Abstract
Cancers evolve by a reiterative process of clonal expansion, genetic diversification and clonal selection within the adaptive landscapes of tissue ecosystems. The dynamics are complex with highly variable patterns of genetic diversity and resultant clonal architecture. Therapeutic intervention may decimate cancer clones, and erode their habitats, but inadvertently provides potent selective pressure for the expansion of resistant variants. The inherently Darwinian character of cancer lies at the heart of therapeutic failure but perhaps also holds the key to more effective control.
Cancer is a major cause of mortality throughout the world and despite the extraordinary amount of effort and money expended over the past several decades, successful eradication and control of advanced disease remains elusive1. In parallel, our understanding of cancer biology and genetics has changed beyond recognition2. The translation of cancer genomics to cancer therapy needs to accommodate the cellular complexity of the disease and address its dynamic, evolutionary character. The latter provides both barriers to success and opportunities.
In 1976 Peter Nowell3 published a landmark perspective on cancer as an evolutionary process, driven by stepwise, somatic cell mutations with sequential, sub-clonal selection. The implicit parallel was to Darwinian natural selection with cancer equivalent to an asexually reproducing, unicellular, quasi-species. The modern era of cancer biology and genomics has validated the fundamentals of cancer as a complex, Darwinian, adaptive system4,5 (Box 1, and additional references in Supplemental information).
Box 1.
Cancer as a Complex System
Cancers exist in an extraordinary variety of taxonomically, quasi-classes, genera, species, characterised by divergent cells of origin and mutational spectra. Each cancer is individually unique.
Cancers evolve over variable time frames (~1–50 years) and tempos and, in any one patient, the clonal structure, genotype and phenotype shifts over time. Contemporaneously, any one cancer is, in effect, multiply different (sub-clonal) cancers occupying overlapping or distinct tissue habitats.
The number of mutations found in any cancer can vary from a handful (10–20) to (more usually) hundreds of thousands. The great majority are ‘passengers’, a modest but undefined number being functionally relevant ‘drivers’. The mutational processes are very diverse.
Cancers acquire, via mutational (and epigenetic) changes, a variety of critical phenotype traits that compound to empower territorial expansion, via proliferative self-renewal, migration and invasion; properties that are part and parcel of normal developmental, physiological and repair processes.
Advanced, disseminated or very malignant cancers appear to be almost uniquely competent to evade therapy.
Most, if not all, of this complexity can be explained by classical evolutionary principles.
Cancer clone evolution takes place within tissue ecosystem habitats which have themselves evolved over a billion years. Their complex anatomies and networked signals have evolved to optimise and integrate multi-cellular functions whilst restraining renegade clonal expansion. The balance however is delicate as the resilience of multi-cellular and long-lived animals such as ourselves depend upon the very phenotypic properties that, if not tightly regulated, drive or sustain malignancy, i.e. self-renewal coupled with stabilization of telomeres that allows extensive proliferation, angiogenesis, cell migration and invasion6.
Tissues provide the context for cancer cell evolution. The usually protracted time required for the clinical emergence of cancer and the resultant mutational complexity often reflect the sequential and random ‘searches’ for phenotypic solutions to micro-environmental constraints. The evolutionary progression of cancer is more often than not stalled or aborted, as revealed by the high frequencies of clinically covert pre-malignant lesions7–9. Cancer suppressive mechanisms relegate most cancers to old age where they have little effect on the reproductive fitness of their hosts.
Resource limitations and other micro-environmental constraints limit the size of tumours at multiple stages of progression. Even full-blown malignancies often exhibit Gompertzian growth10. The doubling time of cancer cells (~1–2 days) is orders of magnitude faster than the doubling time of tumours (~60–200 days)10, implying that the vast majority of cancer cells die before they can divide11. Thus, natural selection in tumours, like selection among organisms, often takes place through severe competition for space and resources.
Oncologists change cancer clone dynamics dramatically by introducing a new potent source of ‘artificial’ selection – with drugs or radiotherapy. But similar evolutionary principles apply. Massive cell death will usually ensue providing selective pressure for the proliferation of variant cells that can, by one of several mechanisms (see below), resist therapeutic oblivion. To make matters worse, many cancer therapeutics are genotoxic; surviving cells regenerating the cancer may have incurred additional mutational insults, some of which could improve their fitness and malignant potential.
These general considerations suggest that much can be gained by applying the tools and insights of evolutionary biology and ecology to the dynamics of cancer pre- and post-treatment. Here, we provide a portrait of cancer as an evolutionary process and argue that this can both explain our modest therapeutic returns and suggest alternative strategies for effective control.
Mutational drivers and clonal dynamics
A classical or Darwinian evolutionary system embodies a basic principle:purposeless genetic variation of reproductive individuals, united by common descent, coupled with natural selection of the fittest variants. That is, natural selection of those rare individuals that fortuitously express the traits that complement or thwart the contemporary selective pressures or constraints. It’s a process replete with chance. Cancer is a clear example of such a Darwinian system. Most mutational processes have biases at the DNA sequence level and mutational spectra in cancer can reflect or implicate particular error-prone repair processes or particular genotoxic exposures, e.g. cigarette carcinogens, UV light, and chemotherapeutics2. Patterns of genetic instability in cancer (chromosomal or microsatellite) may reflect prior exposure and selective pressure exerted by particular classes of chemical carcinogens2. Nevertheless, with respect to functions encoded in genes, mutagenic processes are essentially blind or non-purposeful. (Intrinsic mutagenic or recombinatorial enzymes preferentially targeting lymphoid Ig or TCR genes are the exception to this generalisation12). What we see as recurrent, adaptive mutations endowing fitness traits in cancer reflect the potent impact of clonal selection.
