to analysis, based on the method of Folch et al. (17), with modifications. Briefly, enterocytes were isolated as described above for RNA preparation. PBS-washed enterocyte cell pellets (∼100–300 mg wet weight) were homogenized in an extraction solution (methanol/Triton X-100/water, 98.3/1.1/0.6 (v/v/v)) in a volume equivalent to ∼10 times the cell weight. The homogenate was further extracted, twice, with chloroform. The combined organic phase from chloroform extraction was dried under nitrogen, and the residue was dissolved in the cholesterol reaction buffer from the cholesterol assay kit prior to analysis.
Extraction and GC/MS Analysis of 24-DHL
PBS-washed enterocyte cell pellets (∼40 mg wet weight) were homogenized in 1 ml of water, and 5 μg of cholestane was added per sample as an internal standard. Extraction and derivatization was performed as described by Li and Porter (18), except that the samples were dried under a gentle stream of nitrogen gas rather than by centrifugal evaporation. The trimethylsilyl derivative of 24-DHL (24-DHL-TMS) was prepared by reacting the dried extracts with N,O-bis(trimethylsilyl)trifluoroacetamide containing 1% (v/v) trimethylchlorosilane (Thermo Fisher Scientific, Rockford, IL) at 60 °C for 30 min. GC/MS analyses were performed using an Agilent 7890A GC system with an Agilent HP-5ms 30 m × 0.25-mm (0.25-μm film thickness) column interfaced with an Agilent 5975C Inert XL EI/CI MSD equipped with a triple axis detector. Samples (1 μl) were injected in splitless mode with an injector temperature of 260 °C. The oven temperature was initially at 100 °C for 5 min and was then increased at 20 °C/min to 200 °C, held at 200 °C for 10 min, and increased again at 5 °C/min to 300 °C, which was followed by a final hold at 300 °C for 8 min. The carrier gas was helium, flowing at 1 ml/min. Electron-ionization mass spectra were recorded at 70 eV over a range of m/z 40–640, with an ion source temperature of 226 °C. Authentic 24-DHL (Steraloids, Newport, RI) was used as the standard.
Analysis of FPP and GGPP by HPLC with fluorescence Detection
Extraction of FPP and GGPP from mouse enterocytes was based on the method of Tong et al. (19) with modifications. PBS-washed enterocytes (∼200–400 mg wet weight) were homogenized in 2 ml of an ice-cold extraction solvent (75% ethanol, 0.5% aqueous NH4OH, 3:1) containing 100 μl of PhosStop phosphatase inhibitor (Roche Applied Science). The homogenates were heated at 70 °C for 15 min, vortexed for 2 min, and centrifuged at 1500 × g for 10 min. The supernatants were saved, and the pellets were re-extracted with an additional 2 ml of ice-cold extraction solvent. The two supernatant fractions were combined and were extracted twice with 3-ml portions of hexane. The aqueous layers were combined with 17 ml of 50 mm NH4HCO3. Five-ml portions of these samples were fractionated on 200-mg C18 BondElute SPE columns (Agilent). The columns were washed with 2-ml portions of 50 mm NH4HCO3, followed by 2 ml of 20% methanol, 50 mm NH4HCO3. FPP and GGPP were eluted in 2 ml of 75% methanol, 0.5% NH4OH. The sample eluates were dried at 50 °C under nitrogen. The residue was dissolved in 40 μl of 50 mm Tris-HCl (pH 7.5), containing 5 mm dithiothreitol, 5 mm MgCl2, 10 μm ZnCl2, and 1.0% octyl-β-d-glucopyranoside. Four μl of 125 μm dansyl-Gly-Cys-Val-Leu-Ser (Biosynthesis, Lewisville, TX), 4 μl of 125 μm dansyl-Gly-Cys-Val-Leu-Leu (Biosynthesis), 2.5 μl of 100 ng/μl farnesyltransferase (Jena Bioscience, Jena, Germany), and 2.5 μl of 100 ng/μl geranylgeranyltransferase I (Jena Bioscience) were then added to the reconstituted extracts, and the mixtures were incubated at 37 °C for 2 h in the dark. The derivatization reaction was terminated by the addition of 50 μl of acetonitrile and 5 μl of 10% HCl. The mixture was centrifuged (1500 × g), and 90 μl of the supernatant was injected for HPLC analysis.
HPLC analysis was carried out on an Agilent 1100 system with a model G1321A fluorescence detector. The samples were subjected to chromatography on a Luna C18 250 × 3.0-mm (5-μm particle size) column (Phenomenex, Torrance, CA). The chromatographic conditions were those described by Tong et al. (19). FPP and GGPP were monitored by detection of fluorescence at 528 nm, elicited by excitation at 335 nm. The amounts of FPP and GGPP were determined using authentic standards (Sigma-Aldrich).
Determination of Heme and Bilirubin Contents in Plasma and Enterocytes
Heme content was measured using a QuantiChrom heme assay kit (BioAssay Systems). Plasma and enterocyte homogenates, prepared as described by Mingone et al. (20), were used in the assay at a concentration of ∼5 mg of protein/ml. For bilirubin determination, PBS-washed enterocytes (∼400–500 mg wet weight) were homogenized in 0.5 ml of deionized water containing 1 mg of butylated hydroxytoluene, and bilirubin was extracted as described (21). Mesobilirubin was used as an internal standard. All steps were performed under dim light. Bilirubin analysis was carried out using an Agilent 1100 HPLC system with a diode array detector (set to monitor 405 nm) and a Vydac C8 column (250 mm × 4.6-mm inner diameter, 5 μm) (Discovery Sciences, Deerfield, IL), using a solvent system consisting of solvents A (10 mm ammonium acetate) and B (methanol/100 mm ammonium acetate, 90:10, v/v). A 20-min linear gradient from 60% B to 80% B was applied at a flow rate of 0.5 ml/min for sample elution; the column was maintained at 40 °C. Authentic bilirubin (Sigma) was used as the standard.
Real-time RNA-PCR
Total RNA was isolated from enterocytes with TRIzol as described under “RNA Preparation and Microarray Hybridization.” Real-time RNA-PCRs were performed according to a general protocol described previously (22), with minor modifications. A full list of the primers used and the optimal annealing temperatures is included in supplemental Table 1. All primers were used at 0.1 μm, except for GAPDH, of which primers were used at 0.4 μm. At the end of the PCR cycles, melting curve analysis was performed to assess the purity of the PCR products. The levels of target gene mRNAs in various total RNA preparations were normalized by the level of GAPDH mRNA in a given sample. All reactions were performed in duplicates. Negative control reactions (no template) were routinely included. Identities of PCR products were confirmed by electrophoretic analysis on agarose gels.
Immunoblot Analyses
The basic procedures used for immunoblot analysis were as described (22). The following were used as the primary antibodies: a rat monoclonal antibody to the polymorphic determinant shared by multiple mouse MHC II alloantigens (BD Biosciences, San Jose, CA), a rabbit monoclonal antibody to human STAT1 (42H3, Cell Signaling Technology, Danvers, MA), a rabbit polyclonal antibody to human phospho-Ser-727-STAT1 (Cell Signaling Technology), a rabbit monoclonal antibody to human β-actin (13E5, Cell Signaling Technology), and a goat polyclonal antibody to rabbit GAPDH (GenScript, Piscataway, NJ). Enterocytes were isolated using the same protocol as described above for RNA preparation, except for immunoblot analysis of STAT1 expression, in which the enterocytes were isolated using an elution method described by Ware et al. (23). Whole-cell lysates were prepared using radioimmune precipitation assay buffer (Thermo Fisher Scientific) according to the manufacturer's instructions. Protein concentrations were determined by using the bicinchoninic acid method (Fisher) with bovine serum albumin as the standard. For immunoblot quantitation, a model GS-710 calibrated imaging densitometer (Bio-Rad) was used.
RESULTS
Microarray Analysis
By using relatively conservative criteria in fold change (≥1.75 or ≤0.57) and a p value of ≤0.01, we identified a number of mouse SI genes that were differentially expressed in the enterocytes of the IE-Cpr-null mice, compared with their expression levels in WT mice (34 up-regulated and 17 down-regulated) (Table 1). Genes showing significant differences in expression changes were grouped into functional categories based on GenMAPP, UniProt (24), and additional literature searches. The categories included biotransformation, lipid metabolism, transporters, growth factors, and antigen processing and presentation. Notably, numerous other genes in these and other categories were also found to have altered expression, albeit with smaller fold change or greater p values. For example, although many Cyp genes appeared to be up-regulated in the IE-Cpr-null mice, only Cyp1a1, Cyp1a2, and Cyp51 met the criteria for inclusion in Table 1. In this regard, 27 of the 68 unique mouse P450 genes represented (by 83 probe sets) on the GeneChip were detected in both IE-Cpr-null and WT mice (supplemental Table 2).
