Abstract
Structural analysis of human MRP1-NBD1 revealed that the Walker A S685 forms a hydrogen bond with the Walker B D792 and interacts with the Mg2+ cofactor and the β-phosphate of the bound Mg•ATP. We have found that substitution of the S685 with an amino acid that potentially prevents the formation of the hydrogen bond resulted in misfolding of the protein and significantly affect the ATP-dependent leukotriene C4 (LTC4) transport. In this report we tested whether the corresponding substitution in NBD2 would also result in misfolding of the protein. In contrast to the NBD1 mutations, none of the mutations in NBD2, including S1334A, S1334C, S1334D, S1334H, S1334N, and S1334T, caused misfolding of the protein. However, elimination of the hydroxyl group at S1334 in mutations including S1334A, S1334C, S1334D, S1334H, and S1334N drastically reduced the ATP binding and the ATP-enhanced ADP trapping at the mutated NBD2. Due to this low efficient ATP binding at the mutated NBD2, the inhibitory effect of ATP on the LTC4 binding is significantly decreased. Furthermore, ATP bound to the mutated NBD2 cannot be efficiently hydrolyzed, leading to almost completely abolishing the ATP-dependent LTC4 transport. In contrast, S1334T mutation, which retained the hydroxyl group at this position, exerts higher LTC4 transport activity than the wild-type MRP1, indicating that the hydroxyl group at this position plays a crucial role for ATP binding/hydrolysis and ATP-dependent solute transport.
Structural analyses of human MRP1-NBD1 (1),1 mouse cystic fibrosis transmembrane-conductance regulator (CFTR)-NBD1 (2), and Escherichia coli HlyB-NBD (3) revealed that the Walker A serine residue forms a hydrogen bond with the Walker B aspartic acid and interacts with the β- and γ-phosphate groups of the bound Mg•ATP (1). In addition, the γ-oxygen of the Walker A serine residue and the δ-oxygen of the Walker B aspartate residue interact, together with other four oxygen atoms from β- and γ-phosphate of the bound ATP and water molecules, with Mg2+ cofactor to form the octahedral coordination geometry (1, 2, 4–7). These interactions play very important roles for ATP binding, hydrolysis, and ATP-dependent solute transport by these ATP-binding cassette (ABC) transporters.
We have found that substitution of the Walker A S685 or Walker B D792 in NBD1 with a residue that abolishes the hydrogen bond formation between them caused misfolding of the mutated MRP1 protein (8–10). In contrast, however, substitution of the corresponding Walker B D1454 in NBD2 with the hydrophobic residue leucine (D1454L/E1455L) did not cause misfolding of the mutated MRP1 protein (8), suggesting distinct structures in NBD1 and NBD2 of MRP1. Thus, we speculated that substitution of the Walker A S1334 in NBD2 with a residue that eliminates the hydroxyl group at this position will also not cause misfolding of the protein. However, since substitution of the Walker A S685 with a residue that eliminates the hydroxyl group at this position significantly affected the ATP binding/hydrolysis and ATP-dependent LTC4 transport (10), we speculated that substitution of the Walker A S1334 in NBD2 with a residue that eliminates the hydroxyl group at this position might have more severe effects on the ATP-dependent LTC4 transport than that of the corresponding mutations in NBD1. In order to test this hypothesis, we have substituted the Walker A S1334 with an A (S1334A), a C (S1334C), a D (S1334D), an H (S1334H), an N (S1334N), or a T (S1334T). Indeed, all of these mutants, except S1334T, almost completely abolished the ATP-dependent LTC4 transport, indicating that the interaction of the hydroxyl group at S1334 with Mg•ATP plays a crucial role for ATP binding/hydrolysis and ATP-dependent LTC4 transport.
EXPERIMENTAL PROCEDURES
Materials
Sodium orthovanadate, EGTA, and ATP were purchased from Sigma. [14,15,19,20-3H]Leukotriene C4 was from PerkinElmer Life Sciences. [α-32P]-8-N3ADP, [α-32P]-8-N3ATP, and [γ-32P]-8-N3ATP were from Affinity Labeling Technologies. Fetal bovine serum was from Gemini Bio-Products. The QuikChange site-directed mutagenesis kit was from Stratagene. Anti-mouse Ig conjugated with horseradish peroxidase was from Amersham Biosciences. Chemiluminescent substrates for Western blotting were from Pierce.
Site-Directed Mutagenesis of MRP1 Protein
Wild-type human MRP1 cDNA cloned into pNUT expression vector (11) was used as a template for the in vitro mutagenesis. The serine residue at position of 1334 in the Walker A motif of NBD2 was mutated to either alanine (A), cysteine (C), aspartic acid (D), histidine (H), asparagine (N), or threonine (T) by using the forward/reverse primers and the QuikChange site-directed mutagenesis kit from Stratagene (8). The forward and reverse primers used to introduce these mutations are as follows: S1334A/forward, 5′-CG GGA GCT GGG AAG GCG TCC CTG ACC CTG GGC-3′; S1334A/reverse, 5′-GCC CAG GGT CAG GGA CGC CTT CCC AGC TCC CG-3′; S1334C/forward, 5′-CG GGA GCT GGG AAG TGC TCC CTG ACC CTG GGC-3′; S1334C/reverse, 5′-GCC CAG GGT CAG GGA GCA CTT CCC AGC TCC CG-3′; S1334D/forward, 5′-CG GGA GCT GGG AAG GAC TCC CTG ACC CTG GGC-3′; S1334D/reverse, 5′-GCC CAG GGT CAG GGA GTC CTT CCC AGC TCC CG-3′; S1334H/forward, 5′-CG GGA GCT GGG AAG CAC TCC CTG ACC CTG GGC-3′; S1334H/reverse, 5′-GCC CAG GGT CAG GGA GTG CTT CCC AGC TCC CG-3′; S1334N/forward, 5′-CG GGA GCT GGG AAG AAC TCC CTG ACC CTG GGC-3′; S1334N/reverse, 5′-GCC CAG GGT CAG GGA GTT CTT CCC AGC TCC CG-3′; S1334T/forward, 5′-CG GGA GCT GGG AAG ACG TCC CTG ACC CTG GGC-3′; S1334T/reverse, 5′-GCC CAG GGT CAG GGA CGT CTT CCC AGC TCC CG-3′. The underlined sequences are codons for mutated residues. The serine residue at position1334 in the Walker A motif in pDual/N-half/C-half (12, 13) was mutated to either alanine (A) or threonine (T) by using the corresponding forward/reverse primers and the QuikChange site-directed mutagenesis kit from Stratagene (8). Recombinant viral DNA preparation and viral particle production were carried out according to the procedures described (12). Regions containing these mutations were sequenced to confirm that the correct clones were obtained.