Clonal evolution involves the interplay of selectively advantageous or ‘driver’ lesions, selectively neutral or ‘passenger’ lesions, and deleterious lesions, as well as lesions that increase the rate of other genetic changes (‘mutator’ lesions)13,14, and changes to the micro-environment15 that change the fitness effects of those lesions. (Note that a “hitchhiker” mutation in evolutionary biology is equivalent to a “passenger” mutation in cancer biology.) ‘Driver’ candidature is supported by independent observation in multiple neoplasm beyond what would be expected by the background mutation rate, association with clonal expansions16,17, and type of mutation (missense, nonsense, frame shift, splice-site, phosphorylation sites, double deletions18, etc.)19,20, particularly if the gene involved has a known role in cellular processes relevant to oncogenesis. Evidence for the importance of a mutation in carcinogenesis from genetic studies of human tumours should be corroborated with functional tests and animal models. ‘Passenger’ status may also be ambiguous or context-dependent as, for example, with mono-allelic loss that only impacts on function when the second allele is lost, mutations that only generate a phenotypic effect when another locus mutates, or with mutants that only have selective potency in the face of particular therapeutic challenges.
There has been little work attempting to quantify the selective advantage conferred by driver mutations. Using a non-spatial population genetics model of sequential, exponential clonal expansions, Bozic et al. derived a formula for the proportion of expected neutral passenger mutations versus selectively advantageous driver mutations as a function of the selective advantage of the driver mutations. Fitting this equation to both glioblastoma and pancreatic cancer resequencing data, they estimated that driver mutations confer an average fitness advantage of only 0.4%21. Direct measurements of the selective advantage of a mutant clone would require longitudinal samples of a neoplasm and estimation of the clone sizes at each time point.
The dynamics of somatic evolution depend crucially on the interaction of rates of mutation and clonal expansion. Mutation rates vary substantially for different genomic regions22 and for different types of abnormalities, e.g. single base sequence changes versus balanced chromosomal rearrangements and gene fusions, and will be greatly enhanced by the instigation of genetic instability23–25. The rate of epigenetic changes has been estimated to be orders of magnitude higher than genetic changes26 and could be a major determinant of clonal evolution. Natural selection acts on epigenetic variation within neoplasms27 as well as genetic variation, because epigenetic changes are heritable at cell division and can affect cell phenotypes. Evolutionary biology has generated tools for addressing many of these mutation rate complexities (see Supplementary Information for references) but these remain under-utilised in cancer biology28. The traditional model of clonal evolution posits a series of clonal expansions that grow to dominate the neoplasm (‘selective sweeps’)16,21,29. However, this can only occur if the time until the next driver mutation is longer than the time required for a clone to sweep through the neoplasm. Also, if the second mutation occurs in a competitor clone, then the expansion of both clones is restrained by mutual competition (called ‘clonal interference’)30. Given the large population sizes and high mutation rates typical of neoplasms, clonal competition may be common31–33. Serial sampling is the appropriate way to assess this issue and limited data suggests that parallel clonal expansions may precede dominance of sub-clones early in cancer development34,35. Initial evidence also suggests that large clonal expansions after transformation may be rare26. Selective sweeps are predicted to originate from pre-existing genetic variants or sub-clones. Direct evidence for this comes from serial sampling of oncogenic mutations in advanced disease36 or metastasis37 and, similarly, in post-chemotherapy relapses with oncogenic mutations or with mutations in the drug target itself (see Supplementary information for references).
Punctuated equilibrium versus gradualism
A longstanding debate in species evolution pitting gradualism versus punctuated equilibrium38 has recently emerged in considerations of clonal evolution of neoplasms. Do malignant clones, with their dramatically altered genomes, evolve gradually through a sequence of many genetic alterations and clonal expansions, or are there few, large scale punctuated changes, possibly prompted by acute insult, a single, catastrophic mitosis that generates multiple lesions across the genome or on a single chromosome (chromothripsis)39, or through the accumulation of many lesions over time in a rare, undetected subclone? Evidence of 10’s of non-synonymous mutations in cancers was interpreted as the result of 10’s of clonal expansions29. Reconstruction of genealogies of neoplastic clones, based on genetic heterogeneity within neoplasms, suggests that clones with ancestral genomes are not driven extinct by later clonal expansions31–33. This allows for reconstruction of the history of a neoplasm. Data in breast cancer32 show that clones with intermediate genotypes are difficult to detect and that each clone generates a cloud of genetic variants around it, which may either be neutral or non-viable sub-clones. Work in B-cell chronic lymphocytic leukemia40 suggests that intermediate clones can be detected but at a frequency of < 0.001, which was below the detection threshold of the breast cancer study32. Those rare intermediate clones may be rare because they had limited potential to expand or they may have once been common but lost in competition to more recent clones.
Long periods of stasis of cancer clones are clearly implied by the frequency of pre-malignant clonal lesions (or CIS) that substantially exceed clinical cancer rates7–9 by cancer dormancy41 and by genetic reconstitution of clonal histories37. But cancer clone evolution probably passes a point of no return, perhaps at the juncture of metastatic growth. If unlimited proliferative capacity is guaranteed by telomere stabilization25 then clonal expansion may be curtailed only by the resultant morbidity and lifespan of the patient. And, rather dramatically, when provided (albeit rarely) with the requisite routes of dissemination and immuno-selection, cancer cells are capable of parasite-like immortality and reestablishment in other individuals6,42,43.
The cancer ecosystem
Cancer clone genetic diversification and sub-clonal selection occurs within tissue ecosystems, the latter providing both the venue and the determinants of fitness selection, i.e. the adaptive landscape44. Tissue micro-environments are themselves complex, dynamic states with multiple components that can influence cancer clone evolution (Fig. 1). TGFβ is one instructive example of a cancer ecosystem regulatory molecule45. Other components of inflammatory lesions are potent and common modulators of cancer cell ecosystems25.
Figure 1.
The complexity of tissue ecosystems of cancer cells. Exposures, the constitutive genetics of the host cells, systemic regulators, local regulators and architectural constraints all impinge upon and constrain the evolution of somatic cells.