TABLE 1.
Genes that were differentially expressed in enterocytes of WT and IE-Cpr-null mice
Genes with significantly different expression (p ≤ 0.01), and with at least a 75% difference between the IE-Cpr-null and WT mice (i.e. with fold change ≥1.75 or ≤0.57), in at least one probe set, are shown. For genes represented by multiple probe sets, the results for all probe sets are included, although all probe sets may not meet the selection criteria. For each entry, a reference sequence transcript identification number (RefSeq transcript ID) is given, along with the gene symbol and gene name (according to Affymetrix). The genes selected are grouped according to functional categories (defined in GenMAPP or UniProt or through a literature search).
| Gene symbol | Ref Seq transcript ID | Fold change (IE-Cpr-null/WT) | Gene name |
|---|---|---|---|
| Biotransformation | |||
| Adh4 | NM_011996 | 0.53 | Alcohol dehydrogenase 4 |
| Akr1c14 | NM_134072 | 0.47 | Aldo-keto reductase family 1, member C14 |
| Cyp1a1 | NM_001136059 | 3.8 | Cytochrome P450 1a1 |
| Cyp1a2 | NM_009993 | 44 | Cytochrome P450 1a2 |
| Cyp51 | NM_020010 | 2.4/1.9/1.0 | Cytochrome P450 51 |
| Por | NM_008898 | 0.56 | Cytochrome P450 oxidoreductase |
| Lipid metabolism | |||
| Acer1 | NM_175731 | 2.4 | Alkaline ceramidase 1 |
| Fdft1 | NM_010191 | 1.9/1.7 | Farnesyl diphosphate farnesyl transferase 1 |
| Idi1 | NM_145360 | 2.0/1.5 | Isopentenyl-diphosphate δ isomerase |
| Lss | NM_146006 | 2.0a/1.8/1.0a | Lanosterol synthase |
| Mvd | NM_138656 | 2.7/2.4 | Mevalonate (diphospho) decarboxylase |
| Pla2g7 | NM_013737 | 0.24 | Phospholipase A2, group VII |
| Pmvk | NM_026784 | 1.9 | Phosphomevalonate kinase |
| Pnpla7 | NM_146251 | 2.4 | Patatin-like phospholipase domain containing 7 |
| Scd2 | NM_009128 | 7.1/3.4 | Stearoyl-coenzyme A desaturase 2 |
| Sqle | NM_009270 | 2.5 | Squalene epoxidase |
| Growth factors | |||
| Btc | NM_007568 | 0.76a/0.47 | Betacellulin, EGF family member |
| Caprin2 | NM_181541 | 0.43 | Caprin family member 2 |
| Ereg | NM_007950 | 1.9 | Epiregulin |
| Nrg4 | NM_032002 | 0.41 | Neuregulin 4 |
| Transporters | |||
| mfsd7c/Flvcr2 | NM_145447 | 6.2 | Major facilitator superfamily domain-containing 7C |
| Slc23a2 | NM_018824 | 0.54/0.43 | Solute carrier family 23, member 2 |
| Antigen processing and presentation | |||
| Ciita | NM_007575 | 4.5/4.4 | Class II transactivator |
| H2-Aa | NM_010378 | 3.0/2.2b | Histocompatibility 2, class II antigen A, α |
| H2-Ab1 | NM_207105 | 2.3/2.1/1.9 | Histocompatibility 2, class II antigen A, β1 |
| H2-Dma | NM_010386 | 2.3 | Histocompatibility 2, class II, locus DMa |
| H2-Dmb1/Dmb2 | NM_010387/NM_010388 | 2.9/3.3 | Histocompatibility 2, class II, locus DMb1/2 |
| H2-Eb1 | NM_010382 | 1.9 | Histocompatibility 2, class II antigen E β |
| H2-gs10 | NM_001143689 | 1.8 | MHC class I like protein GS10 |
| 0610037M15Rik | XM_903697 | 1.8 | RIKEN cDNA 0610037M15 gene |
| Other | |||
| 6330442E10Rik | NM_178745 | 0.34 | RIKEN cDNA 6330442E10 gene |
| Acta1 | NM_009606 | 4.1 | Actin, α1, skeletal muscle |
| Apcdd1 | NM_133237 | 1.0a/0.86a/0.79b/0.48 | APC-down-regulated 1 |
| Gbp2 | NM_010260 | 2.2b/2.1 | Guanylate nucleotide-binding protein 2 |
| Gm7120 | NM_001039244 | 1.0a/0.49 | Predicted gene 7120 |
| Gphn | NM_172952 | 0.61b/0.47 | Gephyrin |
| Greb1 | NM_015764 | 0.28 | Gene regulated by estrogen in breast cancer protein 1 |
| Hlf | NM_172563 | 0.88a/0.31 | Hepatic leukemia factor |
| Jdp2 | NM_030887 | 2.1 | Jun dimerization protein 2 |
| Map3k6 | NM_016693 | 2.6 | MAP kinase kinase kinase 6 |
| Mt1 | NM_013602 | 160/8.0 | Metallothionein 1 |
| Mup1 | NM_001045550 | 440/8.0a | Major urinary protein1 (and 2/7/8/10/12/17) |
| Pdk4 | NM_013743 | 0.32 | Pyruvate dehydrogenase kinase isoform 4 |
| Pfkfb3 | NM_133232 | 1.9/1.5a | 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| Rps6ka2 | NM_011299 | 1.0a/0.57 | Ribosomal protein S6 kinase, polypeptide 2 |
| Secisbp2l | NM_177608 | 1.8/1.2a | SECIS binding protein 2-like |
| Susd2 | NM_001162913 | 1.9 | Sushi domain-containing 2 |
| Tmem184c | NM_145599 | 2.5 | Transmembrane protein 184c |
| Tmigd1 | NM_025655 | 4.2 | Transmembrane and immunoglobulin domain-containing 1 |
| Tubb2b | NM_023716 | 0.54 | Tubulin, β2b |
| Ubd | NM_023137 | 7.5 | Ubiquitin D |
a p > 0.05.
b 0.01 ≤ p ≤ 0.05.
To gain a broader view of the genomic changes in the SI, we used a lower stringency for individual gene expression changes (i.e. ≥25% in fold change and a p value of ≤0.05). The selection of this lowered stringency was based on a published method (15) and on the consideration that accumulation of small changes in a biological pathway might cause observable biological effects. By using this criterion, we identified 153 significantly induced and 247 significantly suppressed genes in the enterocytes of the IE-Cpr-null mice, compared with WT mice. Through use of MAPPFinder, we further identified biological processes, molecular functions, and cellular components as GO terms that contain the most significantly altered gene expression, using the criteria of z score ≥4.0, percentage change ≥10%, and change number ≥4 (non-redundant GO terms are shown in Table 2). Genes matching the criteria for changed expression in each of these pathways are shown in supplemental Tables 3 and 4.
TABLE 2.
Gene ontology terms that showed the most significant gene expression alterations in the enterocytes of IE-Cpr-null mice
Pathway analysis was performed using GenMAPP 2.1, MAPP Finder 2.0, and the Mm-std_20070817.gdb database (available at the GenMAPP Web site). The criteria for identification of genes with significantly increased or decreased expression were as follows: fold change ≥ 25% (≥1.25 or ≤0.80) and p ≤ 0.05. GO terms are sorted into three types: biological process, molecular function, and cellular component. For each GO term, the number of genes that meet the criteria for a significant increase or decrease was determined (No. changed). This number was compared with the number of genes in the GO term that are measured by the MOE 430A chip (No. of genes measured), for the calculation of the percentage of genes measured in the GO term that meet the criteria for a significant increase or decrease (% changed, in parentheses). z score, a standardized difference score for comparison of the relative extents of gene expression changes in various GO nodes, was also shown. The pathways shown were filtered using the following criteria: percent changed ≥ 10%, z score ≥ 4.0, and number changed ≥ 4. Redundant pathways were excluded. The specific genes meeting the criteria for changed expression in each GO term are shown in supplemental Tables 3 and 4.