Cell Culture and Cell Lines Expressing MRP1
Baby hamster kidney (BHK-21) cells were cultured in DMEM/F12 medium supplemented with 5% fetal bovine serum at 37 °C in 5% CO2. Subconfluent cells were transfected with pNUT-MRP1/His in the presence of 20 mM HEPES (pH 7.05), 137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, 6 mM dextrose, and 125 mM CaCl2 (11). Surviving colonies in media containing 200 μM methotrexate were mixed and amplified. Cells for membrane vesicle preparation were grown in DMEM/F12 media containing 5% fetal bovine serum and 200 μM methotrexate in roller bottles (on a roller machine from BELLCO).
Spodoptera frugiperda 21 (Sf21) cells were cultured in Grace’s insect cell medium supplemented with 5% heat-inactivated fetal bovine serum at 27 °C. Viral infection was performed according to Invitrogen’s recommendation.
Identification of MRP1 Protein
Western blot was performed according to the method described previously (8). 42.4 monoclonal antibody was used to identify the NBD1-containing fragments of MRP1, whereas 897.2 monoclonal antibody was used to detect the NBD2-containing fragments (8, 13). The secondary antibody used was anti-mouse Ig conjugated with horseradish peroxidase. Chemiluminescent film detection was performed according to the manufacturer’s recommendations (Pierce).
Membrane Vesicle Preparation
Membrane vesicles were prepared according to the procedure described previously (8). The membrane vesicle pellet was resuspended in a solution containing 10 mM Tris-HCl (pH 7.5), 250 mM sucrose, and 1× protease inhibitor cocktail (2 μg/mL aprotinine, 121 μg/mL benzamidine, 3.5 μg/mL E64, 1 μg/mL leupeptin, and 50 μg/mL Pefabloc). Aliquots of the membrane vesicles, after passage through a Liposofast vesicle extruder (Avestin, Ottawa, Canada), were stored in −80 °C.
Membrane Vesicle Transport
ATP-dependent LTC4 transport was assayed by a rapid filtration technique (14, 15). The assays were carried out in a 30 μL solution containing 3 μg of membrane proteins, 50 mM Tris-HCl (pH 7.5), 250 mM sucrose, 10 mM MgCl2, 200 nM LTC4 (17.54 nCi of 3H-labeled LTC4), and 4 mM AMP (used as negative control) or 4 mM ATP. After incubation at 37 °C for 1 min, the samples were brought back to ice and diluted with 1 mL of ice-cold 1× transport buffer (50 mM Tris-HCl, pH 7.5, 250 mM sucrose, and 10 mM MgCl2) and trapped on nitrocellulose membranes (0.2 μm) that had been equilibrated with 1× transport buffer. The filter was then washed with 10 mL of ice-cold 1× transport buffer, air-dried, and placed in a 10 mL of biodegradable counting scintillant (Amersham Pharmacia Biotech). The radioactivity bound to the nitrocellulose membrane was determined by liquid scintillation counting (Beckman LS 6000SC).
Photoaffinity Labeling of MRP1 Protein
Vanadate preparation (from sodium orthovanadate) and photoaffinity labeling of MRP1 protein were performed according to procedures described previously (8, 16, 17). Briefly, photolabeling experiments were carried out in a 10 μL solution containing 10 μg of membrane proteins, 40 mM Tris-HCl (pH7.5), 2 mM ouabain, 10 mM MgCl2, 0.1 mM EGTA, 800 μM vanadate, and either 5 μM [α-32P]-8-N3ADP (Figure 6), 10 μM [α-32P]-8-N3ATP, or 10 μM [γ-32P]-8-N3ATP (Figure 4). After incubation at 37 °C for 10 (Figure 6) or 2 min (Figure 4), the samples were brought back to ice, washed with 0.5 mL of ice-cold Tris–EGTA buffer (0.1 mM EGTA and 40 mM Tris, pH7.5), resuspended in 10 μL of ice-cold Tris–EGTA buffer, and UV-irradiated for 2 min. The labeled proteins were separated by polyacrylamide gel (7%) electrophoresis and electroblotted to a nitrocellulose membrane.
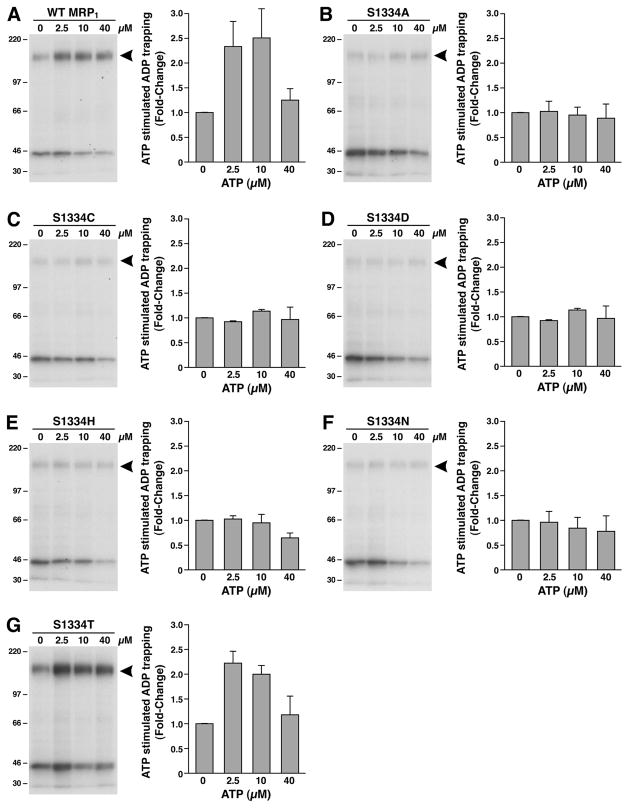
Figure 6.
Substitution of S1334 in the Walker A motif of NBD2 with an amino acid residue that eliminates the hydroxyl group abolished the ATP-enhanced vanadate-dependent ADP trapping at the mutated NBD2. The photolabeling was carried out in 10 μL of solution containing 10 mM MgCl2, 800 μM vanadate, 5 μM [α-32P]-8-N3ADP, varying concentrations of ATP as indicated on top of the gel, and 10 μg of MRP1-containing membrane vesicles. The reaction mixture was incubated at 37 °C for 10 min, brought back to ice, washed with 500 μL of ice-cold Tris–EGTA buffer (0.1 mM EGTA and 40 mM Tris-HCl, pH 7.5), and UV-irradiated (λ = 254 nm) on ice for 2 min. The labeled proteins were separated on a 7% polyacrylamide gel and electroblotted to a nitrocellulose membrane. The arrowhead on the right of the gel indicates the photolabeled MRP1 protein. The counts in each MRP1 band were measured by Instant Imager, and the amount of labeling in the absence of ATP was considered as 1 (n = 3).