Interactions between cancer cells and their tissue habitats are reciprocal and not infrequently cancer cells remodel tissue micro-environments and specialised niches to their competitive advantage46. Architectural constraints or barriers, such as sequestration of stem cells into crypts in the gastrointestinal tract47, as well as the requirement for external signals for proliferation and cell survival, impose restraints on cancer clone expansion. However, other inducible components of the micro-environment can have a positive or promotional impact on neoplastic cells, for example infiltrating macrophages and neo-vascularisation in response to anoxia may support neoplastic cell survival and proliferation. As endorsed by mathematical modelling, cancer clone evolutionary selection for more robust or malignant phenotype is less likely in more stable or homogeneous micro-environments48. Spatial heterogeneity of resources selects for cell migration and emigration from the primary tumour, which may explain why there is selection for metasasis49. Pre-clinical models suggest that normalizing the resources across the primary tumour can suppress metastasis50. As cancer clones and sub-clones expand, migrant cells invade novel habitats or territories within and between tissues, and so experience novel selective pressures that can cause further diversification of the cancer cells. It is this malignant feature and its associated morbidity that characterises end stage, clinically intransigent cancer.
Tissue ecosystems, serving as cancer cell habitats, are not closed systems. In addition to their dynamic regulation by systemic factors (nutrients, hormones) or invasion by inflammatory or endothelial cells, they can be modified by external exposures. In effect, the relevant ecosystem for each cancer includes not only the tissue sites of residence but the environment, lifestyle and associated aetiological exposures of the patient. Genotoxic exposures (e.g. smoking, UV light), infection and chronic dietary/exercise habits impacting on calorie or hormonal levels can have a profound impact on tissue micro-environments as well as directly on cancer cells themselves (Fig. 1). Indeed it is in this context that many exposures are aetiologically linked to initiation or promotion of disease. We assume that without such modulating exposures, the risk of cancer clone initiation and evolution would be much reduced.
The landscape of tissue ecosystems of cancer can also be radically altered following toxic chemotherapy or radiotherapy. Wholesale clearance may decimate most cancer cells but the remodelled landscapes that result then provide novel selective pressures, as well as new resources and opportunities, for the emergence of any pre-existing variant cancer cells that were able to survive therapy. Additionally, and possibly critically, stroma or specialized habitat niches may protect cancer cells from therapy51.
Cancer genomics and clonal architectures
Sequencing of cancer genomes, facilitated by the advent of second generation whole genome sequencing, has opened a new window to the complexity of cancer cell genetics and its evolutionary biology2. Since both transformation and metastases are probably clonal in most cases2, deriving from single cells, identification of mutations present in all cells of a tumour can reconstruct the genotype of the founder cell. These founder events constrain the genetic and clonal complexity of tumors. We already had a long list of recurrent driver mutations (with gain or loss of function) via the fine mapping of chromosome breaks, candidate gene sequencing and functional screening of bulk samples from tumors. What has now emerged in genomic screens is a portrait of just how complex cancer genomes usually are. Individual cancers can contain hundreds or hundreds of thousands of mutations and chromosomal alterations2, the great majority of which are assumed to be neutral mutations arising via genetic instability. Chromosomal instability (amplifications, deletions, translocations and other structural changes) is a common feature of most cancers, however it is not clear if the rate of point mutations is increased in cancer2,21,23,52. Evolutionarily neutral alterations presumably register in the screens as the result of hitchhiking on clonal expansions driven by selectively advantageous alterations or by drift. Additionally, the data vividly confirms that each cancer in each patient has an individually unique genomic profile. It may be that only a modest number of phenotypic traits are required to negotiate all constraints and evolve to full malignant or metastatic status25 but the inference is that this can be achieved by an almost infinite variety of evolutionary trajectories and with multiple, different combinations of driver mutations44.
Paradoxically, genome profiles under-estimate complexity. They are mostly, to date, one-off snapshots taken from a single sample at a single diagnostic time point. Serial or parallel sampling we know, via more conventional genetic analysis, uncovers genetic diversity. Whole genome sequencing of paired primary tumour versus metastasis samples has been limited to date but has revealed that individual metastatic lesions are clonal in origin and genetically unique yet with clonal ancestries traceable back to the primary tumour2. Descriptions of “the genome” of a cancer are perhaps misleading in one other respect. Genetic variants are commonly identified in 5%–50% of reads, suggestingsub-clonal distribution of most mutations53. But, critically, the pattern of segregation of mutations within sub-clones is lost when DNA is extracted from the total cell population. This feature is important if patient-specific genomic profiles are to provide a platform for selecting therapeutic targets. Sub-clonal genetic diversity is arguably a key determinant of therapeutic failure. This limitation in cancer genomicsis broadly recognised and represents a considerable challenge, technically and bioinformatically. Understanding the genetic diversity within neoplasms, and how it changes in response to interventions, will require deep sequencing40 and interrogation of the genomes of single cells for patterns of segregation of mutations.
Sub-clonal segregation of mutations and clonal architecture
The classical model of clonal evolution, derived from Nowell3 envisages, sequential acquisition of mutations with concomitant, successive sub-clonal dominance or ‘selective sweeps’. Histopathological, time-ordered correlates of disease progression, i.e. adenoma, carcinoma, metastases, encourage this view. At all stages of this evolution, however, individual cells and their progeny (i.e., subclones) are vying for space and other limiting resources. Multiplexed, single cell mutational analysis (ideally in serial samples) is the appropriate way to interrogate clonal architecture. There are only a few examples of this to date10,32,33 though they do provide evidence for complex patterns of sub-clonal segregation of mutations, consistent with Nowell’s model. Collectively, a large body of data from tissue section, small biopsy and the more recent single cell analyses33 testifies to the fact that the evolutionary trajectories that emerge are complex and branching, exactly as envisioned by Nowell, providing a striking parallel with Charles Darwin’s iconic evolutionary speciation tree (Fig. 2). Attempts to shoehorn this complexity into a simple linear sequence of mutational events, based on cross-sectional data, have probably been misleading54. However, through a comparison of mutational genomes of sub-clones, it is possible to infer their evolutionary or ancestral relationships as well as the order of events during the development of that neoplasm32,33,37,53,54 (Fig. 4B). Clonal evolution from common ancestral cancer cells is vividly illustrated in identical twin children with concordant acute leukaemia55,56, in metastatic lesions2,10 and, by inference, in some cases of bilateral testicular cancer57 (Fig. 3). Divergent cancer clone genotypes and phenotypes in this context parallel allopatric speciation in separated natural habitats – as with finches in different Galapagos Islands58.
