Despite the importance, abundance, and diversity of bacteria, we are just beginning to understand their evolutionary biology. Sex, understood as genetic exchange, is one of the keystone issues in bacterial evolution. Most bacteria reproduce by binary fission, and genetic exchange is independent of reproduction (1). If there is little or no genetic exchange among clones, each strain evolves as an independent lineage, and standard population genetics concepts, such as allelic frequencies and changes of these frequencies in populations, are not applicable (2). On the other hand, if recombination is common among related bacterial lineages (i.e., within bacterial species), we may analyze bacterial populations like most other organisms using standard population genetics methods (2). Recently, Maynard-Smith et al. (3), using detailed analyses based on multilocus linkage disequilibrium, have shown that there is a wide range of bacterial sexualities, ranging from lineages with little or no recombination (like Salmonella) to others that are almost panmictic (like Neisseria gonorrhoeae), and some with intermediate (and more complicated) sexualities: localized recombination among closely related strains (Rhizobium meliloti) or apparently very clonal populations, due to few very successful strains, have a structure called “epidemic” (e.g., Neisseria meningitidis) (3). These analyses are mainly based on multilocus enzyme electrophoresis data from chromosomal genes. But bacteria have a fascinating complication; they usually present accessory genes, in the form of smaller chromosomes, known as plasmids (4, 5). Plasmids usually encode specific functions, such as conjugation, antibiotic resistance, sugar utilization, colicin activity, and nitrogen fixation (5–8). Some plasmids are small and cryptic (i.e, without any known function, if any), whereas others are large and more complex (5, 9). Plasmidic genes are not only dynamic, due to rearrangements and duplications within and among them (9, 10), they are also capable of moving among strains, among related bacterial species, or even among different genera (9, 11–15). This process is called “horizontal” or “lateral” transfer of plasmids (14). The mobility of plasmidic genes is relevant for the evolutionary ecology of bacteria, since they may confer instantaneous adaptation in changing environments, or they may be costly to the bacteria carrying them (16). If plasmids are only inherited in a “vertical” fashion (from ancestor to their descendants by binary fission), then there should be a close linkage between chromosome and plasmid, and they would evolve together as one evolutionary unit. But if plasmids are very mobile, they would have evolutionary and populational dynamics of their own, and they may behave in a selfish way, similar to pathogens. In this case, they would be found in different chromosomal backgrounds, and their numbers would depend on the advantages they confer to the host, their cost, and their rate of transmission (17).
In this issue of the Proceedings, Wernegreen et al. (5) give us some insight on the dynamics and patterns of plasmid evolution by analyzing Rhizobium associated to four sympatric species of wild clover (Trifolium) growing together in two natural populations from nondisturbed meadows of the Sierra Nevada, in California. To interpret their results from a qualitative perspective, following Maynard-Smith et al. (3) ideas on the genetic structure of bacteria, we propose the following plasmid/chromosome evolutionary patterns (Fig. 1) (considering that the true phylogenies of both chromosome and plasmid are know):
Figure 1.
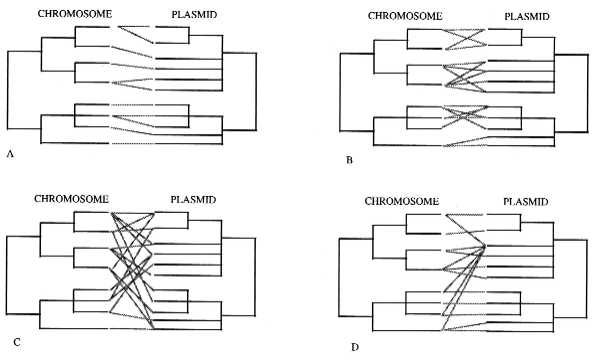
Possible chromosome and plasmid phylogenies (see text). (A) The clonal plasmid/chromosome pattern. (B) The limited plasmid transfer pattern. (C) The “panmictic” plasmid pattern. (D) The “epidemic” plasmid pattern.
(i) The clonal plasmid/chromosome pattern: If the chromosome phylogeny is perfectly mirrored in the plasmid phylogeny, there may be strong coevolution between both plasmid and host, with very limited plasmid transfer among bacteria (Fig. 1A).
(ii) The limited plasmid transfer pattern: There is some plasmid transfer, but only among closely related bacterial strains. The movement is either limited by molecular mechanisms that mediate the transfer and recognition of plasmids, by natural selection which eliminates the “wrong” combinations, or by ecological constraints (e.g., limited dispersal capabilities, different spatial population structures and distributions) (Fig. 1B).
(iii) The “panmictic” plasmid pattern: The plasmids are freely dispersed among chromosomal backgrounds, without any restrictions other than natural selection acting upon the advantages the plasmids may confer to the bacteria and costs of carrying and expressing the plasmid genome (Fig. 1C).
(iv) The “epidemic” plasmid pattern: A very successful plasmid is dispersed in a wide range of chromosome backgrounds. This could be the case of plasmids that carry resistance genes to antibiotics (Fig. 1D).