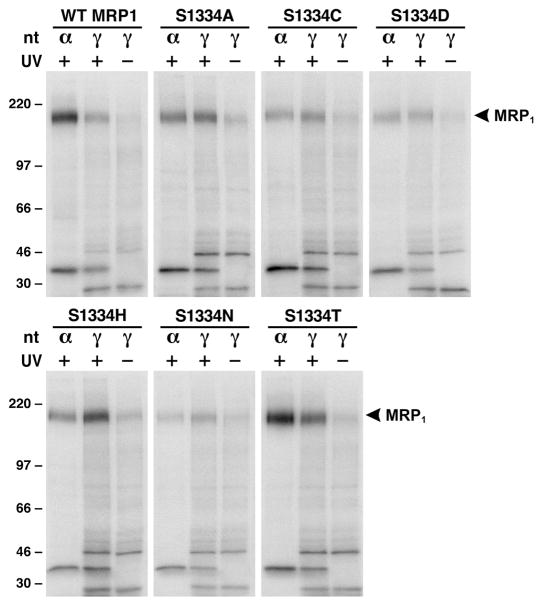
Figure 4.
Substitution of S1334 with an amino acid that eliminates the hydroxyl group at this position impaired ATP binding/hydrolysis at the mutated NBD2. Photolabeling experiments were carried out in 10 μL of solution containing 10 μg of membrane vesicle proteins (the amount of MRP1 protein determined in Figure 2A was adjusted to a similar amount by adding varying amounts of membrane vesicles prepared from parental BHK cell), 10 mM MgCl2, 800 μM vanadate, and 10 μM [α-32P]-8-N3ATP (α) or 10 μM [γ-32P]-8-N3ATP (γ) at 37 °C for 2 min. The reaction mixture was subjected to UV irradiation (+) or without UV irradiation (−) on ice for 2 min, separated by SDS–PAGE (7%), and electroblotted to a nitrocellulose membrane. Molecular mass markers (kDa) are indicated on the left. MRP1 on the right of the gel indicates the 32P-nucleotide-labeled MRP1 protein which was confirmed in Western blot by employing the NBD1-specific monoclonal antibody 42.4 (8).
Membrane vesicles containing MRP1 (25 μg of membrane proteins) were incubated with 3H-LTC4 (0.2 μCi, 61 nM) in 25 μL of solution containing 50 mM Tris-HCl (pH 7.5), 250 mM sucrose, and 10 mM MgCl2 in the presence (+) or absence (−) of 1 mM ATP and 200 μM vanadate at 37 °C for 10 min. The reaction mixture was frozen in liquid nitrogen and UV-irradiated for 2 min. This was repeated 15 times. Radiolabeled proteins were resolved by SDS–PAGE (7% gel), electroblotted to a nitrocellulose membrane, and autoradiographed on a Kodak BioMax MR film.
Endoglycosidase H or PNGase F Treatment of Proteins
The core-glycosylated oligosaccharides were removed by digestion with endoglycosidase H. Cells were lysed with 2% SDS and sonicated for 20 pulses to break the genomic DNA. Ten micrograms of the cell lysates was digested overnight at 37 °C with 200 units (New England BioLabs) of endoglycosidase H in 200 μL of solution containing 50 mM sodium acetate buffer (pH5.3), 0.5% NP-40, 1% β-mercaptoethanol, 0.2% SDS (final concentration), 10 μg of bovine serum albumin, and 1× protease inhibitors (9). The coreglycosylated and the complex-glycosylated oligosaccharides were removed by digestion with PNGase F. Ten micrograms of the cell lysates was digested overnight at 37 °C with 100 units (New England BioLabs) of PNGase F in 200 μL of solution containing 50 mM sodium phosphate buffer (pH 7.5), 0.5% NP-40, 1% β-mercaptoethanol, 0.2% SDS (final concentration), 10 μg bovine serum albumin, and 1× protease inhibitors (18). Following digestion, 800 μL of cold ethanol was added to precipitate the proteins. The pellets were collected by centrifugation at 4 °C for 15 min. The proteins were resolved by SDS–PAGE (7%), electroblotted to nitrocellulose membrane, and probed with monoclonal antibodies against human MRP1 protein (8).
Statistical Analysis
The results in Table 1 were presented as the means ± SD from three independent experiments. The two-tailed P values were calculated based on the unpaired t test from GraphPad Software Quick Calcs. By conventional criteria, if the P value is less than 0.05, the difference between two samples is considered to be statistically significant.
Table 1.
Km (Mg•ATP) and Vmax (LTC4) Values of Wild-Type and Mutant MRP1s
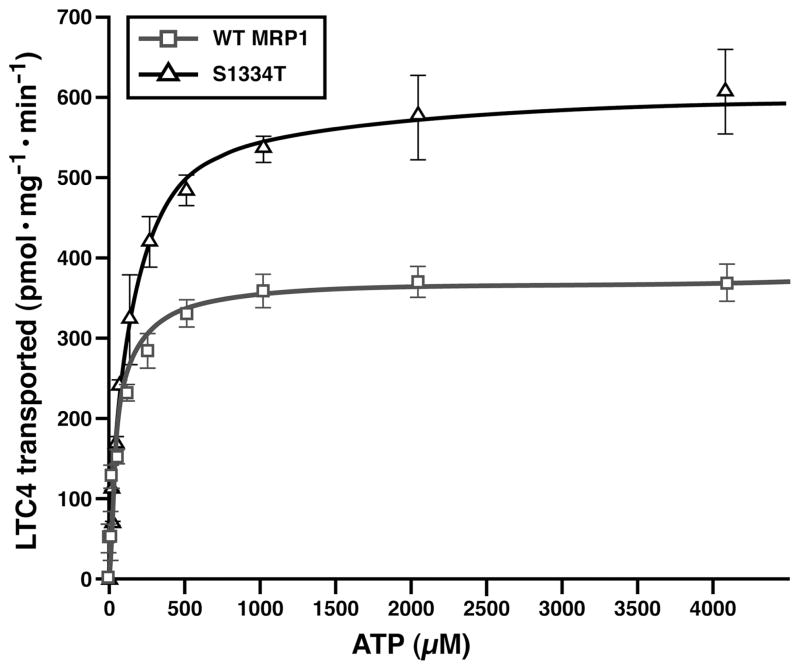
Km (Mg•ATP) and Vmax (LTC4) values (n = 3) for wild type and S1334T were derived from the corresponding Michaelis–Menten curves shown in Figure 3. The P values for comparison of Vmax (LTC4) and Km (Mg•ATP) of wild type versus S1334T are 0.0016 and 0.0024.