Figure 2.
(a) Branching clonal architecture of clonal evolution in cancer. Selective pressures allow some mutant sub-clones to expand while others go extinct.
(b) Darwin’s evolutionary tree of speciation (from his 1837 notebook B). Eco 1–4 (red boxes) different tissue ecosystems/habitats. Tx, therapy. CIS, carcinoma in situ.
Figure 4.
Topography of cancer sub-clones.
A. Tissue section of prostate to detect TMPRSS2/ERG fusion (ERG via rearrangement) and PTEN loss.
B. Presumed sequence of clonal events.
Modified from Clark et al61.
Figure 3.
Divergent (branching) clonal evolution of cancer with topographical separation.
In each example, a clonal (single cell) ancestry is indicated by shared acquired mutations, e.g. ETV6-RUNX1 fusion for the leukaemias, c-kit mutation for the testicular cancers. The time at which the two subclones evolve (T1, T2) can be temporarily synchronous or develop several years apart37,55–57. The probabilities of sub-clones emerging as shown are independent and different (p1, p2). In most cases (90% for monozygotic twins), only one twin develops overt leukaemia. The penetrance of bilateral testicular cancer having a common origin57 is unknown.
Profiling of sub-clones within a neoplasm allows the inference of molecular clocks to time events in histories of neoplasms. DNA methylation changes and point mutations have been used to infer clonal expansion dynamics26, the time between initiation, invasion, and metastasis17,37,52 and even the relative timing of events during progression, based on deep sequencing data from a single sample59.
Sub-clones may be topographically mixed in the primary tissue37,60 but given their single cell origin and bifurcating pathways, it is not surprising that they can also occupy distinctive territories35,37,61,62 (Fig. 4A). This intra-tumour diversity can also be seen in cellular phenotypes that have therapeutic relevance63. The fact that cancer clone evolution involves contemporaneous sub-clones with distinctive mutational profiles, and that may be territorially segregated, has considerable practical implications for biopsy-based diagnosis and prognosis as well as for targeted therapy. Whether all sub-clonal diversification reflects the impact of ‘driver’ mutations and selective advantage or additionally reflects some degree of genetic drift, of selectively neutral mutations, or even epigenetic alterations remains unclear. Sub-clonal structure can be quantified by diversity measures35,64,65, which have been shown to be robust biomarkers for predicting progression to malignancy in Barrett’s esophagus65 and are associated with tumor stage and subtype in breast cancer64.
Units of selection and cancer stem cells
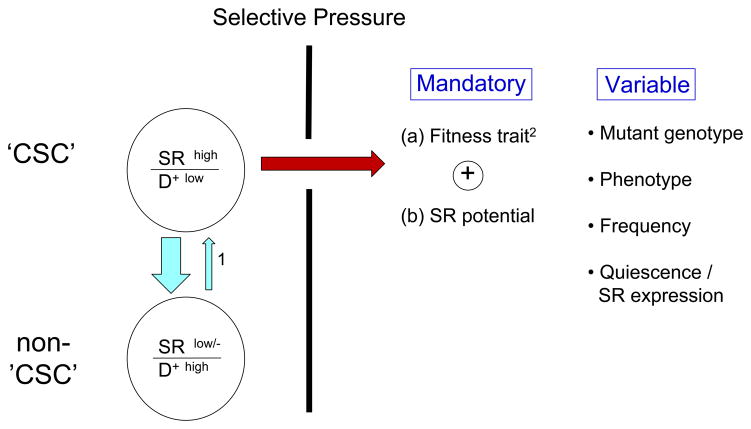
Evolutionary theory posits that natural selection operates in any system that has components with varying reproductive potential4. In the evolutionary progression of cancer or its resurgence post-therapy, the primary unit of selection is the cell. Not any cell but rather a cell that has extensive replicative potential. This equates to the so-called cancer stem cell (a.k.a. cancer initiating or propagating cell) (Fig. 5).
Figure 5.
SR, self-renewal. D+, differentiation. CSC, cancer stem cell.
Selective pressures may include environmentally-derived genotoxicity, natural/physiological restraints, cancer therapy, etc..