Recent detailed studies are helping us to understand how common these patterns are. For instance, Boyd and Hartl (11) found that the F plasmid (a large, low copy number conjugational plasmid) from E. coli is also present in some strains of Salmonella enterica, in an almost identical form. Both lineages are thought to have diverged approximately 140 million years ago. They also detected recombination among different genes in the plasmids. This case may suggest either a panmictic (Fig. 1C) or an epidemic plasmid pattern (Fig. 1D).
The most studied plasmid–chromosome system is the one formed by the nitrogen-fixing bacteria of the genus Rhizobium, and their sym plasmid. In these bacteria, the genetic information that enables them to interact with plant roots, to form nitrogen-fixing nodules, is coded in the large sym plasmid (9). Population genetics analyses indicated extensive plasmidic horizontal exchange in rhizobia associated with beans (13, 15, 18) and clover (12, 19), suggesting that most Rhizobium present a panmictic recombination pattern, where plasmids are intensively transferred among strains.
But how can we evaluate the amount of plasmid transfer? Valdés and Piñero (14) proposed a method, based on simulation analyses involving different levels of horizontal plasmid transmission, to estimate the number of recombination events, looking for differences in the phylogenetic reconstruction (called distortion index) both for the plasmid and for the chromosome. Using Young and Wexler (13) data on Rhizobium leguminosarum biovar phaseoli, they estimated that between 16 and 30% of all genetic types in the population were the source or the target of a plasmid transfer event, while they detected plasmid recombination in 2%.
These data and analyses, both in enterobacteria and in Rhizobium, suggest that there is little association between chromosomal and plasmid genotypes in bacteria, thus indicating that in general terms the panmictic model (Fig. 1C) is the dominant pattern in plasmid evolution. In this case, plasmids should be analyzed as independent genomes, using tools like the ones developed to understand the host/pathogen interaction models (17, 20). However, most of the bacterial population genetics studies are based on either pathogenic bacteria (associated to humans and domesticated animals) or on bacteria associated to cultivated plants. Plasmids free of human selection have been studied in Bacillus cryptic plasmids and the conjugative colicins plasmids in Escherichia coli. In both cases, the evidence suggests a more limited horizontal plasmid transfer (6–8).
Nevertheless, we still do not know if the panmictic plasmid pattern is an artifact due to human induced changes in the environment and the following intense selection, or if it is the normal pattern found in most bacterial populations. Thus, the question remains: How important is plasmid horizontal transfer in natural populations of bacteria?
The data obtained by Wernegreen et al. (5) are relevant to this question. They used restriction fragment length polymorphism analyses with one probe for the chromosome and two for the sym plasmid in 69 R. leguminosarum strains from wild clover populations. What they found was surprising: a close relationship between chromosome and plasmid genotypes. For illustration purposes, we generated the trees shown in Fig. 2. In general terms, it would correspond to the limited plasmid transfer pattern (Fig. 1B), although Wernegreen et al. (5) interpret it as more closely to the clonal pattern (Fig. 1A). With the present data, is not possible to decide to which chromosome/plasmid pattern their data adjust. A larger sample size and more chromosomal genetic markers would be needed to analyze if these populations have a clonal chromosomal genetic structure (high rates of chromosomal recombination would confuse the phylogeny) and to better resolve their phylogenetic relationships. Nevertheless, not all the plasmids and chromosomes seem to behave in the same way. In only one instance did they find one chromosome with one plasmid (the 1d chromosome and HA3 plasmid). On the other extreme, they found a very “virulent” plasmid, B1, present in all four different chromosomes of group 2, and a very promiscuous chromosome, the 1a, which has five different plasmid types. However, they never found any plasmid from a given group, associated with chromosomes from the other group (Fig. 2).
Figure 2.
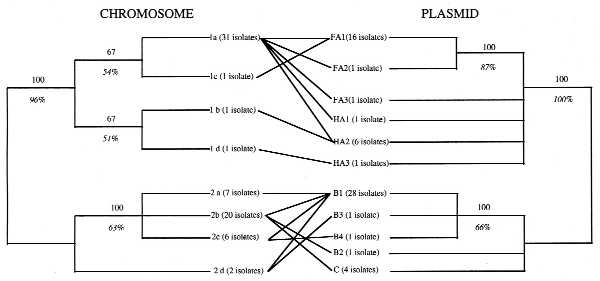
A graphic representation of the results of Wernegreen et al. (5). Both phylogenetic representations were derived from data in Table 2, considering each band of a given size as homologous and considering the presence of the bands as character states. The analyses were done using paup3.0s (21) and are the consensus trees of the most parsimonious trees, obtained using the Branch and Bound method (the 3 most parsimonious trees in the case of the chromosome, the 16 most parsimonious trees for the plasmids). The number of strains with a given chromosome or plasmid type is indicated in parentheses. The numbers above the lines indicate the 50% majority rule consensus of the most parsimonious trees. The numbers under the lines in italics are the bootstrap values, in percentage, obtained with 1,000 subreplicates, done using the Branch and Bound search method.
We consider that another important contribution by Wernegreen et al. (5) is the analysis of Rhizobium strains from several wild host species at the same place and time. A design considering this has never been done before, and it enables us to evaluate the true extent of plasmid movement, as different plant hosts may have different bacterial lineages.
