Abstract
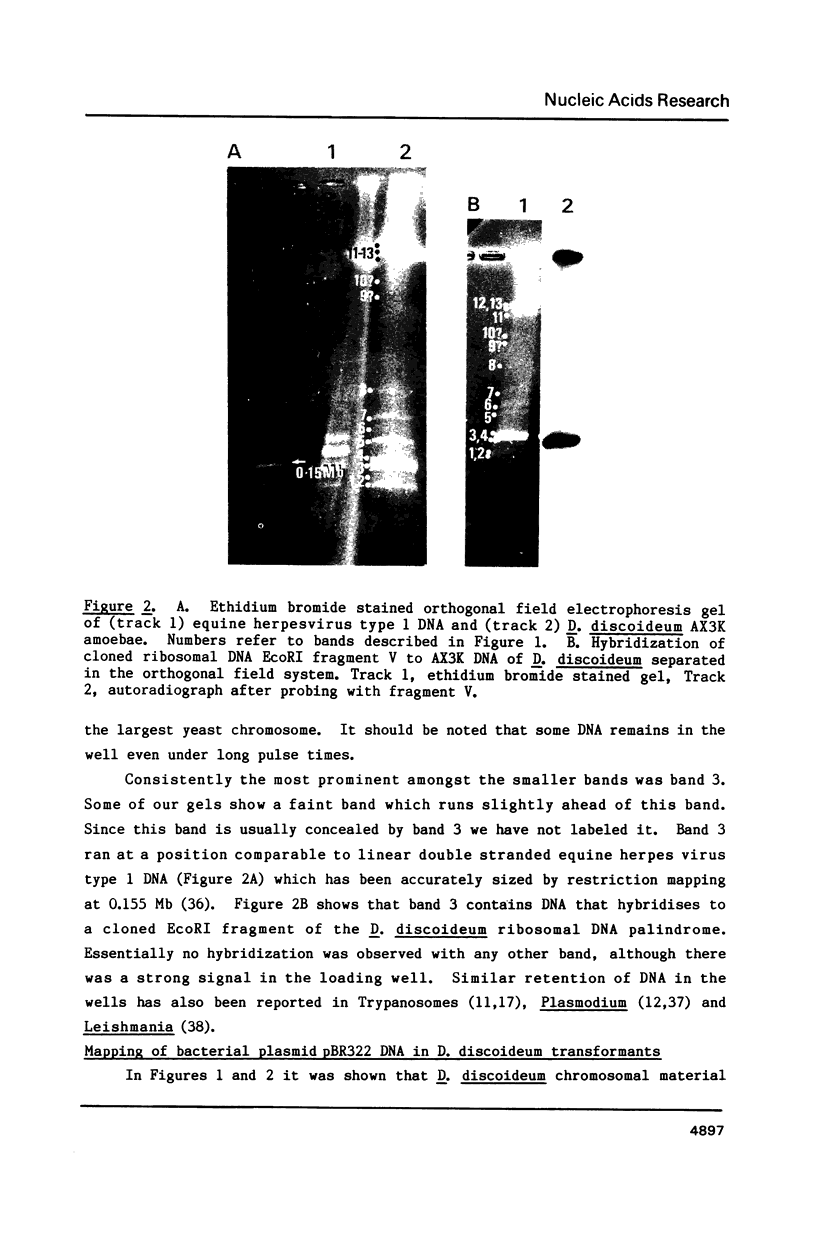
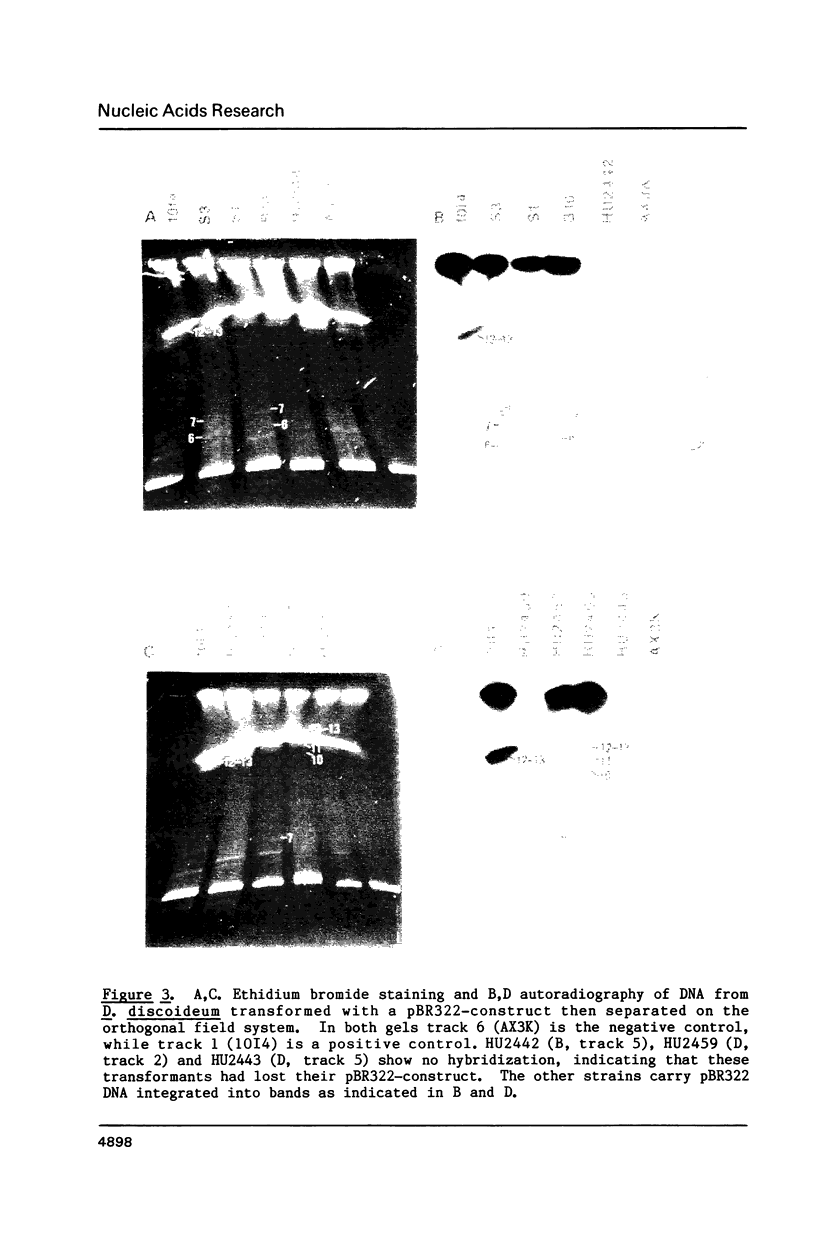
Chromosomes of the cellular slime mold Dictyostelium discoideum were fractionated on three pulse field gel electrophoresis systems (pulse field, orthogonal field and C.H.E.F. (Contour-clamped Homogeneous Electric Fields] into a series of 13 bands ranging from 0.1 Mb to over 2 Mb in size. Since this organism has only seven chromosomes (estimated to be 1-10 Mb), and -90 copies of an 88-kilobase linear ribosomal DNA molecule (14% of genome), it was apparent that not all of these bands were whole chromosomes. However these bands were reproducibly obtained with the cell preparation used. They fell into three categories: i) four large poorly resolved DNA molecules (-2 Mb in size) which represent very large fragments or intact chromosomes, ii) eight faint bands ranging from 0.1 Mb to 2 Mb, iii) a prominent band in the apparent size range of about 0.15 Mb. Cloned Fragment V of an EcoR1 digest of the ribosomal DNA, hybridized to the 0.15 Mb band indicating it contained the linear ribosomal DNA. This chromosomal banding pattern was used to examine the stability and location of vector DNA in 16 transformed strains of D. discoideum. Each transformed strain was initially selected on the basis of G418 resistance with an integrating vector containing pBR322 sequences. Eleven transformants still carried pBR322 sequences after more than 60 generations of growth without selection on G418. All four strains transformed with constructs containing regions of the D. discoideum plasmid Ddp1 had lost their pBR322 insert, indicating that integration of Dictyostelium plasmid DNA into chromosomes leads to instability. Orthogonal field electrophoresis of the eleven strains still carrying pBR322 sequences revealed at least seven different integrating sites for the transforming DNA. We conclude that these vectors have many possible sites of integration in the D. discoideum genome.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschuler M. I., Yao M. C. Macronuclear DNA of Tetrahymena thermophila exists as defined subchromosomal-sized molecules. Nucleic Acids Res. 1985 Aug 26;13(16):5817–5831. doi: 10.1093/nar/13.16.