| GO term | No. of genes in GO term | No. of genes measured | No. of genes changed (% changed) | z score |
|---|---|---|---|---|
| Significantly increased | ||||
| Biological process | ||||
| Antigen processing and presentation | 73 | 44 | 11 (25.0%) | 14.1 |
| Isoprenoid biosynthetic process | 18 | 10 | 6 (60/0%) | 16.6 |
| Negative regulation of transferase activity | 54 | 40 | 4 (10.0%) | 4.9 |
| Steroid metabolic process | 145 | 92 | 15 (16.3%) | 12.9 |
| Cellular component | ||||
| Multivesicular body | 10 | 7 | 4 (70.0%) | 13.2 |
| MHC protein complex | 47 | 24 | 8 (33.3%) | 14.0 |
| Significantly decreased | ||||
| Biological process | ||||
| Hormone metabolic process | 82 | 41 | 6 (14.6%) | 5.6 |
| Molecular function | ||||
| Serine esterase activity | 36 | 31 | 4 (12.9%) | 4.2 |
| Oxidoreductase activity, incorporation, or reduction of molecular oxygen | 143 | 84 | 9 (10.7%) | 5.5 |
| Cellular component | ||||
| Apical part of the cell | 69 | 50 | 6 (12%) | 4.8 |
Three biological processes that showed the most significant increases in gene expression in the enterocytes of IE-Cpr-null mice were: antigen processing and presentation, isoprenoid biosynthetic process, and steroid metabolic process (with z scores of 14.1, 16.6, and 12.9, respectively) (Table 2). In the steroid metabolic process, the majority of the differentially expressed genes were related to cholesterol metabolism, a result similar to what was previously found in the livers of the liver-Cpr-null mice (9). Significant increases in gene expression were also found in the enterocytes of IE-Cpr-null mice in two GO terms associated with cellular components: multivesicular body and major histocompatibility complex protein complex (with z scores of 13.2 and 14.0, respectively); these two GO terms contain a large set of overlapping genes. MHC protein complex contained both MHC I and II genes, whereas multivesicular body contained only MHC II genes as well as the MHC II transmembrane chaperone, CD74.
Significant decreases in gene expression in the enterocytes of IE-Cpr-null mice, when compared with the corresponding levels in WT mice, were also observed. These decreases occurred in the biological process GO terms of hormone metabolic process (with a z score of 5.6), the molecular functions of serine esterase activity and oxidoreductase activity (incorporation or reduction of molecular oxygen) (with z scores of 4.2 and 5.5, respectively), and the cellular component of the apical part of the cell (including several transporters) (with a z score of 4.8).
Cholesterol Synthesis
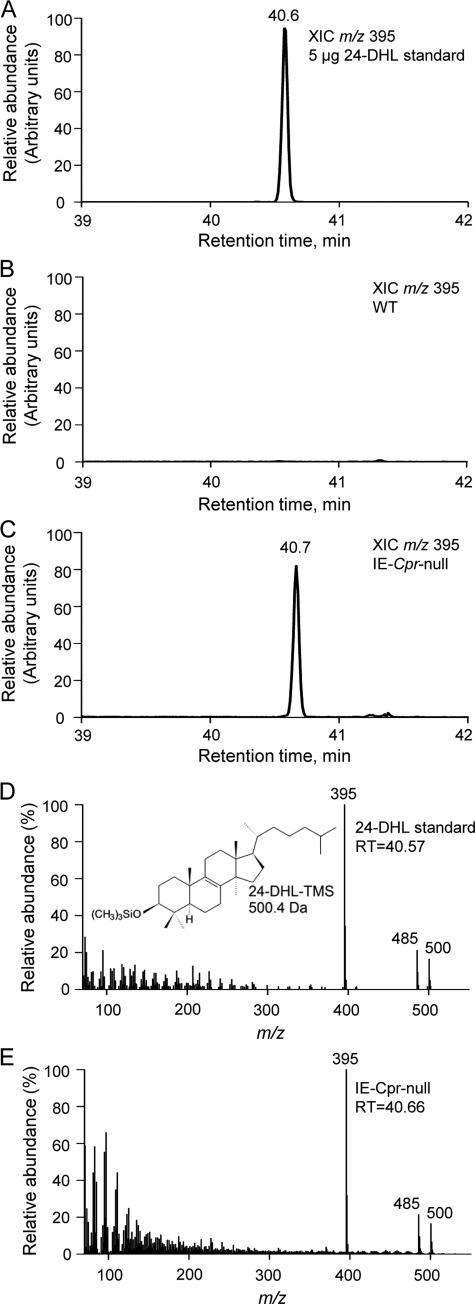
We hypothesized that the induction of expression of multiple genes involved in cholesterol synthesis in the enterocytes of the IE-Cpr-null mice was in response to a blockage of de novo cholesterol biosynthesis that would result from a functional loss of the POR-dependent enzyme CYP51 (9, 10). To confirm the absence of cholesterol synthesis in the enterocytes of the IE-Cpr-null mice, we measured the levels of 24-DHL (a metabolite of the cholesterol precursor, lanosterol), which has been reported to accumulate in the liver of the liver-Cpr-null mice (18). As shown in Fig. 1, 24-DHL, which was readily analyzed as its trimethylsilyl derivative (24-DHL-TMS) by using GC/MS, was not detected in enterocytes from WT mice, but it was abundant in enterocytes of IE-Cpr-null mice. The identity of 24-DHL-TMS was confirmed by comparisons with the results from the analysis of the trimethylsilyl derivative of authentic 24-DHL standard with respect to GC retention times and electron impact mass spectra (Fig. 1). Indistinguishable mass spectra and GC retention times were observed. The mass spectrum of 24-DHL-TMS shows a molecular ion at m/z 500 and a peak at m/z 485 attributed to the loss of a methyl radical (15 Da) from the molecular ion. The base peak in the mass spectrum of 24-DHL-TMS is at m/z 395, which is attributed to the neutral losses of HOSi(CH3)3 (90 Da) and a methyl radical from the molecular ion. The tissue level of 24-DHL was determined by quantitative GC/MS analysis to be 260 pmol/mg enterocytes in the IE-Cpr-null mice (Table 3), a level that is at least 8 times greater than the levels in WT mice (based on a detection limit of 30 pmol/mg). This result confirms that cholesterol synthesis was blocked at CYP51 in the enterocytes of IE-Cpr-null mice.
FIGURE 1.
Accumulation of 24-DHL in the enterocytes of IE-Cpr-null mice. Enterocytes from two 4-month-old male WT or IE-Cpr-null mice were pooled. Sample extraction, preparation of trimethylsilyl derivatives, and GC/MS analysis were as described under “Experimental Procedures.” The data shown represent typical results from one of three independent experiments. Shown are extracted ion chromatograms (XIC) of m/z 395 from the analysis of 5 μg of 24-DHL standard, derivatized to form 24-DHL-TMS (A); a derivatized lipid extract from WT mice (B); and a derivatized lipid extract from IE-Cpr-null mice (C). The mass spectrum for the 24-DHL-TMS peak at 40.7 min (D) from the analysis of the lipid extract from IE-Cpr-null enterocytes is identical to the mass spectrum for the peak at 40.6 min (E) for the 24-DHL-TMS standard. A peak corresponding to 24-DHL-TMS was not detected in the derivatized extract from WT mice (B).
TABLE 3.
Levels of various endogenous compounds in enterocytes of WT and IE-Cpr-null mice
Enterocytes were isolated from male, age-matched (2–4-month-old) WT and IE-Cpr-null mice. The values presented are means ± S.D. (n ≥ 3).
| Strain | Cholesterol (n = 3) | 24-Dihydrolanosterol (n = 9) | FPP (n = 9) | GGPP (n = 9) | Heme (n = 3) | Bilirubin (n = 5) |
|---|---|---|---|---|---|---|
| nmol/mg tissue | pmol/mg tissue | fmol/mg tissue | fmol/mg tissue | nmol/mg protein | fmol/mg tissue | |
| WT | 4.4 ± 0.6 | NDa | 209 ± 28 | 23 ± 5 | 3.3 ± 0.5 | 31.7 ± 18.3 |
| IE-Cpr-null | 4.0 ± 0.3 | 260 ± 18 | 260 ± 41b | 39 ± 6c | 3.6 ± 0.3 | 29.3 ± 5.2 |
a ND, not detected; detection limit was ∼30 pmol/mg tissue.
b p < 0.01, compared with WT value.
c p < 0.001, compared with WT value.
The loss of hepatic cholesterol synthesis in the liver-Cpr-null mice resulted in drastic decreases in plasma total cholesterol (6, 7). Given the relatively large metabolic capacity of the SI, we questioned whether the loss of enterocyte cholesterol synthesis would impact either circulating or else local tissue levels of cholesterol. As shown in Table 3, the levels of total cholesterol in the enterocytes of IE-Cpr-null and WT mice were not significantly different (both at ∼4 nmol/mg enterocytes). Plasma total cholesterol levels were also similar between the mouse strains (at ∼120 ng/ml, or 3.1 mm; data not shown). Thus, de novo cholesterol synthesis in the SI did not noticeably influence cholesterol levels in the plasma or in enterocytes.
