Background: Crystal structures of 6-phosphofructokinases revealed nucleotide binding sites with unknown functional relevance.
Results: Function of two allosteric nucleotide binding sites was determined by mutagenesis and kinetic studies and revealed reciprocal linkage of both.
Conclusion: Activity of mammalian Pfk is regulated by structurally linked new allosteric sites.
Significance: Reciprocal linkage between allosteric binding sites evolved convergent in prokaryotes and eukaryotes.
Keywords: Allosteric Regulation, Enzyme Kinetics, Enzyme Mechanisms, Enzyme Structure, Mutagenesis, 6-Phosphofructokinase
Abstract
6-Phosphofructokinases (Pfk) are homo- and heterooligomeric, allosteric enzymes that catalyze one of the rate-limiting steps of the glycolysis: the phosphorylation of fructose 6-phosphate at position 1. Pfk activity is modulated by a number of regulators including adenine nucleotides. Recent crystal structures from eukaryotic Pfk revealed several adenine nucleotide binding sites. Herein, we determined the functional relevance of two adenine nucleotide binding sites through site-directed mutagenesis and enzyme kinetic studies. Subsequent characterization of Pfk mutants allowed the identification of the activating (AMP, ADP) and inhibitory (ATP, ADP) allosteric binding sites. Mutation of one binding site reciprocally influenced the allosteric regulation through nucleotides interacting with the other binding site. Such reciprocal linkage between the activating and inhibitory binding sites is in agreement with current models of allosteric enzyme regulation. Because the allosteric nucleotide binding sites in eukaryotic Pfk did not evolve from prokaryotic ancestors, reciprocal linkage of functionally opposed allosteric binding sites must have developed independently in prokaryotic and eukaryotic Pfk (convergent evolution).
Introduction
The ATP-dependent 6-phosphofructokinase (EC 2.7.1.11, phosphofructokinase-1, ATP: d-fructose-6-phosphate-1-phosphotransferase; Pfk)2 catalyzes the phosphorylation of fructose 6-phosphate (Fru-6-P) to fructose 1,6-bisphosphate (Fru-1,6-P2). This irreversible reaction is considered to be one of the rate-limiting steps of glycolysis (1–3). In eukaryotes, the Pfk activity is modulated by a number of allosteric regulators, e.g. ATP, AMP, NH4+, fructose 2,6-bisphosphate, citrate, and acyl-CoA (4). Most eukaryotic Pfk are homo- and heterooligomeric enzymes consisting of subunits that evolved from a single ancestor prokaryotic Pfk through gene duplication and mutational events (5, 6).
In eukaryotes, the N-terminal half of a Pfk subunit mediates the catalytic function, whereas in the C-terminal half allosteric ligand binding sites have evolved from former catalytic and regulatory sites (5, 7, 8). It is assumed that specific amino acid residues involved in catalytic and regulatory functions of Pfk from Escherichia coli (9, 10) are conserved in yeast and mammalian Pfk genes. For example, citrate inhibits the enzyme through interaction with sites that have evolved from a duplicated allosteric site (phosphoenolpyruvate binding site) (11, 12), whereas sites for activators, e.g. fructose 2,6-bisphosphate, have evolved from the catalytic site (Fru-6-P) of the ancestral precursor (7, 13). In contrast, the ATP substrate binding site from the ancestral prokaryotic Pfk did not evolve to a new inhibitory ATP binding pocket (13, 15), and it remains unclear as to how adenine nucleotides implement their allosteric inhibitory (ATP) and activating (AMP) effects. Note that ATP serves as a substrate and as an allosteric inhibitor of eukaryotic Pfk. ADP can act as an activator at μm concentrations but inhibits eukaryotic Pfk activity at mm concentrations (16). Until recently, no high resolution structure of eukaryotic Pfk was available to prove the conservation and the functional relevance of allosteric ligand binding sites.
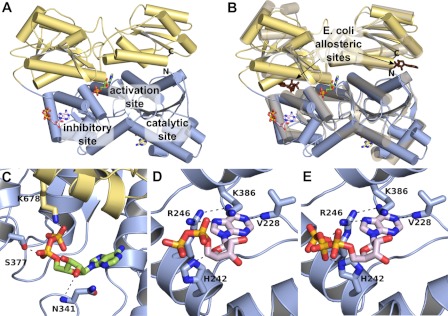
In 2011, crystal structures of three eukaryotic Pfk from the yeasts Pichia pastoris (15) and Saccharomyces cerevisiae (13) and from rabbit muscle (13) were determined. In the crystal structure of P. pastoris Pfk, an additional ATP binding pocket was found in each β chain, which was presumed to be an inactivating site (15). In the structure of rabbit muscle Pfk, this ATP binding site was also discovered in the N-terminal domain of each subunit with either ADP or ATP bound to it. An additional novel ADP binding site was found in the center of each subunit (13). This was proposed as an activating (ADP binding) site because replacement by ATP failed (see Fig. 1). Neither of these two binding sites evolved from a regulatory or catalytic binding site in bacterial Pfk. Fig. 1B shows a superposition of the E. coli Pfk with its two equivalent effector binding sites at the subunit interfaces. The superposition of rabbit and E. coli Pfk suggests that the effector binding sites of the bacterial protein are maintained in the mammalian enzyme but only a phosphate ion is bound to this site. Because ADP is found in at least two binding sites, the functional relevance of the individual binding sites is still unclear. Furthermore, it is unknown whether additional binding sites exist in Pfk mediating the activating and inhibitory effects of adenine nucleotides. There is no crystal structure with AMP bound to Pfk available yet.