RESULTS
Substitution of S1334 in the Walker A Motif with an Amino Acid That Eliminates the Hydroxyl Group at This Position Does Not Affect the Protein Folding and Processing
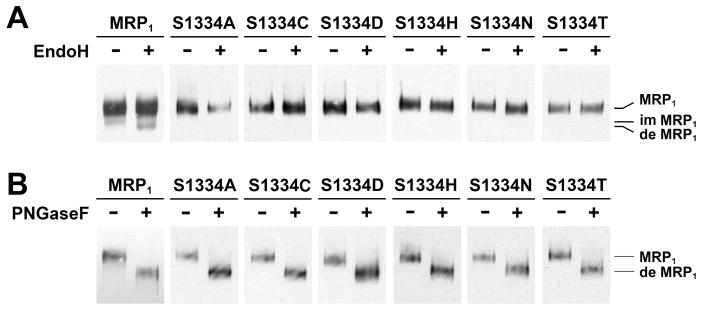
The results in Figure 1A showed that the 190 kDa wild-type MRP1 protein is resistant to endoglycosidase H digestion whereas the minor band is not, indicating that the majority of wild-type MRP1 protein is complex-glycosylated in vivo. All of the mutants, including S1334A, S1334C, S1334D, S1334H, S1334N, and S1334T, have a major band resistant to the endoglycosidase H digestion but sensitive to the PNGase F digestion (Figure 1B), indicating that all of these mutants mainly form complex-glycosylated mature proteins at 37 °C in vivo. If these mutants mainly form complexglycosylated mature MRP1 proteins, they should be sorted to the plasma membrane. Indeed, immunofluorescent staining with monoclonal antibody against human MRP1 demonstrated that all of these mutants were sorted to the plasma membrane surface (data not shown).
Figure 1.
Substitution of S1334 in the Walker A motif of NBD2 with an amino acid that eliminates the hydroxyl group at this position has no effect on protein folding. (A) All of the MRP1 mutants mainly form complex-glycosylated mature proteins at 37 °C. Cells grown at 37 °C were lysed with 2% SDS and incubated in the absence (−) or presence (+) of endoglycosidase H according to the method described in Experimental Procedures. (B) The complex-glycosylated MRP1 proteins are sensitive to PNGase F. The same cell lysates in panel A were incubated in the absence (−) or presence (+) of PNGase F according to the method described in Experimental Procedures. MRP1, im MRP1, and de MRP1 on the right indicate the complex-glycosylated mature MRP1, the core-glycosylated immature MRP1, and the deglycosylated MRP1 proteins that were detected in Western blots by employing the monoclonal antibodies 42.4 and 897.2 against MRP1 (8).
Substitution of S1334 in the Walker A Motif in NBD2 with an Amino Acid That Eliminates the Hydroxyl Group at This Position Abolished ATP-Dependent LTC4 Transport
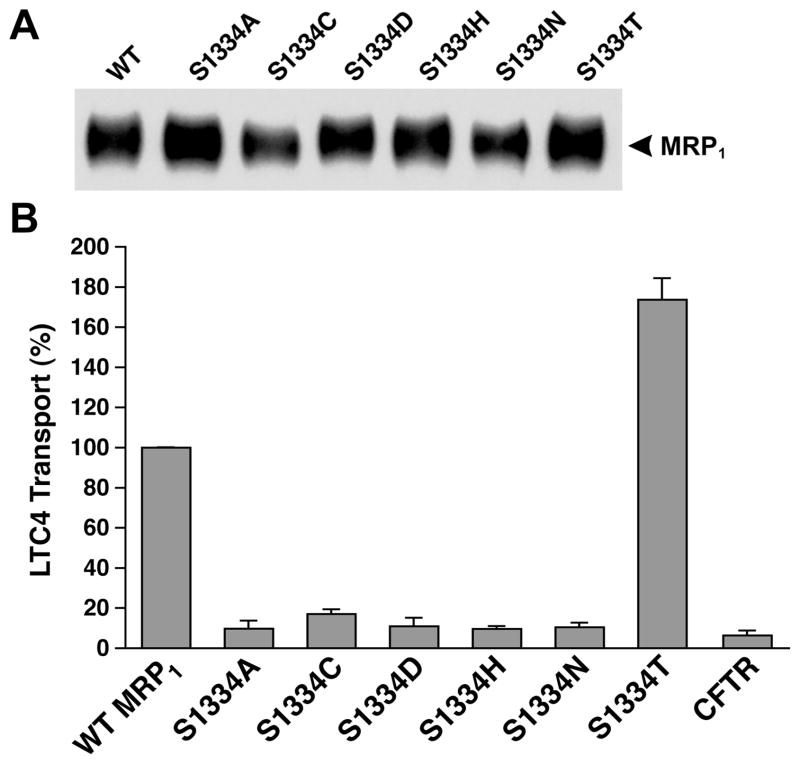
Since S685 in Walker A motif in NBD1 is involved in interacting with the Mg2+ cofactor and the β-phosphate of the bound Mg•ATP (1, 7), thus, substitution of S685 with an amino acid that eliminated the hydroxyl group at this position significantly affected the ATP-dependent LTC4 transport (10). In order to test whether the corresponding mutations in NBD2 also affect the ATP-dependent solute transport, membrane vesicles were prepared from the cells grown at 37 °C (Figure 2A) and used to do the ATP-dependent LTC4 transport assay. As shown in Figure 2B, S1334A-, S1334C-, S1334D-, S1334H-, or S1334N-mutated MRP1 almost completely abolished the ATP-dependent LTC4 transport, whereas S1334T mutation, which retained the hydroxyl group at this position, exerted ~175% of wild-type MRP1 transport activity, suggesting that the hydroxyl group at this position plays a crucial role for the ATP binding/hydrolysis and ATP-dependent solute transport by MRP1.
Figure 2.
Substitution of S1334 with an amino acid that eliminates the hydroxyl group abolishes the ATP-dependent LTC4 transport. (A) MRP1 contents in membrane vesicles. Membrane vesicles were prepared according to the method described in Experimental Procedures. The amounts of proteins loaded in the gel are 400 and 800 ng (only the results derived from 800 ng of protein are shown). MRP1 on the right indicates the complex-glycosylated mature MRP1 that was detected in Western blot by employing the monoclonal antibody 42.4 against NBD1 (8). The intensities of the MRP1 bands were determined by a scanning densitometer. The mean ratios (n = 2, including the results derived from 400 and 800 ng of protein), considering the amount of wild-type MRP1 as 1, of the mutant proteins are as follows: S1334A, 1.46 ± 0.06; S1334C, 0.84 ± 0.14; S1334D, 1.07 ± 0.69; S1334H, 1.17 ± 0.16; S1334N, 0.97 ± 0.08; S1334T, 1.30 ± 0.11. (B) ATP-dependent LTC4 uptake by membrane vesicles prepared from the cells expressing either wild-type or mutant MRP1s. The assays were carried out in a 30 μL solution containing 3 μg of membrane vesicles (the amount of MRP1 protein determined in panel A was adjusted to a similar amount by adding a varying amount of membrane vesicles prepared from parental BHK cells) and 4 mM AMP (used as a control) or 4 mM ATP at 37 °C for 1 min. The amount of LTC4 bound to the membrane vesicles in the presence of 4 mM AMP was subtracted from the corresponding samples in the presence of 4 mM ATP. The data are the means ± SD of three triplicate determinations.