Up-regulation of CIITA and MHC II Genes
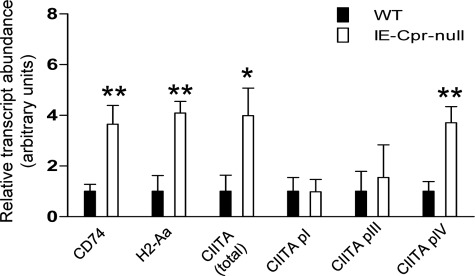
Genes that are up-regulated in expression in the biological process of antigen processing and presentation in the enterocytes of IE-Cpr-null mice (Tables 1 and 2) included MHC I and II genes (1.8–3.3-fold), the class II transactivator (Ciita, the master regulator of MHC II genes; 4.5-fold), and Cd74 (the MHC II invariant chain, a specialized chaperone; 1.6-fold, p = 0.02, not included in Table 1). These changes were confirmed by RNA-PCR analysis of mRNA expression for Ciita, Cd74, and H2-Aa (as a representative MHC II gene) in enterocytes of IE-Cpr-null and WT mice (Fig. 2); the extents of increase in gene expression determined by RNA-PCR were comparable with or greater than the extents revealed by the microarray data. In addition, the mRNA expression of cathepsin E, a gene known to be down-regulated by CIITA (25), also appeared to be decreased in enterocytes of IE-Cpr-null mice, compared with WT mice, as indicated by the microarray data (fold change, 0.21; p = 0.09) and by RNA-PCR analysis (by 85%; data not shown).
FIGURE 2.
Differential expression of genes related to antigen presentation and processing in the enterocytes of WT and IE-Cpr-null mice. RNA samples prepared from enterocytes of 2.5–3-month-old male mice (n = 3) were used for quantitative RNA-PCR analysis. The levels of various target transcripts were normalized to the level of GAPDH mRNA in the same RNA sample. Relative levels of each transcript in the two mouse strains were determined, and the results are shown in arbitrary units obtained by setting the GAPDH-normalized values for the WT samples to 1. The values represent mean ± S.D. (error bars). *, p ≤ 0.05; **, p ≤ 0.01. Data represent typical results from two experiments.
The mouse Ciita gene has three distinct promoters (pI, pIII, and pIV) that are utilized in a cell-specific manner, each producing a unique mRNA. pI and pIII are used exclusively by immune cells, whereas pIV is utilized in multiple cell types (26), including enterocytes (27). The two probe sets for Ciita on the GeneChip are homologous to regions shared by mRNAs from all three promoters. However, RNA-PCR (Fig. 2) analysis revealed increased expression from pIV, but not from pI or pIII, in the enterocytes of IE-Cpr-null mice, compared with expression in WT mice. Furthermore, the magnitude of the increase in Ciita pIV expression was similar to that of the increase of total Ciita transcripts, determined by use of either a general CIITA primer or by the microarray analysis (both 4–5-fold). Thus, the induction of CIITA occurred in enterocytes, rather than in immune cells that conceivably could have been co-isolated with the enterocytes.
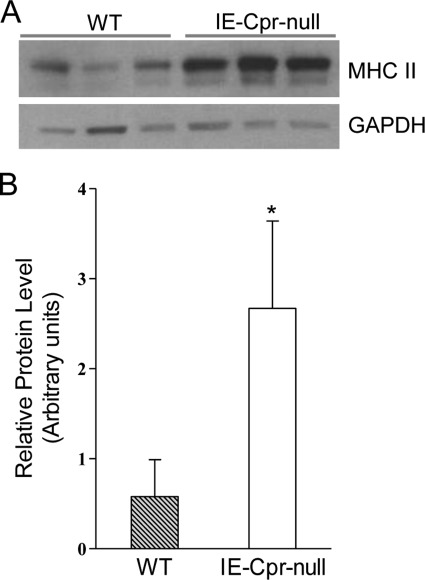
Microarray analysis revealed up-regulation of multiple MHC II genes, including H2-Aa, H2-Ab1, and H2-Eb1, in IE-Cpr-null enterocytes (Table 1). Using an antibody that recognizes both H2-A and H2-E MHC II proteins, we confirmed up-regulation of MHC II protein expression in whole cell lysates from IE-Cpr-null enterocytes (∼4-fold, compared with WT mice) (Fig. 3). The two bands detected on immunoblots represent the α and β subunits of MHC II proteins (28).
FIGURE 3.
Immunoblot analysis of MHC II protein expression in enterocytes of WT and IE-Cpr-null mice. A, mice were fasted overnight, and whole-cell lysates (30 μg/lane) were prepared from enterocytes of three individual, 3-month-old male WT or IE-Cpr-null mice and analyzed on immunoblot with an anti-mouse MHC II antibody. As a loading control, the same samples were also analyzed with an anti-GAPDH antibody. B, results from densitometry analysis. The two bands were combined for determination. Data represent typical results (normalized to GAPDH) from three experiments. *, p < 0.05. Error bars, S.D.
Potential Mechanistic Link between Por Gene Deletion and MHC II Up-regulation
The expression from CIITA pIV can be induced by IFN-γ in multiple cell types (26), including enterocytes (27). Evidence in support of the involvement of IFN-γ or its downstream mediators in the up-regulation of CIITA in the IE-Cpr-null mice was obtained in the analysis of the microarray data, which indicated 1.5-fold or greater increases in the expression of several other genes known to be inducible by IFN-γ. These included Cd74, guanylate-binding protein 2, MHC I, MHC II, metallothionein 1, and ubiquitin D (Table 1) (29–31). The positive regulatory effects of IFN-γ on the expression of these genes are likely to be mediated through activation of STAT1 (29). This contention is supported indirectly by our microarray data showing decreased expression (fold change = 0.50, p = 0.02) of Skp2 (S phase kinase-associated protein 2), which can result from increased STAT1 expression (32), in the enterocytes of IE-Cpr-null mice.
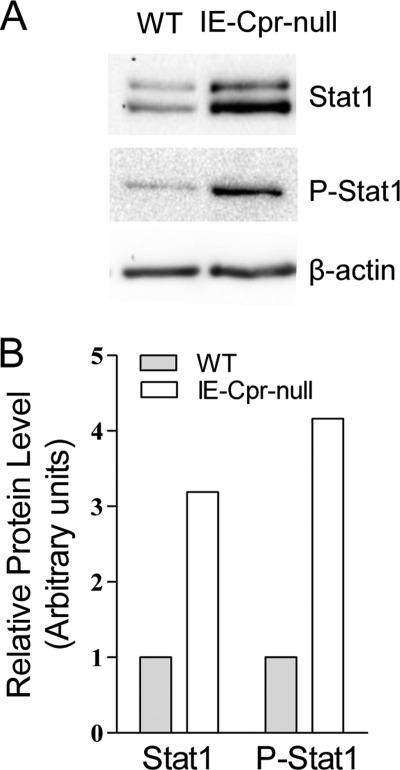
Direct evidence for increased expression of STAT1 protein in IE-Cpr-null enterocytes was obtained by immunoblot analysis, using an anti-STAT1 antibody (Fig. 4), which detected the two bands representing the STAT1 α and β isoforms (33). The levels of both STAT1 isoforms were increased (∼3.2-fold) in whole-cell lysates from IE-Cpr-null enterocytes, compared with the levels in WT enterocytes. Evidence was also obtained for increased activation of STAT1 via phosphorylation at serine 727, a signaling event that is known to be required for maximal activity of the transcription factor (34). As shown in Fig. 4, the level of phosphorylated STAT1 was increased (∼4.2-fold) in enterocytes from IE-Cpr-null mice in comparison with the levels in enterocytes from WT mice.
FIGURE 4.
Immunoblot analysis of STAT1 protein expression and phosphorylation in enterocytes of WT and IE-Cpr-null mice. A, whole cell lysates (40 μg/lane), prepared from enterocytes of individual 3-month-old male WT or IE-Cpr-null mice, were subjected to immunoblot analysis using either anti-STAT1 or anti-phosphorylated STAT 1. As a loading control, the same samples were also analyzed using an anti-β-actin antibody. B, results from densitometry analysis. For STAT1, the two bands were combined for determination. Data represent typical results (normalized to β-actin) from three experiments.