FIGURE 1.
Adenine nucleotide binding sites in crystal structure of rabbit muscle Pfk. A, the crystal structure of rabbit skeletal muscle Pfk (13). One monomer of the dimer found in the crystals is shown. The N-terminal half is colored in blue, and the C-terminal half is in yellow. Three different ADP binding sites were identified in the crystal structure: the catalytic center and the putative inhibitory and activating allosteric sites. B, superposition of rabbit skeletal muscle Pfk (colored as in A) with E. coli Pfk (depicted as a transparent graphic, PDB ID 1PFK (14)). One monomer of the homotetrameric bacterial enzyme superimposes on the N-terminal half, and one superimposes onto the C-terminal half. The two prokaryotic effector binding sites of the dimer are occupied by ADP (brown). The bacterial effector binding sites are located at the subunit interfaces, i.e. between the N- and C-terminal halves in the mammalian Pfk. C, in the activating allosteric sites, the diphosphate moiety of ADP interacts via hydrogen bonds with Ser377 and Lys678, whereas the ribose contacts Asn341. D and E, in the potential inhibitory allosteric site, ADP (D) and ATP (E) are coordinated by Val228, His242, Arg246, and Lys386.
Here, we experimentally addressed the functional relevance of the canonical activating and inhibitory adenine nucleotide binding sites through mutagenesis and kinetic studies. We functionally qualified the adenine nucleotide binding sites and identified a reciprocal linkage between these allosteric binding sites.
EXPERIMENTAL PROCEDURES
Materials
If not otherwise stated, all chemicals and standard substances were purchased from Sigma-Aldrich (Taufkirchen, Germany), Carl Roth GmbH (Karlsruhe, Germany), Roche Applied Science (Mannheim, Germany), Fermentas GmbH (St. Leon-Rot, Germany), and BD Biosciences. Restriction enzymes and primers were purchased from New England Biolabs (Frankfurt/Main, Germany) and Invitrogen (Karlsruhe, Germany), respectively.
Methods
Strains and Growth Conditions
The E. coli strain DH5α was used for the cloning experiments. Transformants were selected on LB medium containing 100 μg/ml ampicillin. The yeast strain for expressing human wild type and mutant muscle Pfk was S. cerevisiae HD114-8D (MATα Scpfk1::HIS3 Scpfk2::HIS3 his3-11,15 leu2-3,112 ura3-52) carrying deletion in both yeast Pfk genes (17, 18). Preparation of competent cells was performed as described (17). Selection for transformation of wild type and mutant Pfk was performed at 30 °C in yeast extract peptone medium (1% yeast extract, 2% Bacto peptone) containing 2% glucose as a carbon source.
Selection of Target Residues for Mutagenic Analysis
Asn341, Ser377, Lys678, Val228, His242, Arg246, and Lys386 were selected based on the crystal structure of rabbit skeletal muscle Pfk (Protein Data Bank (PDB): IDs 3O8N and 3O8L). These residues participate in the proposed ADP activator and the ATP inhibitor binding sites (13, 15) and are conserved between the human and rabbit muscle Pfk.
Construction of Mutant Pfk
All mutants were constructed using PCR-based site-directed mutagenesis, and the fragments were cloned into the plasmid pJJH71PFK (kindly provided by Prof. Dr. Jürgen J. Heinisch, University of Osnabrück (18)). The resulting plasmids were sequenced to ensure sequence correctness.
Expression and Purification of Recombinant Enzymes
Transformation of yeast strain HD114-8D was performed with LiAc/ssDNA/PEG (19). Transformants were grown in liquid medium with shaking at 30 °C. After 24 h, the cells were harvested by centrifugation at 5700 × g for 15 min at 4 °C and washed twice with water and once with a 50 mm potassium phosphate buffer, pH 7.2, containing 1 mm EDTA, 5 mm 2-mercaptoethanol, and 0.5 mm PMSF (buffer A). Cell disruption was performed according to Ref. 20 in buffer A with 50 mm NaF, 50 μm Fru-1,6-P2, and protease inhibitor mixture (Roche Applied Science). The following steps were carried out at 4 °C unless described otherwise. The mixture was centrifuged at 75 000 × g for 30 min. The supernatant was subjected to protamine sulfate precipitation (0.2% w/v) (21) with stirring for 5 min. After centrifugation, the pellet was discarded, and the supernatant was precipitated with PEG 6000 (5% w/v) for 30 min with stirring. After centrifugation as before, the pellet was dissolved in buffer B (50 mm Tris/HCl, pH 8.0, 50 μm Fru-1,6-P2, 0.1 mm EDTA, 5 mm 2-mercaptoethanol, 50 mm NaF, 0.5 mm PMSF; according to Ref. 21). Cibacron Blue F3GA-SephadexTM G75 (prepared according to Ref. 22), equilibrated with buffer B, was added to the solution and stirred for 30 min (binding capacity: 15 units/g of gel wet weight). The gel was washed on a funnel with buffer B. Elution was done with buffer A, pH 8.0, containing 2 mm Fru-6-P and 5 mm ATP. The enzyme was concentrated by ultrafiltration (Omega 100-kDa membrane, Pall Life Sciences, Dreieich, Germany). Glycerol was added to a final concentration of 20% (v/v), and the solution can be stored at −20 °C for weeks without loss of activity. For ion exchange chromatography, the concentrated enzyme sample (without glycerol) was diluted with 0.8 volumes of 0.5× buffer A, pH 8.0, and loaded on a Resource Q-Colum (6 ml, GE Healthcare, Munich, Germany), equilibrated with buffer A, pH 8.0. After washing with buffer A, pH 8.0, the enzyme was eluted with a linear gradient of KCl (0–200 mm) in buffer A, pH 8.0. Fractions of Pfk activity were pooled and supplemented with 2 mm ATP and 50 μm Fru-1,6-P2. After concentration by ultrafiltration (Pall Life Sciences, Omega 100-kDa membrane) and Vivaspin 6 (Sartorius, Göttingen, Germany), 20% glycerol (v/v) was added, and the mixture was stored at −20 °C. Gel permeation chromatography was done on a BioSep SEC-S4000 (600 × 21.2 mm; Phenomenex, Aschaffenburg, Germany). The column was equilibrated with buffer A containing 50 μm Fru-1,6-P2, pH 7.5. Fractions with Pfk activity were pooled, 2 mm ATP was added, and the enzyme solution was concentrated as before. The purified enzyme can be stored with 30% glycerol (v/v) at −20 °C for 2 weeks without loss of activity. For kinetic studies of wild type and mutant Pfk, the partially purified enzyme after affinity chromatography with F3GA-SephadexTM was used.