1 Mutation in progenitor cells may convert these cells ‘back’ to a self-renewing population72.
2 Any phenotypic feature that allows cells to continue to survive and proliferate in the face of particular constraints.
The cancer stem cell (CSC) hypothesis was developed via transplantation experiments with leukemic cells and although promulgated as a general feature of all cancers, it has proven contentious66. There has been no consensus view on whether CSCs are rare or high frequency cells or whether they have fixed, hierarchical or variable phenotypic properties. A consideration of evolutionary progression in cancer suggests that cells with extensive propagating activity are unlikely to be fixed entities67. As cellular drivers of sub-clonal expansion, they are more likely to vary in frequency and phenotypic features. The only obligatory feature they must possess is the potential for extensive self-renewal (Fig. 5). In this respect the clinical relevance of CSCs is endorsed by the finding that quantitative measures of stem cell activity or self-renewal (via xenotransplantation or gene expression signatures) are predictive of clinical outcome in several cancer types68. Self-renewal will be underpinned by aberrant genotype and possibly by other epigenetic features. Several testable predictions follow from this. First, CSCs are highly likely to evolve and change in genotype and phenotype as each cancer evolves pre-and post-therapy67,69,70. Some therapies may even provide strong selection for CSC survival and proliferation71. Second, there may be selective pressure, during cancer progression, for cells with the most extensive self-renewing capacity – at the expense of cells with intact differentiation competence. This has been observed in CML72 and mouse models73,74. A higher probability of symmetrical self-renewing proliferative cycles would be expected to result in an increase in numbers and frequency of CSCs72. It is therefore of some consequence that loss of the p53 DNA damage checkpoint, a highly frequent correlate of cancer progression and clinical intransigence75 appears to ‘release’ stem cell-like transcriptional signatures76 and leads to enhanced self-renewal, in mammosphere culture systems77. The frequency of CSCs could then evolve from low to very high frequency with progression of disease78,79. Third, for selection to operate via micro-environmental or therapeutic pressures, there should be contemporaneous genetic variation in CSCs. This has been born out in leukemias80,81. These considerations have significant clinical implications. Clearly, whatever the frequency and phenotype, if self-renewing CSCs drive and sustain cancer clonal evolution then they are the repository of all functionally relevant mutational events underpinning clonal selection pre-and post-therapy. This then reinforces the view that their restraint or elimination should be the ultimate target of any therapy. But if they are genetically (and epigenetically) diverse, as evolutionary considerations and initial experiments80,81 clearly imply, then a basis for therapeutic failure is immediately apparent. In fact, the expansion of resistant clones under therapy shows that the therapy was able to differentially eliminate CSCs lacking the resistance lesion (see Supplemental information). The adaptability of CSCs afforded by genetic diversity is compounded by what appears to be their intrinsically lowered susceptibility to drugs and irradiation82. This may be the consequence of stromal cell associations83 and quiescence (of CSC sub-populations) along with associated properties of enhanced DNA repair and elevated expression of drug efflux pumps, possibly all evolved contingencies to protect vital normal stem cells.
Sub-clonal genetic heterogeneity is a common, if not universal, feature of cancers84 but it cannot be assumed that all sub-clones are sustained by CSCs; some could be evolutionary dead-ends generated by cells with only limited propagating potential. It is partly to accommodate this fact that the in vivo assay for CSCs involves sequential transplants66. Ideally, the genomes of single CSCs would be interrogated. The issue of genetic heterogeneity can be effectively addressed by comparing sub-clonal diversity or clonal architecture pre- and post-transplant. In one experiment, quadrant sections of glioblastoma were shown to have divergent but related genotypes, but all contained cells reading out in the in vivo (intra-cerebral) CSC assay85. More definitive data comes from a comparison of sub-clonal genetic profiles pre-and post-transplant that were interrogated at the single cell level and/or by SNP arrays in B cell precursor ALL. Multiple sub-clones from each patient’s diagnostic sample registered in the in vivo CSC transplant assays albeit with variable competitive potency33,66,80,81. We await experimental confirmation of the prediction that such genetic diversity of CSCs is a common feature of cancer but, assuming it is, then some important therapeutic implications follow.
Cancer escape: need for a Darwinian bypass?
‘More research should be directed towards understanding and controlling the evolutionary process in tumours before it reaches the late stage seen in clinical cancer.’
P Nowell, 1976
Cancer therapeutics has had its successes but the reality remains that very few advanced or metastatic malignancies are amenable to effective control or eradication. Genetic variation in CSCs, particularly if fermented by genetic instability, provides the substrate for selective escape. In addition, other mechanisms of positive selection by therapy exist that are non-genetic; these can involve signalling plasticity (or oncogene bypass)86, quiescence87 or epigenetic changes88, though many of these may depend on heritable and thus selectable epigenetic variation. There has been great expectation that the audit of cancer genomes, by identifying recurrent and drug able mutations, will herald in a new phase of highly specific or targeted small molecule inhibitors and personalised medicine89. Oncogene addiction appeared to be the Achilles heel for cancer in this respect90. The success of imatinib and derivative ABL kinase inhibitors in chronic myeloid leukaemia (CML)90 was encouraging. But CML is an atypical cancer. It is essentially a pre-malignant (albeit ultimately lethal) condition driven, most probably, by a single founder mutation (BCR-ABL1 fusion), which provides a universal target. Even in this most favourable of circumstances, escape occurs either via quiescence (and coupled resistance) of CSC91 or via mutation of the ABL kinase target. Once CML evolves to overt malignancy or blast crisis, with more genetic complexity, ABL1 kinase-directed therapy is often ineffective.
Other specific small molecule inhibitors directed at mutant products produce initially encouraging results in patients with advanced disease but the benefits have turned out to be transitory as cancer clones re-emerge with resistant features. When targets selected are non-founder mutations, even if they dominate the neoplasm, therapy can be anticipated to select for sub-clones lacking the mutant target70. Alternatively, sub-clones may have additional mutations that enable signalling bypass of the drug target, as with MET amplification in EGFR mutant lung cancer treated with EGFR kinase inhibitors92.
What is a way out of this impasse? Champions of targeted therapy and personalised medicine argue that the problem can be solved by artful combinations of drugs targeting components of networked signalling and tailored to the individual patient’s cancer genome. Synthetic lethal strategies hold promise in this regard93.
Self-renewing cancer cells are the ultimate target so the development of high throughput screening for selective inhibitors is encouraging71. Targeting components of the self-renewing programme itself (independent of specific mutant genotype) deserve exploration, especially if a distinction can be made with normal adult stem cells (see Supplemental information). The problem of intrinsically resistant (and quiescent?) stem cells, in the case of CML, has been addressed by combining selective kinase (ABL1) inhibitors with inhibitors for histone deacetylase94 or BCL695. But ultimately it may prove difficult to thwart the plasticity or adaptability of cancer cells (or CSC) that is an inherent evolutionary feature of advanced disease. A Darwinian bypass may be required. A clear implication of cancer’s evolutionary diversity is that prevention (e.g. cessation of smoking, avoidance of sunburn, prophylactic vaccines, etc) makes a great deal of sense as does early detection and intervention, i.e. prior to extensive genetic diversification and dissemination.