The observed up-regulation of STAT1 and CIITA in the IE-Cpr-null enterocytes is in contrast with the known inhibitory effects of statins on IFN-γ-mediated induction of Ciita transcription in macrophages and microglia (35, 36). The statins inhibit the production of intermediates in the cholesterol synthetic pathway (including FPP and GGPP) through inhibition of the pivotal upstream enzyme, HMG-CoA reductase (37). The molecular mechanism responsible for the inhibitory effects of statins on CIITA transcription is thought to involve decreases in cellular levels of the isoprenoids and consequent reduction in prenylation of Rho family GTPases; the latter event may lead to decreased expression of both STAT1 and CIITA (cf. Ref. 36). We reasoned that, if the same mechanistic link were involved, but in the opposite direction, this could explain the increases in STAT1 and CIITA expression in the IE-Cpr-null enterocytes; the POR loss-related blockage of cholesterol synthesis at CYP51 would result in accumulation of FPP and GGPP. As shown in Table 3, the levels of enterocyte FPP and GGPP were both increased in the IE-Cpr-null mice, compared with the levels in WT mice; the relative extent of increase for FPP (51 fmol/mg, a 24% increase) was lower than that for GGPP (16 fmol/mg; a 70% increase), given the much higher levels of FPP than GGPP in the WT mice.
Heme Metabolism and Homeostasis
HO activity is involved in the breakdown of heme, producing CO, bilirubin, and free iron (2). The absence of POR expression in the IE-Cpr-null enterocytes was accompanied by a ∼90% decrease in total HO activity in enterocyte microsomal preparations, compared with the activity in WT mice (data not shown), consistent with the known dependence of HO on POR (2, 6). However, no significant difference was found in enterocyte heme levels between IE-Cpr-null and WT mice (Table 3). Moreover, enterocyte levels of bilirubin, a heme metabolite capable of regulating STAT1 and MHC II gene expression (38, 39), were likewise not significantly different between IE-Cpr-null and WT mice (Table 3). These results suggest that enterocyte HO activity does not play an essential role in controlling enterocyte levels of heme or its metabolite, bilirubin. Notably, although the biochemical mechanisms involved in heme export from SI are not fully understood (40), preliminary analysis (data not shown) of the expression of genes potentially related to heme transport revealed compensatory changes that may at least partially explain why the heme content in the IE-Cpr-null enterocytes was not different from that in WT mice.
DISCUSSION
Numerous changes in gene expression were observed in the enterocytes of IE-Cpr-null mice in comparison with expression levels in WT mice. However, in contrast to the obvious pathological changes seen in the liver of the liver-Cpr-null mice (enlarged, fatty liver, with necrotic lesions) (6, 7), the changes in gene expression in IE-Cpr-null mice were not accompanied by gross cellular and anatomical changes in the intestine (11). An important reason for the tissue differences between liver and intestine in their response to POR loss may be the fact that the intestinal enterocytes have a short life span, ∼3 days in mice (41); thus, a phenotype (such as lipidosis) that takes considerable time to develop may not be observable. Additionally, some of the POR loss-related metabolic deficiencies in the enterocytes may be compensated for by the availability of substrates and metabolic intermediates (e.g. cholesterol) produced in the liver that are delivered to the intestine via enterohepatic circulation. The influence of the liver on SI gene expression was highlighted in a study of the hepatic POR null mouse (7), in which many changes in gene expression were observed in the intestine (42).
We observed increased expression of genes involved in cholesterol synthesis in the enterocytes of IE-Cpr-null mice in comparison with their expression in WT mice. Similar increases in the expression of genes associated with cholesterol synthesis have been observed in other Por knock-out models and are believed to involve feedback mechanisms mediated by sterol regulatory element-binding proteins (9), which stimulate the expression of genes encoding enzymes of the cholesterol synthetic pathway when cellular oxysterol levels are low (43, 44). Interestingly, we did not observe any changes in serum or enterocyte levels of cholesterol in the IE-Cpr-null mice, despite the blockage in cholesterol synthesis in enterocytes that resulted from loss of POR expression. Our data indicate the lack of a significant contribution from cholesterol synthesis in the intestine to the levels of circulating cholesterol. This observation is consistent with the known function of the liver as the major cholesterol synthesis organ (e.g. see Refs. 6 and 7).
Our data suggest that cholesterol from de novo synthesis in the enterocytes only contributes to a small degree to the total intracellular cholesterol pool. The enterocytes readily obtain cholesterol from extracellular sources in addition to synthesis within the enterocytes; cholesterol in LDL from plasma, cholesterol absorption from the diet, and cholesterol delivered in bile from the liver are all potential sources of enterocyte cholesterol (reviewed in Refs. 45 and 46). Although both endogenously synthesized and absorbed cholesterol can be found in multiple cellular compartments, it has been proposed that, in the enterocytes, absorbed cholesterol is primarily converted to cholesterol esters for transport to the liver in chylomicrons, whereas locally synthesized cholesterol is primarily used for metabolism (47). These same authors suggested that the enterocytes respond differently to increases in cholesterol synthesis versus cholesterol absorption. In this regard, our gene expression data strongly support the concept that enterocytes respond to specific changes in a specific pool of cholesterol (i.e. decreased cholesterol synthesis) despite the absence of changes in total cholesterol levels.
Notably, we cannot rule out the possibility that the IE-Cpr-null enterocytes also compensated for the loss of cholesterol synthesis through changes in cholesterol uptake or export. These changes might also explain, in part, our observation that a loss of de novo cholesterol synthesis in the enterocytes did not result in changes in the levels of total cellular cholesterol. However, this scenario seems unlikely, given that we did not observe an increase in the expression of LDL receptor (based on the cDNA microarray data), which mediates LDL uptake (37), or in the expression of NPC1l1 (Niemann-Pick C1-like protein 1; based on RNA-PCR data (not shown)), a brush border protein critical for cholesterol absorption in enterocytes (48). In addition, cholesterol efflux via ABCA1 (ATP-binding cassette A1) to HDL plays a key role in reverse cholesterol transport (49). The genes encoding major cholesterol efflux transporters (Abca1 and Abcg5/g8) as well as several others encoding enzymes and transporters involved in cholesterol metabolism and disposition are regulated by the liver X receptor (LXR), an oxysterol activated nuclear receptor (50). The loss of POR/P450 activity could potentially lead to decreased levels of oxysterol (9) and thus decreased activation of LXR. However, our cDNA microarray data did not show decreases in the expression of Abca1, Abcg5, Abcg8, or any other LXR target genes, in either SI or liver of the IE-Cpr-null mice, which suggests that a compensatory response, if present, was not initiated via a reduced activation of LXR. Nonetheless, further studies to directly measure rates of intestinal cholesterol absorption and efflux in the IE-Cpr-null mice are warranted in order to detect any potential impact of alterations in de novo cholesterol synthesis on intestinal absorption of dietary cholesterol or on intestinal cholesterol excretion.
The elimination of enterocyte cholesterol synthetic activity in the IE-Cpr-null mice was accompanied by increases in cellular levels of FPP and GGPP in the enterocytes. It has been shown in yeast that overexpression of genes involved in the mevalonate pathway prior to or at the branch point of isoprenoid biosynthesis results in increased levels of farnesol and geranylgeraniol, metabolites of FPP and GGPP, respectively (51, 52). Moreover, studies with rat liver showed that blockage of cholesterol synthesis at SQLE, an enzyme located after the branch point of isoprenoid synthesis, results in increased levels of FPP and GGPP (53). Notably, in the IE-Cpr null enterocytes, blockage of cholesterol synthesis is at CYP51, which is downstream of the branch point of isoprenoid synthesis, and thus can lead to increased accumulation of isoprenoids.