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards A., Kooter J. M., Michels P. A., Moberts R. M., Borst P. Pulsed field gradient electrophoresis of DNA digested in agarose allows the sizing of the large duplication unit of a surface antigen gene in trypanosomes. Gene. 1986;42(3):313–322. doi: 10.1016/0378-1119(86)90235-0. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Frank M., Olson M. V. Electrophoretic separations of large DNA molecules by periodic inversion of the electric field. Science. 1986 Apr 4;232(4746):65–68. doi: 10.1126/science.3952500. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. Separation of chromosomal DNA molecules from yeast by orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res. 1984 Jul 25;12(14):5647–5664. doi: 10.1093/nar/12.14.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Cockburn A. F., Taylor W. C., Firtel R. A. Dictyostelium rDNA consists of non-chromosomal palindromic dimers containing 5S and 36S coding regions. Chromosoma. 1978 Dec 21;70(1):19–29. doi: 10.1007/BF00292212. [DOI] [PubMed] [Google Scholar]
- Dawkins H. J. Molecular weight separation of very large DNA. Parasitol Today. 1987 Feb;3(2):60–62. doi: 10.1016/0169-4758(87)90218-3. [DOI] [PubMed] [Google Scholar]
- De Lozanne A., Spudich J. A. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987 May 29;236(4805):1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- Garvey E. P., Santi D. V. Stable amplified DNA in drug-resistant Leishmania exists as extrachromosomal circles. Science. 1986 Aug 1;233(4763):535–540. doi: 10.1126/science.3726545. [DOI] [PubMed] [Google Scholar]
- Gibson W. C., Miles M. A. The karyotype and ploidy of Trypanosoma cruzi. EMBO J. 1986 Jun;5(6):1299–1305. doi: 10.1002/j.1460-2075.1986.tb04359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger R. M., Maizels N. Dictyostelium ribosomal RNA is processed during transcription. Cell. 1980 Jul;20(3):619–623. doi: 10.1016/0092-8674(80)90308-6. [DOI] [PubMed] [Google Scholar]