An alternative therapeutic strategy directs attention away from the cancer cells and towards their micro-environmental habitats. There are many opportunities here for so-called ‘ecological’ therapy, directed at changing the essential habitat and dependencies of cancer cells96. Anti-angiogenesis is a prime example of this tactic and may provide a potent restraint on CSCs97. Other examples include interference with bone remodelling with bisphosphonates in prostate cancer, aromatase inhibitors in breast cancer, exploiting hypoxia, inhibitors of inflammation or tumour infiltrating macrophages and blockage of CSC interactions with essential stromal or niche components96,98.
A further alternative is to seek to control cancer, rather than eradicate it, turning cancer into a chronic disease. Since the speed of evolution is proportional to the fitness differential between cells, cytotoxic drugs are predicted to rapidly select for resistance5. They likely cause competitive release99, by removing all the competitors of the resistant cells. In contrast, cytostatic drugs should delay progression and mortality longer than cytotoxic drugs, because sensitive competitor cells remain to occupy space and consume resources that would otherwise benefit the resistant clones. In addition, by suppressing cell division, cytostatic drugs also suppress the opportunities for new mutations. Intriguingly, Gate by and colleagues have recently shown that by treating an aggressive ovarian cancer (OVCAR-3) xenograft tumour to maintain a stable size, rather than eradicate it, they were able to keep the host mice alive indefinitely. Moreover, the dose of carboplatin necessary to keep the tumour in check declined over time100. We should be asking what phenotypes can we select for that would make neoplasm less deadly and more clinically manageable?
The evolutionary theory of cancer has survived 35 years of empirical observation and testing and so may be considered a bona fide scientific theory today. While the basic components of somatic evolution are well understood, the dynamics of somatic evolution remain largely opaque. Fortunately, there are tools from evolutionary biology that may be applied to neoplasms in order to address many of the fundamental questions in cancer biology, such as the order of events in progression, distinguishing driver mutations from passengers, as well as understanding and preventing therapeutic resistance. The dynamics of clonal diversification and selection are critical to understanding neoplastic progression and response to therapy. There are exciting clinical opportunities in directly addressing the evolutionary adaptability of neoplasms and designing interventions to slow, direct or otherwise control that evolution so as to delay or prevent cancer mortality.
Supplementary Material
Contributor Information
Mel Greaves, Email: mel.greaves@icr.ac.uk.
Carlo C. Maley, Email: carlo.maley@ucsf.edu.
References
- 1.Jemal A, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 3.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. The foundational paper establishing the evolutionary theory of cancer. [DOI] [PubMed] [Google Scholar]
- 4.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 5.Pepper J, Scott Findlay C, Kassen R, Spencer S, Maley C. Cancer research meets evolutionary biology. Evolutionary Applications. 2009;2:62–70. doi: 10.1111/j.1752-4571.2008.00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greaves M. Cancer: The Evolutionary Legacy. Oxford University Press; 2000. [Google Scholar]
- 7.Sakr WA, Haas GP, Cassin BF, Pontes JE, Crissman JD. The frequence of carcinoma and intraepithelial neoplasia of the prostate in young male patients. J Urol. 1993;150:379–385. doi: 10.1016/s0022-5347(17)35487-3. [DOI] [PubMed] [Google Scholar]
- 8.Mori H, et al. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc Natl Acad Sci USA. 2002;99:8242–8247. doi: 10.1073/pnas.112218799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reid BJ, Li X, Galipeau PC, Vaughan TL. Barrett’s oesophagus and oesophageal adenocarcinoma: time for a new synthesis. Nat Rev Cancer. 2010;10:87–101. doi: 10.1038/nrc2773. nrc2773 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein CA. Parallel progression of primary tumours and metastases. Nature reviews. Cancer. 2009;9:302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- 11.Malaise EP, Chavaudra N, Tubiana M. The relationship between growth rate, labelling index and histological type of human solid tumours. Eur J Cancer. 1973;9:305–312. doi: 10.1016/0014-2964(73)90099-6. [DOI] [PubMed] [Google Scholar]
- 12.Tsai AG, et al. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135:1130–1142. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bardelli A, et al. Carcinogen-specific induction of genetic instability. Proc Natl Acad Sci USA. 2001;98:5770–5775. doi: 10.1073/pnas.081082898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic Instability and Darwinian Selection in Tumors. Trends in Cell Biology. 1999;9:M57–M60. [PubMed] [Google Scholar]
- 15.Barcellos-Hoff MH, Park C, Wright EG. Radiation and the microenvironment -tumorigenesis and therapy. Nature reviews. Cancer. 2005;5:867–875. doi: 10.1038/nrc1735. [DOI] [PubMed] [Google Scholar]
- 16.Maley CC, et al. Selectively advantageous mutations and hitchhikers in neoplasms: p16 lesions are selected in Barrett’s esophagus. Cancer Res. 2004;64:3414–3427. doi: 10.1158/0008-5472.CAN-03-3249. [DOI] [PubMed] [Google Scholar]
- 17.Tao Y, et al. Rapid growth of a hepatocellular carcinoma and the driving mutations revealed by cell-population genetic analysis of whole-genome data. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12042–12047. doi: 10.1073/pnas.1108715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. nature08768 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Youn A, Simon R. Identifying cancer driver genes in tumor genome sequencing studies. Bioinformatics. 2011;27:175–181. doi: 10.1093/bioinformatics/btq630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenman C, Wooster R, Futreal PA, Stratton MR, Easton DF. Statistical analysis of pathogenicity of somatic mutations in cancer. Genetics. 2006;173:2187–2198. doi: 10.1534/genetics.105.044677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bozic I, et al. Accumulation of driver and passenger mutations during tumor progression. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz M, Zlotorynski E, Kerem B. The molecular basis of common and rare fragile sites. Cancer Lett. 2006;232:13–26. doi: 10.1016/j.canlet.2005.07.039. [DOI] [PubMed] [Google Scholar]