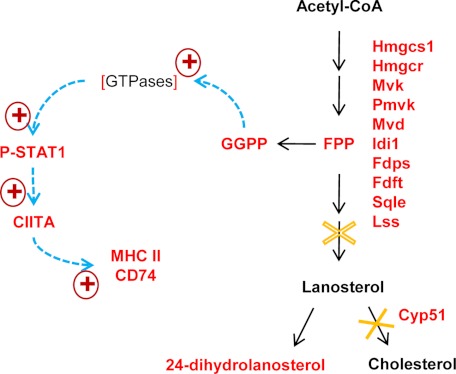
The increased accumulation of isoprenoids, particularly GGPP, in enterocytes may serve as a mechanistic link between the blockage of cholesterol synthesis and the up-regulation of genes related to antigen processing and presentation, including MHC II genes and Ciita, which controls MHC II expression, in the enterocytes of IE-Cpr-null mice. Specifically, we assert that increased GGPP accumulation causes increased prenylation of cellular proteins, including the Rho GTPases, resulting in greater activation of the GTPases, leading in turn to sequential activation of Stat1, Ciita pIV, and the MHC II genes (Fig. 5) in the enterocytes of IE-Cpr-null mice. This mechanism is based on our present observation of the up-regulation of STAT1/PSTAT1, CIITA pIV, and the MHC IIs in the IE-Cpr-null enterocytes and on previously reported evidence (26, 36, 54, 55), including the involvement of IFN-γ, the effects of statins, and the finding that a decrease in protein prenylation of Rho GTPases by GGPP leads to decreased activation of STAT1 and CIITA (36). However, the increased activation of STAT1 in the enterocytes of IE-Cpr-null mice was unlikely to be mediated by IFN-γ, given the fact that we did not observe an increased level of IFN-γ in the serum or any signs of intestinal inflammation upon histological examination of the IE-Cpr-null mice (data not shown).
FIGURE 5.
Proposed mechanistic link between Por deletion and up-regulation of MHC II expression in enterocytes. Metabolic pathways are indicated by black arrows, whereas signaling pathways are represented by blue dashed arrows. Genes found to have increased expression, at either mRNA or protein level, and metabolites found to have increased levels in the IE-Cpr-null enterocytes are shown in red. The GTPase is expected to have increased activity, due to increased prenylation (as indicated by square brackets). The criteria for inclusion of genes in the cholesterol biosynthesis pathway were p < 0.05 and fold change > 25%. The metabolic step catalyzed by CYP51 is completely blocked (solid ×), whereas the step catalyzed by SQLE is partially inhibited (dashed ×), by the loss of POR, leading to accumulation of 24-dihydrolanosterol as well as the isoprenoids. Abbreviations and gene symbols not already listed in Table 1 or mentioned elsewhere include the following: Hmgcs1, 3-hydroxy-3-methylglutaryl-coenzyme A synthase 1; Hmgcr, 3-hydroxy-3-methylglutaryl-coenzyme A reductase; Mvk, mevalonate kinase; FDPS, farnesyl diphosphate synthetase.
Although the available data support a link between GGPP accumulation and increased CIITA and MHC II expression in the IE-Cpr-null enterocytes, it remains possible that other mechanisms also contribute. Of particular relevance are HOs, which also depend on POR for function (2, 6). HO1 was reported to play a role in regulating STAT1 and MHC II expression; studies with dendritic cells show that silencing or inhibition of HO1 results in up-regulation of MHC II expression through induction of CIITA and increased phosphorylation of STAT1 (38). Conversely, bilirubin, a product of heme metabolism, was shown to suppress MHC II expression by reducing expression of CIITA mRNA and reducing STAT1 phosphorylation in endothelial cells (39). Nonetheless, in the enterocytes of IE-Cpr-null mice, the loss of HO activity (data not shown), as a result of the POR loss, was not accompanied by a significant decrease in bilirubin levels. This latter result is explainable by the presence of an alternative source of enterocyte bilirubin, derived from the liver via enterohepatic recirculation (56). Therefore, it is unlikely that the loss of HO activity contributed to the observed increase in MHC II genes in the enterocytes of IE-Cpr-null mice.
In summary, we have explored the potential biological functions of POR-dependent enzymes in the SI through genomic and biochemical analyses of the enterocytes of the IE-Cpr-null mouse. Our findings revealed a novel mechanistic link between POR-dependent enzymes in cholesterol synthesis and the expression of MHC II genes in the enterocytes. This finding, which defines a new physiological/pathological role of intestinal POR/P450 enzymes in modulating the expression of regulators of intestinal immunity, may have important clinical significance. POR is a direct target of inhibition by various drugs and other xenobiotic compounds (e.g. cyclophosphamide (57), ellipticine (58), and cadmium (59)). Furthermore, numerous genetic polymorphisms of the human POR gene that affect either POR expression or POR activity have been identified (60–63). It is conceivable that a decrease in POR activity in the human intestine, either as a result of chemical inhibition or because of POR genetic variations, would also lead to significant increases in the expression of the MHC II genes. Alterations in antigen processing and presentation in the intestine can potentially alter immune responses to antigens. Studies in mice have shown that enterocytes release exosomes that, through MHC II molecules, present antigens to cells of the immune system, resulting in either tolerance (64) or the stimulation of immune responses (65). Furthermore, MHC II expression has been implicated as a factor involved in disease states of the intestine, such as celiac disease (66) and inflammatory bowel disease (67). Additionally, an increase in protein prenylation, resulting from decreases of POR activity and consequent increases of levels of GGPP and FPP, may lead to increased activation of Ras, an oncoprotein, which requires isoprenylation for activation (37, 68) and has been implicated in the development of a large fraction of cancers of the intestine (69, 70). Therefore, any drug that targets cholesterol synthetic enzymes below the branch point of isoprenoid synthesis or has a potential to inhibit POR itself should be monitored for its potential to elicit changes in intestinal antigen processing and presentation or to enhance tumorigenesis in the intestine. This is especially true for drugs that are given orally (as most drugs are) because these would be exposed directly to the intestine.
Supplementary Material
Acknowledgments
We gratefully acknowledge the use of the Microarray Core of the Wadsworth Center. We thank Dr. Bruce Herron for helpful discussions and Weizhu Yang for assistance with animal breeding and genotyping.
This work was supported, in whole or in part, by National Institutes of Health Grant GM082978.
The full microarray dataset reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession numbers(s) GSE35293.

This article contains supplemental Tables 1–4.
- CPR or POR
- NADPH-cytochrome P450 reductase
- P450
- cytochrome P450
- 24-DHL
- 24-dehydrolanosterol
- 24-DHL-TMS
- trimethylsilyl derivative of 24-DHL
- FPP
- farnesyl pyrophosphate
- GGPP
- geranylgeranyl pyrophosphate
- GO
- gene ontology
- HO
- heme oxygenase
- IE
- intestinal epithelium
- SI
- small intestine
- SQLE
- squalene epoxidase
- LXR
- liver X receptor.