Pfk Activity Assay
During preparation, Pfk activity was measured spectrophotometrically at 340 nm and 25 °C according to Ref. 20 except that pH was 8.5 and 1.2 mm ATP was used. The reaction was started with the addition of a 2–5-μl enzyme sample.
Kinetic studies were performed in 50 mm HEPES, 100 mm KCl, 5 mm MgCl2, pH 7.0, 0.2 mm NADH, 0.45 units/ml aldolase, 4.5 units/ml triosephosphate isomerase, and 1.5 units/ml glycerol phosphate dehydrogenase. ATP, Fru-6-P, AMP, and ADP were used as indicated. In the case of the ATP inhibition experiments, the MgCl2 concentration was 15 mm. Auxiliary enzymes were dialyzed before use (Micro Bio-Spin 6, Bio-Rad Laboratories, Munich, Germany). The reaction was started by the addition of a 5-μl enzyme sample appropriately diluted with 50 mm sodium phosphate buffer, pH 7.2, containing 10% glycerol. Curve fittings for kinetic parameters were generated by either Michaelis-Menten or Hill equations using Prism (GraphPad Software, Inc., La Jolla, CA). Thus, the hyperbolic and the sigmoid parts of the curves (Vmin to Vmax) were fit to Michaelis-Menten and Hill equations, respectively. To exclude artifacts in the determination of the kinetic parameters using partially purified enzymes, wild type and two mutants (N341A, R246A/K386A) were purified to homogeneity. Because there were no differences in the kinetic parameters, all mutant Pfk were partially purified (see above) and tested in kinetic assays.
RESULTS
Identification of Activating Site
In the crystal structure of rabbit skeletal muscle Pfk, the diphosphate moiety of ADP in the allosteric activation site is mainly coordinated by Ser377 and Lys678 (Fig. 1C). Mutation of these residues to Ala significantly increased K0.5 for ADP (Table 1). The K0.5 for ADP of the double mutant S377A/K678A was further increased (Table 1), indicating that this ADP binding site is indeed the allosteric activating site in Pfk. The AMP activation of S377A and K678A was less pronounced (Table 1), probably because of weaker or absent interaction between these residues and the monophosphate moiety of AMP.
TABLE 1.
Effect of AMP, ADP, and ATP on kinetic properties of the wild type human muscle Pfk and Pfk mutated at the activating allosteric site
Kinetic properties of wild type and mutant Pfk were determined. Thus, the hyperbolic and the sigmoid parts of the curves (Vmin to Vmax) were fit to Michaelis-Menten (Kd) and Hill (K0.5, Ki) equations, respectively. For ATP, assays were performed with 2 mm Fru-6-P and either with 1 mm AMP or 0.82 mm ADP, respectively. The kinetic parameters for AMP and ADP were determined at 0.5 mm Fru-6-P and 1 mm ATP. Residual Pfk activity was defined as the percentage of enzyme activity at 2.3 mm ADP and maximal activity for each enzyme under these conditions. The values are means ± S.E. of three independent experiments.