The Km (Mg•ATP) Value of S1334T Is Significantly Higher than That of Wild-Type MRP1
The γ-oxygen of the Walker A serine residue may interact with the β-phosphate of the bound ATP and the Mg2+ cofactor, along with five other oxygen atoms from the δ-oxygen of the Walker B aspartate residue, the β- and γ-phosphate of the bound ATP, and water molecules to form the octahedral coordination geometry (1, 2, 4–7). Thus, substitution of this serine with a threonine residue having an extra methyl group at this position might alter the kinetic parameters of the protein. In order to test this possibility, varying concentrations of ATP were utilized to do the ATP-dependent LTC4 transport in the presence of 10 mM MgCl2 (Figure 3). As shown in Figure 3 and Table 1, the Km(Mg•ATP) of S1334T (104 μM Mg•ATP) is significantly higher (with a P value of 0.0024) than that of wild-type MRP1 (61 μM Mg•ATP). In addition, the Vmax (LTC4) value of S1334T (618 pmol mg−1 min−1) is also significantly higher (with a P value of 0.0016) than that of wild-type MRP1 (342 pmol mg−1 min−1), consistent with the results derived from a solution containing 4 mM ATP and 10 mM MgCl2 (Figure 2B).
Figure 3.
S1334T mutation increases the Km (ATP) for ATPdependent LTC4 transport. Membrane vesicles prepared in Figure 2 were used to do the ATP-dependent LTC4 transport, according to the methods described in Experimental Procedures. Briefly, the assays were carried out in a 30 μL solution containing 3 μg of membrane vesicles, 10 mM MgCl2, and varying concentrations of ATP at 37 °C for 1 min. Since the amounts of MRP1 proteins determined in Figure 2A were different, the amounts of membrane vesicles used in these experiments were adjusted to a similar amount by adding varying amounts of membrane vesicles prepared from parental BHK cells: 3 μg of wild-type MRP1; 2.308 μg of S1334T plus 0.692 μg of BHK. The data are the means ± SD of triplicate determinations.
Elimination of the Hydroxyl Group in Walker A S1334 Significantly Affects ATP Hydrolysis at the Mutated NBD2
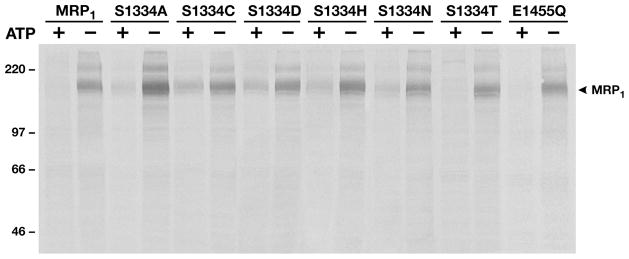
Since substitution of S1334 with a threonine residue having an extra methyl group at this position altered the kinetic parameters of the protein, owing to affecting ATP binding/hydrolysis and release of the hydrolysis products, ADP and phosphate, elimination of the hydroxyl group at this position, by substitution of S1334 with an alanine or other amino acids, should have more severe effects on the ATP binding/hydrolysis and ATP-dependent substrate transport. Indeed, all of the mutants, except S1334T, almost completely abolished the ATP-dependent LTC4 transport (Figure 2B), perhaps owing to affecting the ATP binding/hydrolysis. In order to prove this hypothesis, a similar amount of MRP1 protein in 10 μg of membrane protein (see Figure 4 legend) was used to label them with either [α-32P]-8-N3ATP or [γ-32P]-8-N3ATP (with the same specific activity as [α-32P]-8-N3ATP) at 37 °C in the presence of vanadate. As shown in Figure 4, MRP1 protein, regardless whether it is wild type or mutant, is labeled by [γ-32P]-8-N3ATP without UV irradiation, indicating that MRP1 protein is phospharylated under this condition. However, the labeling intensity of wildtype MRP1 or S1334T with [α-32P]-8-N3ATP, including the intact [α-32P]-8-N3ATP bound at NBD1 and the vanadate-trapped ATP hydrolysis product [α-32P]-8-N3ADP at NBD2 (8), is stronger than that of [γ-32P]-8-N3ATP labeling including the intact [γ-32P]-8-N3ATP bound at NBD1 and NBD2 and the 32P-phospharylated products (8), implying that the majority of the bound ATP at NBD2 is hydrolyzed. The labeling intensity of MRP1 mutants, including S1334A, S1334C, S1334D, S1334H, and S1334N, with [α-32P]-8-N3ATP is weaker than that of wild type or S1334T (Figure 4), indicating that these mutants without the hydroxyl group at this position affect ATP binding, hydrolysis, and vanadatedependent [α-32P]-8-N3ADP trapping. Comparison of labeling intensities with [α-32P]-8-N3ATP and [γ-32P]-8-N3ATP further supports this conclusion: (1) The labeling intensity of MRP1 mutants, including S1334A, S1334C, S1334D, S1334H, and S1334N, with [α-32P]-8-N3ATP is either similar to or weaker than that of [γ-32P]-8-N3ATP labeling (Figure 4), indicating that the bound ATP in these mutants is not efficiently hydrolyzed. (2) The intensity of [γ-32P]-8-N3ATP labeling on the mutants, including S1334A, S1334C, S1334D, S1334H, and S1334N, is either stronger than wild type or similar to wild type (Figure 4), implying that most of those [γ-32P]-8-N3ATP labeling might occur at the unmutated NBD1 (8).
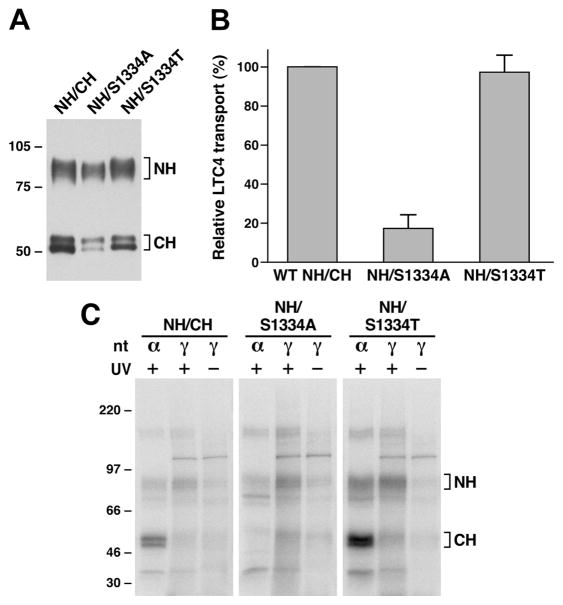
In order to prove this hypothesis, S1334A and S1334T mutations were introduced into pDual/N-half/C-half expression plasmid (12, 13) and expressed in Sf21 cells. Membrane vesicles prepared from Sf21 cells (Figure 5A) were used to do ATP-dependent LTC4 transport and the photoaffinity labeling with either [α-32P]-8-N3ATP or [γ-32P]-8-N3ATP. As shown in Figure 5B, the ATP-dependent LTC4 transport activity of wild type N-half plus S1334T-mutated C-half is more or less similar to that of wild-type, whereas the transport activity of the wild type N-half plus S1334Amutated C-half is significantly reduced. The [γ-32P]-8-N3ATP labeling of wild type NBD1-containing N-half fragment is much higher than the corresponding labeling in wild type or S1334T-mutated NBD2-containing C-half fragment (Figure 5C), whereas the [α-32P]-8-N3ATP labeling of wild type or S1334T-mutated NBD2-containing C-half fragment in the presence of vanadate is much higher than the corresponding labeling in NBD1-containing N-half fragment (Figure 5C), implying that ATP bound to wild type or S1334T-mutated NBD2 is efficiently hydrolyzed. In contrast, the [γ-32P]-8-N3ATP or [α-32P]-8-N3ATP labeling of the unmutated NBD1-containing N-half fragment is stronger than the corresponding labeling on the S1334A-mutated NBD2-containing C-half (Figure 5C), suggesting that most of the [γ-32P]-8-N3ATP or [α-32P]-8-N3ATP labeling occurred at the unmutated NBD1. In addition, the labeling intensity of the S1334A-mutated NBD2-containing C-half with [γ-32P]-8-N3ATP is stronger than that of corresponding labeling with [α-32P]-8-N3ATP (Figure 5C), implying that the trace amount of ATP bound to the S1334A-mutated NBD2 is not efficiently hydrolyzed.