- 23.Loeb LA. Human cancers express mutator phenotypes: origin, consequences and targeting. Nature reviews. Cancer. 2011;11:450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weisenberger DJ, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j. cell.2011.02.013. A consolidation of the common phenotypes that evolve in neoplastic cells of all types. [DOI] [PubMed] [Google Scholar]
- 26.Siegmund KD, Marjoram P, Woo YJ, Tavare S, Shibata D. Inferring clonal expansion and cancer stem cell dynamics from DNA methylation patterns in colorectal cancers. Proc Natl Acad Sci U S A. 2009;106:4828–4833. doi: 10.1073/pnas.0810276106. 0810276106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varley KE, Mutch DG, Edmonston TB, Goodfellow PJ, Mitra RD. Intra-tumor heterogeneity of MLH1 promoter methylation revealed by deep single molecule bisulfite sequencing. Nucleic Acids Res. 2009;37:4603–4612. doi: 10.1093/nar/gkp457. gkp457 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aktipis CA, Kwan VSY, Johnson KA, Neuberg SL, Maley CC. Overlooking evolution: A systematic analysis of cancer relapse and therapeutic resistance research. PLoS One. 2011 doi: 10.1371/journal.pone.0026100. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beerenwinkel N, et al. Genetic progression and the waiting time to cancer. PLoS Comput Biol. 2007;3:e225. doi: 10.1371/journal.pcbi.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Visser JA, Rozen DE. Clonal interference and the periodic selection of new beneficial mutations in Escherichia coli. Genetics. 2006;172:2093–2100. doi: 10.1534/genetics.105.052373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leedham SJ, et al. Individual crypt genetic heterogeneity and the origin of metaplastic glandular epithelium in human Barrett’s oesophagus. Gut. 2008;57:1041–1048. doi: 10.1136/gut.2007.143339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. Single cell sequencing reveals the clonal structure of two breast cancers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson K, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. Single cell genetic analyses and xenografts revealed the clonal architecture within acute lymphoblastic leukemia stem cell populations and demonstrated repeated independent acquisition of copy number changes within the same neoplasm. [DOI] [PubMed] [Google Scholar]
- 34.Tsao JL, et al. Colorectal adenoma and cancer divergence. Evidence of multilineage progression. American Journal of Pathology. 1999;154:1815–1824. doi: 10.1016/S0002-9440(10)65437-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maley CC, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 36.Sidransky D, et al. Clonal expansion of p53 mutant cells is associated with brain tumour progression. Nature. 1992;355:846–847. doi: 10.1038/355846a0. [DOI] [PubMed] [Google Scholar]
- 37.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. nature09515 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gould SJ, Eldredge N. Punctuated equilibrium comes of age. Nature. 1993;366:223–227. doi: 10.1038/366223a0. [DOI] [PubMed] [Google Scholar]
- 39.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell PJ, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13081–13086. doi: 10.1073/pnas.0801523105. Deep sequencing reveals rare (frequency < 0.001) intermediate genotypes between the common clones in leukemias. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007;7:834–846. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Isoda T, et al. Immunologically silent cancer clone transmission from mother to offspring. Proc Natl Acad Sci USA. 2009;106:17882–17885. doi: 10.1073/pnas.0904658106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welsh JS. Contagious cancer. The oncologist. 2011;16:1–4. doi: 10.1634/theoncologist.2010–0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nat Rev Cancer. 2008;8:56–61. doi: 10.1038/nrc2255. [DOI] [PubMed] [Google Scholar]
- 45.Bierie B, Moses HL. TGFβ: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:206–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 46.Lathia JD, Heddleston JM, Venere M, Rich JN. Deadly teamwork: neural cancer stem cells and the tumor microenvironment. Cell Stem Cell. 2011;8:482–485. doi: 10.1016/j.stem.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cairns J. Mutation Selection and the Natural History of Cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. Identified natural selection as a driving force in carcinogenesis, tissue architecture as a cancer suppressor and posited an immortal strand of DNA in tissue stem cells. [DOI] [PubMed] [Google Scholar]
- 48.Anderson AR, Weaver AM, Cummings PT, Quaranta V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell. 2006;127:905–915. doi: 10.1016/j. cell.2006.09.042. S0092-8674(06)01348-1 [pii] [DOI] [PubMed] [Google Scholar]
- 49.Chen J, Sprouffske K, Huang Q, Maley CC. Solving the puzzle of metastasis: the evolution of cell migration in neoplasms. PLoS One. 2011;6:e17933. doi: 10.1371/journal. pone.0017933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mazzone M, et al. Heterozygous deficiency of PHD2 restores tumor oxygenation and inhibits metastasis via endothelial normalization. Cell. 2009;136:839–851. doi: 10.1016/j. cell.2009.01.020. S0092-8674(09)00068-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gilbert LA, Hemann MT. DNA damage-mediated induction of a chemoresistant niche. Cell. 2010;143:355–366. doi: 10.1016/j. cell.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones S, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sprouffske K, Pepper JW, Maley CC. Accurate reconstruction of the temporal order of mutations in neoplastic progression. Cancer prevention research. 2011;4:1135–1144. doi: 10.1158/1940-6207.CAPR-10-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greaves MF, Maia AT, Wiemels JL, Ford AM. Leukemia in twins: lessons in natural history. Blood. 2003;102:2321–2333. doi: 10.1182/blood-2002-12-3817. [DOI] [PubMed] [Google Scholar]
- 56.Bateman CM, et al. Acquisition of genome-wide copy number alterations in monozygotic twins with acute lymphoblastic leukemia. Blood. 2010;115:3553–3558. doi: 10.1182/blood-2009-10-251413. [DOI] [PubMed] [Google Scholar]
- 57.Oosterhuis JW, Looijenga LH. Testicular germ-cell tumours in a broader perspective. Nature reviews. Cancer. 2005;5:210–222. doi: 10.1038/nrc1568. [DOI] [PubMed] [Google Scholar]