Future studies should not only consider detailed analysis of the plasmid/chromosome relationships, but also the level of chromosome recombination, since the observed patterns in plasmids could be just a result of the general recombination of the bacteria. For example, in Rhizobium etli associated to beans in the state of Morelos, Central Mexico, it was found that the less disturbed populations had less chromosomal recombination (they are more clonal), whereas the more intensively managed populations, have substantially more chromosomal recombination (2, 22); similar patterns were found in other populations in the state of Puebla, also in Central Mexico (23). If these ideas are right, and plasmids travel with the rest of the chromosome, in the less managed populations there should be both very specific plasmid/chromosome association and high linkage disequilibirum among chromosomal loci (Fig. 1A), whereas the more human related populations may approach the panmictic pattern both for plasmid and chromosomal loci (Fig. 1C).
We are still far from understanding what bacteria are in evolutionary terms, and this is a very important issue given their abundance and diversity. In particular, it is important to understand the evolutionary biology of the plasmids, since they carry important genes both from medical (e.g., antibiotic resistance and pathogenic genes) and agronomic (e.g., nitrogen fixation) perspectives. Current molecular genetic analysis and evolutionary theory are helping us to learn more about them.
References
- 1.Levin B R. In: The Evolution of Sex. Michod R E, Levin B R, editors. Sunderland, MA: Sinauer; 1988. pp. 194–211. [Google Scholar]
- 2.Souza V, Eguiarte L, Avila G, Cappello R, Gallardo C, Montoya F, Piñero D. Appl Environ Microbiol. 1994;60:1260–1268. doi: 10.1128/aem.60.4.1260-1268.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maynard-Smith J, Smith N H, O’Rourke M, Spratt B G. Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eberhard W. Q Rev Biol. 1990;65:3–22. doi: 10.1086/416582. [DOI] [PubMed] [Google Scholar]
- 5.Wernegreen J J, Harding E E, Riley M A. Proc Natl Acad Sci USA. 1997;94:5483–5488. doi: 10.1073/pnas.94.10.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zawadzaki P, Riley M A, Cohan F M. J Bacteriol. 1996;178:191–198. doi: 10.1128/jb.178.1.191-198.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riley M A, Gordon D M. J Gen Microbiol. 1992;138:1345–1352. doi: 10.1099/00221287-138-7-1345. [DOI] [PubMed] [Google Scholar]
- 8.Mercer A A, Morelli G, Heuzenroeder M, Kamke M, Achtman M. Infect Immun. 1984;46:649–657. doi: 10.1128/iai.46.3.649-657.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martínez-Romero E, Romero D, Palacios R. Crit Rev Plant Sci. 1990;9:17–29. [Google Scholar]
- 10.Brom S, García A, Girard M L, Davila G, Palacios R, Romero D. J Bacteriol. 1991;173:1344–1346. doi: 10.1128/jb.173.3.1344-1346.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyd E F, Hartl D L. J Bacteriol. 1997;179:1622–1627. doi: 10.1128/jb.179.5.1622-1627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schofield P, Gibson A, Dudman W, Watson J. Appl Environ Microbiol. 1987;53:2942–2947. doi: 10.1128/aem.53.12.2942-2947.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young J, Wexler M. J Gen Microbiol. 1988;134:2731–2739. [Google Scholar]
- 14.Valdés A M, Piñero D. Evolution. 1992;46:641–656. doi: 10.1111/j.1558-5646.1992.tb02072.x. [DOI] [PubMed] [Google Scholar]
- 15.Geniaux E, Laguerre G, Amarger N. Mol Ecol. 1993;2:295–302. [Google Scholar]
- 16.Lenski R E, Bouma J. Nature (London) 1990;335:351–352. doi: 10.1038/335351a0. [DOI] [PubMed] [Google Scholar]
- 17.Lenski R E, May R M. J Theor Biol. 1994;169:253–265. doi: 10.1006/jtbi.1994.1146. [DOI] [PubMed] [Google Scholar]
- 18.Segovia L, Piñero D, Palacios R, Martinez-Romero E. Appl Environ Microbiol. 1991;57:426–433. doi: 10.1128/aem.57.2.426-433.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demezas D H, Reardon T B, Strain S R, Watson J M, Gibson A H. Mol Ecol. 1995;4:209–220. [Google Scholar]
- 20.Frank S A. Trends Genet. 1992;8:213–219. doi: 10.1016/0168-9525(92)90236-w. [DOI] [PubMed] [Google Scholar]
- 21.Swofford D L. paup: Phylogenetic Analysis Using Parsimony. Champaign, IL: Ill. Nat. Hist. Survey; 1991. [Google Scholar]
- 22.Souza V, Nguyen T T, Hudson R R, Piñero D, Lenski R E. Proc Natl Acad Sci USA. 1992;89:8389–8393. doi: 10.1073/pnas.89.17.8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Souza, V., Silva, C., Bouchet, V., Valera, A., Bain, J., Marquez, E. & Eguiarte, L. E. (1997) J. Ethnobiol., in press.