Figure 5.
Substitution of S1334 with an alanine residue significantly impaired ATP binding/hydrolysis at the mutated NBD2. (A) MRP1 contents in membrane vesicles prepared from Sf21 cells. Membrane vesicles were prepared according to the method described in Experimental Procedures. The amounts of proteins loaded in the gel are 300 ng of membrane proteins. NH and CH on the right indicate the N-terminal half (detected by the mAb 42.4 against NBD1) and C-terminal half (detected by the mAb 897.2 against NBD2). The intensities of the MRP1 bands were determined by a scanning densitometer. The mean ratios (n = 2, including the results derived from 200 and 300 ng of protein), considering the amount of wild type MRP1 as 1, of the mutant proteins are as follows: S1334A, 0.68 ± 0.06; S1334T, 0.90 ± 0.06. (B) ATP-dependent LTC4 uptake by membrane vesicles prepared from wild type and mutant MRP1s. The assays were carried as described in Figure 2B. (C) S1334A-mutated MRP1 affects both ATP binding and hydrolysis at the mutated NBD2. Wild type MRP1 or S1334A- or S1334T-mutated MRP1 was introduced into the pDual/N-half/C-half and expressed in Sf21 insect cells (12, 25). The membrane vesicles prepared from the viral particle infected Sf21 cells were used to do photolabeling as described in Figure 4. NH and CH on the right indicate the 32P-nucleotide-labeled NBD1-containing N-half and NBD2-containing C-half.
Walker A Serine Mutations Impaired the ATP-Enhanced Vanadate-Dependent ADP Trapping at the Mutated NBD2
The results in Figure 4 implied that substitution of the S1334 with a residue that eliminates the hydroxyl group significantly impaired ATP binding at the mutated NBD2. Limited trypsin digestion (8) of the [α-32P]-8-N3ATP-labeled MRP1 may confirm this conclusion. However, based on the labeling intensities of the S1334A mutant in Figure 5C, the labeling of the mutated NBD2 fragments, after the partial trypsin digestion, might not be detected. Thus, we have to find an alternative way to confirm this conclusion. We have found that ATP binding at the NBD1 induces conformational changes of the protein and enhances nucleotide binding at the NBD2 (16, 17). However, if a mutant significantly affected the ATP binding, such a mutant, such as the Walker A mutation of K1333L, would abrogate the ATP-enhanced nucleotide binding at NBD2 (16). Therefore, we decided to determine whether these S1334 mutants would abrogate the ATP-enhanced nucleotide binding at the mutated NBD2. As shown in Figure 6, 2.5 or 10 μM ATP significantly enhanced the vanadate-dependent [α-32P]-8-N3ADP trapping in wild type MRP1 (Figure 6A) or in S1334T (Figure 6G), whereas there is no enhancement effect in S1334A (Figure 6B), S1334C (Figure 6C), S1334D (Figure 6D), S1334H (Figure 6E), or S1334N (Figure 6F), suggesting that these mutations significantly impaired the nucleotide binding at the mutated NBD2.
Walker A Serine Mutations That Eliminate the Hydroxyl Group at This Position Diminished the Ability to Inhibit the LTC4 Binding
Occupancies of NBD1 and NBD2 by ATP strongly attenuated the substrate, such as LTC4, binding to the MRP1 protein (19–21). Mutations that affect ATP binding may either diminish or abolish this ATP-dependent inhibitory effect (19–21). Thus, if these Walker A serine mutations that eliminated the hydroxyl group significantly impaired the nucleotide binding at the mutated NBD2, these mutations might either diminish or abolish this ATP-dependent inhibitory effect. In order to test this hypothesis, membrane vesicles containing these mutants were used to label them with 3H-LTC4 in the presence or absence of 1 mM ATP and vanadate. As shown in Figure 7, the 3H-LTC4 labeling on wild type MRP1 and S1334T as well as E1455Q is almost completely inhibited by the presence of ATP and vanadate. In contrast, the labeling on S1334A, S1334C, S1334D, S1334H, or S1334N is only partially diminished, implying that ATP binding or vanadate-dependent ATP hydrolysis product ADP trapping at the mutated NBD2 is impaired.
Figure 7.
Walker A serine mutations that eliminate the hydroxyl group at this position diminished the ability to inhibit LTC4 binding. Photolabeling of MRP1 protein with 3H-LTC4 was performed according to the method described in Experimental Procedures in the presence (+) or absence (−) of 1 mM ATP and 200 μM vanadate.
DISCUSSION
We have found that substitution of the Walker A serine S685 in NBD1 of MRP1 with an amino acid residue, such as cysteine, that eliminates the hydroxyl group at this position resulted in misprocessing of the protein (10), indicating that the interactions of the hydroxyl group in S685 with the surrounding residues play a crucial role for MRP1 protein folding in vivo. In contrast, mutation of the corresponding residue in NBD2 of MRP1, such as S1334A, S1334C, S1334D, S1334H, or S1334N, has no effect on the protein folding and processing (Figure 1), implying that the stereo structure surrounding the hydroxyl group of S1334 in NBD2 might be different from that of NBD1. However, molecular modeling of the MRP1 NBD1•ATP•ATP•NBD2 sandwich structure (the model is not shown), based on the crystal structure of wild-type MRP1-NBD1 (1) and MJ0796-E171Q (7), indicates that the Walker A serine S1334 might potentially form a hydrogen bond with the Walker B aspartic acid D1454. Thus, even if the Walker A serine S1334 in NBD2 may form a hydrogen bond with the Walker B aspartic acid D1454, this hydrogen bond formation is not crucial for the protein folding and processing. Indeed, elimination of the carboxyl group at D1454, such as D1454L/E1455L (8), had no effect on the protein folding and processing. Additionally, substitution of the Walker B aspartic acid D555 in NBD1 or D1200 in NBD2 of P-glycoprotein (ABCB1) also did not cause misfolding of the protein (22). However, substitution of the Walker B aspartic acid D572 in NBD1 of CFTR (or ABCC7) with an asparagine resulted in misprocessing of the protein (23). Thus, it might be a unique feature of NBD1 of ABCC subfamily members of ABC transporters that eliminations of the Walker A serine hydroxyl group and/or the Walker B aspartic acid carboxyl group resulted in misprocessing of the protein.