- Kemp D. J., Corcoran L. M., Coppel R. L., Stahl H. D., Bianco A. E., Brown G. V., Anders R. F. Size variation in chromosomes from independent cultured isolates of Plasmodium falciparum. Nature. 1985 May 23;315(6017):347–350. doi: 10.1038/315347a0. [DOI] [PubMed] [Google Scholar]
- Kinashi H., Shimaji M., Sakai A. Giant linear plasmids in Streptomyces which code for antibiotic biosynthesis genes. 1987 Jul 30-Aug 5Nature. 328(6129):454–456. doi: 10.1038/328454a0. [DOI] [PubMed] [Google Scholar]
- Kurtz M. B., Cortelyou M. W., Miller S. M., Lai M., Kirsch D. R. Development of autonomously replicating plasmids for Candida albicans. Mol Cell Biol. 1987 Jan;7(1):209–217. doi: 10.1128/mcb.7.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langsley G., Sibilli L., Mattei D., Falanga P., Mercereau-Puijalon O. Karyotype comparison between P. chabaudi and P. falciparum: analysis of a P. chabaudi cDNA containing sequences highly repetitive in P. falciparum. Nucleic Acids Res. 1987 Mar 11;15(5):2203–2211. doi: 10.1093/nar/15.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee B. B., Magee P. T. Electrophoretic karyotypes and chromosome numbers in Candida species. J Gen Microbiol. 1987 Feb;133(2):425–430. doi: 10.1099/00221287-133-2-425. [DOI] [PubMed] [Google Scholar]
- McPeek F. D., Jr, Coyle-Morris J. F., Gemmill R. M. Separation of large DNA molecules by modified pulsed field gradient gel electrophoresis. Anal Biochem. 1986 Aug 1;156(2):274–285. doi: 10.1016/0003-2697(86)90254-x. [DOI] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae, edition 9. Microbiol Rev. 1985 Sep;49(3):181–213. doi: 10.1128/mr.49.3.181-213.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellen W., Firtel R. A. High-copy-number transformants and co-transformation in Dictyostelium. Gene. 1985;39(2-3):155–163. doi: 10.1016/0378-1119(85)90309-9. [DOI] [PubMed] [Google Scholar]
- Poole S. J., Firtel R. A. Genomic instability and mobile genetic elements in regions surrounding two discoidin I genes of Dictyostelium discoideum. Mol Cell Biol. 1984 Apr;4(4):671–680. doi: 10.1128/mcb.4.4.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Matsumoto T., Niwa O., Klco S., Fan J. B., Yanagida M., Cantor C. R. An electrophoretic karyotype for Schizosaccharomyces pombe by pulsed field gel electrophoresis. Nucleic Acids Res. 1987 Jun 11;15(11):4481–4489. doi: 10.1093/nar/15.11.