REFERENCES
- 1. Taniguchi H., Imai Y., Sato R. (1984) Role of electron transfer system in microsomal drug monooxygenase reaction catalyzed by cytochrome P450. Arch. Biochem. Biophys. 232, 585–596 [DOI] [PubMed] [Google Scholar]
- 2. Schacter B.A., Nelson E. B., Marver H. S., Masters B. S. (1972) Immunochemical evidence for an association of heme oxygenase with the microsomal electron transport system. J. Biol. Chem. 247, 3601–3607 [PubMed] [Google Scholar]
- 3. Ono T., Bloch K. (1975) Solubilization and partial characterization of rat liver squalene epoxidase. J. Biol. Chem. 250, 1571–1579 [PubMed] [Google Scholar]
- 4. Shen A. L., O'Leary K. A., Kasper C. B. (2002) Association of multiple developmental defects and embryonic lethality with loss of microsomal NADPH-cytochrome P450 oxidoreductase. J. Biol. Chem. 277, 6536–6541 [DOI] [PubMed] [Google Scholar]
- 5. Flück C. E., Tajima T., Pandey A. V., Arlt W., Okuhara K., Verge C. F., Jabs E. W., Mendonça B. B., Fujieda K., Miller W. L. (2004) Mutant P450 oxidoreductase causes disordered steroidogenesis with and without Antley-Bixler syndrome. Nat. Genet. 36, 228–230 [DOI] [PubMed] [Google Scholar]
- 6. Gu J., Weng Y., Zhang Q. Y., Cui H., Behr M., Wu L., Yang W., Zhang L., Ding X. (2003) Liver-specific deletion of the NADPH-cytochrome P450 reductase gene. Impact on plasma cholesterol homeostasis and the function and regulation of microsomal cytochrome P450 and heme oxygenase. J. Biol. Chem. 278, 25895–25901 [DOI] [PubMed] [Google Scholar]
- 7. Henderson C. J., Otto D. M., Carrie D., Magnuson M. A., McLaren A. W., Rosewell I., Wolf C. R. (2003) Inactivation of the hepatic cytochrome P450 system by conditional deletion of hepatic cytochrome P450 reductase. J. Biol. Chem. 278, 13480–13486 [DOI] [PubMed] [Google Scholar]
- 8. Wu L., Gu J., Cui H., Zhang Q. Y., Behr M., Fang C., Weng Y., Kluetzman K., Swiatek P. J., Yang W., Kaminsky L., Ding X. (2005) Transgenic mice with a hypomorphic NADPH-cytochrome P450 reductase gene. Effects on development, reproduction, and microsomal cytochrome P450. J. Pharmacol. Exp. Ther. 312, 35–43 [DOI] [PubMed] [Google Scholar]
- 9. Weng Y., DiRusso C. C., Reilly A. A., Black P. N., Ding X. (2005) Hepatic gene expression changes in mouse models with liver-specific deletion or global suppression of the NADPH-cytochrome P450 reductase. J. Biol. Chem. 280, 31686–31698 [DOI] [PubMed] [Google Scholar]
- 10. Wang X. J., Chamberlain M., Vassieva O., Henderson C. J., Wolf C. R. (2005) Relationship between hepatic phenotype and changes in gene expression in cytochrome P450 reductase (POR) null mice. Biochem. J. 388, 857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Q. Y., Fang C., Zhang J., Dunbar D., Kaminsky L., Ding X. (2009) An intestinal epithelium-specific cytochrome P450 (P450) reductase-knockout mouse model. Direct evidence for a role of intestinal P450s in first-pass clearance of oral nifedipine. Drug Metab. Dispos. 37, 651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhu Y., D'Agostino J., Zhang Q. Y. (2011) Role of intestinal cytochrome P450 (P450) in modulating the bioavailability of oral lovastatin. Insights from studies on the intestinal-epithelium-specific P450 reductase knockout mouse. Drug Metab. Dispos. 39, 939–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fang C., Zhang Q. Y. (2010) The role of small-intestinal P450 enzymes in protection against systemic exposure of orally administered benzo[a]pyrene. J. Pharmacol. Exp. Ther. 334, 156–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Q. Y., Dunbar D., Kaminsky L. S. (2003) Characterization of mouse small intestinal cytochrome P450 expression. Drug Metab. Dispos. 31, 1346–1351 [DOI] [PubMed] [Google Scholar]
- 15. Doniger S. W., Salomonis N., Dahlquist K. D., Vranizan K., Lawlor S. C., Conklin B. R. (2003) MAPPFinder. Using gene ontology and GenMAPP to create a global gene expression profile from microarray data. Genome Biol. 4, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dahlquist K. D., Salomonis N., Vranizan K., Lawlor S. C., Conklin B. R. (2002) GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat. Genet. 31, 19–20 [DOI] [PubMed] [Google Scholar]
- 17. Folch J., Lees M., Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 18. Li L., Porter T. D. (2007) Hepatic cytochrome P450 reductase-null mice reveal a second microsomal reductase for squalene monooxygenase. Arch. Biochem. Biophys. 461, 76–84 [DOI] [PubMed] [Google Scholar]
- 19. Tong H., Wiemer A. J., Neighbors J. D., Hohl R. J. (2008) Quantitative determination of farnesyl and geranylgeranyl diphosphate levels in mammalian tissue. Anal. Biochem. 378, 138–143 [DOI] [PubMed] [Google Scholar]
- 20. Mingone C. J., Gupte S. A., Chow J. L., Ahmad M., Abraham N. G., Wolin M. S. (2006) Protoporphyrin IX generation from δ-aminolevulinic acid elicits pulmonary artery relaxation and soluble guanylate cyclase activation. Am. J. Physiol. Lung Cell Mol. Physiol. 291, L337–L344 [DOI] [PubMed] [Google Scholar]
- 21. Zelenka J., Lenicek M., Muchova L., Jirsa M., Kudla M., Balaz P., Zadinova M., Ostrow J. D., Wong R. J., Vitek L. (2008) Highly sensitive method for quantitative determination of bilirubin in biological fluids and tissues. J. Chromatogr. B 867, 37–42 [DOI] [PubMed] [Google Scholar]
- 22. D'Agostino J., Zhang X., Wu H., Ling G., Wang S., Zhang Q. Y., Liu F., Ding X. (2008) Characterization of CYP2A13*2, a variant cytochrome P450 allele previously found to be associated with decreased incidences of lung adenocarcinoma in smokers. Drug Metab. Dispos. 36, 2316–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ware J. A., Graf M. L., Martin B. M., Lustberg L. R., Pohl L. R. (1998) Immunochemical detection and identification of protein adducts of diclofenac in the small intestine of rats. Possible role in allergic reactions. Chem. Res. Toxicol. 11, 164–171 [DOI] [PubMed] [Google Scholar]
- 24. UniProt Consortium (2012) Reorganizing the protein space at the Universal Protein resource (UniProt). Nucleic Acids Res. 40, D71–D75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yee C. S., Yao Y., Li P., Klemsz M. J., Blum J. S., Chang C. H. (2004) Cathepsin E. A novel target for regulation by class II transactivator. J. Immunol. 172, 5528–5534 [DOI] [PubMed] [Google Scholar]
- 26. Reith W., LeibundGut-Landmann S., Waldburger J. M. (2005) Regulation of MHC class II gene expression by the class II transactivator. Nat. Rev. Immunol. 5, 793–806 [DOI] [PubMed] [Google Scholar]
- 27. Waldburger J. M., Suter T., Fontana A., Acha-Orbea H., Reith W. (2001) Selective abrogation of major histocompatibility complex class II expression on extrahematopoietic cells in mice lacking promoter IV of the class II transactivator gene. J. Exp. Med. 194, 393–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bhattacharya A., Dorf M. E., Springer T. A. (1981) A shared alloantigenic determinant on Ia antigens encoded by the I-A and I-E subregions. Evidence for I region gene duplication. J. Immunol. 127, 2488–2495 [PubMed] [Google Scholar]
- 29. Schroder K., Hertzog P. J., Ravasi T., Hume D. A. (2004) Interferon-γ. An overview of signals, mechanisms, and functions. J. Leukoc. Biol. 75, 163–189 [DOI] [PubMed] [Google Scholar]
- 30. De S. K., McMaster M. T., Andrews G. K. (1990) Endotoxin induction of murine metallothionein gene expression. J. Biol. Chem. 265, 15267–15274 [PubMed] [Google Scholar]
- 31. Lukasiak S., Schiller C., Oehlschlaeger P., Schmidtke G., Krause P., Legler D. F., Autschbach F., Schirmacher P., Breuhahn K., Groettrup M. (2008) Proinflammatory cytokines cause FAT10 up-regulation in cancers of liver and colon. Oncogene 27, 6068–6074 [DOI] [PubMed] [Google Scholar]
- 32. Wang S., Raven J. F., Koromilas A. E. (2010) STAT1 represses Skp2 gene transcription to promote p27Kip1 stabilization in Ras-transformed cells. Mol. Cancer Res. 8, 798–805 [DOI] [PubMed] [Google Scholar]
- 33. Najjar I., Schischmanoff P. O., Baran-Marszak F., Deglesne P. A., Youlyouz-Marfak I., Pampin M., Feuillard J., Bornkamm G. W., Chelbi-Alix M. K., Fagard R. (2008) Novel functions of STAT1β in B cells. Induction of cell death by a mechanism different from that of STAT 1α. J. Leukoc. Biol. 84, 1604–1612 [DOI] [PubMed] [Google Scholar]
- 34. Wen Z., Zhong Z., Darnell J. E., Jr. (1995) Maximal Activation of Transcription by STAT1 and STAT3 requires both tyrosine and serine phosphorylation. Cell 82, 241–250 [DOI] [PubMed] [Google Scholar]