- 58.Grant PR, Grant BR. How and why species multiply. Princetown University Press; 2008. [Google Scholar]
- 59.Durinck S, et al. Temporal Dissection of Tumorigenesis in Primary Cancers. Cancer Discovery. 2011;1:OF1–OF7. doi: 10.1158/2159-8290.CD-11-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gonzalez-Garcia I, Sole RV, Costa J. Metapopulation dynamics and spatial heterogeneity in cancer. Proc Natl Acad Sci U S A. 2002;99:13085–13089. doi: 10.1073/pnas.202139299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clark J, et al. Complex patterns of ETS gene alteration arise during cancer development in the human prostate. Oncogene. 2008;27:1993–2003. doi: 10.1038/sj.onc.1210843. [DOI] [PubMed] [Google Scholar]
- 62.Navin N, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20:68–80. doi: 10.1101/gr.099622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Allred DC, et al. Ductal carcinoma in situ and the emergence of diversity during breast cancer evolution. Clin Cancer Res. 2008;14:370–378. doi: 10.1158/1078-0432.CCR-07-1127. [DOI] [PubMed] [Google Scholar]
- 64.Park SY, Gonen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120:636–644. doi: 10.1172/JCI40724. 40724 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Merlo LM, et al. A Comprehensive Survey of Clonal Diversity Measures in Barrett’s Esophagus as Biomarkers of Progression to Esophageal Adenocarcinoma. Cancer Prev Res (Phila) 2010;3:1388–1397. doi: 10.1158/1940-6207.CAPR-10-0108. 1940-6207.CAPR-10-0108 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 67.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 68.Greaves M. Cancer stem cells renew their impact. Nature medicine. 2011;17:1046–1048. doi: 10.1038/nm.2458. [DOI] [PubMed] [Google Scholar]
- 69.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Greaves M. Cancer stem cells: back to Darwin? Sem Cancer Biol. 2010;20:65–70. doi: 10.1016/j.semcancer.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 71.Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jamieson CH, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. The New England journal of medicine. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 73.Akala OO, et al. Long-term haematopoietic reconstitution by Trp53-/-p16Ink4a-/-p19Arf-/-multipotent progenitors. Nature. 2008;453:228–232. doi: 10.1038/nature06869. [DOI] [PubMed] [Google Scholar]
- 74.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 75.Olivier M, Taniere P. Somatic mutations in cancer prognosis and prediction: lessons from TP53 and EGFR genes. Current opinion in oncology. 2011;23:88–92. doi: 10.1097/CCO.0b013e3283412dfa. [DOI] [PubMed] [Google Scholar]
- 76.Mizuno H, Spike BT, Wahl GM, Levine AJ. Inactivation of p53 in breast cancers correlates with stem cell transcriptional signatures. Proc Natl Acad Sci USA. 2010;107:22745–22750. doi: 10.1073/pnas.1017001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cicalese A, et al. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 78.Quintana E, et al. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. nature07567 [pii] New xenograft methods reveal that cancer stem cells are common cell types in melanomas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pece S, et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 80.Notta F, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–367. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 81.Schmitz M, et al. Xenografts of highly resistant leukemia recapitulate the clonal composition of the leukemogenic compartment. Blood. 2011;118:1854–1864. doi: 10.1182/blood-2010-11-320309. [DOI] [PubMed] [Google Scholar]
- 82.Frank NY, Schatton T, Frank MH. The therapeutic promise of the cancer stem cell concept. J Clin Invest. 2010;120:41–50. doi: 10.1172/JCI41004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ishikawa F, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotech. 2007;25:1315–1321. doi: 10.1038/nbt1350. [DOI] [PubMed] [Google Scholar]
- 84.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Piccirillo SGM, et al. Distinct pools of cancer stem-like cells coexist within human glioblastomas and display different tumorigenicity and independent genomic evolution. Oncogene. 2009;28:1807–1811. doi: 10.1038/onc.2009.27. [DOI] [PubMed] [Google Scholar]
- 86.Solit D, Sawyers CL. How melanomas bypass new therapy. Nature. 2010;468:902–903. doi: 10.1038/468902a. [DOI] [PubMed] [Google Scholar]
- 87.Goff D, Jamieson C. Cycling toward elimination of leukemic stem cells. Cell Stem Cell. 2010;6:296–297. doi: 10.1016/j.stem.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 88.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 90.Sawyers CL. Shifting paradigms: the seeds of oncogene addiction. Nat Med. 2009;15:1158–1161. doi: 10.1038/nm1009-1158. [DOI] [PubMed] [Google Scholar]
- 91.Graham SM, et al. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99:319–325. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- 92.Turke AB, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145:30–38. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 94.Zhang B, et al. Effective targeting of quiescent chronic myelogenous leukemia stem cells by histone deacetylase inhibitors in combination with imatinib mesylate. Cancer Cell. 2010;17:427–442. doi: 10.1016/j.ccr.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duy C, et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature. 2011;473:384–388. doi: 10.1038/nature09883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pienta KJ, McGregor N, Axelrod R, Axelrod DE. Ecological therapy for cancer: defining tumors using an ecosystem paradigm suggests new opportunities for novel cancer treatments. Translational Oncol. 2008;1:158–164. doi: 10.1593/tlo.08178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Calabrese C, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 98.Bissell MJ, Hines WC. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wargo AR, Huijben S, de Roode JC, Shepherd J, Read AF. Competitive release and facilitation of drug-resistant parasites after therapeutic chemotherapy in a rodent malaria model. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19914–19919. doi: 10.1073/pnas.0707766104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gatenby RA, Silva AS, Gillies RJ, Frieden BR. Adaptive therapy. Cancer Res. 2009;69:4894–4903. doi: 10.1158/0008-5472. 69/11/4894 [pii] CAN-08-3658. Dosing to maintain tumor size prolongs survival far better than high dose therapy in a mouse xenograft model. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.