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell R. G., Wilkins R. J. Separation of chromosomal DNA molecules from C.albicans by pulsed field gel electrophoresis. Nucleic Acids Res. 1986 Jun 11;14(11):4401–4406. doi: 10.1093/nar/14.11.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M., Anand R., Brown W. R., Fletcher D. S. A model for the separation of large DNA molecules by crossed field gel electrophoresis. Nucleic Acids Res. 1987 Aug 11;15(15):5925–5943. doi: 10.1093/nar/15.15.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spithill T. W., Samaras N. The molecular karyotype of Leishmania major and mapping of alpha and beta tubulin gene families to multiple unlinked chromosomal loci. Nucleic Acids Res. 1985 Jun 11;13(11):4155–4169. doi: 10.1093/nar/13.11.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ploeg L. H., Cornelissen A. W., Michels P. A., Borst P. Chromosome rearrangements in Trypanosoma brucei. Cell. 1984 Nov;39(1):213–221. doi: 10.1016/0092-8674(84)90207-1. [DOI] [PubMed] [Google Scholar]
- Van der Ploeg L. H., Schwartz D. C., Cantor C. R., Borst P. Antigenic variation in Trypanosoma brucei analyzed by electrophoretic separation of chromosome-sized DNA molecules. Cell. 1984 May;37(1):77–84. doi: 10.1016/0092-8674(84)90302-7. [DOI] [PubMed] [Google Scholar]
- Vollrath D., Davis R. W. Resolution of DNA molecules greater than 5 megabases by contour-clamped homogeneous electric fields. Nucleic Acids Res. 1987 Oct 12;15(19):7865–7876. doi: 10.1093/nar/15.19.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts D. J., Ashworth J. M. Growth of myxameobae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem J. 1970 Sep;119(2):171–174. doi: 10.1042/bj1190171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalley J. M., Robertson G. R., Davison A. J. Analysis of the genome of equine herpesvirus type 1: arrangement of cleavage sites for restriction endonucleases EcoRI, BglII and BamHI. J Gen Virol. 1981 Dec;57(Pt 2):307–323. doi: 10.1099/0022-1317-57-2-307. [DOI] [PubMed] [Google Scholar]