| Parameter | Wild type | N341A | S377A | K678A | S377A/K678A |
|---|---|---|---|---|---|
| Without effectors | |||||
| KmATP (μm) | 42.5 ± 2.0 | 23.2 ± 1.9 | 37.0 ± 3.0 | 32.6 ± 1.4 | 27.5 ± 1.7 |
| KiATP (mm) | 2.0 ± 0.1 | 1.4 ± 0.1 | 2.2 ± 0.1 | 1.9 ± 0.1 | 1.6 ± 0.02 |
| 1 mm AMP | |||||
| KmATP (μm) | 49.8 ± 2.7 | 52.8 ± 3.6 | 43.0 ± 2.7 | 38.4 ± 1.7 | 41.5 ± 1.7 |
| KiATP (mm) | 8.5 ± 0.1 | 5.2 ± 0.1 | 8.6 ± 0.1 | 8.1 ± 0.1 | 5.8 ± 0.1 |
| 0.82 mm ADP | |||||
| KmATP (μm) | 169 ± 9.9 | 151 ± 13.7 | 168 ± 12.2 | 250 ± 14.0 | 141 ± 8.3 |
| KiATP (mm) | 5.3 ± 0.1 | 2.1 ± 0.1 | 5.2 ± 0.1 | 4.3 ± 0.1 | 3.1 ± 0.1 |
| K0.5ADP (μm) | 83.6 ± 0.9 | 610 ± 13.1 | 454 ± 13.0 | 422 ± 3.4 | 506 ± 14.1 |
| KiADP (mm) | 2.1 ± 0.1 | 1.3 ± 0.1 | 2.4 ± 0.1 | 2.0 ± 0.1 | 1.6 ± 0.1 |
| K0.5AMP (μm) | 11.9 ± 0.4 | 201 ± 2.9 | 19.7 ± 0.5 | 17.2 ± 0.8 | 49.3 ± 2.1 |
| Residual activity (%) | 52.4 ± 1.6 | 6.4 ± 1.0 | 57.5 ± 0.2 | 33.8 ± 1.3 | 26.1 ± 0.9 |
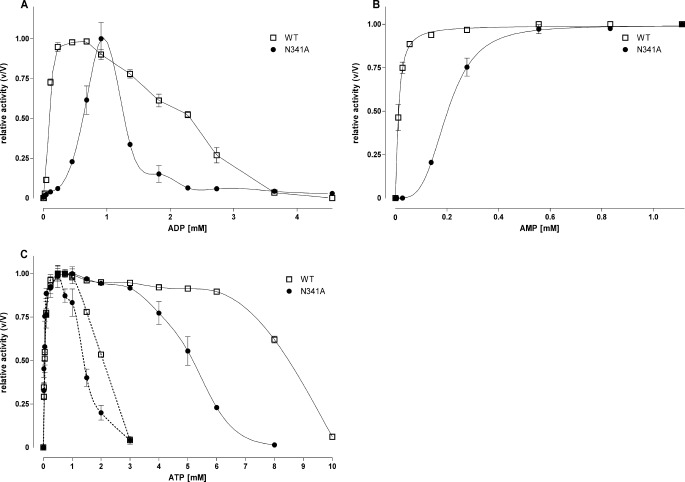
Next, we mutated Asn341 to Ala, which interacts with the ribose of ADP (Fig. 1C) and probably also AMP. In contrast to S377A and K678A, N341A not only showed increased K0.5 values for ADP but also significantly reduced enzyme activation by AMP (Fig. 2, A and B, Table 1).
FIGURE 2.
Effect of ATP, ADP, and AMP on activity of wild type and mutant N341A Pfk. A and B, the effect on the Pfk activity of increasing concentrations of ADP (A) and AMP (B) at the wild type and N341A was determined with 0.5 mm Fru-6-P and 1 mm ATP. Activity is expressed relative to maximal activity (V) for each enzyme under these conditions (V values of wild type were 295 units/ml and 233 units/ml for AMP and ADP, respectively, and 147 units/ml and 57 units/ml for N341A). C, for ATP, assays were performed at 2 mm Fru-6-P. Activity is expressed as a relation between the measured (v) and maximal possible (V) activity for each enzyme under these conditions (V values of wild type were 396 units/ml and 434 units/ml without effectors and 1 mm AMP, respectively, and 147 units/ml and 247 units/ml for N341A). Dotted line, without effectors; continuous line, with 1 mm AMP. Data are means ± S.E. of three independent experiments each performed in duplicate.
ADP and ATP reduced Pfk activity at mm concentrations (Fig. 2, A and C), most likely via binding to the inhibitory nucleotide binding site (Fig. 1, D and E). Strikingly, Ki values of ADP and ATP were significantly reduced in N341A and S377A/K678A (Fig. 2, A and C, Table 1). This effect was found even in the presence of effectors (Fig. 2C, Table 1; data for ADP not shown). In contrast, Ki values of ADP and ATP remained unchanged in S377A and K678A.
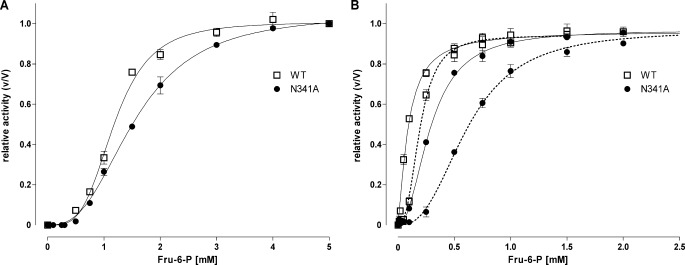
To test whether mutations in the allosteric activator site influence the substrate dependence of the catalytic activity, the influence of Fru-6-P concentrations was tested in the presence of different effectors. As shown in Fig. 3 and Table 2, activities of wild type and mutant enzymes exhibited cooperativity to Fru-6-P without effectors. In the presence of the effectors AMP and ADP, the affinity to this substrate was increased. Specifically, N341A and S377A/K678A displayed significantly increased S0.5 values in comparison with the wild type under these conditions.
FIGURE 3.
Fru-6-P dependence of wild type and mutant N341A Pfk activity. A and B, assays were performed (A) without effectors and (B) with 1 mm AMP (continuous line) and 0.82 mm ADP (dotted line) with fixed concentration of 1 mm ATP and varying concentrations of Fru-6-P (F6P) as indicated. Activity is expressed relative to maximal activity (for wild type, 383, 374, and 359 units/ml without effectors, 1 mm AMP and 0.82 mm ADP, respectively, and 116, 254, and 126 units/ml for N341A). Data are means ± S.E. of three independent experiments each performed in duplicate.
TABLE 2.