Substitution of the Walker A serine S685 in NBD1 of MRP1 with an amino acid residue, such as alanine, that prevents the interactions with the magnesium cofactor and the β-phosphate of the bound Mg•ATP significantly reduced the ATP-dependent LTC4 transport activity (10), indicating that the interactions of the hydroxyl group at the position of 685 with the magnesium cofactor and the β-phosphate of the bound Mg•ATP play an important role for Mg•ATP binding/hydrolysis and ATP-dependent LTC4 transport. If the hydroxyl group of the Walker A serine S1334 in NBD2 of MRP1 plays a similar role as that of S685, elimination of this hydroxyl group at the position of S1334 should also significantly reduce the ATP-dependent LTC4 transport activity. Indeed, substitution of S1334 with an alanine residue (S1334A) reduced the ATP-dependent LTC4 transport activity to ~10% of wild type (Figure 2B). Interestingly, substitution of the hydroxyl group in S1334 with a sulfhydryl group (S1334C) exerted slightly higher transport activity than that of S1334A, S1334D, S1334H, or S1334N (Figure 2B), implying that the sulfhydryl group in S1334C might weakly bind to the magnesium cofactor and the β-phosphate of the bound ATP. In contrast to other mutants, substitution of the S1334 with a threonine (S1334T) increased ATP-dependent LTC4 transport to ~175% of the wild-type MRP1 (Figures 2B and 3 and Table 1), indicating that the interactions of the hydroxyl group at the position of 1334 with the Mg2+ cofactor and the β-phosphate of the bound Mg•ATP(1, 2, 4–7) play a crucial role for ATP binding/hydrolysis at NBD2 and ATP-dependent LTC4 transport. The results shown in Figures 4, 5, 6, and 7 strongly support this conclusion. The labeling intensities of [α-32P]-8-N3ATP in wild type or S1334T are stronger than that of [γ-32P]-8-N3ATP (Figure 4), whereas the labeling intensities of [α-32P]-8-N3ATP in S1334A, S1334C, S1334D, S1334H, or S1334N are slightly weaker than that of [γ-32P]-8-N3ATP (Figure 4), suggesting that even though there might be a trace amount of ATP binding at the mutated NBD2, the bound nucleotide might not be efficiently hydrolyzed. Indeed, a certain amount of [α-32P]-8-N3ATP hydrolysis product [α-32P]-8-N3ADP was trapped in wildtype or S1334T-mutated NBD2, whereas there was no detectable amount of [α-32P]-8-N3ADP trapped in S1334Amutated NBD2 (Figure 5C). All of the S1334 mutants, including S1334A (Figure 6B), S1334C (Figure 6C), S1334D (Figure 6D), S1334H (Figure 6E), and S1334N (Figure 6F), lost the ATP-enhanced vanadate-dependent ADP trapping (16, 17) at the mutated NBD2, suggesting that these mutations significantly decreased the affinity for nucleotide at the mutated NBD2. In contrast, although substitution of the S1334 with a threonine (S1334T) introduced an extra methyl group, but kept the hydroxyl group at the original position, the S1334T mutation retained the ATP-enhanced vanadate-dependent ADP trapping (Figure 6G), indicating that the interactions of the hydroxyl group at position 1334 with the Mg2+ cofactor and the β-phosphate of the bound Mg•ATP (1, 2, 4–7) play a very important role for ATP binding at NBD2. The reduced nucleotide binding at the mutated NBD2, such as S1334A, S1334C, S1334D, S1334H, and S1334N, significantly decreased the ability to inhibit the LTC4 binding (Figure 7), similar to the mutations of K684E, G771A, or K1333E (19). This inhibitory effect is directly related to nucleotide binding, but not hydrolysis, at NBD1 and NBD2 since the E1455Q mutation, which abolished ATP hydrolysis but not binding (12), exerted similar ability as wild type MRP1 to inhibit the LTC4 binding (Figure 7). These results are consistent with the substitutions of H1486 in the H-loop of NBD2 with residues that might potentially form a hydrogen bond with the γ-phosphate of the bound Mg•ATP exerted either full or partial function whereas the substitutions with residues that prevented formation of these hydrogen bonds almost abolished the ATP-dependent solute transport completely (24). Thus, the binding forces between the residues in the Mg•ATP binding pocket and Mg•ATP at NBD2 might be a decisive factor for MRP1 to transport the bound substrate across the biological membranes.
Acknowledgments
We thank Irene Beauvais for preparation of the manuscript and Marv Ruona and Nichole Boruff for preparation of the graphics.
Footnotes
This work was supported by a grant from the National Cancer Institute, National Institutes of Health (CA89078 to X.C.).
Abbreviations: BHK, baby hamster kidney; Sf21, Spodoptera frugiperda 21; ABC, ATP binding cassette; MRP1, multidrug resistance-associated protein 1; CFTR, cystic fibrosis transmembrane-conductance regulator; NBD, nucleotide binding domain; LTC4, leukotriene C4; 8-N3ATP, 8-azidoadenosine 5′-triphosphate; PBS, phosphate-buffered saline; EGTA, ethylene glycol bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid; SDS, sodium dodecyl sulfate.