- 35. Youssef S., Stüve O., Patarroyo J. C., Ruiz P. J., Radosevich J. L., Hur E. M., Bravo M., Mitchell D. J., Sobel R. A., Steinman L., Zamvil S. S. (2002) The HMG-CoA reductase inhibitor, atorvastatin, promotes a Th2 bias and reverses paralysis in central nervous system autoimmune disease. Nature 420, 78–84 [DOI] [PubMed] [Google Scholar]
- 36. Lee S. J., Qin H., Benveniste E. N. (2008) The IFN-γ-induced transcriptional program of the CIITA gene is inhibited by statins. Eur. J. Immunol. 38, 2325–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goldstein J. L., Brown M. S. (1990) Regulation of the mevalonate pathway. Nature 343, 425–430 [DOI] [PubMed] [Google Scholar]
- 38. Cheng C., Noorderloos M., van Deel E. D., Tempel D., den Dekker W., Wagtmans K., Duncker D. J., Soares M. P., Laman J. D., Duckers H. J. (2010) Dendritic cell function in transplantation arteriosclerosis is regulated by heme oxygenase 1. Circ. Res. 106, 1656–1666 [DOI] [PubMed] [Google Scholar]
- 39. Wu J., Ma J., Fan S. T., Schlitt H. J., Tsui T. Y. (2005) Bilirubin derived from heme degradation suppresses MHC class II expression in endothelial cells. Biochem. Biophys. Res. Commun. 338, 890–896 [DOI] [PubMed] [Google Scholar]
- 40. West A. R., Oates P. S. (2008) Mechanisms of heme iron absorption. Current questions and controversies. World J. Gastroenterol. 14, 4101–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Potten C. S., Hendry J. H. (1983) Stem Cells, Churchill Livingstone, Edinburgh, Scotland [Google Scholar]
- 42. Mutch D. M., Klocke B., Morrison P., Murray C. A., Henderson C. J., Seifert M., Williamson G. (2007) The disruption of hepatic cytochrome P450 reductase alters mouse lipid metabolism. J. Proteome Res. 6, 3976–3984 [DOI] [PubMed] [Google Scholar]
- 43. Hua X., Sakai J., Brown M. S., Goldstein J. L. (1996) Regulated cleavage of sterol regulatory element binding proteins requires sequences on both sides of the endoplasmic reticulum membrane. J. Biol. Chem. 271, 10379–10384 [DOI] [PubMed] [Google Scholar]
- 44. Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., Goldstein J. L. (2003) Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. 100, 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chang T. Y., Chang C. C., Ohgami N., Yamauchi Y. (2006) Cholesterol sensing, trafficking, and esterification. Annu. Rev. Cell Dev. Biol. 22, 129–157 [DOI] [PubMed] [Google Scholar]
- 46. Ikonen E. (2008) Cellular cholesterol trafficking and compartmentalization. Nat. Rev. Mol. Cell Biol. 9, 125–138 [DOI] [PubMed] [Google Scholar]
- 47. Stange E. F., Suckling K. E., Dietschy J. M. (1983) Synthesis and coenzyme A-dependent esterification of cholesterol in rat intestinal epithelium. Differences in cellular localization and mechanisms of regulation. J. Biol. Chem. 258, 12868–12875 [PubMed] [Google Scholar]
- 48. Altmann S. W., Davis H. R., Jr., Zhu L. J., Yao X., Hoos L. M., Tetzloff G., Iyer S. P., Maguire M., Golovko A., Zeng M., Wang L., Murgolo N., Graziano M. P. (2004) Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science 303, 1201–1204 [DOI] [PubMed] [Google Scholar]
- 49. Brufau G., Groen A. K., Kuipers F. (2011) ATVB in focus: HDL structure, function, therapeutics, and imaging. Reverse cholesterol transport revisited. Contribution of biliary versus intestinal cholesterol excretion. Arterioscler. Thromb. Vasc. Biol. 31, 1726–1733 [DOI] [PubMed] [Google Scholar]
- 50. Kalaany N. Y., Mangelsdorf D. J. (2006) LXRs and FXR. The Yin and Yang of cholesterol and fat metabolism. Annu. Rev. Physiol. 68, 159–191 [DOI] [PubMed] [Google Scholar]
- 51. Ohto C., Muramatsu M., Obata S., Sakuradani E., Shimizu S. (2009) Overexpression of the gene encoding HMG-CoA reductase in Saccharomyces cerevisiae for production of prenyl alcohols. Appl. Microbiol. Biotechnol. 82, 837–845 [DOI] [PubMed] [Google Scholar]
- 52. Ohto C., Muramatsu M., Obata S., Sakuradani E., Shimizu S. (2010) Production of geranylgeraniol on overexpression of a prenyl diphosphate synthase fusion gene in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 87, 1327–1334 [DOI] [PubMed] [Google Scholar]
- 53. Keller R. K. (1996) Squalene synthase inhibition alters metabolism of nonsterols in rat liver. Biochim. Biophys. Acta 1303, 169–179 [DOI] [PubMed] [Google Scholar]
- 54. Sadeghi M. M., Tiglio A., Sadigh K., O'Donnell L., Collinge M., Pardi R., Bender J. R. (2001) Inhibition of interferon-γ-mediated microvascular endothelial cell major histocompatibility complex class II gene activation by HMG-CoA reductase inhibitors. Transplantation 71, 1262–1268 [DOI] [PubMed] [Google Scholar]
- 55. Ghittoni R., Napolitani G., Benati D., Ulivieri C., Patrussi L., Laghi Pasini F., Lanzavecchia A., Baldari C. T. (2006) Simvastatin inhibits the MHC class II pathway of antigen presentation by impairing Ras superfamily GTPases. Eur. J. Immunol. 36, 2885–2893 [DOI] [PubMed] [Google Scholar]
- 56. Wang X., Chowdhury J. R., Chowdhury N. R. (2006) Bilirubin metabolism: Applied physiology. Curr. Paediatr. 16, 70–74 [Google Scholar]
- 57. Marinello A. J., Berrigan M. J., Struck R. F., Guengerich F. P., Gurtoo H. L. (1981) Inhibition of NADPH-cytochrome P450 reductase by cyclophosphamide and its metabolites. Biochem. Biophys. Res. Commun. 99, 399–406 [DOI] [PubMed] [Google Scholar]
- 58. Guenthner T. M., Kahl G. F., Nebert D. W. (1980) NADPH-cytochrome P-450 reductase. Preferential inhibition by ellipticine and other type II compounds having little effect on NADPH-cytochrome c reductase. Biochem. Pharmacol. 29, 89–95 [DOI] [PubMed] [Google Scholar]
- 59. Trakshel G. M., Kutty R. K., Maines M. D. (1986) Cadmium-mediated inhibition of testicular heme oxygenase activity. The role of NADPH-cytochrome c (P-450) reductase. Arch. Biochem. Biophys. 251, 175–187 [DOI] [PubMed] [Google Scholar]
- 60. Hart S. N., Wang S., Nakamoto K., Wesselman C., Li Y., Zhong X. B. (2008) Genetic polymorphisms in cytochrome P450 oxidoreductase influence microsomal P450-catalyzed drug metabolism. Pharmacogenet. Genomics 18, 11–24 [DOI] [PubMed] [Google Scholar]
- 61. Huang N., Agrawal V., Giacomini K. M., Miller W. L. (2008) Genetics of P450 oxidoreductase. Sequence variation in 842 individuals of four ethnicities and activities of 15 missense mutations. Proc. Natl. Acad. Sci. 105, 1733–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gomes A. M., Winter S., Klein K., Turpeinen M., Schaeffeler E., Schwab M., Zanger U. M. (2009) Pharmacogenomics of human liver cytochrome P450 oxidoreductase. Multifactorial analysis and impact on microsomal drug oxidation. Pharmacogenomics 10, 579–599 [DOI] [PubMed] [Google Scholar]
- 63. Marohnic C. C., Panda S. P., McCammon K., Rueff J., Masters B. S., Kranendonk M. (2010) Human cytochrome P450 oxidoreductase deficiency caused by the Y181D mutation. Molecular consequences and rescue of defect. Drug Metab. Dispos. 38, 332–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Karlsson M., Lundin S., Dahlgren U., Kahu H., Pettersson I., Telemo E. (2001) “Tolerosomes” are produced by intestinal epithelial cells. Eur. J. Immunol. 31, 2892–2900 [DOI] [PubMed] [Google Scholar]
- 65. Van Niel G., Mallegol J., Bevilacqua C., Candalh C., Brugière S., Tomaskovic-Crook E., Heath J. K., Cerf-Bensussan N., Heyman M. (2003) Intestinal epithelial exosomes carry MHC class II/peptides able to inform the immune system in mice. Gut 52, 1690–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mayrhofer G. (1995) Absorption and presentation of antigens by epithelial cells of the small intestine. Hypotheses and predictions relating to the pathogenesis of coeliac disease. Immunol. Cell Biol. 73, 433–439 [DOI] [PubMed] [Google Scholar]
- 67. Brandtzaeg P., Haraldsen G., Rugtveit J. (1997) Immunopathology of human inflammatory bowel disease. Springer Semin. Immunopathol. 18, 555–589 [DOI] [PubMed] [Google Scholar]
- 68. Appels N. M., Beijnen J. H., Schellens J. H. (2005) Development of farnesyltransferase inhibitors. A review. Oncologist 10, 565–578 [DOI] [PubMed] [Google Scholar]
- 69. Bos J. L. (1989) Ras oncogenes in human cancers. A review. Cancer Res. 49, 4682–4689 [PubMed] [Google Scholar]
- 70. Arai M., Shimizu S., Imai Y., Nakatsuru Y., Oda H., Oohara T., Ishikawa T. (1997) Mutations of the KI-Ras, P53, and APC genes in adenocarcinomas of the human small intestine. Int. J. Cancer 70, 390–395 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.