Saturation kinetics for Fru-6-P on the wild type human muscle Pfk and Pfk mutated at the activating allosteric site
Assays were carried out at in the presence of 1 mm ATP and indicated concentrations of effectors. By fitting with the Hill equation, the parameters were determined with S0.5 as the substrate concentration at half-maximal activity and nH as the Hill coefficient. The values are means ± S.E. of three independent experiments.
| Parameter | Wild type | N341A | S377A | K678A | S377A/K678A |
|---|---|---|---|---|---|
| Without effectors | |||||
| S0.5Fru-6-P (mm) | 1.2 ± 0.1 | 1.6 ± 0.1 | 1.2 ± 0.1 | 1.5 ± 0.1 | 1.3 ± 0.1 |
| nHFru-6-P (mm) | 2.6 ± 0.1 | 2.7 ± 0.1 | 2.9 ± 0.2 | 3.0 ± 0.2 | 3.0 ± 0.1 |
| 1 mm AMP | |||||
| S0.5Fru-6-P (mm) | 0.09 ± 0.01 | 0.29 ± 0.01 | 0.07 ± 0.01 | 0.07 ± 0.01 | 0.15 ± 0.01 |
| nHFru-6-P (mm) | 1.4 ± 0.1 | 2.1 ± 0.1 | 0.9 ± 0.1 | 1.3 ± 0.1 | 1.6 ± 0.1 |
| 0.82 mm ADP | |||||
| S0.5Fru-6-P (mm) | 0.19 ± 0.01 | 0.61 ± 0.01 | 0.18 ± 0.01 | 0.42 ± 0.01 | 0.58 ± 0.01 |
| nHFru-6-P (mm) | 2.7 ± 0.2 | 2.6 ± 0.1 | 2.9 ± 0.2 | 2.5 ± 0.1 | 2.1 ± 0.1 |
Identification of Inhibitory Site
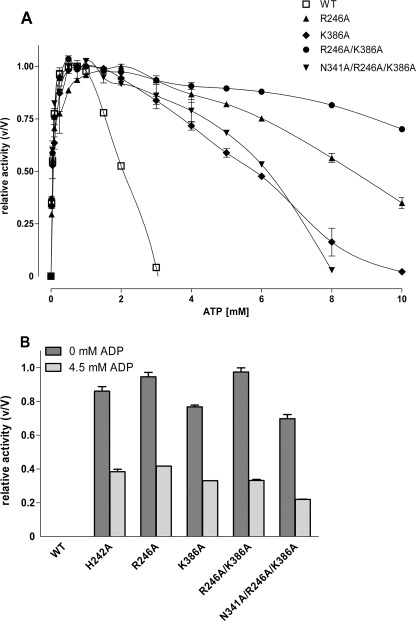
ADP and ATP inhibit Pfk activity by binding to one or more allosteric inhibitory sites. In the rabbit muscle Pfk crystal structure, ADP and ATP in the potential allosteric inhibition site are mainly coordinated by Val228, His242, Arg246, and Lys386 (Fig. 1, D and E). V228A had no significant effects on Pfk activity when compared with the wild type enzyme (data not shown), most probably because of the very conservative structural change. In contrast, mutation of the other three residues to Ala significantly increased Ki for ATP and ADP (Fig. 4, Table 3). Ki values for ATP and ADP of the double mutant R246A/K386A were further increased (Fig. 4, Table 3), indicating that this nucleotide binding site is indeed the allosteric inhibition site of ADP and ATP in mammalian Pfk. Interestingly, all mutants showed significantly higher basal activity in the absence of the activators AMP and ADP (Fig. 4B, Table 3), suggesting that the enzyme is constitutively activated due to mutation in the allosteric inhibitor site. The Fru-6-P dependence of the Ala mutants of His242, Arg246, and Lys386 presented a Michaelis-Menten kinetic indicating a lack of cooperativity (data not shown).
FIGURE 4.
Effect of ATP and ADP on activity of wild type Pfk and various mutations at the inhibitory allosteric site. A, the effect on the Pfk activity of increasing concentrations of ATP at the wild type, R246A, K386A, R246A/K386A, and the enzyme mutated additionally at the activating site (N341A/R246A/K386A) was determined at 2 mm Fru-6-P. Activity is expressed relative to maximal activity for each enzyme under these conditions (V values were 274, 233, 213, and 164 units/ml for R246A, K386A, R246A/K386A, and N341A/R246A/K386A, respectively). To compare data with N341A, see Fig. 2C. B, the activity of wild type and the mutants was measured at 0.5 mm Fru-6-P and 1 mm ATP in the absence and presence of 4.5 mm ADP (inhibitory concentration). Under both conditions, the wild type and N341A displayed no significant activity. Activity is expressed relative to maximal activity (V) for each enzyme under these conditions (V values: wild type, 295 units/ml; H242A, 270 units/ml; R246A, 237 units/ml; K386A, 171 units/ml, R246A/R386A, 250 units/ml; N341A/R246A/K386A, 209 units/ml). Data are means ± S.E. of three independent experiments each performed in duplicate.
TABLE 3.