References
- 1.Ramaen O, Leulliot N, Sizun C, Ulryck N, Pamlard O, Lallemand JY, Tilbeurgh H, Jacquet E. Structure of the human multidrug resistance protein 1 nucleotide binding domain 1 bound to Mg2+/ATP reveals a non-productive catalytic site. J Mol Biol. 2006;359:940–949. doi: 10.1016/j.jmb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Lewis HA, Buchanan SG, Burley SK, Conners K, Dickey M, Dorwart M, Fowler R, Gao X, Guggino WB, Hendrickson WA, Hunt JF, Kearins MC, Lorimer D, Maloney PC, Post KW, Rajashankar KR, Rutter ME, Sauder JM, Shriver S, Thibodeau PH, Thomas PJ, Zhang M, Zhao X, Emtage S. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaitseva J, Jenewein S, Jumpertz T, Holland IB, Schmitt L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 2005;24:1901–1910. doi: 10.1038/sj.emboj.7600657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verdon G, Albers SV, Dijkstra BW, Driessen AJ, Thunnissen AM. Crystal structures of the ATPase subunit of the glucose ABC transporter from Sulfolobus solfataricus: nucleotide-free and nucleotide-bound conformations. J Mol Biol. 2003;330:343–358. doi: 10.1016/s0022-2836(03)00575-8. [DOI] [PubMed] [Google Scholar]
- 5.Lu G, Westbrooks JM, Davidson AL, Chen J. ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation. Proc Natl Acad Sci USA. 2005;102:17969–17974. doi: 10.1073/pnas.0506039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Procko E, Ferrin-O’Connell I, Ng SL, Gaudet R. Distinct structural and functional properties of the ATPase sites in an asymmetric ABC transporter. Mol Cell. 2006;24:51–62. doi: 10.1016/j.molcel.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 7.Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, Hunt JF. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou Y, Cui L, Riordan JR, Chang XB. Allosteric interactions between the two non-equivalent nucleotide binding domains of multidrug resistance protein MRP1. J Biol Chem. 2000;275:20280–20287. doi: 10.1074/jbc.M001109200. [DOI] [PubMed] [Google Scholar]
- 9.Cui L, Hou YX, Riordan JR, Chang XB. Mutations of the Walker B motif in the first nucleotide binding domain of multidrug resistance protein MRP1 prevent conformational maturation. Arch Biochem Biophys. 2001;392:153–161. doi: 10.1006/abbi.2001.2441. [DOI] [PubMed] [Google Scholar]
- 10.Yang R, Scavetta R, Chang XB. The hydroxyl group of S685 in Walker A motif and the carboxyl group of D792 in Walker B motif of NBD1 play a crucial role for multidrug resistance protein folding and function. Biochim Biophys Acta. 2008;1778:454–465. doi: 10.1016/j.bbamem.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang XB, Hou YX, Riordan JR. ATPase activity of purified multidrug resistance-associated protein. J Biol Chem. 1997;272:30962–30968. doi: 10.1074/jbc.272.49.30962. [erratum: (1998) J. Biol. Chem. 273, 7782] [DOI] [PubMed] [Google Scholar]
- 12.Yang R, McBride A, Hou YX, Goldberg A, Chang XB. Nucleotide dissociation from NBD1 promotes solute transport by MRP1. Biochim Biophys Acta. 2005;1668:248–261. doi: 10.1016/j.bbamem.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Q, Chang XB. Mutation of the aromatic amino acid interacting with adenine moiety of ATP to a polar residue alters the properties of multidrug resistance protein 1. J Biol Chem. 2004;279:48505–48512. doi: 10.1074/jbc.M408343200. [DOI] [PubMed] [Google Scholar]
- 14.Leier I, Jedlitschky G, Buchholz U, Keppler D. Characterization of the ATP-dependent leukotriene C4 export carrier in mastocytoma cells. Eur J Biochem. 1994;220:599–606. doi: 10.1111/j.1432-1033.1994.tb18661.x. [DOI] [PubMed] [Google Scholar]
- 15.Loe DW, Almquist KC, Deeley RG, Cole SP. Multidrug resistance protein (MRP)-mediated transport of leukotriene C4 and chemotherapeutic agents in membrane vesicles. Demonstration of glutathione-dependent vincristine transport. J Biol Chem. 1996a;271:9675–9682. doi: 10.1074/jbc.271.16.9675. [DOI] [PubMed] [Google Scholar]
- 16.Hou YX, Cui L, Riordan JR, Chang XB. ATP binding to the first nucleotide-binding domain of multidrug resistance protein MRP1 increases binding and hydrolysis of ATP and trapping of ADP at the second domain. J Biol Chem. 2002;277:5110–5119. doi: 10.1074/jbc.M107133200. [DOI] [PubMed] [Google Scholar]
- 17.Hou YX, Riordan JR, Chang XB. ATP binding, not hydrolysis, at the first nucleotide-binding domain of multidrug resistance-associated protein MRP1 enhances ADP•Vi trapping at the second domain. J Biol Chem. 2003;278:3599–3605. doi: 10.1074/jbc.M210480200. [DOI] [PubMed] [Google Scholar]
- 18.Almquist KC, Loe DW, Hipfner DR, Mackie JE, Cole SP, Deeley RG. Characterization of the Mr 190,000 multidrug resistance protein (MRP) in drug-selected and transfected human tumor cell. Cancer Res. 1995;55:102–110. [PubMed] [Google Scholar]
- 19.Payen L, Gao M, Westlake C, Theis A, Cole SP, Deeley RG. Functional interactions between nucleotide binding domains and leukotriene C4 binding sites of multidrug resistance protein 1 (ABCC1) Mol Pharmacol. 2005;67:1944–1953. doi: 10.1124/mol.104.007708. [DOI] [PubMed] [Google Scholar]
- 20.Payen LF, Gao M, Westlake CJ, Cole SP, Deeley RG. Role of carboxylate residues adjacent to the conserved core Walker B motifs in the catalytic cycle of multidrug resistance protein 1 (ABCC1) J Biol Chem. 2003;278:38537–38547. doi: 10.1074/jbc.M305786200. [DOI] [PubMed] [Google Scholar]
- 21.Qian YM, Qiu W, Gao M, Westlake CJ, Cole SP, Deeley RG. Characterization of binding of leukotriene C4 by human multidrug resistance protein 1: evidence of differential interactions with NH2- and COOH-proximal halves of the protein. J Biol Chem. 2001;276:38636–38644. doi: 10.1074/jbc.M107025200. [DOI] [PubMed] [Google Scholar]
- 22.Hrycyna CA, Ramachandra M, Germann UA, Cheng PW, Pastan I, Gottesman MM. Both ATP sites of human P-glycoprotein are essential but not symmetric. Biochemistry. 1999;38:13887–13899. doi: 10.1021/bi991115m. [DOI] [PubMed] [Google Scholar]
- 23.Gentzsch M, Choudhury A, Chang XB, Pagano RE, Riordan JR. Misassembled mutant ΔF508 CFTR in the distal secretory pathway alters cellular lipid trafficking. J Cell Sci. 2007;120:447–455. doi: 10.1242/jcs.03350. [DOI] [PubMed] [Google Scholar]
- 24.Yang R, Chang XB. Hydrogen-bond formation of the residue in H-loop of the nucleotide binding domain 2 with the ATP in this site and/or other residues of multidrug resistance protein MRP1 plays a crucial role during ATP-dependent solute transport. Biochim Biophys Acta. 2007;1768:324–335. doi: 10.1016/j.bbamem.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao M, Cui HR, Loe DW, Grant CE, Almquist KC, Cole SP, Deeley RG. Comparison of the functional characteristics of the nucleotide binding domains of multidrug resistance protein 1. J Biol Chem. 2000;275:13098–13108. doi: 10.1074/jbc.275.17.13098. [DOI] [PubMed] [Google Scholar]