Functional characterization of human muscle Pfk modified in the inhibitory allosteric site
Kinetic parameters for ATP of wild type and mutant Pfk were determined at 2 mm Fru-6-P and the indicated effector concentrations. Thus, the hyperbolic and the sigmoid parts of the curves (Vmin to Vmax) were fit to Michaelis-Menten (Km) and Hill (K0.5, Ki) equations, respectively. For comparing data of the triple mutant with N341A, see Table 1. For ADP, kinetic properties were determined with 0.5 mm Fru-6-P and 1 mm ATP. Residual Pfk activity was defined as the percentage of enzyme activity at 4.5 mm ADP and maximal activity of each enzyme under these conditions. The values are means ± S.E. of three independent experiments. ND, not detectable.
| Parameter | Wild type | H242A | R246A | K386A | R246A/K386A | N341A/R246A/K386A |
|---|---|---|---|---|---|---|
| Without effectors | ||||||
| KmATP (μm) | 42.5 ± 2.0 | 38.2 ± 2.0 | 43.2 ± 1.3 | 52.4 ± 4.7 | 47.7 ± 2.4 | 35.6 ± 2.0 |
| KiATP (mm) | 2.0 ± 0.1 | 6.8 ± 0.1 | 8.6 ± 0.3 | 5.6 ± 0.1 | >10.0 | 5.9 ± 0.1 |
| 1 mm AMP | ||||||
| KmATP (μm) | 49.8 ± 2.7 | 33.8 ± 2.0 | 50.8 ± 5.0 | 35.6 ± 1.9 | 42.5 ± 1.5 | 63.0 ± 2.3 |
| KiATP (mm) | 8.5 ± 0.1 | >10.0 | >10.0 | >10.0 | >10.0 | 8.8 ± 0.1 |
| 0.82 mm ADP | ||||||
| KmATP (μm) | 169 ± 9.9 | 114 ± 4.4 | 166 ± 7.1 | 168 ± 6.0 | 234 ± 13.6 | 220 ± 7.5 |
| KiATP (mm) | 5.3 ± 0.1 | >10.0 | >10.0 | 9.5 ± 0.1 | >10.0 | 7.6 ± 0.1 |
| K0.5ADP (μm) | 83.6 ± 0.9 | ND | ND | ND | ND | ND |
| KiADP (mm) | 2.1 ± 0.1 | 3.2 ± 0.1 | 3.4 ± 0.1 | 3.2 ± 0.1 | 2.5 ± 0.1 | 2.3 ± 0.1 |
| Residual activity at 4.5 mm ADP (%) | 0 | 38.4 ± 1.2 | 41.7 ± 0.1 | 33.1 ± 0.1 | 33.2 ± 0.5 | 21.9 ± 0.1 |
Finally, we combined mutations of the allosteric activating (N341A) and the inhibitory (R246A/K386A) sites in the triple mutant N341A/R246A/K386A. This mutant was also constitutively activated but with a lower extent (66% of maximum activity). The Ki values for ATP and ADP when compared with mutants of the inhibitory allosteric site alone were significantly decreased (Table 3, Fig. 4).
DISCUSSION
Pfk activity underlies strong allosteric regulation by several effectors including adenine nucleotides. The crystal structures of prokaryotic and eukaryotic Pfk revealed a number of adenine nucleotide molecules bound in the catalytic center as well as in allosteric binding sites (Fig. 1, A and B). In the crystal structure of rabbit skeletal muscle Pfk (13), ADP was found in two binding sites (Fig. 1, C and D), which were different from the well characterized allosteric binding sites in prokaryotic Pfk (Fig. 1B). Because ADP can activate (at μm concentration) and also inactivate (at mm concentration) the enzyme, we approached the functional relevance of these two nucleotide binding sites by site-directed mutagenesis. In one crystal structure of the rabbit skeletal muscle Pfk, ATP was found in one of the two ADP binding sites (Fig. 1E), suggesting this as the inhibitory allosteric site; however, an experimental proof was missing.
The single mutations of binding relevant residues in the potential activating binding site (N341A, S377A, K678A) shifted the K0.5 values of ADP to higher concentrations (Table 1). Increased K0.5 values of AMP were found for S377A/K678A and N341A (Table 1), strongly supporting annotation of this nucleotide binding site as the allosteric activating binding site for AMP and ADP. Interestingly, the K0.5 values for AMP were unchanged in the single mutants (S377A, K678A). This suggests that in contrast to ADP, where the diphosphate moiety of ADP is coordinated by Ser377 and Lys678 (Fig. 1C), binding of the monophosphate residue of AMP may involve additional or other residues. Only a crystal structure of muscle Pfk with AMP bound may clarify detailed coordination of this nucleotide in the activating allosteric site.
Mutation of Asn341 was more efficient in increasing the K0.5 values of both ADP and AMP (Table 1). Because the backbone nitrogen atom of Asn341 interacts with the ribose of ADP, the side chain is probably involved in the transmission of the allosteric signal or necessary for the correct positioning of the backbone chain. Most strikingly, however, Ki values of ADP and ATP were significantly shifted toward lower concentrations, suggesting an increase of the effector affinity at the allosteric inhibitory binding site or an enhanced transduction of the inhibitory effect of ATP and ADP (Fig. 2, A and C).
Single Ala mutations of His242, Arg246, and Lys386 and double mutation (R246A/K386A) in the putative inhibitory binding site reduced the inhibitory potency of ATP and ADP (Table 3), strongly supporting the postulated function as the inhibitory allosteric site. However, Pfk activity was almost at the maximum in the absence of the activator AMP or ADP (Fig. 4B), indicating constitutively activated mutant enzymes. The maximum activity of e.g. R246A/K386A was not significantly different from the wild type, and the specific activity of highly purified Ala mutants did not differ from the wild type enzyme under optimal conditions excluding gross structural changes in the mutants. It appears that mutations of the inhibitory allosteric site “locked” the enzyme in an active conformation mimicking the state in the presence of AMP. This is supported by the fact that Fru-6-P dependence of the Ala mutants (H242A, R246A, K386A) presented a Michaelis-Menten kinetic indicating a lack of cooperativity. However, additional mutation on the activating allosteric site (N341A/R246A/K386A) did not reverse this constitutively activated conformation but improved ATP inhibition when compared with R246A/K386A (Fig. 4, A and B). This indicates a more complex interplay of the allosteric sites or several (dissociable) active states.
From recent studies, we know that the smallest active enzyme form of mammalian Pfk consists of four subunits but that higher oligomeric complexes exist that are also functional (23, 24). Physiologically, AMP and μm concentration of ADP promote enzyme activation, and mm concentrations of ADP and ATP promote enzyme inhibition. This allosteric regulation directly couples cellular energy metabolism to cellular ATP/AMP ratio. Our data now suggest that this ATP/AMP-driven allostery is further fine-tuned by a reciprocal linkage of the two functionally opposed allosteric sites. Mutation of the activating allosteric site reduces the activating effect of AMP and ADP and synergistically increases the inhibitory effect of ATP and ADP. Vice versa, mutation of the inhibitory allosteric site increases Ki values of ATP and ADP and results in a constitutively activated enzyme. One may speculate that mutation or ligand occupation of one allosteric site leads to structural changes followed by changes in the affinity of the opposite allosteric site. For example, occupation of the allosteric activator site by AMP in the wild type Pfk decreases the affinity of ATP at the allosteric inhibitory site. Indeed, early studies with the skeletal muscle Pfk showed that the binding of AMP resulted in an antagonism of ATP inhibition by decreasing the affinity of ATP binding (25). Further evidence for such a mechanistic explanation comes from the crystal structures of the prokaryotic Pfk from E. coli. In the inactive state, the AMP binding site is structurally different when compared with the active state (10). Crystal structures of eukaryotic Pfk clearly showed that the allosteric nucleotide binding sites are different from those in the prokaryotic Pfk (13, 15). Therefore, reciprocal linkage of functionally opposed allosteric binding sites must have developed independently in prokaryotic and eukaryotic Pfk, giving an impressive example of convergent evolution at the molecular level.
Although a reciprocal linkage of functionally opposite allosteric binding sites is in agreement with the extended Monod-Wyman-Changeux model of allostery (26), this allostery model has been shown inappropriate in a certain number of cases. For prokaryotic Pfk, the classical two-state model is not sufficient to describe the allosteric behavior (27), so additional models of allostery were developed (26–29). Because mammalian Pfk are even more complex (in terms of structure and regulation) than prokaryotic Pfk, the appropriate crystal structures of the various states are required to clarify whether the functionally opposed allosteric sites are linked within a subunit or between subunits.
In sum, we assigned two new allosteric adenine nucleotide binding sites to be the activating and inhibitory nucleotide binding sites. The latter is occupied by ATP or ADP. The novel ADP binding site found in the crystal structure of mammalian Pfk is the activating allosteric site and also binds AMP. Our data strongly support that activating and inhibitory allosteric binding sites do not only regulate the catalytic Pfk activity but also modulate the properties of each other in a reciprocal manner. This cross-talk between the allosteric nucleotide binding sites may further fine-tune Pfk activity depending on the ATP/AMP ratio.
Acknowledgments
We thank Prof. Gunther Fischer and Wolfgang Schellenberger and the two anonymous reviewers for critically reading the manuscript and their very helpful comments.
This work was supported by Deutsche Forschungsgemeinschaft Grant Sfb610.
- Pfk
- 6-phosphofructokinase(s)
- Fru-6-P
- fructose 6-phosphate
- Fru-1,6-P2
- fructose 1,6-bisphosphate.
REFERENCES
- 1. Hofmann E. (1976) The significance of phosphofructokinase to the regulation of carbohydrate metabolism. Rev. Physiol. Biochem. Pharmacol. 75, 1–68 [DOI] [PubMed] [Google Scholar]
- 2. Hofmann E., Kopperschläger G. (1982) Phosphofructokinase from yeast. Methods Enzymol. 90, 49–60 [DOI] [PubMed] [Google Scholar]
- 3. Uyeda K. (1979) Phosphofructokinase. Adv. Enzymol. Relat. Areas Mol. Biol. 48, 193–244 [DOI] [PubMed] [Google Scholar]
- 4. Jenkins C. M., Yang J., Sims H. F., Gross R. W. (2011) Reversible high affinity inhibition of phosphofructokinase-1 by acyl-CoA: a mechanism integrating glycolytic flux with lipid metabolism. J. Biol. Chem. 286, 11937–11950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Poorman R. A., Randolph A., Kemp R. G., Heinrikson R. L. (1984) Evolution of phosphofructokinase: gene duplication and creation of new effector sites. Nature 309, 467–469 [DOI] [PubMed] [Google Scholar]
- 6. Heinisch J., Ritzel R. G., von Borstel R. C., Aguilera A., Rodicio R., Zimmermann F. K. (1989) The phosphofructokinase genes of yeast evolved from two duplication events. Gene 78, 309–321 [DOI] [PubMed] [Google Scholar]
- 7. Kemp R. G., Gunasekera D. (2002) Evolution of the allosteric ligand sites of mammalian phosphofructo-1-kinase. Biochemistry 41, 9426–9430 [DOI] [PubMed] [Google Scholar]
- 8. Arvanitidis A., Heinisch J. J. (1994) Studies on the function of yeast phosphofructokinase subunits by in vitro mutagenesis. J. Biol. Chem. 269, 8911–8918 [PubMed] [Google Scholar]
- 9. Evans P. R., Hudson P. J. (1979) Structure and control of phosphofructokinase from Bacillus stearothermophilus. Nature 279, 500–504 [DOI] [PubMed] [Google Scholar]
- 10. Schirmer T., Evans P. R. (1990) Structural basis of the allosteric behavior of phosphofructokinase. Nature 343, 140–145 [DOI] [PubMed] [Google Scholar]
- 11. Li Y., Rivera D., Ru W., Gunasekera D., Kemp R. G. (1999) Identification of allosteric sites in rabbit phosphofructo-1-kinase. Biochemistry 38, 16407–16412 [DOI] [PubMed] [Google Scholar]
- 12. Usenik A., Legis̆a M. (2010) Evolution of allosteric citrate binding sites on 6-phosphofructo-1-kinase. PLoS One 5, e15447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Banaszak K., Mechin I., Obmolova G., Oldham M., Chang S. H., Ruiz T., Radermacher M., Kopperschläger G., Rypniewski W. (2011) The crystal structures of eukaryotic phosphofructokinases from bakers' yeast and rabbit skeletal muscle. J. Mol. Biol. 407, 284–297 [DOI] [PubMed] [Google Scholar]
- 14. Shirakihara Y., Evans P. R. (1988) Crystal structure of the complex of phosphofructokinase from Escherichia coli with its reaction products. J. Mol. Biol. 204, 973–994 [DOI] [PubMed] [Google Scholar]
- 15. Sträter N., Marek S., Kuettner E. B., Kloos M., Keim A., Brüser A., Kirchberger J., Schöneberg T. (2011) Molecular architecture and structural basis of allosteric regulation of eukaryotic phosphofructokinases. FASEB J. 25, 89–98 [DOI] [PubMed] [Google Scholar]
- 16. Mediavilla D., Metón I., Baanante I. V. (2008) Purification and kinetic characterization of 6-phosphofructo-1-kinase from the liver of gilthead sea bream (Sparus aurata). J. Biochem. 144, 235–244 [DOI] [PubMed] [Google Scholar]
- 17. Heinisch J. J. (1993) Expression of heterologous phosphofructokinase genes in yeast. FEBS Lett. 328, 35–40 [DOI] [PubMed] [Google Scholar]
- 18. Raben N., Exelbert R., Spiegel R., Sherman J. B., Nakajima H., Plotz P., Heinisch J. (1995) Functional expression of human mutant phosphofructokinase in yeast: genetic defects in French Canadian and Swiss patients with phosphofructokinase deficiency. Am. J. Hum. Genet. 56, 131–141 [PMC free article] [PubMed] [Google Scholar]
- 19. Gietz R. D., Schiestl R. H. (2007) Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 38–41 [DOI] [PubMed] [Google Scholar]
- 20. Kirchberger J., Bär J., Schellenberger W., Dihazi H., Kopperschläger G. (2002) 6-Phosphofructokinase from Pichia pastoris: purification, kinetic, and molecular characterization of the enzyme. Yeast 19, 933–947 [DOI] [PubMed] [Google Scholar]
- 21. Martínez-Costa O. H., Hermida C., Sánchez-Martínez C., Santamaría B., Aragón J. J. (2004) Identification of C-terminal motifs responsible for transmission of inhibition by ATP of mammalian phosphofructokinase, and their contribution to other allosteric effects. Biochem. J. 377, 77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Böhme H. J., Kopperschläger G., Schulz J., Hofmann E. (1972) Affinity chromatography of phosphofructokinase using Cibacron Blue F3G-A. J. Chromatogr. 69, 209–214 [DOI] [PubMed] [Google Scholar]
- 23. Ferreras C., Hernández E. D., Martínez-Costa O. H., Aragón J. J. (2009) Subunit interactions and composition of the fructose 6-phosphate catalytic site and the fructose 2,6-bisphosphate allosteric site of mammalian phosphofructokinase. J. Biol. Chem. 284, 9124–9131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zancan P., Almeida F. V., Faber-Barata J., Dellias J. M., Sola-Penna M. (2007) Fructose 2,6-bisphosphate counteracts guanidinium chloride-, thermal-, and ATP-induced dissociation of skeletal muscle key glycolytic enzyme 6-phosphofructo-1-kinase: a structural mechanism for PFK allosteric regulation. Arch. Biochem. Biophys. 467, 275–282 [DOI] [PubMed] [Google Scholar]
- 25. Kemp R. G., Krebs E. G. (1967) Binding of metabolites by phosphofructokinase. Biochemistry 6, 423–434 [DOI] [PubMed] [Google Scholar]
- 26. Changeux J. P. (2012) Allostery and the Monod-Wyman-Changeux model after 50 years. Annu. Rev. Biophys. 41, 83–113 [DOI] [PubMed] [Google Scholar]
- 27. Fenton A. W., Reinhart G. D. (2009) Disentangling the web of allosteric communication in a homotetramer: heterotropic inhibition in phosphofructokinase from Escherichia coli. Biochemistry 48, 12323–12328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Peracchi A., Mozzarelli A. (2011) Exploring and exploiting allostery: models, evolution, and drug targeting. Biochim. Biophys. Acta 1814, 922–933 [DOI] [PubMed] [Google Scholar]
- 29. Fenton A. W., Paricharttanakul N. M., Reinhart G. D. (2004) Disentangling the web of allosteric communication in a homotetramer: heterotropic activation in phosphofructokinase from Escherichia coli. Biochemistry 43, 14104–14110 [DOI] [PubMed] [Google Scholar]